Giardia is a deeply branching unicellular eukaryote. Here, the authors provide evidence for a microRNA, miR4, that regulates the expression of variant surface proteins (VSPs). Biogenesis of this miRNA requires Giardia dicer and the miRNA to be associated with the Giardia Argonaute protein. These results suggest a very ancient origin of miRNA regulatory systems.
Keywords: Giardia, microRNA, variant surface protein, gene expression, microRNA biogenesis
Abstract
We have previously shown that a snoRNA-derived microRNA, miR2, in Giardia lamblia potentially regulates the expression of 22 variant surface protein (VSP) genes. Here, we identified another miRNA, miR4, also capable of regulating the expression of several VSPs but derived from an unannotated open reading frame (ORF) rather than a snoRNA, suggesting a canonical miRNA biogenesis pathway in Giardia. miR4 represses expression of a reporter containing two miR4 antisense sequences at the 3′ UTR without causing a corresponding decrease in the mRNA level. This repression requires the presence of the Giardia Argonaute protein (GlAgo) and is reversed by 2′ O–methylated antisense oligo to miR4, suggesting an RNA-induced silencing complex (RISC)–mediated mechanism. Furthermore, in vivo and in vitro evidence suggested that the Giardia Dicer protein (GlDcr) is required for miR4 biogenesis. Coimmunoprecipitation of miR4 with GlAgo further verified miR4 as a miRNA. A total of 361 potential target sites for miR4 were bioinformatically identified in Giardia, out of which 69 (32.7%) were associated with VSP genes. miR4 reduces the expression of a reporter containing two copies of the target site from VSP (GL50803_36493) at the 3′ UTR. Sixteen of the 69 VSP genes were further found to contain partially overlapping miR2 and miR4 targeting sites. Expression of a reporter carrying the two overlapping sites was inhibited by either miR2 or miR4, but the inhibition was neither synergistic nor additive, suggesting a complex mechanism of miRNA regulation of VSP expression and the presence of a rich miRNAome in Giardia.
INTRODUCTION
Giardia lamblia is a deeply branching, minimalistic protozoan causing the diarrheal disease giardiasis in humans (Morrison et al. 2007). Past studies and recent analysis of its genome suggested that Giardia utilizes simplified machineries for many important biological processes, ranging from DNA replication to translation initiation (Morrison et al. 2007). Few consensus promoter sequences or transcription factors have been identified in Giardia (Yee et al. 2007; Wang et al. 2010). An 8-bp AT-rich region was shown to be sufficient to initiate transcription, which may explain the abundance of sterile transcripts in Giardia (Elmendorf et al. 2001a,b). Additionally, Giardia mRNAs typically have short 5′ and 3′ untranslated regions (UTR) of <30 nucleotides (nt), which minimizes translational regulation. The mechanism of ribosome scanning, for instance, was found to be missing from Giardia mimicking that observed in Archaea (Li and Wang 2004). These data suggest that gene expression in Giardia is poorly controlled. However, the expression of variant surface proteins (VSPs) in this organism is tightly regulated. Over 200 VSP genes have been identified in the Giardia genome, of which only one VSP is expressed on the cell surface at any given time (Nash et al. 2001; Adam et al. 2010). The VSP coat is shed from the membrane surface and replaced with a different VSP coat every six to 13 generations during the growth of Giardia (Nash et al. 1990). The mechanism behind this phenomenon is poorly understood. A microRNA (miRNA)-mediated translational repression mechanism was, however, recently found to be involved in regulating VSP expression (Saraiya and Wang 2008), and the importance of Dicer in regulating VSP expression has been shown (Prucca et al. 2008).
miRNAs are major post-transcriptional regulators of gene expression during cell development, differentiation, and maintenance (Pasquinelli and Ruvkun 2002; Miska 2005; Stadler and Ruohola-Baker 2008). In higher eukaryotes, the canonical miRNA biogenesis pathway involves the digestion of a primary-miRNA (pri-miRNA) by the nuclear RNaseIII, Drosha, into a 70-nt pre-miRNA (Lee et al. 2003). The latter is exported by Exportin5 into the cytoplasm and is further processed into a mature miRNA by the cytoplasmic RNaseIII, Dicer (Bernstein et al. 2001; Yi et al. 2003; Lund et al. 2004). The miRNA is then incorporated into the Argonaute protein to form the core RNA-induced silencing complex (RISC), which binds to the target sequence at the 3′ UTR of an mRNA partly complementary to the miRNA to block its translation (Liu et al. 2004; Meister et al. 2004). RNA polymerase II transcripts of noncoding genes are the main source of miRNA precursors, but miRNAs derived from introns, which bypass the canonical biogenesis pathway, have also been described (Brown et al. 2008; Lin et al. 2008).
We previously identified a small nucleolar (sno) RNA-derived miRNA, miR2, in Giardia (Saraiya and Wang 2008). This miRNA is from the 3′ end of a Box C/D snoRNA, GlsR17, and requires Dicer processing for maturation, which bypasses the requirement for Drosha and Exportin5 that are apparently missing from Giardia (Saraiya and Wang 2008). This was a new example of miRNA biogenesis bypassing the canonical biogenesis pathway. A total of 22 VSP genes were found to contain potential target sites for miR2 at their 3′ UTRs (Saraiya and Wang 2008). Translation of a reporter transcript carrying the target site from one of the VSPs at the 3′ UTR was repressed by miR2 in Giardia, suggesting the involvement of a miRNA-mediated mechanism in controlling VSP expression (Saraiya and Wang 2008). Our subsequent studies indicated that more miRNAs are derived from snoRNAs and function in regulating VSP expression in Giardia (Li et al. 2011; W Li, AA Saraiya, and CC Wang, in prep.).
At the same time, Ender et al. (2008) independently reported that a human H/ACA snoRNA, ACA45, could be processed to 20- to 25-nt-long small RNAs that were stably associated with the Argonaute protein and post-transcriptionally repressed the expression of a cellular target mRNA of CDC2L6. Similar to that in Giardia, processing of this snoRNA-derived miRNA is independent of the Drosha/DGCR8 complex but requires Dicer. It thus appears that snoRNAs are one of the sources of functional miRNAs preserved through evolution. The subsequent, systematic analysis of small-RNA deep sequencing libraries from various organisms showed that small RNAs with conserved sizes and positions were derived from many snoRNA loci in human, mouse, chicken, fruit fly, Arabidopsis, and fission yeast (Taft et al. 2009). Some of these small RNAs showed 20- to 100-fold decreases in response to loss of Dicer and were associated with specific members of the Argonaute family (Taft et al. 2009). Recently, it was further reported that 20 known miRNA precursors showed significant sequence similarities to box H/ACA snoRNAs, and five of them could bind to dyskerin, a specific protein component of the functional box H/ACA snoRNP (Scott et al. 2009). Another study showed that the precursors and/or mature form of a number of rat miRNAs could be detected in the nucleolus and that some of them were significantly concentrated there (Politz et al. 2009). It is thus becoming increasingly clear that snoRNAs could be a family of evolutionarily conserved precursors of miRNAs (Scott et al. 2009).
Since our demonstration that snoRNA-derived miRNA-mediated gene regulation is present in Giardia and that it may be involved in controlling VSP expression, two additional questions emerged: (1) Are snoRNAs the only source of miRNAs in Giardia? (2) Are there additional miRNAs that regulate VSP expression? To answer these questions, we created a small RNA library from Giardia and identified a small RNA derived from an unannotated open reading frame (ORF) rather than a snoRNA that was able to repress translation of a reporter transcript carrying the potential target site. This target site is located in the 3′ end of coding region of many VSP genes, thus suggesting a mechanism of VSP expression regulation involving miRNAs derived from precursors other than snoRNAs.
