Ribonuclease J is an essential enzyme, and the Bacillus subtilis ortholog possesses both endoribonuclease and 5′ → 3′ exoribonuclease activities. Chloroplasts also contain RNase J, which has been postulated to participate, as both an exo- and endonuclease, in the maturation of polycistronic mRNAs. Virus-induced gene silencing of RNase J expression caused surprisingly few disruptions in the cleavage of polycistronic rRNA and mRNA precursors. In contrast, antisense RNAs accumulated massively, suggesting that the failure of chloroplast RNA polymerase to terminate effectively leads to extensive symmetric transcription products that are normally eliminated by RNase J. Accordingly, a major role of chloroplast RNase J is RNA surveillance to prevent overaccumulation of antisense RNA, which would otherwise exert deleterious effects on chloroplast gene expression.
Keywords: antisense RNA, RNase J, chloroplast, exoribonuclease
Abstract
Ribonuclease J is an essential enzyme, and the Bacillus subtilis ortholog possesses both endoribonuclease and 5′ → 3′ exoribonuclease activities. Chloroplasts also contain RNase J, which has been postulated to participate, as both an exo- and endonuclease, in the maturation of polycistronic mRNAs. Here we have examined recombinant Arabidopsis RNase J and found both 5′ → 3′ exoribonuclease and endonucleolytic activities. Virus-induced gene silencing was used to reduce RNase J expression in Arabidopsis and Nicotiana benthamiana, leading to chlorosis but surprisingly few disruptions in the cleavage of polycistronic rRNA and mRNA precursors. In contrast, antisense RNAs accumulated massively, suggesting that the failure of chloroplast RNA polymerase to terminate effectively leads to extensive symmetric transcription products that are normally eliminated by RNase J. Mung bean nuclease digestion and polysome analysis revealed that this antisense RNA forms duplexes with sense strand transcripts and prevents their translation. We conclude that a major role of chloroplast RNase J is RNA surveillance to prevent overaccumulation of antisense RNA, which would otherwise exert deleterious effects on chloroplast gene expression.
INTRODUCTION
The chloroplast is a semi-autonomous organelle derived from a cyanobacterial ancestor. The chloroplast combines bacterial and acquired eukaryotic traits, and among chloroplast functions that have particularly complex origins are the various steps of gene expression (Stern et al. 2010; Barkan 2011). For example, chloroplasts have a eubacterial RNA polymerase, prokaryotic-type operons, and bacterial-like ribosomes. On the other hand, a second, phage-like, RNA polymerase was probably acquired through horizontal gene transfer; polycistronic transcripts are generally processed to give rise to smaller species, and chloroplasts possess numerous introns and frequently modify transcripts by C-to-U RNA editing. These characteristics raise the question of how retained prokaryotic mechanisms are intertwined with eukaryotic ones.
Chloroplast RNA metabolism is an aspect of gene expression that possesses mechanisms of dual origins. RNA splicing, RNA editing, and RNA stability all are heavily dependent on a variety of RNA-binding proteins that are not found in prokaryotes. These include members of the penta-tricopeptide repeat (PPR) family, which is massively expanded in plants and primarily regulates organellar genes (for review, see Schmitz-Linneweber and Small 2008), as well as other families. On the other hand, the catalytic enzymes of RNA metabolism, both endo- and exoribonucleases, are mainly recognizable as prokaryotic.
The major chloroplast endoribonucleases so far described are RNase E, which is also found in prokaryotes, and CSP41, which appears to be plant-specific. The RNase E catalytic domain has a similar activity to its Escherichia coli counterpart (Schein et al. 2008), but the protein also contains a long and variable plant-specific domain of unknown function. RNase E is essential for photosynthesis, and null mutants are stunted and chlorotic (Mudd et al. 2008). Arabidopsis rne mutants accumulate some transcript precursors, and its phenotype has been proposed to result from poor expression of one or more ribosomal protein transcripts (Walter et al. 2010). CSP41 is a heterodimer of two related proteins, and double mutants have a mild growth defect and few RNA aberrations (Bollenbach et al. 2009), although CSP41 has strong endonucleolytic activity in vitro (Yang et al. 1996).
