RNA editing converts hundreds of cytidines into uridines in plant organelles. Recognition of editing sites requires nuclear encoded RNA-binding pentatricopeptide repeat (PPR) proteins. Previous studies have documented the presence of PPR proteins encoded by the genome of the protist Naegleria gruberi, raising the possibility that plant-like RNA editing might occur in this organism. This paper shows that this is, indeed, the case. Because Naegleria and plants diverged ∼1.5 billion years ago, these findings suggest either horizontal gene transfer or that plant-type editing is much more ancestral than previously believed.
Keywords: DYW-type PPR proteins, C-to-U RNA editing, plant mitochondria, protists, Heterolobosea, Naegleria gruberi
Abstract
RNA editing converts hundreds of cytidines into uridines in plant mitochondrial and chloroplast transcripts. Recognition of the RNA editing sites in the organelle transcriptomes requires numerous specific, nuclear-encoded RNA-binding pentatricopeptide repeat (PPR) proteins with characteristic carboxy-terminal protein domain extensions (E/DYW) previously thought to be unique to plants. However, a small gene family of such plant-like PPR proteins of the DYW-type was recently discovered in the genome of the protist Naegleria gruberi. This raised the possibility that plant-like RNA editing may occur in this amoeboflagellate. Accordingly, we have investigated the mitochondrial transcriptome of Naegleria gruberi and here report on identification of two sites of C-to-U RNA editing in the cox1 gene and in the cox3 gene, both of which reconstitute amino acid codon identities highly conserved in evolution. An estimated 1.5 billion years of evolution separate the heterolobosean protist Naegleria from the plant lineage. The new findings either suggest horizontal gene transfer of RNA editing factors or that plant-type RNA editing is evolutionarily much more ancestral than previously thought and yet to be discovered in many other ancient eukaryotic lineages.
INTRODUCTION
RNA editing of mitochondrial and chloroplast transcripts of land plants converts numerous cytidines into uridines. The majority of these site-specific, post-transcriptional sequence changes serve to reconstitute evolutionarily conserved codons in messenger RNAs. Approximately 200–500 RNA editing sites are present in flowering plant mitochondria (Giegé and Brennicke 1999; Notsu et al. 2002; Handa 2003; Mower and Palmer 2006; Sloan et al. 2010), but RNA editing frequencies vary even more widely in other plant clades. Less than a dozen sites are affected in the mitochondrial transcriptomes of funariid mosses (Rüdinger et al. 2009, 2011), whereas more than 2000 RNA editing events have been found in the spikemoss Selaginella moellendorffii, a lycophyte representing an ancient vascular plant lineage (Hecht et al. 2011).
Following the seminal discovery of the first nuclear-encoded proteins that target specific RNA editing sites in chloroplasts (Kotera et al. 2005) and mitochondria (Zehrmann et al. 2009), numerous similar editing factors have been identified (see, e.g., Chateigner-Boutin and Small 2010; Fujii and Small 2011). All of them are so-called pentatricopeptide repeat (PPR) proteins, which are characterized by arrays of tandemly repeated, loosely conserved 35-amino acid motifs that likely mediate sequence-specific RNA-binding. PPR proteins are encoded by vastly extended nuclear gene families in plants containing hundreds of members, most of which are targeted to chloroplasts and mitochondria (Lurin et al. 2004; O'Toole et al. 2008; Schmitz-Linneweber and Small 2008). To date, all plant PPR proteins specific for RNA editing are characterized by additional carboxy-terminal protein domains, the E/E+ “extensions,” and in many cases, these are followed by an additional “DYW” domain (Fig. 1A). The highly conserved 100-amino acid DYW domain is named for the conserved tripeptide motif at its end. The weak similarity of the DYW domain to deaminases has received particular attention (Salone et al. 2007), because the C-to-U conversions effected by plant organelle RNA editing may be a simple deamination process (Yu and Schuster 1995). Equally important, the presence of DYW-type PPR protein genes correlates perfectly with the occurrence of RNA editing in the plant kingdom, both of which seem to be entirely absent in green algae and the marchantiid liverworts (Salone et al. 2007; Rüdinger et al. 2008). The lack of RNA editing in the marchantiid liverworts obviously reflects a secondary loss (Groth-Malonek et al. 2007), and it is thus parsimonious to assume that RNA editing originated together with DYW-type PPR proteins at the origin of land plant life. However, the extreme degree of sequence conservation in the DYW domain across 500 million years of land plant diversification made its evolutionary origin quite mysterious. No sequence homology of the DYW domain with any other protein sequences had been identified other than the very weak similarity with deaminases, which is largely restricted to a few conserved histidine and cysteine residues (see Fig. 1A).