RESULTS
Identification of additional potential miRNAs in Giardia
In an effort to identify additional miRNAs, a cDNA library was constructed from ∼20–35 nt, small RNAs size-fractionated from Giardia WB isolate trophozoites. The library was cloned into a pGEM-T vector and sequenced. One of the small RNAs thus identified was a 26-nt small RNA (5′-UCUGCACCAAGGAGCUGAUCCAGACC-3′), which is of the same size as that of a previously identified miRNA, miR2, in Giardia (Saraiya and Wang 2008). This small RNA was subsequently identified in a small RNA library by Illumina sequencing, thus confirming its presence in Giardia WB isolate (data not shown). A BLAST analysis of the Giardia WB genome database (www.giardiadb.org, v 1.1) identified the small RNA in a 972-bp ORF encoding a hypothetical protein (GL50803_92663), with the small RNA sequence positioned from nucleotides 215–240 (Fig. 1A; Aurrecoechea et al. 2009). It suggested that this small RNA could be derived from a precursor other than snoRNA through a probable canonical biogenesis pathway. Comparison of this ORF among the genome databases of the three different Giardia isolates, WB, P15, and GS, however, showed no conservation of this ORF in P15 and GS, suggesting that this ORF and the small RNA are present only in Giardia WB. Interestingly, the first approximately 177 amino acids in ORF GL50803_92663 are highly similar to the first approximately 177 amino acids in another putative DNA polymerase gene in both the WB and P15 isolates (GL50803_137688 and GLP15_4358, respectively). The potential significance in this coincidence is not yet clear.
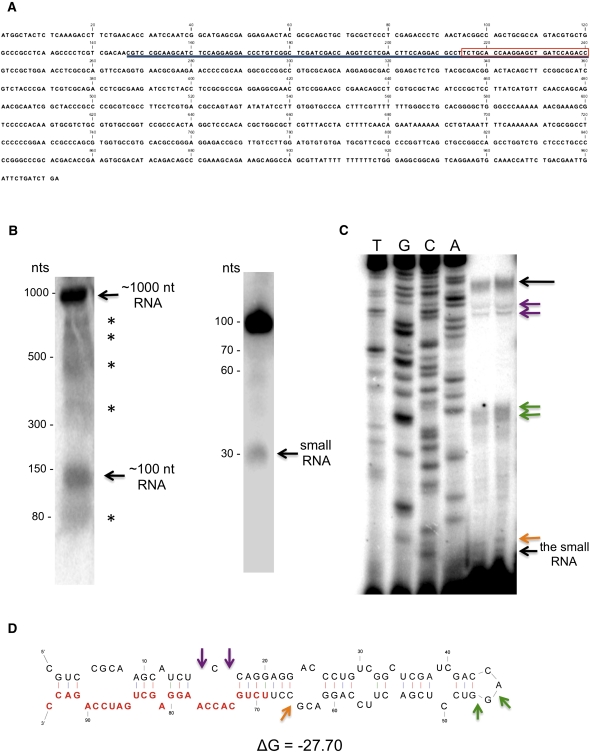
FIGURE 1.
Identification of a small RNA and its precursor in Giardia. (A) Sequence of the 972-nt ORF from which the small RNA and its precursor are derived. The small RNA is boxed in red, while the precursor is underlined. (B) Northern blot analysis of total RNA (left panel) showed the presence of an ∼1000-nt RNA that corresponds to the GL50803_92663 ORF transcript, suggesting that it is expressed. An ∼100-nt band and an ∼70-nt band that correspond to bands seen on the Northern blot of size-fractionated small RNA (<200 nt) were also observed. Additional bands may represent GlDcr intermediate products or degradation. Northern blot analysis of size-fractionated small RNA (<200 nt) identified an ∼30-nt band corresponding to the small RNA and an ∼100-nt band that may correspond to its precursor (right panel). Additional faint bands were also observed at ∼60 nt and ∼70 nt. (C) Primer extension of size-fractionated (<200 nt) RNA from Giardia trophozoites identified a 94-nt fragment and a 26-nt band corresponding to the small RNA. Additional bands shown by color-coded arrows have their corresponding locations indicated on the pre-miR4 secondary structure below. (D) MFOLD secondary structure of the 94-nt RNA identified by primer extension. The small RNA is indicated in red.
Further characterization of the small RNA
The presence of this small RNA in Giardia was confirmed by Northern blot analysis using size-fractionated small RNA (<200 nt) from Giardia trophozoites probed with the sequence complementary to that of the 26-nt small RNA. The Northern blot showed the presence of a major ∼100-nt band, a minor ∼30-nt band, and two faint bands of ∼60 and 70 nt, suggesting that the ORF transcript is discretely processed instead of being randomly degraded in Giardia (Fig. 1B, right panel). The larger RNAs identified in this Northern blot could be precursors of the ∼30-nt RNA.
The sequence at the 5′ end of the ∼100-nt RNA was determined by primer extension on size-fractionated small RNAs (<200 nt) from Giardia using a 20-nt 5′ end radiolabeled primer complementary to nucleotides 4–23 in the 26-nt small RNA sequence (5′-CTGGATCAGCTCCTTGGTGC-3′). The results showed a 94-nt band at the top of the gel, which could correspond to the ∼100-nt band seen in the Northern (Fig. 1C). The sequence of this 94-nt RNA corresponds to the sequence between nucleotides 147–240 in the ORF (see Fig. 1A) with its 3′ end corresponding to the sequence of the 26-nt small RNA. Presence of the 26-nt small RNA was also confirmed by the data from primer extension (Fig. 1C). Several additional bands in the 50- to 70-nt range were also identified in primer extension, which could represent either the degradation products of the 94-nt RNA, the intermediary precursors of the 26-nt RNA, or premature terminations due to secondary structure of the 94-nt RNA. The minimal free energy structure identified by MFOLD modeling of the 94-nt RNA sequence showed a hairpin structure with the 5′ end of the 26-nt small RNA portion located in the stem (Fig. 1D). The colored arrows in Figure 1D, pointing to the bulges in the secondary structure, agree well with the additional bands observed in Figure 1C, suggesting premature terminations during primer extension. This could be a likely substrate structure for Dicer processing to generate the 26-nt small RNA. The latter thus could be a miRNA in Giardia.
We then confirmed the presence of the full-length ORF transcript of 972 nt in Giardia with RT-PCR. PCR amplification of a Giardia poly (T) reverse transcribed (RT) cDNA library using two ORF-specific primers generated an amplified DNA band with the anticipated size (data not shown). This data confirmed that the full-length ORF is transcribed as a polyadenylated RNA. Furthermore, a Northern blot of total Giardia RNA probed with a 94-nt antisense RNA corresponding to the potential small RNA precursor showed the presence of a major ∼1000-nt RNA band, which could be the 972-nt ORF (Fig. 1B, left panel).
To further verify whether the 94-nt RNA is related to snoRNAs in Giardia, fluorescence in situ hybridization was performed on Giardia trophozoites as previously described (Saraiya and Wang 2008). The results indicated that both the 94-nt and 26-nt small RNAs are evenly distributed in the nucleus and cytoplasm without any sign of specific localization in the nucleolus (Supplemental Figure 1). The 94-nt RNA is thus probably unrelated to snoRNAs.
