Pre-mRNA splicing and transcription are thought to be coupled by the C-terminal domain (CTD) of the large subunit of RNA polymerase II (Pol II). In yeast, the U1 snRNP subunit Prp40 has been proposed to mediate cotranscriptional recruitment of early splicing factors through binding of its WW domains to the Pol II CTD. In this article, the role of Prp40’s WW domains was investigated. The Prp40 WW domains were dispensable for yeast viability. In their absence, no defect in splicing in vivo, U1 or U2 snRNP recruitment in vivo, or early splicing complex assembly in vitro was detected. Accordingly, the WW domains of Prp40 do not mediate essential coupling between U1 snRNP and Pol II.
Keywords: U1 snRNP, commitment complexes, RNA polymerase II, protein interaction
Abstract
Complex cellular functions involve large networks of interactions. Pre-mRNA splicing and transcription are thought to be coupled by the C-terminal domain (CTD) of the large subunit of RNA polymerase II (Pol II). In yeast, the U1 snRNP subunit Prp40 was proposed to mediate cotranscriptional recruitment of early splicing factors through binding of its WW domains to the Pol II CTD. Here we investigate the role of Prp40 in splicing with an emphasis on the role of the WW domains, which might confer protein–protein interactions among the splicing and transcriptional machineries. Affinity purification revealed that Prp40 and Snu71 form a stable heterodimer that stably associates with the U1 snRNP only in the presence of Nam8, a known regulator of 5′ splice site recognition. However, the Prp40 WW domains were dispensable for yeast viability. In their absence, no defect in splicing in vivo, U1 or U2 snRNP recruitment in vivo, or early splicing complex assembly in vitro was detected. We conclude that the WW domains of Prp40 do not mediate essential coupling between U1 snRNP and Pol II. Instead, delays in cotranscriptional U5 snRNP and Prp19 recruitment and altered spliceosome formation in vitro suggest that Prp40 WW domains assist in late steps of spliceosome assembly.
INTRODUCTION
Splicing, the removal of noncoding introns from pre-mRNA and joining together of exons, is an essential event in eukaryotic gene expression. This process is performed effectively and precisely by the spliceosome, which assembles on each intron from five small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP factors (Wahl et al. 2009). In the yeast Saccharomyces cerevisiae, U1 snRNP first recognizes the 5′ splice site and then associates with Mud2p and Msl5/SF1/BBP (hereafter BBP) bound to the branchpoint, forming, respectively, the commitment complexes CC1 and CC2 (CC) (Ruby and Abelson 1988; Séraphin and Rosbash 1989, 1991; Berglund et al. 1997). Pre-spliceosomes result from the ATP-dependent joining of U2 snRNPs. Subsequent association of the U5/U4•U6 tri-snRNP forms the mature spliceosome. Several rearrangements occur, including displacement of the U1 snRNP from the 5′ splice site and release of the U4 snRNP. These changes allow the formation of new RNA–RNA interactions, such as pairing of the U5 and U6 snRNAs with the 5′ splice site (Newman and Norman 1992; Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993; Staley and Guthrie 1998). These changes, promoted by factors joining the spliceosome such as RNA helicases or the Nineteen Complex (NTC), signal spliceosome activation and catalysis of the two transesterification steps to form mature mRNA (Tarn et al. 1993; Staley and Guthrie 1998).
In vivo, spliceosome assembly occurs while the nascent RNA is still attached to chromatin via RNA polymerase II (Pol II), i.e., cotranscriptionally (Görnemann et al. 2005; Lacadie and Rosbash 2005; Tardiff et al. 2006). Recently, it was shown that transcriptional pausing is linked to cotranscriptional splicing (Alexander et al. 2010; Carrillo Oesterreich et al. 2010; Oesterreich et al. 2011). However, the extent to which recruitment of spliceosomal components is influenced by direct interactions with Pol II remains unclear. Intron recognition mediated by the U1 snRNP plays a crucial role in triggering subsequent steps of spliceosome assembly and intron removal. U1 snRNP accumulates cotranscriptionally on intron-containing genes in yeast shortly downstream from the 5′ splice site within the intron, indicating rapid interactions with 5′ splice sites sequences in vivo (Kotovic et al. 2003; Görnemann et al. 2005; Lacadie and Rosbash 2005; Tardiff and Rosbash 2006). Thus, the recruitment of the U1 snRNP to the 5′ splice site, the first step in committing the nascent pre-mRNA to splicing, is a potential site for communication between transcription and spliceosome assembly.
The core of the U1 snRNP, consisting of the U1 snRNA, seven Sm proteins, U1-A/Mud1p, U1-C/Yhc1p, and U1-70k/Snp1p, is conserved among yeast and metazoans (Hochleitner et al. 2005; Pomeranz Krummel et al. 2009). Seven additional proteins are stably associated with the U1 snRNP in yeast: Prp39p, Prp42p, Luc7p, Snu71p, Nam8p, Snu56p, and Prp40p (Neubauer et al. 1997; Gottschalk et al. 1998; Fortes et al. 1999; Rigaut et al. 1999). Some of these U1 snRNP proteins have been implicated in splice site selection, a task that in multicellular organisms is regulated by splicing factors that are not stably snRNP-associated. Metazoan orthologs of the yeast U1 snRNP proteins (e.g., yLuc7p/hsLuc7, Nam8p/TIA-1) interact weakly with the U1 snRNP (Puig et al. 1999, 2007; Förch et al. 2000). All U1 snRNP components, with the exception of Mud1p and Nam8p, are essential in yeast, and it is likely that the less well-characterized yeast U1 snRNP proteins also have orthologs in mammals (e.g., Fortes et al. 2007).
