Nonsense-mediated mRNA decay (NMD) is traditionally portrayed as a quality control mechanism that degrades mRNAs with truncated open reading frames (ORFs). However, it is clear that NMD also contributes to the post-transcriptional gene regulation of numerous physiological mRNAs. To identify endogenous NMD substrate mRNAs and analyze the features that render them sensitive to NMD, the authors performed transcriptome profiling of human cells depleted of the NMD factors UPF1, SMG6, or SMG7. It revealed that mRNAs up-regulated by NMD abrogation had a greater median 3′-UTR length compared to that of the human mRNAome and were also enriched for 3′-UTR introns and uORFs. Intriguingly, most mRNAs coding for NMD factors were among the NMD-sensitive transcripts, implying that the NMD process is autoregulated.
Keywords: autoregulation, mRNA, microarray, NMD, 3′-UTR length
Abstract
Nonsense-mediated mRNA decay (NMD) is traditionally portrayed as a quality-control mechanism that degrades mRNAs with truncated open reading frames (ORFs). However, it is meanwhile clear that NMD also contributes to the post-transcriptional gene regulation of numerous physiological mRNAs. To identify endogenous NMD substrate mRNAs and analyze the features that render them sensitive to NMD, we performed transcriptome profiling of human cells depleted of the NMD factors UPF1, SMG6, or SMG7. It revealed that mRNAs up-regulated by NMD abrogation had a greater median 3′-UTR length compared with that of the human mRNAome and were also enriched for 3′-UTR introns and uORFs. Intriguingly, most mRNAs coding for NMD factors were among the NMD-sensitive transcripts, implying that the NMD process is autoregulated. These mRNAs all possess long 3′ UTRs, and some of them harbor uORFs. Using reporter gene assays, we demonstrated that the long 3′ UTRs of UPF1, SMG5, and SMG7 mRNAs are the main NMD-inducing features of these mRNAs, suggesting that long 3′ UTRs might be a frequent trigger of NMD.
INTRODUCTION
To maintain a healthy homeostasis, cells have to constantly ensure sufficient accuracy of the numerous complex biochemical processes. This requirement led to the evolution of a variety of quality-control, checkpoint, and proofreading mechanisms. Among these, nonsense-mediated mRNA decay (NMD) represents a quality-control mechanism that acts on the level of mRNA translation. Historically, NMD was discovered as a process that degrades mRNAs with premature termination codons (PTC) (Losson and Lacroute 1979; Maquat et al. 1981). However, subsequent research during the last decade revealed that NMD also regulates the steady-state levels of many “normal” physiological mRNAs that encode functional full-length proteins (Lelivelt and Culbertson 1999; Mendell et al. 2004; Rehwinkel et al. 2005; Taylor et al. 2005; Wittmann et al. 2006). These findings, together with new insights into the mechanism of NMD substrate recognition, led to a broader definition of NMD as a translation-dependent process that recognizes and degrades mRNAs with a termination codon (TC) that is not in a favorable environment for efficient translation termination (Amrani et al. 2006; Nicholson et al. 2010).
According to these new mechanistic working models, normal translation termination involves an interaction between poly(A) binding protein (PABP) and ribosome-bound eukaryotic release factor 3 (eRF3), which promotes fast polypeptide release, disassembly of the ribosomal subunits, and recycling of the ribosomal subunits to the start codon (Hoshino et al. 1999; Amrani et al. 2004; Behm-Ansmant et al. 2007). When the ribosome reaches a TC located in a messenger ribonucleoprotein (mRNP) environment where it fails to receive the PABP-mediated termination-stimulating signal, the ribosome stalls for a prolonged period of time, allowing binding of UPF1, a key factor in NMD, to eRF3 (Singh et al. 2008). This event marks the mRNA for degradation; however, at this point the commitment to the NMD pathway is thought to still be reversible. UPF1 is a group 1 RNA helicase, a nucleic acid–dependent ATPase, and a phosphorylation substrate of SMG1 kinase (Czaplinski et al. 1995; Bhattacharya et al. 2000; Yamashita et al. 2001). Two other core NMD factors, UPF2 and UPF3, are thought to promote SMG1-mediated phosphorylation of UPF1 (Kashima et al. 2006). Following phosphorylation and possibly induced by ATP hydrolysis, UPF1 undergoes a conformational change that increases its affinity for RNA. After that, UPF1 is ready for interaction with SMG5, SMG6, and SMG7 proteins, which initiate mRNA degradation. At this point the mRNA is irreversibly committed to NMD. SMG6 has been shown to be an endonuclease that cleaves NMD substrate mRNAs near the stop codon (Huntzinger et al. 2008; Eberle et al. 2009), whereas SMG5 and SMG7 are thought to promote mRNA deadenylation followed by decapping and exonucleolytic decay (Unterholzner and Izaurralde 2004).
According to the unified working model of NMD, several different features of an mRNP can trigger NMD (for review, see Rehwinkel et al. 2006; Nicholson et al. 2010). The experimental evidence from several different model organisms suggests, for example, that long 3′ UTRs can elicit NMD (Muhlrad and Parker 1999; Amrani et al. 2004; Kertesz et al. 2006; Behm-Ansmant et al. 2007; Longman et al. 2007; Eberle et al. 2008; Kerenyi et al. 2008; Singh et al. 2008; Hansen et al. 2009; Kebaara and Atkin 2009). A greater distance between the TC and PABP would impede transmission of the PABP-mediated termination-stimulating signal to the ribosome stalled at the TC, thus tilting the balance from efficient termination and ribosome release toward NMD. Another well-documented NMD-eliciting feature is the presence of intron(s) >50 nt downstream from the termination codon (Nagy and Maquat 1998). Sites of intron removal get marked on the spliced mRNPs with exon junction complexes (EJC). The EJC is a dynamic structure with a heterogeneous protein composition that, among other roles, serves as an anchoring point for UPF2 and UPF3 (Le Hir et al. 2001). When an EJC is situated downstream from a TC, the interaction between UPF1 bound to the termination complex and UPF2 and/or UPF3 is greatly facilitated by virtue of their close proximity, resulting in efficient NMD (Nagy and Maquat 1998; Stalder and Muhlemann 2008). A further element potentially resulting in NMD is an open reading frame (ORF) upstream of the main coding sequence. If a uORF (upstream ORF) is translated by the ribosome, its TC could be regarded as premature due to the bulk of the downstream sequences that separate stop codon position from poly(A) tail as well as due to possible downstream intron(s). Furthermore, programmed frameshifts, selenocysteine-encoding UGA codons, and leaky scanning initiation codons can target transcripts for NMD (Moriarty et al. 1998; Welch and Jacobson 1999; Plant et al. 2004).