Two chloroplast exoribonucleases have been studied, RNase R and polynucleotide phosphorylase (PNPase). RNase R mutants have a similar growth phenotype to rne mutants and have defects in the processing of some rRNAs (Kishine et al. 2004; Bollenbach et al. 2005). The rnr mutant also overaccumulates an antisense RNA (asRNA) complementary to 5S rRNA precursors, which we have proposed leads to depletion of 5S rRNA and inefficient translation (Sharwood et al. 2011). PNPase has both 3′ → 5′ exonucleolytic activity and 5′ → 3′ polymerization activity (Yehudai-Resheff et al. 2003) and is involved both in polyadenylation, which destabilizes chloroplast transcripts, and in transcript maturation. PNPase mutants have chlorotic young leaves but are fertile (Marchive et al. 2009) and have multiple defects in 3′-end maturation as well as intron degradation (Walter et al. 2002; Germain et al. 2011).
A chloroplast enzyme whose function remains enigmatic is RNase J, which has been identified through proteomics (Kleffmann et al. 2004) and whose absence is known to cause embryo lethality (Tzafrir et al. 2004). RNase J was discovered in Bacillus subtilis (Even et al. 2005), which lacks RNase E and is found as a heterotetramer of RNase J1 and the related RNase J2 (for review, see Condon 2010). RNase J contains a β-CASP metallo-β-lactamase fold and is essential for viability in B. subtilis, although the molecular basis for this is still unclear. RNase J is not found in E. coli; however, it is present in cyanobacteria and Archaea (Clouet-d'Orval et al. 2010; Hasenohrl et al. 2011).
RNase J from B. subtilis was found to have both endonucleolytic and exonucleolytic activities in vitro, with the exonucleolytic activity being more robust (Deikus et al. 2008). Bacterial RNase J catalyzes 5′ → 3′ degradation (Mathy et al. 2007), which is most efficient with a 5′ monophosphate or hydroxyl due to the structure of the catalytic site (de la Sierra-Gallay et al. 2008). Thus, primary triphosphorylated transcripts are thought to be largely impervious to RNase J exonucleolytic attack. Chloroplasts are known to have a net 5′ → 3′ RNA degradation pathway (Drager et al. 1999; Hicks et al. 2002), and RNase J has also been proposed to catalyze 5′ end trimming of monocistronic RNAs and polycistronic transcript derivatives, being stalled by gene-specific RNA binding proteins (see Fig. 2 in Barkan 2011). Because the endonucleolytic activity of RNase J requires high enzyme concentrations in vitro, it is unclear whether this activity is relevant to its in vivo function. We show here that the recombinant plant enzyme catalyzes both 5′ → 3′ exonucleolytic and significant endonucleolytic activities.
To deduce the role of RNase J in chloroplast RNA metabolism, we have used Virus-Induced Gene Silencing (VIGS) to repress expression of the encoding gene. While we did detect modest changes in the patterns of rRNAs and mRNAs, most striking was a massive accumulation of asRNA. Our data suggest that this asRNA sequesters sense transcripts in dsRNA duplexes, preventing efficient translation and ultimately causing the observed lethality of RNase J null mutants.
RESULTS
In vitro activity of chloroplast RNase J
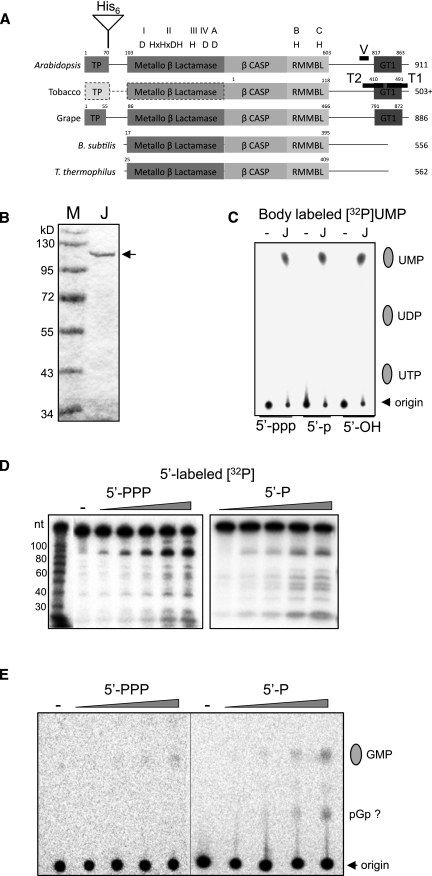
Figure 1A compares the domain structures of RNase J from B. subtilis with several predicted plant sequences including the two studied here, Arabidopsis thaliana and Nicotiana benthamiana. Each protein includes the signature domains of RNase J as a metallo-β-lactamase protein, denoted motifs I–IV and A–C. The plant proteins in addition possess short N-terminal extensions, which, in the case of the Arabidopsis protein, was demonstrated by transient expression of YFP fusions in protoplasts to confer chloroplast targeting (Supplemental Fig. S1). In our experiments no evidence for mitochondrial localization was observed, nor has RNase J been reported in mitochondrial proteomes. One intriguing feature of the plant enzymes is a putative C-terminal GT-1-like DNA-binding domain (Lam 1995), whose properties remain to be tested.