The small RNA functions as a miRNA
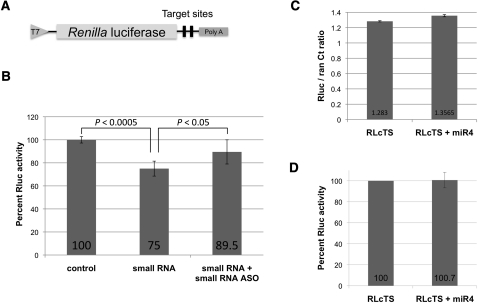
To test whether the 26-nt small RNA functions as a miRNA in Giardia, two copies of the antisense sequence were inserted into the 3′ UTR of a Renilla luciferase (RLuc) reporter gene to make the construct RLcTS (Fig. 2A). A capped in vitro transcript of RLcTS was electroporated into Giardia WB trophozoites either with or without the chemically synthesized 26-nt small RNA. The transfected cells were incubated for 5 h at 37°C, lysed, and assayed for RLuc activity with the cells transfected with RLcTS mRNA alone set at 100% RLuc expression. Cotransfection of RLcTS mRNA with the synthetic small RNA resulted in a ∼25% decrease in RLuc activity (Fig. 2B). This decrease could be reversed back to ∼90% activity when the cells were also cotransfected with an equivalent amount of 2′ O-methyl small RNA antisense oligo (Fig. 2B). These data suggested that the RLuc activity is specifically repressed by the 26-nt small RNA. The latter was thus tentatively designated as miRNA 4 (miR4) and the 94-nt RNA pre-miR4.
FIGURE 2.
The small RNA–mediated repression of RLuc expression. (A) Diagram of the RLuc reporter construct RLcTS containing two copies of the small RNA antisense sequence at the 3′ UTR. (B) Percentage of RLuc activity of Giardia trophozoites transfected with capped in vitro transcript of RLcTS along with or without the synthetic small RNA (1 μg). The presence of the synthetic small RNA resulted in a ∼25% decrease in RLuc activity. This loss could be restored to ∼90% by cotransfecting the cells with a 2′-O methylated small RNA antisense oligo (1 μg). These results represent the average and standard deviations from at least three independent experiments and the Students t-test was used to determine P-values. Comparison of single or double antisense target sites using this assay resulted in a similar outcome (Li et al. 2011). (C) Total RNA from Giardia WB cells transfected with RLcTS mRNA alone or with miR4 was isolated. The RLcTS mRNA level was assayed by real-time PCR with Ran mRNA as a control. The ratios of RLcTS/Ran show little difference between the two samples, suggesting that the inhibition in RLuc expression is not due to mRNA degradation. (D) The presence of miR4 did not affect the expression of RLuc in the GlAgo knockdown cells, suggesting that GlAgo is required for the miR4-mediated inhibition.
To verify if the observed miR4-mediated decrease in RLuc activity is due to translational repression, total RNA was isolated from the RLcTS mRNA and miR4 cotransfected Giardia cells, and the RLcTS mRNA level was determined using quantitative PCR (qPCR). There was no detectable difference in the CT values between cells transfected with RLcTS mRNA alone and cells transfected with RLcTS mRNA plus synthetic miR4 (Fig. 2C), thus suggesting translational repression by miR4. This lack of mRNA degradation suggests that the Giardia Argonaute (GlAgo) does not have the ability to cleave mRNA, which may explain the absence of observable RNA interference (RNAi) in Giardia (data not shown).
We have previously constructed a plasmid of GlAgo antisense-hammerhead ribozyme and used it for knocking down the expression of GlAgo in Giardia (Saraiya and Wang 2008). These GlAgo knockdown cells were transfected with RLcTS mRNA and synthetic miR4. The results showed that the RLuc expression was not affected by introducing miR4 into the cells (Fig. 2D), suggesting that GlAgo is required for the miR4-mediated reduction of Rluc expression.
Is miR4 associated with GlAgo in Giardia?
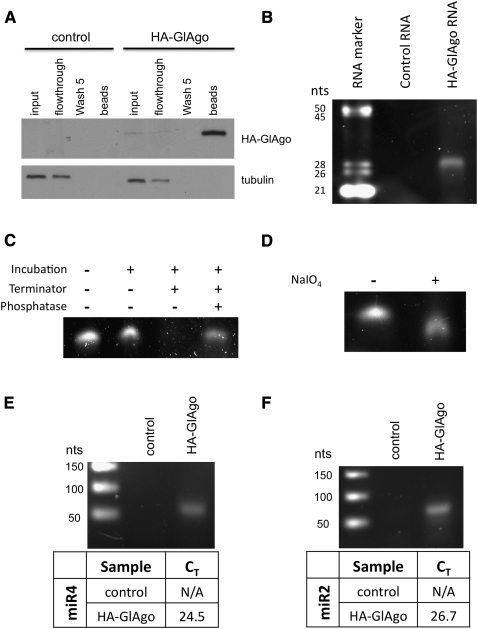
It is well known that miRNAs need to be associated with Argonaute proteins to mediate translation repression (Liu et al. 2004; Meister et al. 2004). To see if miR4 is associated with GlAgo in Giardia, an N-terminal 2xHA-tagged GlAgo (HA-GlAgo) was cloned into a tetracycline inducible overexpression vector and introduced into Giardia trophozoites (Sun and Tai 2000). Lysate of the tetracycline-induced cells was treated with α-HA beads to immunoprecipitate HA-GlAgo followed by Trizol isolation of the coimmunoprecipitated RNA. Analysis of the immunoprecipitate by SDS-PAGE followed by SYPRO ruby staining showed the presence of a number of protein bands that could be associated with GlAgo and could represent other potential subunit proteins in RISC (Supplemental Fig. 2A). Western blot using an anti-HA antibody confirmed that HA-GlAgo was pulled down by the beads (Fig. 3A). A well-defined ∼26- to 30-nt RNA band was coimmunoprecipitated with HA-GlAgo (Fig. 3B) together with a few other larger RNA bands that were also observed in the control (Supplemental Fig. 2B). The presence of the HA-GlAgo–associated small RNAs with the size of known miRNAs in Giardia (Saraiya and Wang 2008) suggests that most, if not all, of the RNA molecules in the ∼26- to 30-nt band could be miRNAs. It also suggests that the tagged GlAgo is functional and capable of interacting with miRNAs. The immunoprecipitation of HA-GlAgo also allowed us to examine the ends of the coimmunoprecipitated small RNAs. During incubation with the Terminator 5′-3′ exonuclease, which requires a 5′ phosphate in the substrate RNA for degradation, the small RNAs were significantly digested. A pretreatment of the small RNAs with Antarctic phosphatase to remove the 5′ phosphates effectively protected them from Terminator 5′-3′ exonuclease degradation (Fig. 3C). Most, if not all, of the small RNAs thus carry a 5′ phosphate. The 3′ ends of the HA-GlAgo coimmunoprecipitated small RNAs were examined by β-elimination (Zhang et al. 2008). Denaturing PAGE analysis of the reaction product showed a decrease in the size of RNAs following β-elimination (Fig. 3D), suggesting that the immunoprecipitated small RNAs have 2′- and 3′-OH groups. The presence of a 5′ phosphate and 2′,3′-OH in the small RNAs is characteristic of the Dicer product, thus providing further supporting evidence that they are miRNAs.
FIGURE 3.
Immunoprecipitation of HA-GlAgo. (A) The HA-GlAgo immunoprecipitation was analyzed by Western blot using an anti-HA antibody. The HA-GlAgo was significantly enriched in the beads indicating that HA-GlAgo was effectively immunoprecipitated. (B) Analysis of the RNAs coimmunoprecipitated with HA-GlAgo showed an associated ∼26- to 30-nt RNA band not seen in the control, indicating that these small RNAs are specifically associated with HA-GlAgo. (C) Terminator exonuclease was able to degrade the untreated small RNAs but not the phosphatase pretreated RNAs, indicating that the GlAgo-associated small RNAs have 5′ phosphates. (D) The small RNAs treated with periodate showed an increased gel mobility, indicating the loss of a base due to β-elimination. This suggests that the GlAgo-associated small RNAs have free 2′,3′ hydroxyls. (E) The small RNA sample was isolated from the immunoprecipitated HA-GlAgo and reverse transcribed using miRNA specific primers and assayed by qPCR. The qPCR showed the presence of miR4 (CT = 24.5) and (F) miR2 (CT = 26.7) in the GlAgo sample but not in the control (N/A indicates no amplification). The amplified miRNA cDNAs were analyzed in 2% agarose gel to confirm the presence of a specific 76- to 77-nt band.