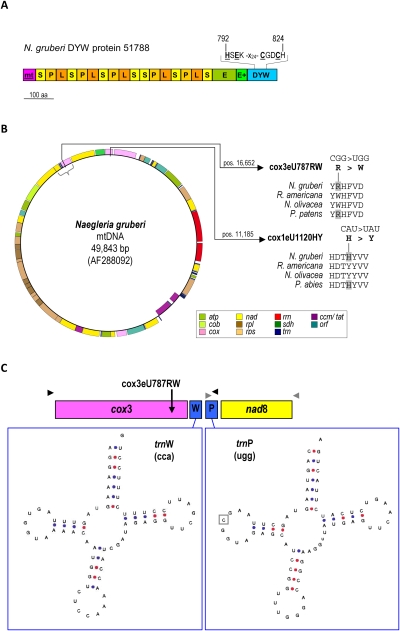
FIGURE 1.
(A) A candidate mitochondrially targeted DYW protein in Naegleria gruberi. Mitochondrial (mt) targeting is predicted (http://ihg.gsf.de/ihg/mitoprot.html) for one Naegleria gruberi 860-amino acid DYW-type PPR protein (NCBI accession XP_002673292). Like the other nine Naegleria DYW proteins, it shares all elements conserved among the plant homologues: length-variable PPR repeats (canonical 35-aa “P,” long 35–36- aa “L,” and short 31-aa “S” variants) and carboxy-terminal domains E, E+, and DYW. The conserved deaminase motif is shown above the DYW domain. (B) Two sites of RNA editing in Naegleria gruberi mitochondria. A gene map of Naegleria gruberi mtDNA (accession AF288092) was drawn using the OGDRAW service at http://ogdraw.mpimp-golm.mpg.de (Lohse et al. 2007) with genes color-coded as indicated (see Materials and Methods for details). Transcriptome sequencing identified one site of C-to-U RNA editing in each cox1 and cox3 (positions in the chondrome sequence entry indicated), which reconstitute widely conserved amino acid codons illustrated by alignments including the jakobid protist Reclinomonas americana and the green alga Nephroselmis olivacea. Editing event cox3eU787RW is conserved in the moss Physcomitrella patens and cox1eU1120HY in Picea abies and other gymnosperms. The curly brace indicates the cox3-trnW-trnP-nad8 region investigated for cotranscription (see C). (C) RNA editing in cotranscripts. Two pairs of PCR primers (arrowheads) were designed to amplify predicted cotranscription products of the mitochondrially encoded cox3-trnW-trnP-atp8 gene continuity. Cotranscripts were detected and found to be edited only at the cox3eU787RW site. Editing of a candidate cytidine in the pseudouridine loop of trnP (boxed) was not detected. The tRNA graphics were made using tRNAscan-SE (Lowe and Eddy 1997).
Surprisingly, we recently found the first DYW protein homologues outside of land plants in the genome of the heterolobosean protist Naegleria gruberi (Fritz-Laylin et al. 2010; Knoop and Rüdinger 2010). The N. gruberi nuclear genome encodes a small gene family encoding ten DYW-type PPR proteins, quite similar to its counterpart in the moss Physcomitrella patens. Aside from the ten plant-like PPR proteins of the DYW-type, the Naegleria genome encodes 32 “pure” PPR proteins lacking carboxy-terminal domain additions. Although Naegleria is separated from land plants by some 1.5 billion years of evolution, the correlation of DYW-type PPR proteins with RNA editing among land plants led us to investigate whether plant-type mitochondrial RNA editing may also exist in Naegleria.