FIGURE 1.
Recombinant AtRNase J has both exo- and endoribonuclease activities. (A) Alignment of RNase J proteins from Arabidopsis (At5g63420), tobacco (Unigene U431192), grape (XM_002279762.1), B. subtilis, and Thermus thermophilus. (TP) predicted chloroplast transit peptide; (His6) site of histidine tag insertion for recombinant protein. V, T1, and T2 mark regions of the gene targeted by VIGS constructs. The plant C-terminal regions include homology with the GT1 DNA-binding domain. The N-terminal portion of the tobacco protein marked with a dashed line and lighter shades is unsequenced and thus a prediction. The conserved domains of the β-lactamase superfamily (I–IV; A–C) are indicated along with the diagnostic amino acid residues. (B) Coomassie-stained gel of bacterially expressed and purified Arabidopsis RNase J lacking the transit peptide (arrow). (C) RNase J was incubated for 4 h with body-labeled RNAs with 5′ ends as indicated at the bottom of the figure, and analyzed by TLC. (Lane –) Incubation without protein. (D) Recombinant RNase J was incubated with tri- or monophosphorylated 5′-end-labeled RNA. Following incubation for 0, 10, 30, 60, or 120 min, the RNA was analyzed by denaturing PAGE. (Lane –) Incubation without protein for 120 min. (E) The samples shown in panel C were analyzed by TLC. The migration of GMP was determined by using a standard. The second radioactive species appearing in the reaction where monophosphorylated RNA was analyzed migrated similarly to a GDP standard and therefore could be pGp.
To explore the catalytic properties of chloroplast RNase J, the recombinant Arabidopsis protein, minus the chloroplast transit peptide, was expressed and purified (Fig. 1B). To determine whether RNase J could catalyze exonucleolytic digestion, a uniformly [α-32P]UTP-labeled synthetic transcript corresponding to a portion of the chloroplast petD gene was incubated with the enzyme, and the products were analyzed by thin layer chromatography (Fig. 1C). Because bacterial RNase J poorly digests triphosphorylated substrates exonucleolytically, we compared activity on triphosphorylated, monophosphorylated, and 5′-hydroxylated versions. The results (Fig. 1C) demonstrated that RNase J generated similar amounts of UMP from each substrate after a 4-h incubation, confirming its exonucleolytic activity. Since the archaeal and bacterial enzymes discriminate for monophosphate at the 5′ end (e.g., Hasenohrl et al. 2011), there are two ways to interpret these results. The first one is that Arabidopsis RNase J, unlike the other RNase J enzymes characterized so far, degrades exonucleolytically 5′-end mono- and triphosphorylated RNAs at a similar rate. The second possibility is that although the exoribonucleolytic activity is sensitive to the nature of the 5′ end, the Arabidopsis enzyme has strong endonucleolytic activity that produces 5′-monophosphorylated molecules that are then rapidly degraded by the exonuclease activity. This would lead to similar apparent exonucleolytic degradation rates for the two substrates when incubated for longer periods.