Prp40p is a potential integrator of splicing and transcription, because it was reported to bind directly to the Pol II CTD (Morris and Greenleaf 2000). Sequence analyses reveal that Prp40p is composed of several domains, namely, two N-terminal WW domains and four FF domains (Fig. 1A; Bedford and Leder 1999; Sudol et al. 2001). Pol II CTD binding to Prp40 was ascribed to the WW domains and was proposed to promote intron recognition through the juxtaposition of splice sites (Morris and Greenleaf 2000). Prp40 homologs and the Pol II CTD have been reported to interact in other animal and plant species, suggesting that this event is part of a conserved mechanism (Lin et al. 2004; Kang et al. 2009). However, analyses of the yeast Prp40p WW-domain structure and binding specificities questioned whether the Prp40 WW domains are capable of binding directly to the Pol II CTD (Wiesner et al. 2002). Interestingly, the WW domains have also been proposed to bind BBP and the N-terminal part of the U5 snRNP factor Prp8p in yeast, through PPxY motifs present in these two proteins (Abovich and Rosbash 1997; Wiesner et al. 2002), possibly bridging interactions across the intron during spliceosome formation. Interaction of branchpoint bound BBP with the Prp40 subunit of U1 snRNP might lead to the formation of the first spliceosomal complex that involves the 5′ and the 3′ splice sites, namely, commitment complex 2 (CC2) (Abovich and Rosbash 1997). The interaction of Prp40p with Prp8p may help recruit U5 snRNP/tri-snRNP to the pre-mRNA, thereby promoting full spliceosome assembly. Thus, it is currently unclear whether Prp40p plays a role in U1 snRNP recruitment to nascent transcripts via CTD interactions and/or whether it plays a later role in spliceosome maturation.
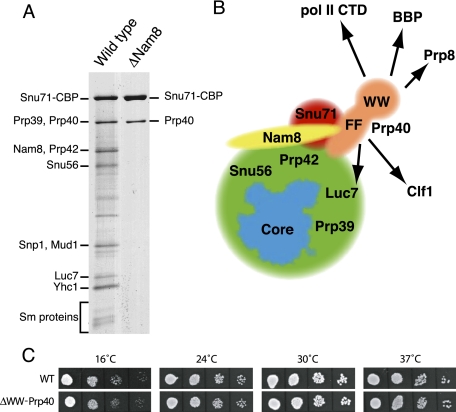
FIGURE 1.
Organization of the protein subunits of the yeast U1 snRNP and growth phenotype of the ΔWW-Prp40 mutant. (A) Profiles of the proteins purified from wild-type and ΔNam8 strains carrying a TAP-tagged Snu71 gene following the TAP strategy. Proteins were fractionated on denaturing exponential gradient gels and stained with silver. Stronger bands represent subunits (names indicated), while fainter bands are likely to represent degradation products. (B) Schematic representation of the yeast U1 snRNP organization. (Blue) The core conserved between yeast and human and containing the U1 snRNA, U1C, U1-70k, U1A, and the Sm subunits is schematized following the contour of the human particle (Pomeranz Krummel et al. 2009). The Prp40 (orange), Nam8 (yellow), and Snu71 (red) are indicated individually, while the other yeast U1 snRNP subunits are not differentiated (green). Factors that have been proposed to interact with the FF or WW domains of Prp40 are indicated (arrows). (C) Growth of wild-type and Prp40Δ strains at different temperatures: Serial dilutions of liquid cultures were spotted on solid medium and incubated at the indicated temperatures for 2 d (4 d at 16°C).
The U1 snRNP protein Nam8 was previously shown to be an important regulator of intron recognition through its binding to nonconserved sequences located downstream from the 5′ splice site (Puig et al. 1999). During the course of our analysis of the role of Nam8p in splicing, we noticed that the stable association of Prp40p and Snu71p with the U1 snRNP required the presence of Nam8p. This prompted us to investigate whether these factors, and particularly the WW domain of Prp40, could somehow also be involved in regulating intron recognition, e.g., through an interaction with the Pol II CTD or BBP. Our analyses demonstrate that the WW domains of Prp40 are not essential for viability. This allowed us to evaluate their in vivo and in vitro roles in U1 snRNP function, spliceosome assembly, and splicing.
RESULTS
An Snu71–Prp40 module interacts with the remainder of the U1 snRNP in an Nam8-dependent manner
While investigating the role of Nam8 in pre-mRNA splicing, we sought to compare the U1 snRNP protein composition in the presence and absence of Nam8. The U1 snRNP was purified using an Snu71-TAP tag fusion in either wild-type or Nam8Δ backgrounds. To our surprise, only two protein bands were reproducibly detected from Snu71-TAP purifications performed in the absence of Nam8 (Fig. 1A, lane 2). This effect was specific, because the normal U1 snRNP protein profile was recovered when Snu71-TAP was purified from a strain expressing Nam8 (Fig. 1A, lane 1) or from a strain lacking Mud1 (data not shown). The absence of the other U1 snRNP subunits cannot be attributed to their instability in the absence of Nam8. Indeed, U1 snRNP is still functional in the absence of Nam8, and native gel analyses indicate that its mobility is not significantly altered (Puig et al. 1999).
The larger of the proteins recovered in the absence of Nam8 comigrated with the largest subunit of the U1 snRNP purified in the presence of Nam8. This protein has been previously identified as Snu71-CBP. Its presence was expected because this factor carries the TAP tag that was used for the purification. The second band migrated at the size of Prp39 and Prp40. Mass spectrometry analyses revealed that the second band corresponded to Prp40. No peptides originating from Prp39 were identified. This result is consistent with the description of a direct interaction between Snu71 and Prp40 (Ester and Uetz 2008) and the previous report that Snu71 integration in U1 snRNP is impaired in an Nam8 mutant strain (Gottschalk et al. 1998). However, the latter study suggested that, in the absence of Nam8, Prp40 was only released from U1 snRNP at 450 mM salt, while we observed that Snu71 and Prp40 were simultaneously released at 150 mM salt. Our results are consistent with the presence of an Snu71–Prp40 heterodimer that stably interacts with the remainder of the U1 snRNP only in the presence of Nam8. While we cannot exclude that Nam8 might induce a long-distance conformational modification of the U1 snRNP favoring the docking of Snu71/Prp40, the simplest explanation of our data is that Snu71 and/or Prp40 contact several subunits of the U1 snRNP, including Nam8; in the absence of Nam8, the remaining interactions are too weak to withstand the TAP purification procedure (Fig. 1B). Interestingly, this suggests that Prp40 and Snu71 are located close to Nam8 and could, in a manner similar to the latter factor, modulate intron recognition. This possibility suggested that the Prp40 WW domains—proposed to interact with the CTD of Pol II, BBP and Prp8—could mediate networks of interactions during cotranscriptional splicing.