To test if miR4 is associated with GlAgo in Giardia, the immunoprecipitated 26- to 30-nt small RNA band was excised and extracted from the gel and reverse transcribed using miR4-specific primers and was analyzed by Taqman qPCR. The result showed that miR4 is present in the GlAgo pull-down RNA sample (CT = 24.51) (Fig. 3E). The qPCR was also able to identify our previously identified miRNA, miR2 (Saraiya and Wang 2008), in the GlAgo pull-down (CT = 26.67) (Fig. 3F), thus providing a strong indication that both miR4 and miR2 are bona fide miRNAs in Giardia.
The Dicer-mediated processing of pre-miR4
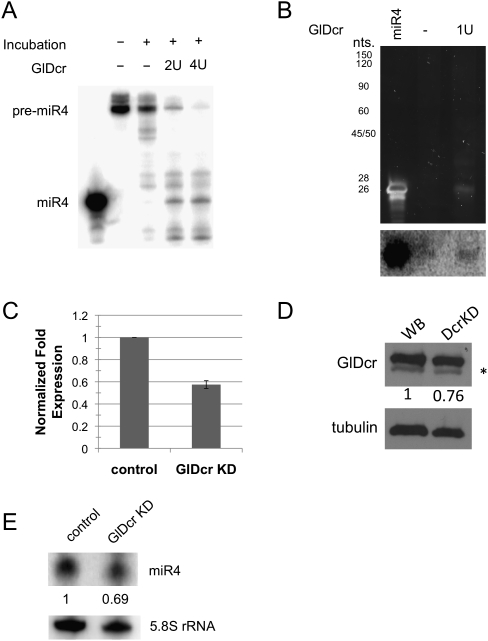
Since the Northern blot in Figure 1B suggests that miR4 could be a processed small RNA from the 94-nt RNA, we tested if the Giardia Dicer (GlDcr) is responsible for the processing. In vitro transcribed, radiolabeled pre-miR4 was incubated with a commercial sample of GlDcr (Powercut Dicer, Finnzyme) for 16 h at 37°C as suggested by the supplier. Denaturing PAGE analysis of the reaction products showed a significant decrease in pre-miR4 along with the accumulation of a 26-nt band corresponding to the size of miR4 (Fig. 4A). It is thus likely that GlDcr is capable of digesting the pre-miR4 to generate mature miR4. In the absence of an identifiable Drosha homolog in Giardia, the mechanism for pre-miRNA production from the 972-nt pri-miR4 is unclear. One possibility is that GlDcr could be responsible for processing both pri-miR4 as well as pre-miR4. To test such a possibility, in vitro transcribed pri-miR4 was incubated with the commercial GlDcr, and the product was analyzed on an 8% polyacrylamide gel (Fig. 4B). A small RNA band of ∼26 nt was observed after GlDcr digestion. The gel was blotted onto a membrane and probed with radiolabeled pre-miR4 (Fig. 4B, bottom). The result indicates that the 26-nt RNA is likely miR4, suggesting that GlDcr is capable of processing pri-miR4 into pre-miR4 and then into miR4.
FIGURE 4.
GlDcr is required for processing the mature miR4. (A) Digestion of radiolabeled pre-miR4 by GlDcr was analyzed by denaturing PAGE. The decrease in pre-miR4 level and the emergence of a 26-nt RNA band suggest that GlDcr is capable of processing pre-miR4 into mature miR4. (B) Analysis of in vitro transcribed pri-miR4 digested by GlDcr also shows the emergence of a 26-nt RNA band that is not observed in the control. Northern blot analysis of the digest presented at the bottom suggests the identity of the 26-nt RNA band to be miR4. (C) Quantitative PCR of GlDcr mRNA in ribozyme-mediated GlDcr knockdown cells shows a ∼40% decrease. (D) Western blot analysis of the GlDcr level in GlDcr knockdown cells shows a ∼25% decrease. The * represents a nonspecific band. (E) Northern blot analysis of the miR4 level in GlDcr knockdown cells show a 30% decrease. The 5.8S ribosomal RNA was used as a loading control.
To further verify that GlDcr is responsible for the maturation of miR4, we performed a giardiavirus-mediated antisense ribozyme knockdown of GlDcr in Giardia (Saraiya and Wang 2008). qRT-PCR of RNA from the knockdown cells showed that the GlDcr mRNA was reduced by ∼42% (Fig. 4C), and Western blot indicated a ∼25% decrease in GlDcr protein (Fig. 4D). Northern blot on size-fractionated (<200 nt) small RNAs from the knockdown cells with an antisense miR4 showed a 30% decrease of miR4 (Fig. 4E). This result suggests that the GlDcr is responsible for maturation of miR4 in vivo.
Identification of putative miR4 target sites in Giardia
A bioinformatic search for miR4 target sites was performed using the miRanda program (version 3; score threshold, 120; energy threshold, −20 kcal/mol; and scaling parameter, 4) (John et al. 2004). To limit the number of potential target sites, we confined our search to within a 150-nt region corresponding to 50 nt upstream of and 100 nt downstream from the stop codon from each ORF in the Giardia genome database. The identified target sites were further refined by requiring at least five consecutive base pairs (one G:U base pair allowed) within two bases from the miRNA 5′ end to form the seed sequence. Of the 4969 ORFs in the Giardia genome database, 361 were found to have potential miR4 target sites at their 3′ ends. Of the 361 ORFs, 211 were annotated, among which 69 (32.7%) were VSP genes. Furthermore, 54 of the 69 VSP target sites are located at the same position (nucleotides 15–39 in the 150-nt ORF fragment) in the VSP gene. This significant enrichment of potential miR4 targets with similar localization among the VSP genes is reminiscent of the previous identification of target sites for miR2, for which 22 VSP genes were found to carry the target site (Saraiya and Wang 2008). Since miR4 is likely missing from the two other Giardia isolates P15 and GS (see above), we used miRanda analysis on the genomic databases of these two isolates and found no enrichment of VSP targets for miR4 (two out of 163 potential GS target sites and 10 out of 163 potential P15 target sites), suggesting that miR4 is a Giardia WB-specific miRNA.
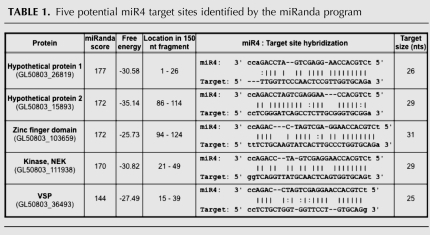
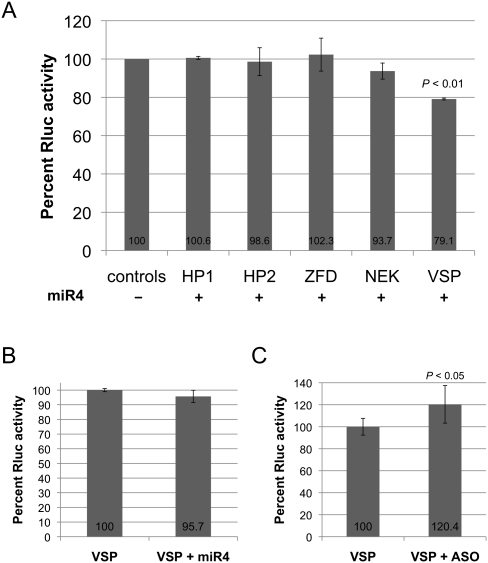
Five potential target sites were selected for further analysis based on their miRanda scores and free energy of binding (Table 1). They were derived from genes encoding two hypothetical proteins, a zinc finger domain containing protein, a NEK kinase, and a VSP. To test if these targets could interact with miR4 and repress translation in Giardia, two copies of each target site (similar to the number of target sites in RLcTS to allow comparison) were incorporated into the 3′ UTR of RLuc. The ability of miR4 to repress the expression of the reporter mRNA containing the target sites was assayed in Giardia. The results presented in Figure 5A indicate that only the expression of RLuc carrying the VSP target site was significantly repressed by miR4 (∼20%). The data also suggest that the miRanda score and free energy are not the sole predictors of potential miRNA target sites in Giardia, However, the program does allow us to limit the number of potential targets for testing in Giardia. Additional non-VSP target sites in the program were not tested and cannot be ruled out as potential miR4 targets at the present time.