To check for endonucleolytic activity, we analyzed the activity of RNase J on a 5′ [32P]-labeled petD substrate harboring one or three phosphates at their 5′ ends, reasoning that only endonucleolytic cleavage products would retain the [32P] label, since any 5′ → 3′ exonucleolytic activity would remove it. As shown in Figure 1D, recombinant RNase J generated discrete endonucleolytic 5′-labeled cleavage products, and the kinetics of their appearance were similar for RNA with mono- or triphosphates at the 5′ end. Hence, chloroplast RNase J appears to harbor both exo- and endonuclease activity under in vitro conditions, and the endonuclease activity is insensitive to the number of phosphates at the 5′ end. To analyze if the exonucleolytic activity is sensitive to the number of phosphates, the initial degradation rates of mono- and triphosphorylated RNAs, labeled at their 5′ ends, were analyzed by TLC by detecting the released 5′-end mononucleotide (Fig. 1E). Although the amount of signal obtained by analysis at these short incubation times was low as compared to the 4-h time point shown in Figure 1C, a higher accumulation of GMP from the monophosphorylated RNA was repeatedly observed. Together, these results show that the Arabidopsis RNase J is active both as a 5′ → 3′ exonuclease and an endonuclease, and that as for other RNase J enzymes, the exonucleolytic activity prefers 5′-monophosphorylated molecules. However, due to the relatively robust endonucleolytic activity that is insensitive to the nature of the 5′ end and produces 5′-monophosphorylated cleavage products, the apparent degradation rates of uniformly labeled molecules harboring one or three phosphates at the 5′ end is similar.
Repression of RNase J expression leads to chlorosis and more diffuse RNA patterns
Because RNase J null mutants cannot be propagated, we used VIGS to silence RNase J partially. To control for any off-target effects, we initially compared results for Arabidopsis and N. benthamiana (tobacco). We also used two different constructs for tobacco, targeting different segments (T1 and T2) of the RNJ transcript (see Materials and Methods). These constructs gave indistinguishable results; therefore, only data from T1-infiltrated plants are shown here.
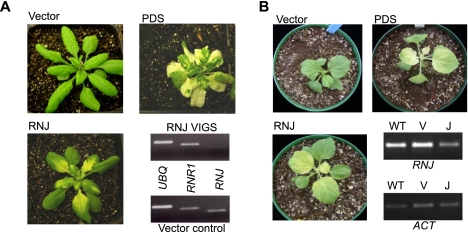
Figure 2A shows the phenotypes of Arabidopsis plants 25 d after infiltration with Agrobacterium carrying the VIGS vector only, a construct targeting the carotenoid biosynthesis enzyme phytoene desaturase (PDS), or the RNase J T1 construct. As expected, plants infiltrated with only the vector showed no phenotypic effects, and the expected patchy chlorosis was observed when carotenoid synthesis was blocked (PDS). For RNase J, a slightly milder chlorosis was observed in the basal portion of emerging leaves, indicating that the expression of RNJ is required for photosynthetic competence. To verify that the RNJ gene was being silenced, RT-PCR was used. This confirmed that RNJ mRNA was reduced by ∼70%, when compared with two control nuclear genes. We do not yet possess an antibody that can measure the RNase J protein level; however, it is reasonable to assume that RNJ transcript depletion leads to a reduction of the enzyme level, which is, in turn, responsible for the plant phenotype. When the experiment was repeated in tobacco, comparable results were obtained, as shown in Figure 2B, although chlorosis eventually spread to the entire leaf. We conclude that partial silencing of RNase J expression leads to chlorosis, suggesting an impact on chloroplast gene expression.
FIGURE 2.
Generation of RNase J–deficient plant material. (A) Arabidopsis plants 25 d post-infiltration and analysis of silencing using RT-PCR. “Vector” plants were infiltrated with the empty pYL170 plasmid; “PDS” plants were silenced for phytoene desaturase gene expression; and in “RNJ” plants, RNase J was targeted. For the RT-PCR analysis, UBQ (ubiquitin) and RNR1 (chloroplast RNase R) were used as nuclear gene controls. (B) Tobacco plants 23 d post-infiltration and analysis of silencing using RT-PCR. Labeling is as for panel A, except for RT-PCR; (V) vector-only control; (J) plants silenced for RNase J expression.