RESULTS
The Naegleria gruberi mitochondrial genome is a circular, gene-rich mtDNA (Gray et al. 2004), encoding 43 proteins, two rRNAs, and 21 tRNAs (Fig. 1B). We sequenced the mitochondrial transcriptome including all coding regions and also resequenced selected coding regions in the mtDNA to avoid false positives due to potential sequencing errors or polymorphisms between our biological material and the published mtDNA sequence (NCBI accession AF288092). All protein coding mitochondrial mRNAs in Naegleria gruberi were found to be identical to the corresponding genomic sequences in the mitochondrial DNA with two exceptions. Position 1120 of the cox1 cDNA and position 787 of the cox3 cDNA contained uridines instead of the genomically encoded cytidines (Fig. 1B). These C-to-U RNA editing events, designated as cox1eU1120HY and cox3eU787RW following recent nomenclature recommendations (Lenz et al. 2009), restore evolutionarily conserved amino acids in the two cytochrome c oxidase subunits vital for respiration: a histidine codon is converted into a tyrosine codon in cox1 and an arginine codon is converted into a tryptophan codon in the cox3 coding sequence. Strikingly, such C-to-U editing events at the corresponding sites have also been observed in phylogenetically distant land plants. RNA editing site cox1eU1120HY is present among gymnosperms (Lu et al. 1998), and editing event cox3eU787RW is conserved at the homologous position in the moss Physcomitrella patens (Rüdinger et al. 2009). Coincidentally, the DYW-type PPR protein recognizing the latter editing site in Physcomitrella patens has recently been identified, and the growth retardation of the corresponding knockout strain demonstrates the vital importance of this RNA editing event (Ohtani et al. 2010).
RNA editing of such essential sites in mRNAs is an efficient post-transcriptional process in plant mitochondria, frequently faster than (and mostly independent of) other steps of RNA maturation such as splicing, endonucleolytic processing, or RNA end trimming. Only some RNA editing events, mostly at “silent” sites that leave the encoded amino acid unchanged, are often found only partially edited to varying degrees in the steady state transcript pool of plant mitochondria (e.g., Mower and Palmer 2006; Grewe et al. 2011).
We wished to test whether RNA editing similarly precedes other steps of RNA maturation in Naegleria gruberi mitochondria. As no introns are present in the N. gruberi mitochondrial genome, we examined the order of maturation events of a predicted cotranscribed region, cox3-trnW-trnP-nad8 (Fig. 1B) that would require endonucleolytic processing for maturation of the two tRNAs encoded between cox3 and nad8. This gene arrangement was additionally interesting, given the distant possibility that a cytidine in position 17 of the trnP dihydrouridine arm may first undergo C-to-U editing before being modified to dihydrouridine (Fig. 1C). RT-PCR products corresponding to cox3 and the downstream tRNAs and, conversely, to nad8 and the upstream trnP, were easily retrieved from cDNA, demonstrating their cotranscription and incomplete processing in the steady-state mitochondrial RNA pool (Fig. 1C). Sequencing of the corresponding cDNAs confirmed that editing of the cox3eU787RW site already took place in these unprocessed transcripts. However, no other editing events, including the weak candidate site in trnP, were identified. Likewise, exhaustive sequencing of all other mitochondrial genes on transcriptome level, including the large and small subunit ribosomal RNAs and other tRNAs showing candidate positions similar to trnP, did not identify any RNA editing sites other than cox1eU1120HY and cox3eU787RW in Naegleria gruberi.
DISCUSSION
After the seminal discoveries of nuclear-encoded RNA editing factors in model plants like Arabidopsis and rice, the moss Physcomitrella patens has emerged as an interesting alternative organism to study RNA editing in plants. The very few RNA editing sites in the moss—only eleven in mitochondria and only two in chloroplasts—and its amenability to targeted nuclear gene knockouts make Physcomitrella an attractive complementary model system. Knockout analyses in Physcomitrella patens have so far clearly assigned six of its ten DYW-type PPR proteins to nine of its eleven mitochondrial editing sites (Ohtani et al. 2010; Tasaki et al. 2010; Rüdinger et al. 2011; Uchida et al. 2011). Three of these DYW proteins address two editing sites simultaneously, possibly due to the sequence similarities around those editing sites.