Previous gene expression profiling studies in Saccharomyces cerevisiae, Drosophila melanogaster, and human cells showed that 3%–10% of all mRNAs were up-regulated in NMD-deficient cells (Lelivelt and Culbertson 1999; He et al. 2003; Mendell et al. 2004; Rehwinkel et al. 2005; Rodriguez-Gabriel et al. 2006; Wittmann et al. 2006). Although some of them were PTC-containing mRNAs, such as inefficiently or alternatively spliced mRNA isoforms and transcripts from pseudogenes, the majority were normal physiological transcripts. These NMD targets were mainly non-orthologous between different species and also differed between distinct studies of the same human cell line (Mendell et al. 2004; Rehwinkel et al. 2006; Wittmann et al. 2006; Viegas et al. 2007). Overrepresented cellular processes among the identified NMD substrates are also different from study to study, with the exceptions of telomere maintenance, NMD, and DNA repair, processes in which NMD factors might have a conserved role (Rehwinkel et al. 2006).
To get further insights into how NMD regulates physiological transcripts, we have conducted microarray experiments on RNA from HeLa cells that were depleted of UPF1, SMG6, or SMG7. The analysis of the NMD-sensitive transcriptome shows a clear enrichment for transcripts with longer than average 3′ UTRs, genes that contain at least one transcript variant harboring 3′-UTR intron(s), as well as moderate enrichment for mRNAs with uORFs. Given the large number of physiological transcripts that presumably are under NMD control, the NMD pathway can be considered as an important post-transcriptional gene regulation mechanism. To keep the levels of physiological NMD targets constant, the efficiency of NMD itself would have to be tightly regulated. Indeed, we identified a feedback regulation of the NMD process whereby mRNAs coding for most known NMD factors are targeted themselves by the NMD pathway. Intriguingly, these NMD-sensitive NMD factor-coding mRNAs all possess relatively long 3′ UTRs, and for UPF1, SMG5, and SMG7, we demonstrated that their 3′ UTRs are sufficient to elicit NMD. This autoregulation has a potential to quickly establish balanced levels of NMD factors and subsequently regulated levels of a bulk of NMD substrates in the cell.
RESULTS
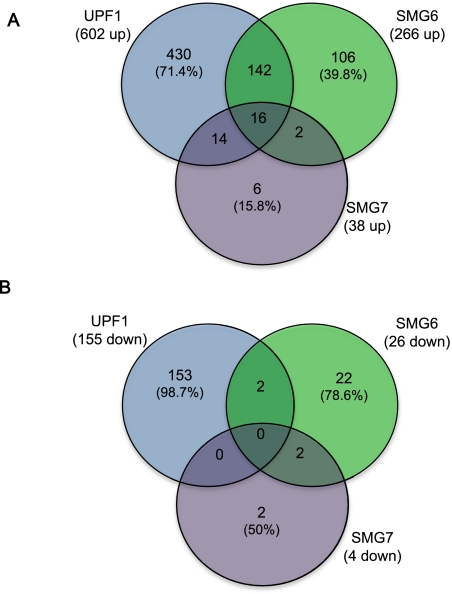
To broaden our understanding of the role of NMD in physiological processes of the cell, we performed transcriptome profiling of HeLa cells depleted of UPF1, SMG6, or SMG7 using Affymetrix exon arrays. The depletion was achieved through shRNA-mediated knockdown (KD) using two different shRNA target sequences for each NMD factor in independent experiments (Supplemental Fig. 1). To minimize possible off-target effects, only the genes differentially expressed upon both shRNA treatments were followed further in our study. Consistent with the role of UPF1, SMG6, and SMG7 in the decay of mRNAs, the majority of differentially expressed genes on the microarray were up-regulated (Fig. 1; Supplemental Table I). Interestingly, most of the genes up-regulated in SMG6 and SMG7 KD were also induced in UPF1 depletion, whereas upon UPF1 KD the majority of the changes in mRNA levels were unique, possibly due to widespread non-NMD roles of UPF1 (Fig. 1A). Similarly, 153 out of 155 genes down-regulated in UPF1 depletion were not significantly changed in SMG6 or SMG7 knockdowns (Fig. 1B).
FIGURE 1.
Venn diagrams illustrating the genes differentially expressed in UPF1, SMG6, and SMG7 knockdowns (>1.5-fold, false discovery rate ≤ 0.05). (A) Up-regulated sets of genes. (B) Down-regulated sets of genes. The total number of differentially expressed genes in a specific knockdown is depicted in parentheses below the respective NMD factor, and the percentage values indicate the genes that were differentially expressed only in one of the three knockdown conditions.
To validate our microarray data, we tested several mRNAs that are stabilized in all tested KD conditions by quantitative real-time reverse transcription PCR (hereafter RT-qPCR) and could reproduce the microarray results for the majority of the transcripts with a general tendency of a higher fold change in RT-qPCR assays (Supplemental Fig. 2).