Construction of a yeast strain lacking the Prp40 WW domains
We used a pop-in/pop-out strategy to replace one PRP40 allele of a wild-type diploid strain by a version lacking amino acids 2–76 of Prp40. After sporulation and dissection of the resulting strain, haploid spores containing the ΔWW-Prp40 were recovered at normal frequencies and showed no strong growth phenotypes. Because Prp40 is essential for yeast viability, this result indicated that the Prp40 WW domains are not required for cell growth. To assess whether deletion of the WW domains of Prp40 produces a subtle phenotype, we compared the growth of the mutant and an isogenic wild-type strain at different temperatures (Fig. 1C). This failed to reveal any significant difference associated with the removal of the Prp40 WW domains. Because the WW domains deletion was introduced at the chromosomal PRP40 locus, the absence of phenotype does not result from the overexpression of a partially active protein. These results suggest that the Prp40 WW domains are not essential for expression of spliced transcripts in cells; nevertheless, Prp40 WW domains might modulate splicing efficiency or specificity in a nonessential manner.
The WW domain is not required for U1 snRNP association
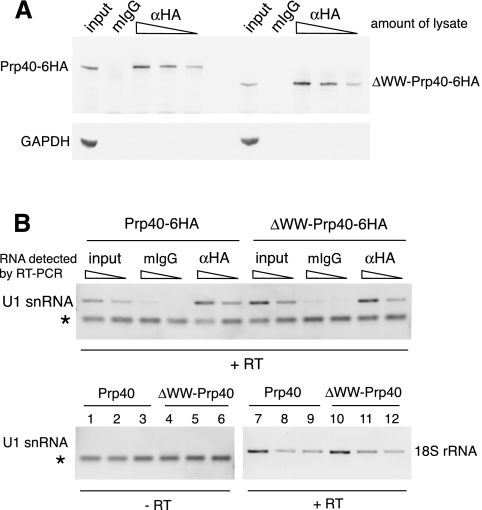
To determine whether Prp40 lacking the WW domain is incorporated into the U1 snRNP, we introduced an HA-tag at the C terminus of the truncated protein and, as a control, to the C terminus of its wild-type counterpart. HA-tagged Prp40 was immunoprecipitated, and samples were analyzed for the coprecipitation of U1 snRNA (Fig. 2). The amount of protein in the lysate from the ΔWW-Prp40 strain and its isogenic wild type was similar, and comparable amounts of Prp40 were pulled down through the HA-epitope. This demonstrates that the stability of ΔWW-Prp40 was unchanged compared with the full-length protein. Both full-length and ΔWW-Prp40 pulled down substantial, comparable amounts of U1 snRNA. We conclude that ΔWW-Prp40 is efficiently and stably incorporated in the U1 snRNP, to an extent similar to the full-length protein. This is consistent with recent findings (Ester and Uetz 2008), suggesting that Prp40 interacts with other U1 snRNP components, namely the proteins Snu71 and Luc7, through its FF domains.
FIGURE 2.
Prp40 and U1 snRNA are found in the same complex in wild-type and ΔWW-Prp40 cells. (A) Full-length and truncated Prp40 have comparable expression levels. C-terminally HA-tagged Prp40 was immunoprecipitated from whole cell extracts and analyzed by Western blotting. (Upper panel) Detection of Prp40 with mouse-anti-HA antibody (12CA5). (Lower panel) Detection of GAPDH in the input samples as a loading control. The changing amounts of immunoprecipitated Prp40 from different amounts of lysate used demonstrate that the lysates do not appear to contain the same amounts of Prp40 due to saturation of binding sites. The input is 1:20 volume of the highest input, fraction of lysate in the samples: 2:3, 1:3, 1:9, unspecific antibody: 2:3. (B) Full-length and ΔWW-Prp40 coimmunoprecipitate similar amounts of U1 snRNA. Coimmunoprecipitated RNAs were reverse-transcribed and analyzed by PCR using specific primers for U1 snRNA and 18S rRNA. Lanes in the lower panel: 1,4,7,10: input; 2,5,8,11: mIgG (unspecific antibody); 3,6,9,12: αHA antibody. Amount of cDNA used in the PCR reactions: input: 0.3 μL and 0.1 μL (U1 snRNA), 0.03 μL (18S RNA); IP samples: 1 μL and 0.3 μL (U1 snRNA), 0.1 μL (18S RNA). For the αHA samples, only those originating from the IP using 1:9 volume of lysate are shown. Of −RT samples, 1 μL was used in the PCR. 1:5 of each reaction was loaded on the gel. PCR with 18S rRNA primers based on −RT templates did not amplify detectable amounts of DNA (data not shown). The starred band is a nonspecific by-product of the U1 primers (primer dimer).
Analysis of commitment complex and spliceosome formation
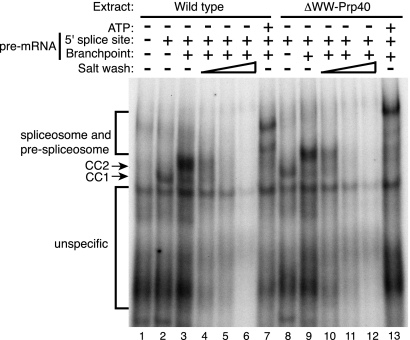
The activity of U1 snRNP in yeast splicing complex formation can be assayed by following the formation of two commitment complexes, CC1 and CC2, using native gels. We tested the formation of commitment complexes in extracts prepared from the wild-type strain and from the ΔWW-Prp40 strain. CC1 formation occurred similarly in both extracts when using a substrate lacking a functional branchpoint (Fig. 3). Interestingly, both extracts were competent for the formation of CC2 when incubated with a wild-type pre-mRNA substrate in the absence of ATP (Fig. 3). This result was intriguing because the WW domains of Prp40 have been suggested to bridge the interaction between the U1 snRNP and BBP that occurs in CC2. Similar ratios of CC2 to CC1 were observed in the two extracts (1.06 vs. 1.01), suggesting that the contribution of the Prp40 WW domains to CC2 formation, if any, was modest. To detect a minor contribution of the proposed interaction of the WW domains of Prp40 with BBP to the CC2 conformation, we analyzed whether the stability of CC2 was altered in the absence of the Prp40 WW domain. To do so, CC2 formation was performed as described above, but increasing amounts of salt were added after the reactions had been stopped. As observed previously, commitment complexes were destabilized at high salt concentrations (Séraphin and Rosbash 1989, 1991). However, we did not observe significant differences between the stability of CC2 in the presence or absence of the Prp40 WW domains. These observations demonstrate that the interaction reported between the Prp40 WW domains and BBP was required neither for commitment complexes formation nor the stability of these assemblies. Nevertheless, spliceosome assembly was slightly altered in the ΔWW-Prp40 mutant strains compared with wild type (lane 13 vs. lane 7). In these gels, (pre-)spliceosomes appear as two bands (Séraphin and Rosbash 1989), and the slower migrating band, which still contains U1 snRNP, is over-represented in the mutant extract compared with wild type (the ratio of slower migrating [pre-]spliceosome to faster migrating species is 0.97 in wild type and 1.23 in mutant). This indicates that absence of the WW domains of Prp40 affects the late steps of spliceosome assembly.