TABLE 1.
Five potential miR4 target sites identified by the miRanda program
FIGURE 5.
Examination of five potential miR4 target sites. (A) The capability of miR4 to inhibit expression of the transcript of RLuc reporter gene containing one of the five potential miR4 target sites was tested (see Table 1). HP1 indicates hypothetical protein 1; HP2, hypothetical protein 2; ZFD, zinc finger domain; and NEK, NEK kinase. Expression in the absence of miR4 was set at 100% in all the controls. Only the transcript carrying the VSP target site showed an a ∼20% decrease in RLuc activity, suggesting that miR4 specifically regulates VSP gene expression. The results represent the average and standard deviations from at least three independent experiments, and the Students t-test was used to determine P-values. (B) The miR4-mediated repression of Rluc expression from the transcript carrying the VSP target sites was significantly reduced in the GlAgo knockdown cells, indicating that GlAgo is required for miR4 action. (C) Rluc activity was increased by ∼20% when miR4 2′ O-methyl antisense oligo was introduced into the cells, suggesting that endogenous miR4 represses the reporter gene expression. The results represent the average and standard deviations of at least two independent experiments, and the Students t-test was used to determine the P-values.
The transcript of RLuc carrying the double target sites from the VSP gene was also tested in the GlAgo knockdown cells. The result shows little effect from exogenously introduced miR4 (Fig. 5B), suggesting that the effect of miR4 on the target site from VSP depends on the presence of GlAgo. Additionally, transfection of the reporter along with the 2′ O-methyl miR4 antisense oligo showed a ∼20% increase in Rluc activity, suggesting that endogenous miR4 is actively repressing the translation of the reporter gene containing the VSP target sites (Fig. 5C). Expression of the 69 VSP genes carrying potential miR4 binding sites, most of which carry highly similar sequences in the region, could be also affected by introducing the 2′ O-methyl miR4 antisense into the trophozoites. But the effect may differ in individual cases, because many VSP mRNAs may carry multiple binding sites for multiple miRNAs (see below).
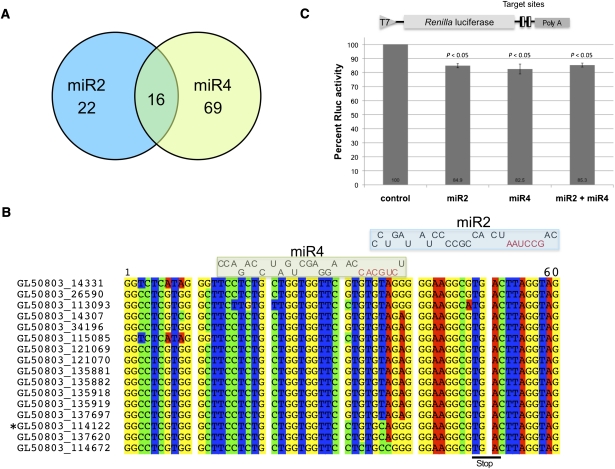
Expression of the VSP genes with target sites for both miR2 and miR4
Two Giardia miRNAs that regulate VSP expression, miR2 and miR4, have now been identified. Of the 22 VSP genes carrying miR2 target sites and the 69 VSP genes with miR4 target sites, 16 of them carry both target sites (Fig. 6A). Invariably, these 16 genes have the miR2 binding site spanning the stop codon with the seed sequence in the 3′ UTR and the miR4 binding site entirely in the coding region with the 3′ end of miR2 overlapping the seed sequence of miR4 (Fig. 6B). This positioning suggests that while both miR2 and miR4 may bind to the same VSP transcript, the translational repression may not be cooperative. To test if this postulation is correct, two copies of the sequence that contains both the miR2 and the miR4 target site from VSP-213 (GL50803_114122) were incorporated into the truncated 3′ end of RLuc (Fig. 6C). Cotransfection of the transcript from this construct with either miR2 or miR4 resulted in a ∼15% decrease in Rluc expression (Fig. 6C). However, when both miR2 and miR4 were present, the RLuc expression also resulted in a ∼15% decrease, suggesting that there is no cooperative action between the two miRNAs (Fig. 6C).
FIGURE 6.
Overlap of miR2 and miR4 VSP target sites. (A) Diagram indicating the number of miRanda-identified target sites for miR2 and miR4 among the VSP genes and the number of VSP genes carrying both miR2 and miR4 target sites. (B) Alignment of the 3′ ends of the 16 VSP genes with both miR4 and miR2 target sites. The two miRNAs are aligned on top of the potential binding sites with their seed sequences colored in red. The miR2 binding site straddles the stop codon with the seed sequence in the 3′ UTR, while the miR4 binding site is completely in the coding region with the 3′ end of miR2 occluding the miR4 seed sequence. (C) Diagram of the reporter construct containing two copies of the VSP-213 gene sequence (GL50803_114122) from nucleotides 12–62 (see Fig. 6B, the gene identified with *) containing both the miR2 and miR4 target sites. Capped RLuc reporter mRNA was transfected into Giardia cells with miR2, miR4, or both miR2 and miR4 and was assayed for RLuc activity. The activity was reduced by ∼15% in the presence of either miR2 or miR4, indicating that both miRNAs can regulate VSP-213 expression. Presence of both miRNAs did not, however, further reduce RLuc expression, suggesting that the inhibitory effect of miR2 and miR4 is not cooperative. The results represent the average and standard deviation of at least three independent experiments, and the Students t-test was used to determine P-values.
DISCUSSION
In the present study, we show that the deeply branching protozoan Giardia has the ability to utilize a gene transcript other than snoRNAs to produce a miRNA, which may play an important role in regulating the expression of a number of VSP genes.
Sources of miRNAs in Giardia
We have previously shown that snoRNA-derived miRNA-mediated translational repression is present in Giardia (Saraiya and Wang 2008). Other than the well-characterized miR2 (Saraiya and Wang 2008), we have also recently identified several additional miRNAs that are derived from snoRNAs and are playing regulatory roles in VSP expression in Giardia (Li et al. 2011; data not shown). The use of snoRNAs as miRNA precursors could bypass the need for Drosha/Pasha and Exportin5, which are apparently absent in Giardia. However, since there are only 24 snoRNAs identified in Giardia thus far (Niu et al. 1994; Yang et al. 2005; Chen et al. 2007), the exclusive use of snoRNAs as miRNA precursors would limit the number of miRNAs in Giardia and the number of mRNAs that could be regulated by miRNAs. The identification of a 94-nt RNA species derived from an unannotated 927-nt ORF as the precursor of miR4 indicates that additional sources of miRNAs are present in Giardia. The 972 nt pri-miR4 is further reduced to the 94-nt pre-miR4 and then the mature miR4 by the action of GlDcr. Thus, a miRNA biogenesis pathway reminiscent of the canonical miRNA biogenesis pathway found in higher organisms is also present in Giardia, except that GlDcr is apparently assuming the functions of both a Drosha and a Dicer. Since the usual sources of miRNAs such as the introns and long intergenic spaces found in higher eukaryotes are largely missing from Giardia (Nixon et al. 2002; Russell et al. 2005; Morrison et al. 2007), it is possible that the latter could utilize ORFs as well as snoRNAs as miRNA precursors. Recently, Ringqvist et al. (2011) identified an ORF that was highly up-regulated during host–Giardia interaction. Northern blot analysis identified the presence of this ORF and also a 24-nt small RNA, suggesting that this ORF may also be a precursor for a miRNA (Ringqvist et al. 2011). The possibility of ORF-derived miRNAs would significantly increase the potential sources of miRNAs in Giardia and make the miRNA-mediated translational repression a potentially major mechanism of gene regulation in this ancient eukaryote. It also raises questions on the potential mechanisms involved in controlling degradation of one mRNA to generate a miRNA for regulating the expression of another mRNA in Giardia.