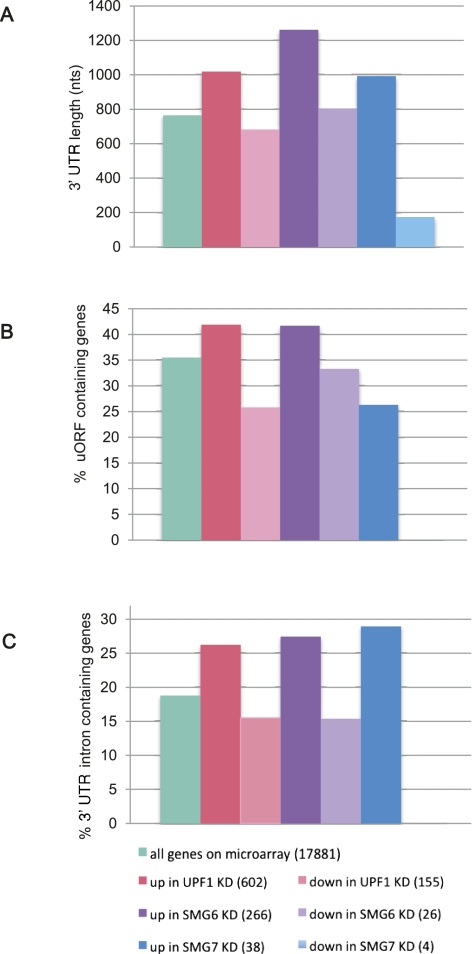
Next, we analyzed the sets of differentially expressed genes for various NMD-inducing features, such as 3′-UTR length and the presence of uORFs and 3′-UTR introns located >50 nt downstream from the TC. First, we calculated the median 3′-UTR length of human mRNAs based on the annotations in the reference sequence (RefSeq) database. The mRNA entries consisting of only predicted ORFs were ignored. The median 3′-UTR length of 33,404 RefSeq mRNAs that contained an annotated 3′ UTR was calculated to be 608 nt, whereas the median 3′-UTR length of mRNAs that are present on the Affymetrix Human Exon 1.0 ST Array was calculated to be 765 nt. A possible reason for this discrepancy is the enrichment of microarray genes for well-annotated transcripts with complete sequence data. Importantly, the median 3′-UTR length for genes up-regulated in UPF1, SMG6, or SMG7 knockdowns was 1019, 1262, and 992 nt, respectively, revealing a substantial enrichment of long-3′-UTR-containing mRNAs among NMD substrates (Fig. 2A). In contrast, the down-regulated genes in the UPF1 and SMG6 KD had a median 3′-UTR length similar to that of the human mRNAome. The exceptionally small median 3′-UTR length for genes down-regulated in SMG7 KD is highly uncertain due to only four genes in this set.
FIGURE 2.
Analysis of NMD-inducing features among the genes on the microarray. (A) The median 3′-UTR length is increased in genes up-regulated by UPF1, SMG6, and SMG7 knockdown (nts, nucleotides). (B) Genes harboring uORF(s) are enriched among the populations up-regulated upon UPF1 and SMG6 depletion. (C) The percentage of the genes that harbor at least one intron >50 nt downstream from the termination codon is elevated among the genes up-regulated by UPF1, SMG6, and SMG7 knockdown. The number of transcripts in each group is indicated in parentheses.
Next we looked at the possible enrichment of uORFs in the same subsets of microarray genes. To do so, first we obtained 5′-UTR sequences from the RefSeq database and identified uORFs. The criteria to define a uORF were the presence of an AUG followed by an in-frame stop codon upstream of the main ORF of the gene. According to this definition, 35.5% of all genes on the microarray do harbor at least one uORF (Fig. 2B). Genes up-regulated in UPF1 and SMG6 knockdown conditions have a higher percentage of such uORF-containing mRNAs. This is not the case for down-regulated genes or the genes up-regulated upon SMG7 depletion (Fig. 2B). It is not known at which of these uORFs the ribosome initiates. We predict that NMD substrate genes would be further enriched among transcripts with uORFs that indeed get translated.
Another extensively documented NMD-inducing feature is the presence of an intron >50 nt downstream from the termination codon. Initially we attempted to identify the genes that harbor such 3′-UTR introns from the RefSeq database. However, we realized that the RefSeq database is curated in such a way that it excludes many transcripts precisely because of the presence of one or more 3′-UTR introns, which makes the transcript a candidate substrate for NMD, and the curators therefore do not regard it as a physiologically meaningful mRNA isoform. Hence, we then turned to the Ensembl database (Flicek et al. 2011), which has a less-stringent curation policy in this respect. Our search revealed that 18.8% of genes on the microarray have at least one transcript variant that harbors one or several introns >50 nt downstream from the stop codon. The level of stabilization of a 3′-UTR-intron-containing transcript variant can be marred on the gene level due to the noise from the probe sets that belong to other, non-NMD substrate transcript variants of the same gene. Nonetheless, we found impressive enrichments for 3′-UTR introns in all subsets of up-regulated genes after KD of NMD factors (Fig. 2C).
It should be noted that secondary effects could greatly contribute to the observed changes since the microarray experiment was performed with RNA isolated from cells 4 d post-transfection of the shRNA-expressing plasmid. However, the overrepresentation of all NMD-inducing features among the genes that are stabilized in the cells depleted of NMD factors suggests that many of these genes are direct substrates for nonsense-mediated mRNA decay. It is possible that a number of up-regulated genes are under the control of a common transcription factor or other regulatory factor and are induced as a consequence of the stabilization of that common regulator. If so, these genes are likely to be functionally related. Such examples are provided in the study of coregulated sets of genes in yeast by Taylor et al. (2005). In addition, the genes of the same biological pathway could have evolved NMD-inducing features enabling a direct regulation through NMD. Using Gene Ontology (GO) annotation, we examined our lists of genes with the aim of identifying statistically significantly overrepresented biological processes. One of the leading GO categories enriched among up-regulated genes under NMD inhibition conditions is alternative splicing (AS). Strikingly, another top GO category was the NMD pathway itself, with most of the NMD factor-encoding mRNAs being up-regulated 1.5- to 3.8-fold. Whereas the link between AS and NMD is well established in that one-third of alternatively spliced transcripts contain PTCs (Lewis et al. 2003) and that many core spliceosomal components are under NMD regulation (Saltzman et al. 2008), the autoregulation of the NMD pathway has been less appreciated so far. It was shown that SMG5 and SMG6 were targets of feedback regulation in Drosophila cells (Rehwinkel et al. 2005), SMG7 and UPF3 were targeted by NMD in plants (Kerenyi et al. 2008; Saul et al. 2009), and SMG5 was induced under UPF1 depletion conditions in HeLa cells (Mendell et al. 2004; Chan et al. 2007), but in a different study of NMD inhibition by UPF2 KD in HeLa cells, the levels of UPF1, UPF3a, UPF3b, SMG5, and SMG6 remained unchanged (Wittmann et al. 2006).