If RNA editing in Naegleria mitochondria uses similar mechanisms, it would obviously require only one or two DYW proteins to recognize the two mitochondrial RNA editing sites that we have identified here. Similar to Physcomitrella, however, the Naegleria nuclear genome encodes ten DYW genes, and all of them carry the conserved cytidine deaminase signature HxExnCxxC. Interestingly, however, only one of these (NCBI accession XP_002673292, protein 51788) has a clear mitochondrial targeting prediction (Fig. 1A). This leaves open the exciting possibility that some DYW proteins may edit nuclear-encoded RNAs in Naegleria. Alternatively, some DYW domains may have acquired novel functionalities in RNA maturation or stabilization as has been shown for the CRR2 protein in Arabidopsis, which participates in processing a chloroplast cotranscript (Hashimoto et al. 2003). Finally, although unlikely given that our transcriptome analysis covered >92% of the Naegleria mitochondrial genome on the transcript level (see Materials and Methods), it cannot be ruled out completely that additional editing sites may have been missed.
Naegleria and plants belong to two different of the six major eukaryotic groups and are, therefore, separated by some estimated 1500 million years of evolution (Fritz-Laylin et al. 2010). The simultaneous coexistence of RNA editing and DYW-type PPR proteins both in land plants and in the phylogenetically distant protist is puzzling. The presence of DYW proteins may possibly be explained by horizontal gene transfer, which would subsequently allow for T-C mutations in the recipient organism to be compensated by RNA editing. Alternatively, it is possible that C-to-U RNA editing and DYW proteins are much older than previously thought, and are ancestral to both lineages. This would imply that many additional examples await discovery in other eukaryotic lineages. The rarity of RNA editing found in Naegleria gruberi mitochondria clearly underlines the risk of overlooking similar processes in mitochondria of other eukaryotic lineages.
MATERIALS AND METHODS
Nucleic acids were prepared from Naegleria gruberi strain NEG-M grown aerobically in axenic medium M7 at 30°C. DNA was isolated using a standard guanidium thiocyanate extraction protocol. RNA was isolated using Trizol Reagent (Invitrogen), according to the manufacturer's instructions. RNA was treated with DNase I (Fermentas Life Sciences) to remove vestiges of DNA. First strand cDNA was synthesized using the RevertAid M-MulV Reverse Transcriptase kit (Fermentas) and a hexanucleotide random primer mix (10 μM; Carl Roth). Oligonucleotide pairs were designed to flank the coding regions of all N. gruberi mitochondrial protein coding genes: five ATP synthase subunits (atp), apocytochrome b (cob), four cytochrome c oxidase subunits (cox), two cytochrome c maturation subunits (ccm), 11 NADH dehydrogenase subunits (nad), six proteins of the large (rpl) and 11 of the small ribosomal subunit (rps), one subunit each of succinate dehydrogenase (sdh) and the twin arginine translocase (tat), four uncharacterized open reading frames (orf), and the two rRNA genes (rrn). These oligonucleotides (sequences available upon request) were successfully used for PCR amplification of the entire coding regions of the DNA and cDNA, except in four cases (atp1, rps4, ccmF, rrnL) where we were unable to amplify the terminal 40–70 nucleotides of the cDNA coding sequences. Furthermore, RT-PCR was performed for 11 tRNA genes (trn), where RNA folding prediction (http://lowelab.ucsc.edu/tRNAscan-SE) suggested potential C-to-U editing to improve base-pairing in the secondary structures. Additionally, we targeted cotranscripts in the cox3-trnW-trnP-atp8 cluster (see Fig. 1C). PCR reactions typically contained 1–3 μL template (10 ng–1 μg), 0.4 mM MgCl2, 0.8 mM dNTPs, 0.4 μM of each primer, 1 U GoTaq DNA Polymerase and 1× recommended PCR buffer (Promega) in a total volume of 25 μL. PCR amplification included an initial 5 min denaturation at 94°C, followed by 30 cycles each of 30 sec denaturation at 94°C, 30 sec annealing at 42–53°C, 1–3 min synthesis at 72°C, and a final step of synthesis for 5 min at 72°C. Control PCRs were performed on RNA not treated with reverse transcriptase to exclude false positives from residual DNA. RT-PCR and PCR products were cloned into the pGEM-T Easy vector (Promega) or sequenced directly (Macrogen Inc. or GATC Biotech AG). DNA and cDNA sequences were aligned to the corresponding gene sequences of the annotated mitochondrial genome of Naegleria gruberi (GenBank accession AF288092) using the MEGA software (Tamura et al. 2007).