FIGURE 3.
Native gel assay of spliceosomal complex formation and stability in wild-type and ΔWW-Prp40 strains. Formation of commitment complexes and spliceosomes was assayed by a native gel using a pre-mRNA substrate mutated in its 5′ splice site and branch point (lane 1), only in its branch point (lanes 2,8) or with a wild-type substrate (all other lanes). Pre-mRNAs were incubated in extracts prepared from wild-type (lanes 1–7) or ΔWW-Prp40 strains in the absence of ATP, except for lanes 7 and 13, where ATP was included to allow the formation of the spliceosome. The various splicing complexes are labeled on the left. To monitor the stability of commitment complexes 2 (CC2), KCl was added after the completion of the reaction (corresponding to lanes 3 and 9) to a final concentration of 100 mM (lanes 4,10), 150 mM (lanes 5,11), or 200 mM (lanes 6,12). Note that the apparent slight difference in complex mobility between the wild-type and ΔWW-Prp40 reactions is in part due to gel smiling (see other background bands) in this particular experiment. However, we cannot exclude that commitment complexes containing the mutant protein have a slightly retarded migration, which may partly be related to the poor efficiency of the latter steps of spliceosome assembly.
Spliceosome assembly in vivo
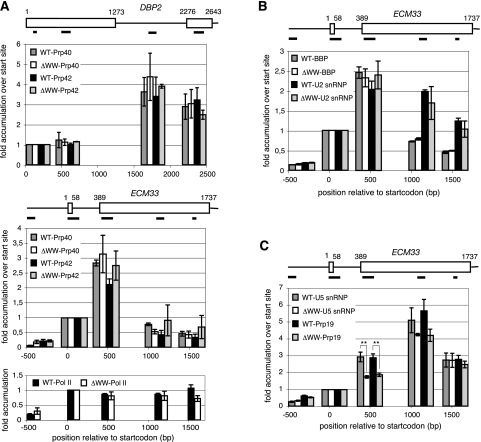
Because our in vitro analyses failed to demonstrate a role for the Prp40 WW domain in early intron recognition events and suggested that later steps were affected, we monitored cotranscriptional spliceosome assembly in vivo. This assay should be sensitive to alterations in the interactions of Prp40 with several of its potential partners including BBP, Prp8, and the CTD of Pol II. To this end, we used a previously established chromatin immunoprecipitation (ChIP) assay, relying on the coprecipitation of target gene regions with cotranscriptionally assembled splicing complexes (Görnemann et al. 2005). The ChIP assay yields temporal resolution of spliceosome assembly, based on the assumption that the complexes are attached to Pol II via the nascent RNA and thus reflect assembly along the transcription unit as Pol II proceeds. We used tagged components of the U1 snRNP (Prp42 and Prp40), the U2 snRNP (Msl1), and the U5 snRNP (Brr2), as well as the non-snRNP splicing factors BBP, Mud2, and Prp19 to monitor the formation of the various splicing complexes. For this purpose, the sequence encoding the HA-epitope was fused to the endogenous genes both in the wild-type and isogenic ΔWW-Prp40 strains. The distribution of these spliceosome components along the intron-containing gene ECM33 was determined. For the U1 snRNP proteins Prp40 and Prp42, we also assayed their presence along the DBP2 split gene.
The patterns of DNA coimmunoprecipitation with the two U1 snRNP proteins were the same in both strain backgrounds (Fig. 4). The signals peak ∼500 bp downstream from the 5′ splice site on both genes, then decline further downstream. This pattern of accumulation reflects the association of U1 snRNP with the transcribed pre-mRNA once the 5′ splice site is available and its later replacement during the final stages of spliceosome assembly (Kotovic et al. 2003). This confirms that in the absence of the Prp40 WW domains, the U1 snRNP is intact (see above). The same accumulation profile is seen for BBP and Mud2 (data not shown) in both yeast strains. Moreover, U2 snRNP accumulation along these genes was not altered in the mutant strain. Thus, deletion of the WW domain from Prp40 does not impair binding of the U1 snRNP to the pre-mRNA, the recruitment of BBP and Mud2 to the branchpoint, or U2 snRNP accumulation. We conclude that the Prp40 WW domain is not required for proper commitment complex and pre-spliceosome formation in vivo.
FIGURE 4.
In vivo spliceosome assembly does not differ between wild-type (WT) and ΔWW-Prp40 strains. (A) The pattern of Prp40 and Prp42 accumulation is indistinguishable in wild-type and ΔWW-Prp40 background on two intron-containing genes. ChIP analysis of the distribution of the U1 snRNP proteins Prp40 and Prp42 along the two intron-containing genes DBP2 and ECM33 reveals no difference of either profile in WT and ΔWW-Prp40 strains. No difference is observed between Pol II profiles in the two strains. DBP2: The data represent the average of three independent experiments, except for Prp40/ΔWWPrp40 with n = 4. ECM33: Prp40/WT: n = 4; Prp40/ΔWWPrp40: n = 6; Prp42/WT: n = 5; Prp42/ΔWWPrp40: n = 4; Pol II/WT and Pol II/ΔWWPrp40: n = 3. (B,C) BBP, the U2 snRNP (Msl1), U5 snRNP (Brr2), and Prp19 display a similar distribution along ECM33 in wild-type and ΔWW-Prp40 background. The binding pattern of BBP is the same as for the U1 snRNP components, with a peak ∼500 bp downstream from the 5′ splice site and then a pronounced decline toward the 3′ end of the gene, whereas the U2 snRNP shows its maximal accumulation after the 3′ splice site is made and then persists. No distinction between the two strains can be seen. The signal of the U5 snRNP and Prp19 is maximal ∼700 bp downstream from the 3′ splice site, to then slowly go down again, but a significant decrease is seen for the factors in the ΔWW-Prp40 background; (**) p < 0.005 by Student's t-test. The same number of independent experiments were done in both backgrounds for each factor with BBP: n = 3; U2 snRNP (Msl1): n = 3; U5 snRNP (Brr2): n = 4; and Prp19: n = 5. Diagrams representing the genes DBP2 and ECM33 show the positions of the PCR products used for analysis. In the panels below these diagrams, histogram bars are placed according to the positions of the PCR products along the gene. The levels of accumulation are given relative to the start site-proximal position. All replicates are biological replicates. Error bars represent SEM.