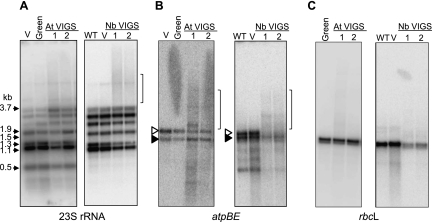
To gain a general picture of the effects of RNase J depletion on chloroplast transcripts, we performed a series of RNA gel blots, and comparisons of Arabidopsis and tobacco for three representative genes are shown in Figure 3. When rRNAs were examined, we observed a decrease in the abundance of mature species for both 16S (Supplemental Fig. S2, left side) and 23S (Fig. 3A) rRNAs, and in the case of Arabidopsis, a concomitant accumulation of precursors. This phenotype is commonly observed in chloroplast gene expression mutants, however, and may be a pleiotropic consequence of ribosome assembly defects (Barkan 1993; Yu et al. 2008 and references therein). We also compared the 5′ termini of 16S and 23S rRNAs between control and RNase J–deficient plants by gel analysis of 5′-RACE products; RNase J has been implicated in maturation of both the 16S and 23S rRNA 5′ ends in bacteria (Britton et al. 2007; Madhugiri and Evguenieva-Hackenberg 2009). We found that RNase J deficiency resulted in a series of 5′ extensions of 16S rRNA and the presence of a single 5′-extended species for 23S rRNA (Supplemental Fig. S2, right side), suggestive of a conserved function. Finally, the 23S rRNA probe identified a higher-molecular-weight smear (indicated by a bracket for tobacco), which was not further investigated.
FIGURE 3.
Transcript analysis from RNase J–deficient Arabidopsis and tobacco. Total RNA was analyzed from Arabidopsis (left gel of each panel) or tobacco (right gel of each panel). (WT) Wild-type untransformed; (V) vector only control; (Green) asymptomatic tissue; (At VIGS or Nb VIGS) independent RNase J–deficient Arabidopsis or tobacco plants, respectively. RNA blots were hybridized with [32P]dCTP-labeled DNA probes to detect (A) 23S rRNA; (B) the dicistronic atpBE transcript, which encodes the ATP synthase β and ɛ subunits); and (C) rbcL, which encodes the Rubisco large subunit. In panel B, the open and filled arrowheads represent transcription initiation by chloroplast RNA polymerases PEP and NEP, respectively (Schweer et al. 2006). Brackets in panels A and B mark the positions of heterodisperse RNAs discussed in the text.
When two mRNAs, atpBE (Fig. 3B) and rbcL (Fig. 3C), were examined, a variety of effects were observed. In the case of atpBE, the predominant WT species were still present, but in diminished abundance, particularly in tobacco. At the same time, higher-molecular-weight smears were seen for both species, along with a few discrete transcripts (brackets). For rbcL, no significant change was seen for Arabidopsis, whereas in tobacco the WT species underaccumulated, suggesting that rbcL is less sensitive than atpBE to the RNase J level. In addition, for rbcL, only a faint higher-molecular-weight smear was observed. While low-molecular-weight smearing is generally a sign of RNA degradation artifacts, the longer transcripts observed here do not suggest this interpretation. Instead, this result was consistent with a role of RNase J in maturing these mRNAs.
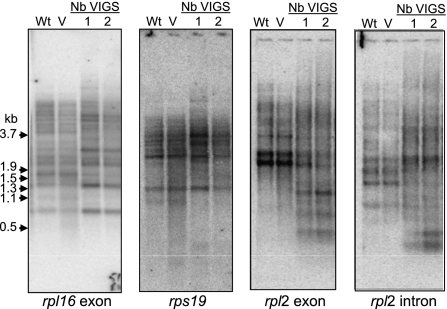
Because the rRNA results led us to infer that ribosome assembly might be perturbed, we examined transcripts encoding ribosomal proteins, as exemplified in Figure 4 (from this point forward, only results from tobacco are shown). The three genes shown are members of the S10 operon, which generates the complex transcript patterns typical of chloroplasts. In all cases, results analogous to those observed for atpBE were obtained. Each probe detected the same bands as seen in WT or vector control material, but at a diminished level, while additional higher- and lower-molecular-weight bands, and a diffuse smear, were also present. It would not be surprising if these altered RNA patterns caused reduced expression of the encoded proteins, although we have not tested this directly. In summary, RNase J depletion similarly affects RNAs emanating from monocistronic genes (atpBE and rbcL) and polycistronic gene clusters. In examining other chloroplast genes, a consistent story emerged (Supplemental Fig. S2). We conclude that RNase J has a global role in chloroplast RNA metabolism, especially where mRNAs are concerned.