ACKNOWLEDGMENTS
The corresponding author thanks the Deutsche Forschungsgemeinschaft for funding of research on RNA editing and PPR proteins in his lab through grant Kn411/7.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.02962911.
REFERENCES
- Chateigner-Boutin AL, Small I 2010. Plant RNA editing. RNA Biol 7: 213–219 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642 [DOI] [PubMed] [Google Scholar]
- Fujii S, Small I 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol 191: 37–47 [DOI] [PubMed] [Google Scholar]
- Giegé P, Brennicke A 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci 96: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G 2004. Mitochondria of protists. Annu Rev Genet 38: 477–524 [DOI] [PubMed] [Google Scholar]
- Grewe F, Herres S, Viehöver P, Polsakiewicz M, Weisshaar B, Knoop V 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res 39: 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V 2007. Evolution of a pseudogene: Exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae. Mol Biol Evol 24: 1068–1074 [DOI] [PubMed] [Google Scholar]
- Handa H 2003. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31: 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T 2003. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol 3: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Rüdinger M 2010. DYW-type PPR proteins in a heterolobosean protist: Plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett 584: 4287–4291 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T 2005. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Lenz H, Rüdinger M, Volkmar U, Fischer S, Herres S, Grewe F, Knoop V 2009. Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Curr Genet 56: 189–201 [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52: 267–274 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MZ, Szmidt AE, Wang XR 1998. RNA editing in gymnosperms and its impact on the evolution of the mitochondrial coxI gene. Plant Mol Biol 37: 225–234 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Palmer JD 2006. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genomics 276: 285–293 [DOI] [PubMed] [Google Scholar]
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268: 434–445 [DOI] [PubMed] [Google Scholar]
- Ohtani S, Ichinose M, Tasaki E, Aoki Y, Komura Y, Sugita M 2010. Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol 51: 1942–1949 [DOI] [PubMed] [Google Scholar]
- O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I 2008. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Polsakiewicz M, Knoop V 2008. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat (PPR) proteins in jungermanniid but not in marchantiid liverworts. Mol Biol Evol 25: 1405–1414 [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V 2009. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics 281: 473–481 [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Szövényi P, Rensing SA, Knoop V 2011. Assigning DYW-type PPR proteins to RNA editing sites in the funariid mosses Physcomitrella patens and Funaria hygrometrica. Plant J 67: 370–380 [DOI] [PubMed] [Google Scholar]
- Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C 2007. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett 581: 4132–4138 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR 2010. Extensive loss of RNA editing sites in rapidly evolving silene mitochondrial genomes: Selection vs. retroprocessing as the driving force. Genetics 185: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tasaki E, Hattori M, Sugita M 2010. The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J 62: 560–570 [DOI] [PubMed] [Google Scholar]
- Uchida M, Ohtani S, Ichinose M, Sugita C, Sugita M 2011. The PPR-DYW proteins are required for RNA editing of rps14, cox1, and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett 585: 2367–2371 [DOI] [PubMed] [Google Scholar]
- Yu W, Schuster W 1995. Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J Biol Chem 270: 18227–18233 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M 2009. A DYW domain–containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]