U5 snRNP and Prp19 accumulation differs significantly between the ΔWW-Prp40 background and the isogenic wild type. While the general shape of the binding pattern is preserved for both spliceosomal complexes with peaks ∼700 bp downstream from the 3′ splice and then a decrease toward the 3′ end of the gene, the onset of binding is delayed in the ΔWW-Prp40 strain. In the wild-type strain, an increase in U5 snRNP signal was observed just downstream from the 3′ splice site (Fig. 4). In the ΔWW-Prp40 background, we also detected U5 snRNP and Prp19 recruitment at this site, but they were significantly reduced compared with wild type. This suggests a delay in U5/tri-snRNP recruitment that, in turn, could delay NTC association. However, the downstream peaks of U5 snRNP and NTC were similarly positioned among the two strains, suggesting that delays in spliceosome assembly steps are compensated during the process of transcription.
The ChIP analysis indicates that the lack of the WW domain of the U1 snRNP protein Prp40 does not dramatically impair spliceosome assembly in vivo under standard conditions of growth at 30°C in rich medium. Despite the proposed role of the direct interaction between BBP and the WW domain of Prp40 in cross-intron bridging and commitment complex formation, binding of CC factors does not differ between the two yeast strains. Apparently, there is also no functional difference, because the spliceosome formation proceeds to addition of the U2 snRNP and beyond. The delay and reduction in signal from the U5 snRNP might result from the impaired interaction between the WW domain and the U5 snRNP protein Prp8. Consistent with this interpretation, ChIP analyses confirm that removal of the WW domain of Prp40 also delays the onset of joining of Prp8 in vivo (data not shown). However, this slow joining appears not to be sufficient to prevent spliceosome assembly. Overall, this model is consistent with the lack-of-growth phenotype that would have been expected if a significant splicing defect had occurred.
Removal of the Prp40 WW domains does not impact on splicing or induce pre-mRNA leakage
We further tested potential effects of the Prp40 WW domain on spliceosome formation and splicing, by analyzing the steady-state levels of several endogenous (pre-)mRNAs. Measurement of steady-state mRNA and pre-mRNA levels is a sensitive indicator of splicing defects. To this end, total RNA was reverse-transcribed using random hexamer primers, and the resulting cDNAs were analyzed by quantitative PCR using primer sets specific for the exon junction (“spliced”), the intron (“unspliced”), and the second exon (“total”). We analyzed several transcripts using U6 snRNA for normalization and found differences in levels of unspliced ECM33, DBP2, and SAC6 mRNAs between ΔWW-Prp40 and wild-type strains of 1.07 ± 0.32, 0.86 ± 0.07, and 1.16 ± 0.10, respectively; values for spliced mRNA were 1.07 ± 0.002, 1.02 ± 0.11, and 1.02 ± 0.10, respectively. Thus, this analysis revealed no significant decrease in splicing of endogenous transcripts due to Prp40 WW-domain deletion at steady state.
To increase the sensitivity of this approach, we introduced reporter plasmids encoding splicing reporters in the wild-type and mutant strains. This allowed a quantitative assessment of splicing through the quantification of β-galactosidase, including in the case of a poorly spliced, and thus sensitized, reporter (Legrain and Rosbash 1989). Moreover, we also analyzed a construct reporting pre-mRNA leakage that would reveal defects and/or delays in splicing complex formation (Legrain and Rosbash 1989). Figure 5 shows that splicing of neither the wild-type RP51A intron nor a poorly spliced synthetic construct was significantly affected by the absence of the Prp40 WW domains. Importantly, leakage of the reporter pre-mRNA to the cytoplasm was not stimulated when the WW domains of Prp40 were missing. Analysis of an additional set of 11 reporters, including sensitized constructs carrying duplicated splice sites and/or introns sensitive to the activity of Nam8 (Puig et al. 1999), failed to uncover an effect of the Prp40 WW domain in splicing (Fig. 5B). This contrasts with previous analyses, which demonstrated that sensitized reporters constitute a powerful tool to detect subtle splicing phenotypes (Luukkonen and Séraphin 1997; Dziembowski et al. 2004).
FIGURE 5.
Analysis of the splicing efficiency or pre-mRNA leakage of different reporters in wild-type or ΔWW-Prp40 strains. (A) Histogram reporting the results of β-galactosidase assays from the reporters carrying no LacZ gene (“background”), the LacZ reporter gene with no intron (“No intron”), the LacZ reporter gene interrupted by a simple intron (“Wild type RP51A intron”), a weak synhetic intron or pre-mRNA leakage expressed either in a wild type yeast strain (hatched bars), or in a ΔWWPrp40 strain (gray bars). The reporter constructs were described earlier (Legrain and Rosbash 1989). (B) Histogram reporting the results of β-galactosidase assays from reporters either in a wild-type yeast strain (hatched bars) or in a ΔWWPrp40 strain (gray bars). Control reporters carry no LacZ coding sequence (“background”) or the LacZ reporter gene with no intron (“No intron”). A first set of reporters described earlier (Séraphin and Rosbash 1990) contains the LacZ reporter gene interrupted by a simple intron with GG at nucleotides −3 and −4 in the upstream exon, combined with either wild-type or mutant 5′ splice site (5′II, GUAUaU) (Jacquier et al. 1985) and either wild-type or frameshifted exon 2 (WT GG, 5′ II GG, WT GG – FS, 5′II GG – FS). A second set of reporters contain the LacZ reporter gene interrupted by an intron carrying duplicated 5′ splice sites (Séraphin and Kandels-Lewis 1993) either with AA at positions −1 and −2 in the upstream exon and combinations of wild-type or mutant 5′ splice site (5′II, GUAUaU) (Jacquier et al. 1985) or with combinations of AG and UC at positions −1 and −2 in the upstream exon. Finally, the last reporter contained the LacZ reporter gene interrupted by an intron with a duplicated branchpoint and 3′ splice site (Goguel and Rosbash 1993). Constructs carrying the 5′II mutant 5′ splice site have previously been shown to be poorly spliced in the absence of Nam8 (Puig et al. 1999; data not shown quoted therein).