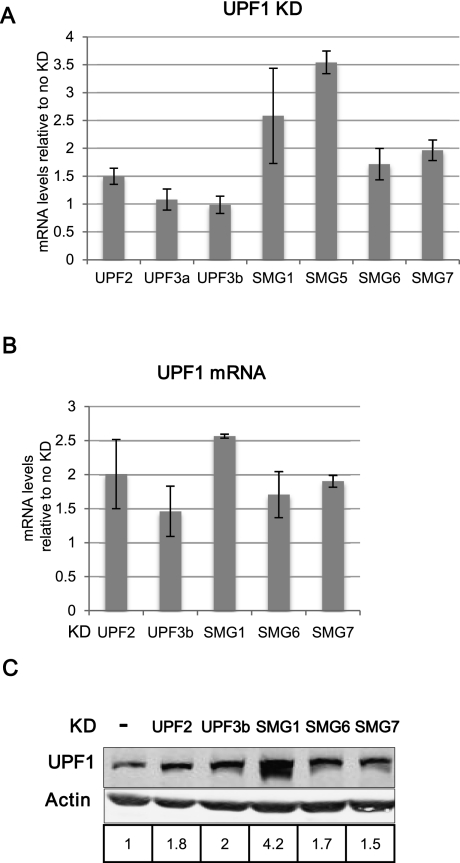
To validate the microarray data with respect to the apparent NMD factor autoregulation, we performed RT-qPCR of an array of NMD factors under UPF1 KD conditions. The mRNA levels of UPF2, SMG1, SMG5, SMG6, and SMG7 were reproducibly increased (Fig. 3A), whereas UPF3a and UPF3b levels remained unchanged in accordance with the microarray data. To assess how the UPF1 mRNA behaves, we abrogated the NMD process by knocking down several NMD factors such as UPF2, UPF3b, SMG6, and SMG7, whereupon a moderate but consistent increase of UPF1 mRNA was observed (Fig. 3B). This increased UPF1 mRNA expression resulted also in a higher intracellular abundance of the UPF1 protein (Fig. 3C).
FIGURE 3.
Autoregulation of NMD factors. The levels of mRNAs are normalized to β-actin, GAPDH mRNA, or 18S rRNA levels. (A) Transcripts encoding UPF2, SMG1, SMG5, SMG6, and SMG7 are up-regulated in UPF1 KD conditions. The average values and standard deviations of three biological replicates represent changes of mRNA upon UPF1 knockdown and were determined by RT-qPCR. (B) UPF1 mRNA is up-regulated when NMD is abrogated by knockdown of other NMD factors. Average values and standard deviations of two independent experiments represent the change of UPF1 mRNA levels upon KD of the indicated factors. (C) UPF1 protein levels increase when NMD is abrogated by knockdown of other NMD factors. The quantification of UPF1 protein amounts, relative to no KD control and normalized to β-actin levels, is displayed below the Western blots.
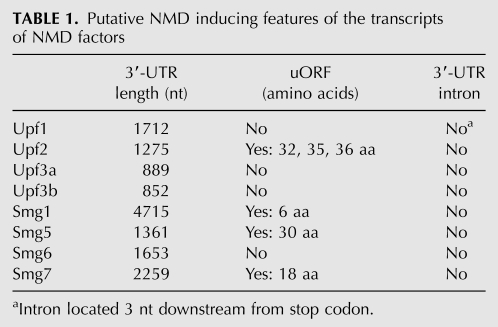
To understand the mechanism of NMD self-regulation, we analyzed the mRNAs of NMD factors for the presence of NMD-inducing features (Table 1). Remarkably, all of them have longer 3′ UTRs compared with the average 3′-UTR length of human mRNAs. The 3′-UTR lengths of the NMD factor-encoding mRNAs tend to correlate with the levels at which they are up-regulated by NMD abrogation (cf. Fig. 3A and Table 1): UPF3a and UPF3b with the shortest 3′ UTRs remained unchanged in UPF1 KD conditions, UPF2 with the next-shortest 3′ UTR of 1275 nt showed a mild increase, and the transcripts with 3′-UTR lengths >1300 nt (encoding UPF1, SMG1, SMG5, SMG6, and SMG7) were up-regulated between 1.5-fold and fourfold upon NMD inhibition. Besides the above average 3′-UTR sizes, UPF2, SMG1, SMG5, and SMG7 mRNAs also contain uORFs, whereas none of the investigated NMD factors harbors an intron >50 nt downstream from the stop codon.
TABLE 1.
Putative NMD inducing features of the transcripts of NMD factors
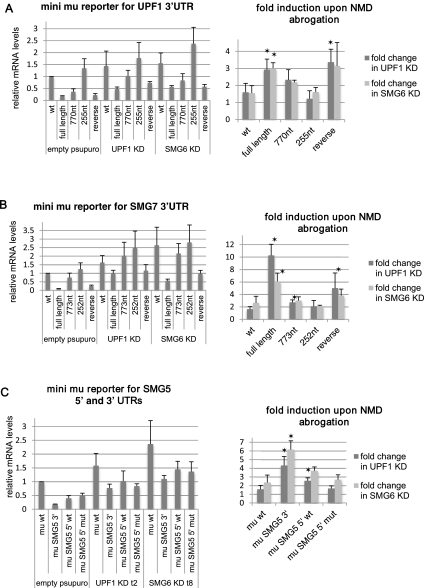
To test if the 3′ UTRs of the NMD factors can convert a reporter gene to a NMD substrate, we cloned the 3′ UTRs of UPF1 and SMG7 into the immunoglobulin mu minigene (mini mu) (Fig. 4; Buhler et al. 2004). The addition of full-length 3′ UTRs of UPF1 or SMG7 greatly reduced the levels of mini mu reporter mRNA, whereas the truncation of the trailers to ∼255 nt (which is similar to the mini mu wt reporter 3′-UTR length) did not influence reporter levels (Fig. 5A,B). An intermediate truncation of the 3′ UTRs to ∼800 nt showed intermediate levels of reporter transcript, consistent with our previous findings of a gradual decline of NMD efficiency with decreasing distance between the TC and the poly(A) tail (Eberle et al. 2008). To further test if the reason for the low reporter mRNA levels was, indeed, the 3′-UTR length rather than the nucleotide sequence, we measured the transcript levels of a reporter construct that contained the reverted full-length 3′-UTR sequence of UPF1 or SMG7. These reporters have identical 3′-UTR lengths compared with their corresponding full-length 3′-UTR reporters, but a different nucleotide sequence (Figs. 4E, 5A,B). Again, the transcript levels were low, comparable to those of the full-length reporter. A recent study has identified the neuron-specific microRNA miR-128 as a regulator of UPF1 and hence of NMD through a target sequence in the 3′ UTR of UPF1 (Bruno et al. 2011). However, the UPF1 3′-UTR-mediated reduction of reporter mRNAs in our HeLa cells occurred independently of the reported miR-128 regulation, since the reverse nucleotide sequence of UPF1 that no longer contains the miR-128 seed sequence still targeted the reporter transcript for degradation. Moreover, the mu reporter with the shortest UPF1 3′-UTR fragment still included the reported miR-128 binding site, yet its mRNA levels were similar to the wt mini mu mRNA. Collectively, our data are in agreement with the unified NMD model (Muhlemann 2008) by identifying the distance between stop codon and PABP as an important regulator of transcript stability. Importantly, the reduced levels of long-3′-UTR-containing mRNAs were increased when NMD was inhibited by knockdown of UPF1 or SMG6 (Fig. 5A,B). Noteworthy, the reference transcript mini mu wt was also slightly stabilized upon NMD inhibition, but the observed stabilization of reporter transcripts with full-length 3′ UTRs was significantly greater. Taken together, these data show that the long 3′ UTRs of the transcripts coding for the NMD factors UPF1 and SMG7 are sufficient to target their mRNAs for UPF1- and SMG6-dependent decay in a sequence-independent but length-dependent manner.