The dependence of miR4 function on GlAgo
In a transfected Giardia cell line where GlAgo is partially depleted, miR4 loses its capability of reducing the expression of a reporter transcript carrying two miR4 antisense sequences at the 3′ UTR (Fig. 2D). This indicates that the function of miR4 is dependent on the presence of GlAgo, most likely through formation of a RISC. This postulated RISC formation was strongly supported by the coimmunoprecipitation of miR4 with HA-tagged GlAgo (Fig. 3E), establishing miR4 as a potentially functional miRNA in Giardia. By pulling down miR4 (Fig. 3E), miR2 (Fig. 3F), and a single RNA band with an estimated size of 26–30 nt from Giardia (Fig. 3B), this immunoprecipitation experiment with HA-GlAgo carries the potential of isolating most, if not all, of the miRNAs from Giardia and could constitute a library for global analysis.
Biogenesis of miR4
The indication that the 972-nt, full-length, unannotated ORF (GL50803_92663) is transcribed to generate a polyadenylated transcript, pri-miR4, suggests that the latter is further processed to produce the 94-nt pre-miR4. In vitro digestion of pri-miR4 and pre-miR4 to generate miR4 and in vivo reduction of miR4 by a partial knockdown of GlDcr indicated that GlDcr could be the sole enzyme responsible for miR4 biogenesis (Fig. 4). This tentative conclusion was further supported by the finding that the small RNAs, including miR4 and miR2, immunoprecipitated with GlAgo possess 5′ phosphates (Fig. 3C) and 2′,3′-OH (Fig. 3D), a specific characteristic of Dicer products.
In the apparent absence of Drosha or other RNaseIII motif–containing homologs in Giardia, it is possible that GlDcr is responsible for processing the pri-miR4 as well as the pre-miR4 to produce miR4 by a mechanism similar to that identified in plants. Unlike metazoa, plants utilize nuclear Dicer-like proteins to process both the pri-miRNA and the pre-miRNA. After generating the pri-miRNA by the action of RNA polymerase II, the RNA product is bound to the RNA-binding protein DAWDLE (DDL) and localizes to the nuclear processing center D-bodies (Voinnet 2009). Dicer-like 1 processes the pri-miRNA into pre-miRNA and then the pre-miRNA into the mature miRNA (Voinnet 2009). A similar mechanism is apparently present in Giardia, even though a DDL homolog has not yet been identified. Whether other RNA-binding proteins are involved in this processing is unclear. It is possible that GlDcr can shuttle between the cytoplasm and the nucleus and can process both pri-miRNAs and pre-miRNAs or that the entire miRNA maturation process occurs in the cytoplasm.
miRNA regulation of VSP expression
The presence of a potential miR4 target site at the 3′ end of the coding region in 69 VSP genes suggests that miR4 could play a role in controlling the expression of these VSPs in Giardia (Fig. 6A). This likelihood was supported by the miR4-mediated inhibition of the expression of a reporter transcript with a potential target site from a VSP gene incorporated into the 3′ UTR (Fig. 5A). The inhibition was significantly reduced in a transfected Giardia cell line partially depleted of GlAgo (Fig. 5B), suggesting the need of RISC formation between miR4 and GlAgo for the inhibitory action.
Of interest was the additional finding that 16 of the 69 VSP genes carry the potential target sites of both miR4 and miR2 (Fig. 6B). The miR2 target site spans the VSP gene stop codon with the seed sequence in the 3′ UTR, whereas the miR4 target site is completely in the coding region, but its seed sequence overlaps the 3′ end of miR2 (Fig. 6B). It is thus unlikely that both miRNAs could simultaneously bind to the same mRNA to exert their repressive effects. This prediction was verified by data from experiments in which the expression of a reporter transcript with both miR4 and miR2 target sites was inhibited by either miR4 or miR2 alone to a similar extent (Fig. 6C). The inhibitory effects, however, were neither synergistic nor cooperative (Fig. 6C), suggesting that bindings of miR4 and miR2 are mutually exclusive. At any given time, different miRNAs may bind and exert their effects on one VSP mRNA. Our data also showed that there are sufficient differences in the conserved VSP 3′ ends for binding of multiple miRNAs. We propose that multiple VSP mRNA-targeting miRNAs exist in Giardia. After transcription of multiple VSP genes, the cytoplasmic VSP mRNAs are targeted by different miRNAs. Translational inhibition by multiple miRNAs may be sufficient to completely inhibit expression of a targeted gene. Differences in the miRNA expression or miRNA concentrations may allow a unique VSP or a subset of VSP genes to be translated in a stochastic manner.
The precise mechanism by which only one VSP gene is selected for expression at a given time in Giardia is still unclear. Other potential regulatory mechanisms in addition to the miRNA machinery remain a possibility at the present time. Initial studies have shown that the upstream histones of a VSP promoter were acetylated, indicating an epigenetic mechanism of gene regulation (Kulakova et al. 2006). More recently, however, nuclear run-on experiments showed that multiple VSP genes are transcribed (Prucca et al. 2008), suggesting that epigenetic mechanisms may not be solely responsible for regulating VSP expression. Prucca et al. (2008) proposed that VSP expression is regulated by a RNAi mechanism. This was based on an antisense knock down of RNA-dependent RNA polymerase (RdRP) or GlDcr, which led to Giardia trophozoite populations expressing more than one VSP (Prucca et al. 2008). The investigators demonstrated that VSP double-stranded (ds) RNA could generate a small RNA band using Giardia cell lysate (Prucca et al. 2008). These observations led to the conclusion that VSP mRNAs are turned into dsRNAs by RdRP, processed by GlDcr, and bound to GlAgo to function in an RNAi pathway (Prucca et al. 2008). It is, however, not possible to identify a specific GlDcr cleaved product in a Giardia lysate due to the presence of many RNase activities. Furthermore, GlDcr is incapable of digesting long dsRNAs in vivo. The dsRNA giardiavirus is stable and multiplying in the cytoplasm of Giardia trophozoites (Miller et al. 1988). A recent study of dsRNAs artificially generated in Giardia trophozoites also showed no degradation of the dsRNAs into small RNAs, suggesting that GlDcr is incapable of processing long dsRNAs (Rivero et al. 2010). Additionally, our investigations of miR2 and miR4 showed that neither miRNA is derived from VSP antisense RNA. Inhibition of mRNA expression by these miRNAs does not lead to reduction of the mRNA level, indicating that GlAgo does not cleave the targeted mRNA even in the presence of a perfectly complimentary target site. This is a standard feature of the miRNA-Argonaute-mediated repression of translation. With all the information available, we conclude that the miRNAs are playing a role in regulating the expression of VSPs in Giardia.