DISCUSSION
Due to previously reported physical interactions with other proteins, the U1 snRNP protein Prp40 and, more specifically, its WW domains have been proposed to play roles in several key steps of spliceosome assembly. These include the initial recruitment of splicing factors to the transcription unit via the CTD of Pol II, commitment complex formation, and cross-intron bridging through interactions with the U5 snRNP protein Prp8. Here, we report that Prp40 and Snu71 form a stable heterodimeric module that associates with U1 snRNP in an Nam8-dependent fashion. While destabilization of Snu71 and Prp40 upon Nam8 deletion had been noticed before, the two proteins were thought to be released independently at different salt concentrations (Gottschalk et al. 1998). The connection of Prp40 with Nam8, a factor that is required for the efficient splicing of a subset of yeast mRNAs (Puig et al. 1999), suggests a regulatory role for Prp40. Based on a yeast strain that expresses ΔWW-Prp40 as the only version of Prp40 at levels that are comparable to Prp40 expression in wild-type strains, we examined viability, in vitro formation of commitment complexes, cotranscriptional assembly of splicing complexes in vivo, and the outcome of splicing of endogenous pre-mRNAs as well as several sensitized reporters, which should reveal subtle requirements for splicing regulation. However, we did not observe defects attributable to the lack of the WW domains on any of these processes. Our observations do not exclude the possibility that there might be conditions under which the Prp40 WW domain is essential for splicing. It is nevertheless surprising that systematic probing for multiple aspects of WW domain function did not reveal a role for the Prp40 WW domain in any splicing activity proposed in the literature.
Our in vitro results indicate that the WW domains do not stimulate the formation of CC1 and CC2. This is supported by in vivo ChIP results, which indicate that the lack of the WW domains of Prp40 does not dramatically impair commitment complex or pre-spliceosome assembly in cells under standard conditions of growth. Despite the proposed role of the direct interaction between BBP and the WW domain of Prp40 in cross-intron bridging and commitment complex formation, binding of CC factors does not differ between the wild-type and mutant strains. Apparently, there is also no functional difference, because the spliceosome formation proceeds to addition of the U2 snRNP and beyond. A significant delay of binding and reduction in signal of the U5 snRNP might result from the missing interaction between the WW domain and the U5 snRNP protein Prp8; however, this delay is overcome during transcription and is not sufficient to prevent maturation of the spliceosome as indicated by the accumulation of Prp19 and subsequent spliceosome disassembly. A mutation in the splicing machinery might have an impact on Pol II elongation, which could mask effects on the apparent timing of spliceosome assembly. However, Pol II density along the genes analyzed was indistinguishable between the two strains, arguing against this possibility. Moreover, the similarity of the snRNP profiles among wild-type and mutant strains is consistent with an unchanged distribution of nascent transcripts along each transcription unit. Thus, while deletion of the Prp40 WW domain affects spliceosome assembly in vitro and in vivo, these effects are too limited to impact on the splicing efficiency in vivo, even for weakly spliced reporter genes.
While the Prp40 WW domains had been proposed to facilitate cotranscriptional spliceosome assembly by tethering splicing factors to elongating Pol II (Morris and Greenleaf 2000), the fact that U1 snRNP is not detectable by ChIP on highly expressed intronless genes where Pol II is abundant argues against direct recruitment via Pol II binding (Kotovic et al. 2003). In the present study, we further show that the accumulation of the U1 snRNP protein Prp42 is unaffected by deletion of the Prp40 WW domain. Importantly, we show that the ΔWW-Prp40 mutant protein associates with U1 snRNA but does not affect U1 snRNP activity with respect to commitment complex formation and splicing; consistent with this, ΔWW-Prp40 protein was itself well recruited to intron-containing genes due to its continued presence in the U1 snRNP. We conclude that the Prp40 WW domain is not required for U1 snRNP association, function, or interactions with the Pol II CTD, in agreement with the predictions of a previous structural study (Wiesner et al. 2002).
Cross-intron bridging, in which the 5′ splice site and the 3′ region of the intron are brought together in the commitment complex, appears not to be crucial for spliceosome formation (Rutz and Séraphin 1999, 2000); yet the occurrence of this interaction is well established by in vitro analyses (Séraphin and Rosbash 1991). The proposed interaction between the Prp40 WW domains and BBP seems to play little, if any, role in cross-intron bridging as shown by our biochemical analysis of commitment complexes, suggesting that other factors mediate this connection. An alternative U1 snRNP subunit candidate for cross-intron bridging is Prp39, which has been reported to interact in yeast two-hybrid assays with the 3′ splice site recognizing CC components BBP and Mud2 (Fromont-Racine et al. 1997) and, like Prp40, with an N-terminal region of the U5 snRNP protein Prp8 (van Nues and Beggs 2001). In addition, an interaction between the U1 snRNP protein Snu56 and Mud2 has been detected by two-hybrid assay (Balzer and Henry 2008). In contrast to Prp40, it has been recently reported that the WW domains of the mammalian splicing factor FBP21 have an impact on splicing (Huang et al. 2009). FBP21 is associated with the U2 snRNP and is thought to act in cross-intron bridging by its WW domains, which interact not only with mammalian SF1/BBP but also with the U1 snRNP component U1C. While overexpression of full-length FBP21 increases the relative amount of spliced product in an in vivo reporter assay, a mutant with alanine substitutions of the signature tryptophans of the WW domains does not. Cross-intron bridging may have an increased importance in mammalian cells that have longer and multiple introns in contrast to yeast.
What is the role of Prp40? One possibility is that the FF domains of Prp40p are involved in U1 snRNP recruitment cotranscriptionally. The FF domains bound the phosphorylated CTD in GST pull-down assays, but this interaction could not be confirmed in complementary assays (Morris and Greenleaf 2000). Moreover, interaction of the FF motif with the Pol II CTD could not be reconstituted using highly purified protein (Gasch et al. 2006). Instead, FF domains were proposed to interact with the U1 snRNA itself (Gasch et al. 2006), other subunits of the U1 snRNP (Ester and Uetz 2008), and to the Clf1 splicing factor (Chung et al. 1999). Interestingly, mutant analyses demonstrated that these interactions occur through the first FF domains, some of which are essential for viability. This contrasts with the C-terminal FF domains, which are dispensable and for which interacting partners remain unknown (Ester and Uetz 2008). While the deletion of the WW domain does not impair growth at any temperature, yeast cells are highly sensitive to the deletion of the FF domains, which leads to slow growth and temperature sensitivity. The physical interaction of the essential central part of Prp40 with the U1 snRNP proteins Luc7 and Snu71 suggests that the crucial role of Prp40 could be to stabilize the U1 snRNP by holding these proteins in place, while interactions outside of the U1 snRNP are not essential.