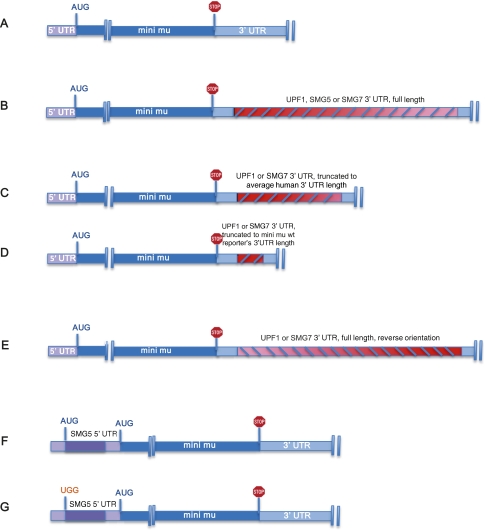
FIGURE 4.
Schematic illustration of UTR reporter constructs used in Figures 5 and 6. (A) Mini mu wild-type (wt) reporter with a 288-bp-long 3′ UTR. (B) Reporters with either UPF1 (1834 bp), SMG5 (1483 bp), or SMG7 (2381 bp) full-length 3′ UTRs. (C) Reporters with truncated UPF1 (770 bp) or SMG7 (773 bp) 3′ UTRs. The length of these 3′-UTR fragments is similar to the average human 3′-UTR length. (D) UPF1 (255 bp) or SMG7 (252 bp) 3′-UTR fragments corresponding to the wt reporter's 3′-UTR length. (E) The inverted sequence of the UPF1 (1834 bp) or SMG7 (2381) full-length 3′ UTR. (F) Reporter with the wt 5′ UTR of SMG5. (G) Reporter with a SMG5 5′ UTR in which the start codon of the uORF was mutated (gAUG to gUGG). The constructs are not drawn to scale.
FIGURE 5.
Untranslated regions of UPF1, SMG7, and SMG5 are sufficient to trigger NMD. The levels of mRNAs are normalized to cotransfected wild-type (wt) β-globin reporter levels. Asterisks indicate that the P-value of the fold change (Student's t-test) is <0.05 compared with the fold induction of the mu wt 3′-UTR reporter. Full-length but not truncated sequences of the UPF1 3′ UTR (A) and of the SMG7 3′ UTR (B) convert the mini mu reporter gene into a strong NMD substrate. Average values and standard deviations of four independent experiments are shown. (C) The 3′ UTR of SMG5 converts mini mu reporter to a strong NMD substrate (average of three experiments).
Next we tested the effect of a uORF on the reporter mRNA stability. In plants it was shown that uORFs can trigger NMD in a length-dependent manner (Nyiko et al. 2009). Therefore, we tested the 5′ UTR of SMG5, which has a 30-amino-acid-long uORF. We cloned the 5′ UTR of SMG5 upstream of the main ORF of the mu reporter gene, either with the intact uORF or with a uORF with a mutated AUG (Fig. 4F,G). In addition, we also created a reporter gene that harbors the 3′ UTR of SMG5 (Fig. 4B). Similar to UPF1 and SMG7 and consistent with the findings by Singh et al. (2008), the 3′ UTR of SMG5 significantly reduced mini mu reporter mRNA levels, and NMD abrogation led to a fourfold to sixfold increase of transcript levels (Fig. 5C). On the other hand, both wt and AUG-mutated 5′ UTRs of SMG5 slightly reduced mini mu reporter levels. The recovery upon NMD abolishment was similar to that of control mini mu transcripts with the wt uORF of SMG5 conferring a somewhat higher increase to the reporter (Fig. 5C). These results show that in the case of SMG5 mRNA, the long 3′ UTR is the main NMD-inducing feature, and the uORF contributes only marginally, if at all, to the NMD sensitivity of this transcript.
To test if the measured changes in steady-state levels of various mini mu reporter transcripts indeed reflect differential decay rates rather than changes at the transcription level, we determined the decay kinetics of these reporters using the Tet-Off Advanced System (Clontech). These assays showed that the wt mini mu reporter with a 3′ UTR of only 288 nt has an average half-life of 7.3 h. The addition of 3′ UTRs of UPF1, SMG5, and SMG7 accelerated the decay of the reporter mRNAs and resulted in half-lives of 3.9, 3.5, and 2.6 h, respectively (Fig. 6).
FIGURE 6.
Reduced half-lives of reporter mRNAs with 3′ UTRs of UPF1, SMG5, and SMG7. To determine the decay kinetics, the tet-off system was used. Relative mRNA levels were measured by RT-qPCR 0, 2, 4, and 8 h after stopping the transcription of the reporter gene by addition of doxycycline. Average values and standard deviations of three independent experiments are shown. The levels of mRNAs are normalized to cotransfected wild-type (wt) β-globin reporter levels.
To test if endogenous mRNAs coding for NMD factors are direct NMD substrates, we performed RNA immunoprecipitation (RIP) experiments.