MATERIALS AND METHODS
RNA isolation
Total RNA was isolated from Giardia trophozoites using Trizol (Invitrogen), while size-fractionated RNA (<200 nt) was isolated using the High Pure miRNA Isolation kit (Roche). Isolation of small RNAs <40 nt was accomplished by gel fractionation of the size-fractionated RNA (<200 nt) and excision of the 26- to 30-nt region. The excised gel fragment was crushed, and the RNA was eluted into 300 μL 1× TE buffer with 1 U/μL SUPERase•In (Ambion) and incubated overnight at 4°C with end-over-end mixing followed by standard ethanol precipitation.
Cloning of the Rluc reporter gene with miR4 target sites
We have previously constructed a plasmid pRL containing the RLuc reporter gene under control of the T7 promoter and a multiple cloning site followed by A50 at the 3′ end (Saraiya and Wang 2008). Two copies of each target site was incorporated into the Rluc 3′ UTR by PCR amplification using the Rluc seq primer (5′-AGAGAAAGGTGAAGTTCGTC-3′) and the appropriate target site primers (RLcTS: 5′-ATCCCGCGGTCTGCACCAAGGAGCTGATCCAGACCGGGCCCTCTGCACCAAGGAGCTGATCCAGACCAGATCTTTATTGTTC-3′; GL50803_26819: 5′-GATCCGCGGATATCTGCACCAACGGAGTTGGGAACCAACCGGTTCTGCACCAACGGAGTTGGGAACCAAAGATCTTTATTGTTC-3′; GL50803_15893: 5′-GATCCGCGGATATCCGCACCCGCAAGAGGCTGATCCCGAGGGTACCTCCGCACCCGCAAGAGGCTGATCCCGAGGAGATCTTTATTGTTC-3′; GL50803_103659: 5′-GATCCGCGGATATCTGCACCAGGGCAAGTGATACTTGCAGACCGGTTCTGCACCAGGGCAAGTGATACTTGCAGAAGATCTTTATTGTTC-3′; GL50803_111938: 5′-GATCCGCGGATAACTGCACCACTGAGTTGCATAACCTGACCCCGGTACTGCACCACTGAGTTGCATAACCTGACCAGATCTTTATTGTTC-3′; GL50803_36493: 5′-GATCCGCGGATACCTGCACAGGAACCACCAGCAGAGGCCGGTCCTGCACAGGAACCACCAGCAGAGGAGATCTTTATTGTTC-3′) and cloned into pGEM-T (Promega). Clones were sequenced and subcloned into pRL using the SacII and BsrGI restriction sites.
The first miR2/miR4 dual target site was incorporated into pRL by PCR amplification using the Rluc seq primer and primer miR2/4 114122 R-1 (5′-AGGAACCAGCTGCACTACCTAAGTCACGCCTTCCCCCTGCACAGGAACCACCAGCAGAGGAAGAGGCGCGCCTTATTG-3′). The resulting PCR product was used as a template to add the second miR2/miR4 target site by PCR amplification using the Rluc seq primer and primer miR2/4 114122 R-2 (5′-AATCCGCGGCACTACCTAAGTCACGCCTTCCCCCTGCACAGGAACCACCAGCAGAGGAAGCAGCTG-3′). The resulting PCR product was cloned into pRL as described above.
miRNA assay
The miRNA assay was performed as previously described using either Giardia WB trophozoites or the GlAgo knockdown cells (Saraiya and Wang 2008).
HA-GlAgo cloning, expression and immunoprecipitation
HA-GlAgo was PCR amplified from Giardia genomic DNA using pNlop4 2HA GlAgo F primer (5′-GATACTAGTATGTATCCTTATGATGTTCCTGATTATGCTTATCCTTATGATGTTCCTGATTATGCTATGGTAGCAGATGTTGTC-3′) and the R primer (5′-ATGCGAATTCAAATATTTACTGCCTAGGTTATATGAAGAATGGTC T-3′). The product was cloned into pGEM-T (Promega), sequenced, and then subcloned into the pNlop4 vector, a derivative of the pNLop3-GtetR vector, using SpeI and EcoRI (Sun and Tai 2000). The resulting construct (pNlop4 HA-GlAgo) was transfected into Giardia trophozoites as previously described, and the transformants were selected with 200 μg/mL G418 (Singer et al. 1998). The selected cells were induced with 10 μg/mL of tetracycline for 24 h at 37°C and assayed for HA-GlAgo by Western blot to confirm its expression (data not shown).
Immunoprecipitation of HA-GlAgo was performed using the ProFound HA Tag IP/Co-IP Kit (Pierce). Briefly, about 108 cells were induced with 10 μg/mL tetracycline for 24 h at 37°C, pelleted, and lysed with 700 μL RIPA buffer (50 mM Tris-HCl at pH 7.5; 150 mM NaCl; 2 mM EDTA at pH 8.0; 1% NP-40) in the presence of 2× Halt protease inhibitor cocktail (Thermo Scientific) and 1 U/μL SUPERase•In (Ambion). The cells were allowed to lyse for 1–2 h at 4°C with end-over-end mixing followed by centrifugation at 16,000g for 20 min at 4°C to pellet cellular debris. The cleared lysate was mixed with 6 μL of anti-HA agarose beads that were preincubated with 150 μg of BSA and 150 μg of Salmon sperm DNA (Invitrogen). After an incubation at 4°C with end-over-end mixing for 16 h, the mixture was pulse centrifuged at 16,000g to remove the supernatant. The agarose beads were then washed five times with 500 μL of Tris buffered saline plus 0.05% Tween-20. The RNA associated with the HA-GlAgo–anti-HA agarose complex was extracted with 500 μL Trizol (Invitrogen) using the manufacturer's protocol and suspended in 20 μL RNase-free water. The extracted RNA was analyzed by 15% denaturing PAGE. An ∼26- to 30-nt band was observed in the HA-GlAgo immunoprecipitation, but not in the control. The ∼26- to 30-nt band was excised from the gel, crushed, and incubated overnight at 4°C with end-over-end mixing in 1× TE buffer with 1U/μL SUPERase•In (Ambion) to extract the RNA followed by ethanol precipitation.
Real-time PCR identification of GlAgo associated miRNAs
The extracted RNA was reverse transcribed with miRNA specific primers (miR2: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGACTAT-3′; miR4: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGTCTG-3′) following the provided protocol for SuperScript III (Invitrogen). The cDNA was used to perform a Taqman assay using PrimeTime qPCR (IDT) primers to detect the cDNAs of miR4 (Probe: 5′-FAM-AGTACTGGCGGGGCCTGAC-Iowa Black 3′; Primer 1: 5′-GGAAACTCCTTAACTCGTTCCG-3′; Primer 2: 5′-GTGTTCCCACGACGTCTC-3′) and miR2 (Probe: 5′-FAM-ACTGGATAC-ZEN-GACGGACTATAG-Iowa Black 3′; Primer 1: 5′-CAGCCTAATCACCGCC-3′; Primer 2: 5′ GTCGTATCCAGTGCAGG-3′) using the Bio-Rad CFX96 Real-Time System.
Examining the ends of small RNAs coimmunoprecipitated with HA-GlAgo
To examine the 5′ ends of the small RNAs coimmunoprecipitated with HA-GlAgo, the small RNAs were treated with the 5′-3′ exonuclease Terminator (Epicentre) with or without an Antarctic phosphatase (NEB) pretreatment to remove potential 5′ phosphates as directed by the suppliers. Products from the reactions were analyzed by 8% denaturing PAGE.
The 3′ ends of the small RNAs coimmunoprecipitated with GlAgo were checked by β-elimination. Briefly, HA-GlAgo coimmunoprecipitated small RNAs were incubated with 100 mM sodium periodate in 10 mM Tris-HCl (pH 7.5) for 30 min at 22°C. An equal volume of formamide loading buffer was then added, and the reaction mixture was incubated for 30 min at 99°C for cleavage of the 3′-terminal nucleotide mediated by the 2′,3′-dialdehyde generated by the periodate treatment. Products from the reactions with an anticipated loss of 3′ end nucleotide were analyzed by 8% denaturing PAGE.