Clues to Prp40 function may also come from orthologs in other species. The splicing factor FBP11 is closely related to Prp40p in higher organisms, sharing the domain organization with two N-terminal WW and several FF domains in the central and C-terminal regions of the protein. Although the FF-domain properties of FBP11 and Prp40p differ significantly (Gasch et al. 2006), both interact with BBP (Abovich and Rosbash 1997; Lin et al. 2004) and phosphorylated CTD peptides (Morris and Greenleaf 2000; Allen et al. 2002). Yet another protein with a related domain structure found in metazoans, the transcription elongation factor CA150, has been proposed to connect transcription and splicing, because it interacts with Pol II and SF1 and has been found in spliceosome preparations (Carty et al. 2000; Goldstrohm et al. 2001; Makarov et al. 2002; Rappsilber et al. 2002). Thus, these related proteins represent prime candidates for coupling transcription and spliceosome assembly by recruitment of splicing factors to the transcription unit. Given the results presented here, it remains to be seen whether Prp40 orthologs have additional roles in spliceosome assembly and splicing in other organisms.
In summary, several groups have proposed models for the function of the Prp40 protein WW domains based on their physical interactions in yeast and other eukaryotic organisms (Abovich and Rosbash 1997; Morris and Greenleaf 2000; Wiesner et al. 2002; Lin et al. 2004; Kang et al. 2009), including association of the splicing machinery with the transcription apparatus or networking of various splicing factors. While these models are essential to form testable hypotheses, the present example demonstrates that physical interactions of even a conserved essential protein do not necessarily predict essential functions. Biochemical and in vivo assays are essential to ascertain the function of proposed interactions. In this case, various assays indicate that the Prp40 WW domains weakly affect spliceosome assembly in vitro and in vivo but that their effects are not sufficient to alter splicing.
MATERIALS AND METHODS
Yeast strains, maintenance
Yeast strains were grown, manipulated, and maintained according to standard procedures. The strains used in this study are based on MGD353-13D (MATa, ade2 arg4 leu2-3,112 trp1-289 ura3-52) and its derivative BSY909 (MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 prp40ΔWW) (Table 1). The latter was constructed by pop-in/pop-out. A PCR-based strategy for epitope tagging of endogenous genes (Knop et al. 1999) was used to generate the strains used for immunoprecipitation and chromatin immunoprecipitation. TAP tagging was performed as described previously (Puig et al. 1998). Growth at different temperatures was determined by spotting 10-fold serial dilutions starting from O.D.600 = 0.06 on solid YPD and SD medium, followed by 2 d of incubation at the respective temperatures (4 d at 16°C).
TABLE 1.
Yeast strains (from this study except otherwise indicated)
TAP purifications
TAP purifications were performed according to the published procedure (Rigaut et al. 1999; Puig et al. 2001). Mass spectrometric identification of Prp40 was performed by LC-MS/MS (Cellzome) following in gel trypsin digestion.
Native gel analysis of splicing complex formation
Splicing extracts and native gels for the analysis of commitment complexes, pre-spliceosome, and spliceosome were prepared and performed as described earlier (Séraphin and Rosbash 1989). RNA substrates were also described (Séraphin and Rosbash 1991).
Chromatin immunoprecipitation and quantitative PCR
ChIP and QPCR analysis were performed as described previously in Kotovic et al. (2003) and Görnemann et al. (2005). In all experiments, RNA-Polymerase II (8WG16 antibody from Neoclone) was analyzed in parallel, as a quality control for each experiment, while unspecific mouse IgG (Sigma-Aldrich) served as background control.
Immunoprecipitation
Yeast cells were grown in 200 mL of YPD to an O.D.600 of 0.6–0.8 and harvested by centrifugation. After washing the pellets with ice-cold water once, cells were resuspended in 800 μL of IP buffer (50 mM Tris at pH 7.4, 125 mM KCl, 0.1% NP-40) and lysed by vigorous shaking with 300 μL of glass beads for 20 min at 4°C. Cleared lysates were obtained by centrifugation for 20 min at 14,000 rpm at 4°C and subsequent incubation of the supernatant with 200 μL of 4CL-B agarose for 1 h at 4°C. Eight micrograms of antibody (pre-immune IgG, 12CA5) was bound to 50 μL of γ-bind beads for 2 h prior to addition of lysate. Total protein content of the pre-cleared lysate was determined by Bradford assay and volumes adjusted accordingly. Immunoprecipitations were performed in a volume of 450 μL; the amount of lysate used is given as a fraction of total volume with the individual experiment. After 2 h of nutating at 4°C, beads were washed with cold buffer three times, transferred to fresh tubes, and washed again. Samples were then split in two parts for analysis of protein and RNA content, respectively. For analysis of immunoprecipitated proteins, the Sepharose beads were boiled in protein sample buffer and subjected to Western blot analysis with 12CA5 antibody (α-HA) beside input samples. Blots were stripped and probed with mouse-α-GAPDH (Novus Biologicals). For RNA analysis, RNA was eluted by addition of 100 μL of TE/1% SDS and incubation for 15 min at 70°C. For input samples, 10 μL of a 1:10 dilution of lysate was incubated in 100 μL of TE/1% SDS in parallel. RNA was extracted by TRIzol, followed by isopropanol precipitation. Samples were DNase I (NEB) treated. 1:25 (2 μL) of each RNA sample was reverse-transcribed in a volume of 20 μL (SuperScript III) using random hexamer primers (Roche). −RT samples were run in parallel omitting the Reverse Transcriptase. cDNAs were amplified by PCR (Taq polymerase, 25 cycles) with primer pairs specific for U1 snRNA and 18S snRNA.