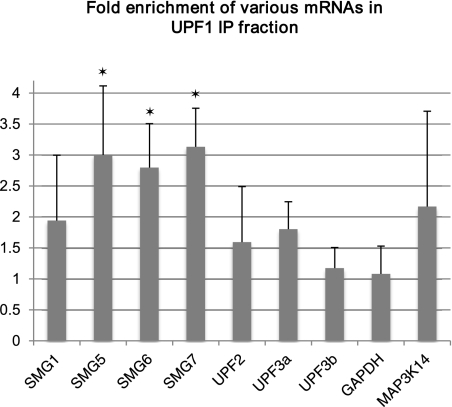
For this analysis, HeLa cells were transfected with Flag-tagged UPF1 protein (FLAG-UPF1) or, as a negative control, with Flag-tagged green fluorescent protein (FLAG-GFP). RNAs associated with tagged UPF1 and GFP proteins were coimmunoprecipitated from cell lysates using anti-Flag antibody, and the relative abundance of various mRNAs in these precipitates was determined. Relative to the GFP IP fraction, the mRNAs of SMG5, SMG6, and SMG7 were substantially enriched (threefold to 3.5-fold) in the UPF1 IP fraction, SMG1, UPF2, and UPF3a mRNAs as well as the mRNA of the well-known NMD substrate MAP3K14 (Mendell et al. 2004; Gardner 2008; Stalder and Muhlemann 2009) were moderately enriched (about twofold), and the mRNA levels of UPF3b and the negative control GAPDH were similar in the UPF1 and GFP IP fractions (Fig. 7).
FIGURE 7.
RNA immunoprecipitation with anti-UPF1 antibody. Fold enrichment of indicated mRNAs in the UPF1 immunoprecipitate fraction relative to negative control (GFP IP fraction) is shown. Relative mRNA levels were determined by RT-qPCR and were normalized to input levels of mRNA. Asterisks signify <0.05 P-value (Student's t-test) for enrichment compared with the enrichment of GAPDH mRNA in UPF1 immunoprecipitates.
Based on these data, we conclude that the NMD process is feedback-regulated in human cells, whereby most of the mRNAs coding for NMD factors are directly recognized as NMD substrates, most likely due to their long 3′ UTRs.
DISCUSSION
Using a genome-wide transcriptome profiling, the goal of this study was to identify and analyze the population of endogenous mRNAs that are substrates for the NMD pathway. Overall, ∼4% of the genes present on the microarray chip were up-regulated in our NMD-deficient HeLa cells, and only 1% were down-regulated. The strong enrichment for NMD-inducing features among the up-regulated transcripts suggests that a substantial fraction of these mRNAs are direct NMD substrates. Comparison of our results with other microarray data performed in HeLa cells either following UPF1 knockdown (Mendell et al. 2004) or stable depletion of UPF2 (Wittmann et al. 2006) showed that among the 197 genes up-regulated ≥1.9-fold in the former study, 35 were also significantly increased in our UPF1 KD (Supplemental Table II), whereas only four genes were in common between our UPF1-targeted transcripts and the 37 genes up-regulated twofold or more in the stable UPF2 KD. The rather low overlap between these different gene sets can be explained by different experimental setups as well as different statistical stringencies used to evaluate the data by the different laboratories. Another reason for the low overlap is the off-target effects when RNAi is used to achieve gene silencing. That was apparent in our microarray experiments where we used two different shRNA target sequences for knocking down each UPF1, SMG6, and SMG7: mRNA data sets that are differentially expressed as a result of the same NMD factor KD show widespread off-target effects. Our choice to work with the set of mRNAs that are differentially expressed in both KDs greatly decreased the number of false positives. Among 173 down-regulated genes in the study of Mendell et al. (2004), only three were found in our list of 155 UPF1-down-regulated genes. Notably, 26 of the transcripts with decreased abundance in our UPF1 depletion data set map to histone genes (Supplemental Table IA). For several histone transcripts, this UPF1-mediated down-regulation was validated in subsequent RT-qPCR assays (Supplemental Fig. 3). Such a profound change of histone levels has not been previously noted in microarrays performed by various laboratories in yeast, fly, and human cells (Lelivelt and Culbertson 1999; He et al. 2003; Mendell et al. 2004; Rehwinkel et al. 2005), possibly because these previous studies selectively detected the polyadenylated transcriptome, whereas our exon arrays detect also non-polyadenylated transcripts due to the use of randomly attaching primers. In contrast to our data, which suggest that UPF1 enhances the expression of histone genes and/or the stability of histone transcripts, UPF1 has previously been implicated in the degradation of histone mRNAs (Kaygun and Marzluff 2005).
The median 3′-UTR length for the 35 genes stabilized by UPF1 in our study and in the study of Mendell et al. (2004) is longer than the median human 3′-UTR length (903 compared to 765 nt, respectively), and it increases to 1298 nt if we consider only the mRNAs categorized under “unknown NMD-inducing feature” in the list of the Mendell paper. This impressive enrichment, together with the finding that (1) the mRNAs up-regulated by UPF1, SMG6, or SMG7 depletion have considerably longer median 3′-UTR lengths than the average human transcript and that (2) long 3′ UTRs of several mRNAs target the reporter transcripts for decay, reinforce the notion that long 3′ UTRs indeed constitute an important NMD-inducing feature of endogenous mRNAs. Consistent with this conclusion, “>2 kb long 3′ UTR” was also the most enriched NMD-inducing feature among the transcripts that were up-regulated by miR-128, which was shown to down-modulate NMD by targeting UPF1 in neuronal cells (Bruno et al. 2011). Moreover, our conclusion is further supported by a recent study demonstrating that UPF1 associated with mRNAs in a 3′-UTR-length-dependent manner and was highly enriched on transcripts containing 3′ UTRs known to elicit NMD (Hogg and Goff 2010). It was suggested that UPF1 preferentially accumulates on long 3′ UTRs, establishing an mRNP state primed for decay (Hogg 2011), although the mechanism that discriminates NMD-sensitive and NMD-immune mRNAs with similarly long 3′ UTRs has yet to be discovered.