In vitro GlDcr digestion of pri-miR4 and pre-miR4
The pri-miR4 was PCR amplified from Giardia genomic DNA with pri-miR4 T7 F (5′-TAATACGACTCACTATAGCCATGGATGGCTACTCTCAAAGACC-3′) and pri-miR4 RNA R (5′-TCAGATCAGAATCAATTCGTCAG-3′) and cloned into pGEM-T and sequenced. The clone was used as a template to PCR amplify the pri-miR4. The amplified DNA was used as template for in vitro transcription (Ambion) of the pri-miR4, which was gel purified and used for the GlDcr digestion assay. The pri-miR4 was incubated with 1 U of the commercial GlDcr for 16 h at 37°C. The reaction was analyzed by denaturing 8% PAGE.
The pre-miR4 was PCR amplified from Giardia genomic DNA with pre-miR4 T7 F (5′-TAATACGACTCACTATAGCGTCCGCAAGCATCTCC-3′) and miR4 full R (5′-GGTCTGGATCAGCTCCTTGGTGCAGA-3′). The ethanol precipitated DNA was used for trace [32P-UTP]-labeled in vitro transcription (Ambion) of pre-miR4. The RNA product was passed through a G-25 Sephadex column (GE Healthcare) to remove unincorporated nucleotides and was incubated with 2 or 4 U of a commercial GlDcr sample (PowerCut Dicer, Finnzymes) for 16 h at 37°C. The reaction products were analyzed by denaturing 10% PAGE.
Analysis of ribozyme-mediated GlDcr knockdown
Total RNA from GlDcr knockdown cells was isolated with Trizol and reverse transcribed with Superscript III (Invitrogen) using an oligo(T)16 primer. GlDcr mRNA levels were observed by qPCR using primers qPCR Dicer F (5′-AAAATACAGCCTGACAGCAAGG-3′) and qPCR Dicer R (5′-TGGTCGCCAGAAGGGTGTA-3′) along with 1 μL of cDNA and the Faststart Universal SYBR Green Master mix (Roche). The Giardia tubulin mRNA [qPCR tubulin F (5′-TACATGGCGTGCTGCATGATG-3′) and qPCR tubulin R (5′-TCGTGTTGGAGATCATGAGGC-3′)] was used as a control. The qPCR was performed using the Bio-Rad CFX96 real-time thermocycler.
The GlDcr protein levels were determined using a rabbit poly-clonal antibody raised against two GlDcr peptides (peptide 1: C-PTGPVASKVLALYEK; peptide 2: C-PSVVSEVIIESHPKI). An anti-tubulin antibody (DM1A; Sigma) was used as a loading control.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Wang laboratory of UCSF for useful suggestions and critical comments throughout this study. The work was supported by an NIH grant R01 AI-30745.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028118.111.
REFERENCES
- Adam RD, Nigam A, Seshadri V, Martens CA, Farneth GA, Morrison HG, Nash TE, Porcella SF, Patel R 2010. The Giardia lamblia vsp gene repertoire: characteristics, genomic organization, and evolution. BMC Genomics 11: 424 doi: 10.1186/1471-2164-11-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. 2009. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res 37: D526–D530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Brown JW, Marshall DF, Echeverria M 2008. Intronic noncoding RNAs and splicing. Trends Plant Sci 13: 335–342 [DOI] [PubMed] [Google Scholar]
- Chen XS, Rozhdestvensky TS, Collins LJ, Schmitz J, Penny D 2007. Combined experimental and computational approach to identify non-protein-coding RNAs in the deep-branching eukaryote Giardia intestinalis. Nucleic Acids Res 35: 4619–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf HG, Singer SM, Nash TE 2001a. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res 29: 4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf HG, Singer SM, Pierce J, Cowan J, Nash TE 2001b. Initiator and upstream elements in the α2-tubulin promoter of Giardia lamblia. Mol Biochem Parasitol 113: 157–169 [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human microRNA targets. PLoS Biol 2: e363 doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova L, Singer SM, Conrad J, Nash TE 2006. Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Mol Microbiol 61: 1533–1542 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Li L, Wang CC 2004. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J Biol Chem 279: 14656–14664 [DOI] [PubMed] [Google Scholar]
- Li W, Saraiya AA, Wang CC 2011. Gene regulation in Giardia lamblia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl Trop Dis 5: e1338 doi: 10.1371/journal.pntd.0001338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Kim H, Ying SY 2008. Intron-mediated RNA interference and microRNA (miRNA). Front Biosci 13: 2216–2230 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miller RL, Wang AL, Wang CC 1988. Purification and characterization of the Giardia lamblia double-stranded RNA virus. Mol Biochem Parasitol 28: 189–195 [DOI] [PubMed] [Google Scholar]
- Miska EA 2005. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926 [DOI] [PubMed] [Google Scholar]
- Nash TE, Banks SM, Alling DW, Merritt JW Jr, Conrad JT 1990. Frequency of variant antigens in Giardia lamblia. Exp Parasitol 71: 415–421 [DOI] [PubMed] [Google Scholar]
- Nash TE, Lujan HT, Mowatt MR, Conrad JT 2001. Variant-specific surface protein switching in Giardia lamblia. Infect Immun 69: 1922–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu XH, Hartshorne T, He XY, Agabian N 1994. Characterization of putative small nuclear RNAs from Giardia lamblia. Mol Biochem Parasitol 66: 49–57 [DOI] [PubMed] [Google Scholar]
- Nixon JE, Wang A, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J 2002. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci 99: 3701–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G 2002. Control of developmental timing by microRNAs and their targets. Annu Rev Cell Dev Biol 18: 495–513 [DOI] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T 2009. MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD 2008. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456: 750–754 [DOI] [PubMed] [Google Scholar]
- Ringqvist E, Avesson L, Soderbom F, Svard SG 2011. Transcriptional changes in Giardia during host-parasite interactions. Int J Parasitol 41: 277–285 [DOI] [PubMed] [Google Scholar]
- Rivero MR, Kulakova L, Touz MC 2010. Long double-stranded RNA produces specific gene downregulation in Giardia lamblia. J Parasitol 96: 815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Shutt TE, Watkins RF, Gray MW 2005. An ancient spliceosomal intron in the ribosomal protein L7a gene (Rpl7a) of Giardia lamblia. BMC Evol Biol 5: 45 doi: 10.1186/1471-2148-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog 4: e1000224 doi: 10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ 2009. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 5: e1000507 doi: 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SM, Yee J, Nash TE 1998. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol 92: 59–69 [DOI] [PubMed] [Google Scholar]
- Stadler BM, Ruohola-Baker H 2008. Small RNAs: keeping stem cells in line. Cell 132: 563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Tai JH 2000. Development of a tetracycline controlled gene expression system in the parasitic protozoan Giardia lamblia. Mol Biochem Parasitol 105: 51–60 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang YT, Pan YJ, Cho CC, Lin BC, Su LH, Huang YC, Sun CH 2010. A novel pax-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J Biol Chem 285: 32213–32226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Zhou H, Luo J, Qu LH 2005. Identification of 20 snoRNA-like RNAs from the primitive eukaryote, Giardia lamblia. Biochem Biophys Res Commun 328: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Yee J, Tang A, Lau WL, Ritter H, Delport D, Page M, Adam RD, Müller M, Wu G 2007. Core histone genes of Giardia intestinalis: genomic organization, promoter structure, and expression. BMC Mol Biol 8: 26 doi: 10.1186/1471-2199-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U 2008. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog 4: e1000219 doi: 10.1371/journal.ppat.1000219 [DOI] [PMC free article] [PubMed] [Google Scholar]