Splicing assays
To analyze the levels of spliced mRNAs and unspliced pre-mRNAs in vivo, yeast cells were grown to log phase, harvested by centrifugation, washed once with ice-cold RNA-buffer (50 mM Tris-Cl at pH 7.4, 100 mM NaCl, 10 mM EDTA) and frozen at −80°C. After thawing, the pellet was resuspended and extracted by vigorous shaking in equal volumes of buffer, glass beads, and phenol–chloroform. Following centrifugation, the aqueous phase was extracted once more with phenol–chloroform, and RNA was precipitated. The resuspended RNA was DNAse I treated (NEB) and reverse-transcribed (SuperScript III; Invitrogen) using random hexamer primers. cDNAs were analyzed with specific primers by quantitative PCR/SYBR Green methodology (Stratagene, Mx3000).
Analyses of pre-mRNA splicing using reporter and β-galactosidase assays were performed as described earlier (Brooks et al. 2009).
ACKNOWLEDGMENTS
We thank Raphael Ramirez-Morales for his input in constructing the original Prp40-ΔWW strain; N. Dreumont for suggestions and the Institut de Génétique et de Biologie Moléculaire et Cellulaire for assistance. We also thank Jaz Woo for his enthusiastic contribution to this study. Work in the laboratory of B. Séraphin is supported by La Ligue contre le Cancer (Equipe Labellisée 2011) and the CNRS. This work was also supported by the European Union 6th Framework program “EURASNET” (LSHG-CT-2005-518238) to B.S. and K.M.N.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.02646811.
REFERENCES
- Abovich N, Rosbash M 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412 [DOI] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Friedler A, Schon O, Bycroft M 2002. The structure of an FF domain from human HYPA/FBP11. J Mol Biol 323: 411–416 [DOI] [PubMed] [Google Scholar]
- Balzer RJ, Henry MF 2008. Snu56p is required for Mer1p-activated meiotic splicing. Mol Cell Biol 28: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Leder P 1999. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem Sci 24: 264–265 [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787 [DOI] [PubMed] [Google Scholar]
- Brooks MA, Dziembowski A, Quevillon-Cheruel S, Henriot V, Faux C, van Tilbeurgh H, Séraphin B 2009. Structure of the yeast Pml1 splicing factor and its integration into the RES complex. Nucleic Acids Res 37: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Carty SM, Goldstrohm AC, Suñé C, Garcia-Blanco MA, Greenleaf AL 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci 97: 9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, McLean MR, Rymond BC 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprévote O, Séraphin B 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 23: 4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester C, Uetz P 2008. The FF domains of yeast U1 snRNP protein Prp40 mediate interactions with Luc7 and Snu71. BMC Biochem 9: 29 doi: 10.1186/1471-2091-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Fortes P, Bilbao-Cortés D, Fornerod M, Rigaut G, Raymond W, Séraphin B, Mattaj IW 1999. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev 13: 2425–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Longman D, McCracken S, Ip JY, Poot R, Mattaj IW, Cáceres JF, Blencowe BJ 2007. Identification and characterization of RED120: A conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett 581: 3087–3097 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Gasch A, Wiesner S, Martin-Malpartida P, Ramirez-Espain X, Ruiz L, Macias MJ 2006. The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J Biol Chem 281: 356–364 [DOI] [PubMed] [Google Scholar]
- Goguel V, Rosbash M 1993. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell 72: 893–901 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Albrecht TR, Suñé C, Bedford MT, Garcia-Blanco MA 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol Cell Biol 21: 7617–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393 [PMC free article] [PubMed] [Google Scholar]
- Hochleitner EO, Kastner B, Fröhlich T, Schmidt A, Lührmann R, Arnold G, Lottspeich F 2005. Protein stoichiometry of a multiprotein complex, the human spliceosomal U1 small nuclear ribonucleoprotein: Absolute quantification using isotope-coded tags and mass spectrometry. J Biol Chem 280: 2536–2542 [DOI] [PubMed] [Google Scholar]
- Huang X, Beullens M, Zhang J, Zhou Y, Nicolaescu E, Lesage B, Hu Q, Wu J, Bollen M, Shi Y 2009. Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21). J Biol Chem 284: 25375–25387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A, Rodriguez JR, Rosbash M 1985. A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell 43: 423–430 [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S, Séraphin B 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039 [DOI] [PubMed] [Google Scholar]
- Kang CH, Feng Y, Vikram M, Jeong IS, Lee JR, Bahk JD, Yun DJ, Lee SY, Koiwa H 2009. Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch Biochem Biophys 484: 30–38 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E 1999. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262: 1982–1988 [DOI] [PubMed] [Google Scholar]
- Lin KT, Lu RM, Tarn WY 2004. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol Cell Biol 24: 9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Séraphin B 1997. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J 16: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Neubauer G, Gottschalk A, Fabrizio P, Séraphin B, Lührmann R, Mann M 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci 94: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AJ, Norman C 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754 [DOI] [PubMed] [Google Scholar]
- Oesterreich FC, Bieberstein N, Neugebauer KM 2011. Pause locally, splice globally. Trends Cell Biol 21: 328–335 [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Rutz B, Luukkonen BG, Kandels-Lewis S, Bragado-Nilsson E, Séraphin B 1998. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast 14: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Séraphin B 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev 13: 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Puig O, Bragado-Nilsson E, Koski T, Séraphin B 2007. The U1 snRNP-associated factor Luc7p affects 5′ splice site selection in yeast and human. Nucleic Acids Res 35: 5874–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Ruby SW, Abelson J 1988. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science 242: 1028–1035 [DOI] [PubMed] [Google Scholar]
- Rutz B, Séraphin B 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B, Séraphin B 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J 19: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Kandels-Lewis S 1993. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell 73: 803–812 [DOI] [PubMed] [Google Scholar]
- Séraphin B, Rosbash M 1989. Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59: 349–358 [DOI] [PubMed] [Google Scholar]
- Séraphin B, Rosbash M 1990. Exon mutations uncouple 5′ splice site selection from U1 snRNA pairing. Cell 63: 619–629 [DOI] [PubMed] [Google Scholar]
- Séraphin B, Rosbash M 1991. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA–pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J 10: 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Kretzner L, Rosbash M 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J 7: 2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Sudol M, Sliwa K, Russo T 2001. Functions of WW domains in the nucleus. FEBS Lett 490: 190–195 [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Rosbash M 2006. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA 12: 968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly post-transcriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Lee KR, Cheng SC 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci 90: 10821–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Stier G, Sattler M, Macias MJ 2002. Solution structure and ligand recognition of the WW domain pair of the yeast splicing factor Prp40. J Mol Biol 324: 807–822 [DOI] [PubMed] [Google Scholar]