It must be emphasized that according to the unified NMD model, a long 3′ UTR will not necessarily trigger NMD, because the physical distance between the termination codon and the PABP rather than the number of nucleotides in between is the crucial parameter (Eberle et al. 2008). Data from different laboratories indicate that a 3′ UTR of >420 nt can trigger NMD (Eberle et al. 2008; Singh et al. 2008). However, the majority of the ∼55% of human mRNAs with a 3′ UTR of >500 nt (Supplemental Table III) are obviously not NMD substrates. Many of these mRNAs probably have evolved means to avoid NMD. For example, spatial arrangements of the 3′ UTR that bring the PABP in proximity to the termination codon can suppress NMD (Eberle et al. 2008; Singh et al. 2008). In addition, mRNAs might possess NMD-inhibitory sequences similar to the stability element in the Rous sarcoma virus (Weil and Beemon 2006). Adding further complexity, long 3′ UTRs can accommodate numerous regulatory sequences, for example, microRNA binding sites (Cheng et al. 2009), which independently of NMD also affect the half-life of an RNA. Along the same lines, the other well-established NMD-inducing features such as downstream introns or uORFs do not always trigger NMD of the transcript (Romao et al. 2000; Singh et al. 2008). In summary, our still limited understanding of the molecular mechanisms resulting in NMD currently prevents reliable bioinformatics predictions of NMD substrates. In particular, the widely used prediction of NMD substrates solely based on the “50 nucleotides rule” (Nagy and Maquat 1998) will only reveal a fraction of the endogenous NMD substrates, and on top of that it will also produce a considerable number of false-positive predictions.
The mRNAs of NMD factors, however, are under direct surveillance of NMD as demonstrated in the reporter gene and RNA immunoprecipitation experiments (Figs. 5–7). Our experiments establish NMD as an autoregulatory process with a potential to swiftly establish the homeostasis of NMD factors. Such self-adjustment of NMD could consequently offer stability on the transcriptome level, given that up to 10% of mRNAs directly or indirectly are under NMD control. For example, metabolic or environmental fluctuations could influence NMD factor levels translating into greater insult at the transcriptome level. Tight control of NMD factor concentrations through autoregulation buffers such undesired consequences. Previously, individual NMD factor-encoding mRNAs have already been reported to be targeted by NMD in different organisms: SMG5 has been identified as an NMD target in human and fruit fly cells (Mendell et al. 2004; Rehwinkel et al. 2005; Chan et al. 2007), SMG7 and UPF3 in plants (Kerenyi et al. 2008; Saul et al. 2009), SMG6 in fruit fly (Rehwinkel et al. 2005), and UPF1 in mouse (Weischenfeldt et al. 2008). Thus the self-regulation of NMD that we demonstrated in this study for human cells likely extends to other organisms.
MATERIALS AND METHODS
Cell culture, transfections, and drug treatments
Plain HeLa cells or HeLa cells with stably integrated Tet-Off Transactivator (tTA-Advanced; Clontech) were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin (full medium). Transfections were generally performed in six-well plates. Two × 105 cells were seeded in 2 mL medium per well and transfected the next day with 100 ng of reporter plasmids, 400 ng of pSUPERpuro plasmid (Paillusson et al. 2005), and 3 μL of DreamFect (OZ Biosciences) according to the manufacturer's protocol. One day later, cells were exposed to 1.5 μg/mL puromycin for selection. After 2 d of selection for antibiotic resistance, cells were cultured in antibiotic-free media for another day before total RNA was isolated and whole cell lysates for immunoblotting were prepared. Total cellular RNA was isolated using either TRI-reagent or the “Absolutely RNA Miniprep Kit” (Stratagene). For transcript half-life determination experiments, the transcription of reporter genes was switched off via addition of doxycyline (Sigma-Aldrich) to a final concentration of 1 μg/mL, and RNA was isolated 0, 2, 4, and 8 h after transcription arrest.
Plasmids
For the knockdown experiments, short hairpin RNAs (shRNA) expressed from the pSUPERpuro plasmid (Paillusson et al. 2005) were used. The shRNA target sequences are provided in the Supplemental Material. The mini mu reporter gene transcripts were driven by the β-actin promoter from pβmini mu vector (Buhler et al. 2004). The 3′ UTRs of the UPF1, SMG5, and SMG7 genes were cloned into the 3′ UTR of mini mu wild type between ClaI and SpeI sites. The 5′ UTR of SMG5 was cloned into the SalI site of mini mu wild-type leader sequence. For the half-life measurements, the entire reporter constructs were inserted into NotI–XbaI of the pTRE-Tight vector (Clontech).
Quantitative real-time reverse transcription PCR (RT-qPCR)
In general, 1 μg of total RNA was reverse-transcribed in 20 μL of StrataScript 6.0 RT buffer containing 0.4 mM dNTPs, 300 ng of random hexamers, and 50 U of StrataScript 6.0 reverse transcriptase (Stratagene) according to the manufacturer's protocol. Reverse-transcribed material corresponding to 24 ng of RNA was amplified using Brilliant II Fast/Brilliant II Fast SYBR Green qPCR Master Mix (Stratagene) or KAPA PROBE FAST/KAPA SYBR FAST Universal qPCR Master Mix (Kapa Biosystems) and corresponding primers in a total volume of 15 μL with the Rotor-Gene 6000 rotary analyzer (Corbett). The primer and probe sequences are listed in the Supplemental Material.
Immunoblotting
Whole cell lysates corresponding to 105 cells per lane were electrophoresed on a 10% SDS-PAGE. Proteins were transferred to Optitran BA-S 85 reinforced nitrocellulose (Schleicher and Schuell) and probed with 1:1000 diluted polyclonal rabbit anti-hUpf1 antibody (sc-48802; Santa Cruz Biotechnology) or 1:3000 diluted polyclonal rabbit anti-actin antibody (A5060; Sigma-Aldrich). One-to-10,000 diluted donkey anti-rabbit IRDye800CW (LI-COR) was used as secondary antibody. The bands were visualized by scanning the membrane on an Odyssey Infrared Imager.
RNA immunoprecipitation
HeLa cells of ∼80% confluency grown in 10 mL of full medium in 75 cm2 flasks were transfected with Flag-tagged UPF1 or Flag-GFP (negative control) and an empty pSUPERpuro plasmid with 18 μL of DreamFect (OZ Biosciences) according to the manufacturer's protocol. The next day, cells were put under 1.5 μg/mL puromycin selection for 2 d. After another day of incubation without antibiotic, the cells were counted and 1.5 to 4 × 107 cells were cross-linked in 0.5% formaldehyde for 15 min at room temperature. After washing with cold PBS, cells were lysed in FA lysis buffer (50 mM HEPES-KOH at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, complete protease inhibitor [Roche]) containing RNase inhibitor (RNAsin, Promega; 80 U/500 μL of buffer), and then nucleic acids were fragmented by sonication. Before immunoprecipitation, the extract was treated with DNase (400–500 units of RNase-free DNase I [Sigma-Aldrich] per 500-μL extract) in 25 mM MgCl2, 5 mM CaCl2, and an additional 100 U of RNAsin for 10 min at 37°C. DNase I digestion was stopped by the addition of EDTA to 20 mM. Reactions were cleared by centrifugation at 16,100g in a microcentrifuge for 20 min. For input RNA, all following steps up to proteinase K digestion were omitted. Immunoprecipitation was performed by adding 80 μL of anti-Flag M2 agarose beads (Sigma-Aldrich) equilibrated in FA lysis buffer to the extracts and incubating head over tail overnight at 4°C. All subsequent precipitate washes were performed using standard ChIP buffers from Millipore supplemented with RNase inhibitor. Following the washes, two-step elution was performed, first using 1% SDS, 0.1 M NaHCO3 buffer at room temperature for 20 min followed by elution using 50 mM Tris/HCL, 20 mM EDTA, and 1.3% SDS buffer for 20 min at 60°C. The eluates were pooled and adjusted to 200 mM NaCl concentration, and 80 μg of proteinase K was added followed by incubation for 1 h at 42°C. Cross-links were reversed by heating for 3 h at 65°C. The reactions were extracted with TRI-reagent according to a standard TRIzol protocol for RNA isolation. The levels of transcripts of interest were determined by RT-qPCR.
Microarray analysis and bioinformatics
HeLa cells were transfected either with empty pSUPERpuro plasmid or pSUPERpuro encoding for short hairpin RNAs targeting two distinct sequences in each UPF1, SMG6, and SMG7 gene. Cells were selected for expression of knockdown plasmids, and 4 d post-transfection total cellular RNA was isolated with the Absolutely RNA Miniprep Kit (Stratagene). All RNA quantities were assessed by a NanoDropND-1000 spectrophotometer, and the RNA quality was assessed using RNA 6000 NanoChips with the Agilent 2100 Bioanalyzer (Agilent). For each sample, 1 μg of total RNA was amplified using the wt sense strand Target Labeling kit (Affymetrix, Cat. no. 900223); 5.5 μg of the resulting sense cDNA was fragmented by UDG (uracil DNA glycosylase) and APE 1 (apurinic/apyrimidic endonuclease 1), and biotin-labeled with TdT (terminal deoxynucleotidyl transferase) using the GeneChip WT Terminal labeling kit (Affymetrix Cat. no. 900671; Santa Clara). Affymetrix Human Exon 1.0 ST arrays (Affymetrix) were hybridized with 2.7 μg of biotinylated target, for 17 h at 45°C, washed, and stained according to the protocol described in the Affymetrix GeneChip Expression Analysis Manual (Fluidics protocol FS450_0001). The arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix), and raw data were extracted from the scanned images and analyzed with the Affymetrix Power Tools software package (Affymetrix). Statistical analyses were performed using the free high-level interpreted statistical language R and various Bioconductor packages (http://www.Bioconductor.org). Hybridization quality was assessed using the Expression Console software (Affymetrix). Normalized expression signals were calculated from Affymetrix CEL files using the RMA normalization method. Differential hybridized features were identified using the Bioconductor package “limma” that implements linear models for microarray data (Smyth 2004). The P-values were adjusted for multiple testing with Benjamini and Hochberg's method to control the false discovery rate (FDR) (Benjamini and Hochberg 1995). Probe sets showing at least 1.5-fold change and an FDR <0.05 were considered significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Sylvain Pradervand, Alexandra Paillusson, and Otto Hagenbüchle from the Genomic Technologies Facility of the University of Lausanne for running the microarrays and statistical analyses. We thank the members of the OM research group for discussions and assistance throughout the study. This work was supported by the European Research Council (ERC StG 207419), the Swiss National Science Foundation (grants 31003A-113878 and 31003A-127614), and by the Canton of Bern.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030247.111.
NOTE ADDED IN PROOF
While this paper was under review, Wilkinson and colleagues (Huang et al. 2011) also reported the autoregulation of NMD factor-encoding mRNAs by NMD.
REFERENCES
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A 2006. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. 2011. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 42: 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Paillusson A, Muhlemann O 2004. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res 32: 3304–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, Wilkinson MF 2007. An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Bhardwaj N, Gerstein M 2009. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics 10: 431 doi: 10.1186/1471-2164-10-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW 1995. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92 doi: 10.1371/journal.pbio.0060092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2011. Ensembl 2011. Nucleic Acids Res 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB 2008. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol 28: 3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. 2009. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet 5: e1000525 doi: 10.1371/journal.pgen.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Hogg JR 2011. This message was inspected by Upf1: 3′UTR length sensing in mRNA quality control. Cell Cycle 10: 372–373 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43:950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kebaara BW, Atkin AL 2009. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 37: 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D 2008. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D 2006. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34: 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 19: 6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 100: 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF 2007. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci 76: 5134–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J 1981. Unstable β-globin mRNA in mRNA-deficient β0 thalassemia. Cell 27: 543–553 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18: 2932–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann O 2008. Recognition of nonsense mRNA: Towards a unified model. Biochem Soc Trans 36: 497–501 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyiko T, Sonkoly B, Merai Z, Benkovics AH, Silhavy D 2009. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol Biol 71: 367–378 [DOI] [PubMed] [Google Scholar]
- Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O 2005. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res 33: e54 doi: 10.1093/nar/gni052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant EP, Wang P, Jacobs JL, Dinman JD 2004. A programmed −1 ribosomal frameshift signal can function as a cis-acting mRNA destabilizing element. Nucleic Acids Res 32: 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E 2005. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E 2006. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P 2006. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol 26: 6347–6356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao L, Inacio A, Santos S, Avila M, Faustino P, Pacheco P, Lavinha J 2000. Nonsense mutations in the human β-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood 96: 2895–2901 [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul H, Elharrar E, Gaash R, Eliaz D, Valenci M, Akua T, Avramov M, Frankel N, Berezin I, Gottlieb D, et al. 2009. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J 60: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111 doi: 10.1371/journal.pbio.0060111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2008. The meaning of nonsense. Trends Cell Biol 18: 315–321 [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2009. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R, Kebaara BW, Nazarenus T, Jones A, Yamanaka R, Uhrenholdt R, Wendler JP, Atkin AL 2005. Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway. Eukaryot Cell 4: 2066–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE 2007. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res 35: 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil JE, Beemon KL 2006. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22: 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Jacobson A 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J 18: 6134–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]