FIGURE 4.
Ribosomal protein transcript analysis in tobacco. Total RNA was analyzed as described in the legend to Figure 3.
Diffuse transcripts and novel bands are mainly antisense RNAs
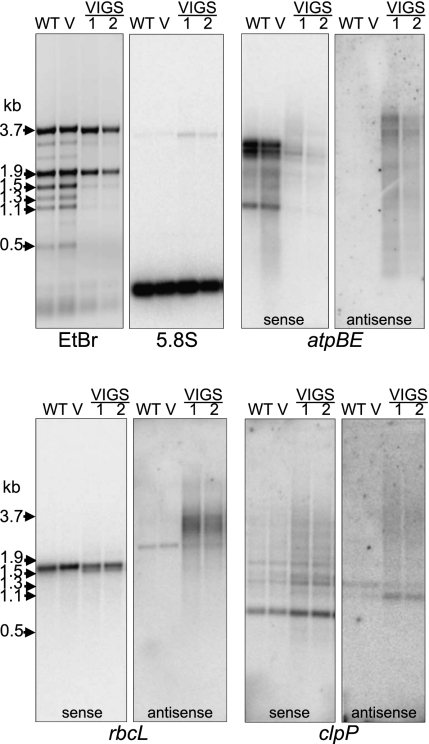
Although our initial hypothesis, as stated above, was that RNase J–deficient material was accumulating maturation and possibly degradation intermediates, we elected to repeat our gel blots using strand-specific probes. We reasoned that several recent reports, including our own, had described the accumulation of chloroplast antisense or non-coding RNAs (Lung et al. 2006; Georg et al. 2010; Hotto et al. 2010), and that these might be substrates for RNase J. If this were the case, they could overaccumulate in mutant material. Figure 5 demonstrates that this is, indeed, the case for atpBE, rbcL, and clpP. When probes were used that specifically detect sense strand transcripts, mainly quantitative differences were observed. The decreased abundance for atpBE and rbcL was consistent with our interpretations from double-stranded probes and also probably reflects a generally decreased level of chloroplast transcripts, as can be seen by the reduced accumulation of chloroplast rRNAs in the ethidium bromide–stained gel (top left panel). In contrast, clpP mRNA abundance appeared to increase, although the transcript pattern did not change.
FIGURE 5.
Accumulation of antisense transcripts in RNase J–reduced leaves. Strand-specific transcript analysis. Total RNA isolated from control and RNase J–reduced tobacco leaves was analyzed by RNA gel blot. Loading is reflected by ethidium bromide staining (top left) and by hybridization to detect nuclear 5.8S rRNA. For atpBE, rbcL, and clpP, pairs of identical gel blots were probed with [32P]UTP uniformly labeled RNAs designed to detect sense or antisense transcripts, as indicated. 1 and 2 are independent VIGS-treated plants.
A very different picture emerged when probes were used to detect asRNAs. For all three genes examined, a disperse set of transcripts accumulated. For atpBE, two relatively distinct species of 3–4 kb were seen, as well as indistinct hybridization down to a size slightly larger than the 164-nt 5.8S rRNA, which was used as a loading control. No signal was seen for the WT or vector control. For rbcL, one distinct species slightly larger than the mature sense RNA was seen in both control and VIGS samples; to our knowledge, this species has not been previously reported. In addition, a higher-molecular-weight smear of transcripts was seen in the VIGS material. Results for clpP asRNAs largely paralleled those obtained for rbcL, with a single discrete and multiple diffuse transcripts accumulating in all or VIGS tissues, respectively. In summary, the results in Figure 5 show that for at least three genes, and likely for many or most chloroplast genes, reduction of RNase J expression leads to the accumulation of asRNAs, suggesting that a major and unanticipated role of RNase J in WT plants is to prevent its accumulation. To test whether this accumulated asRNA might be responsible for the deleterious plant growth phenotype, we examined the cases of atpBE and rbcL in more detail.
Antisense RNAs entirely overlap the corresponding coding regions and lead to formation of stable double-stranded molecules
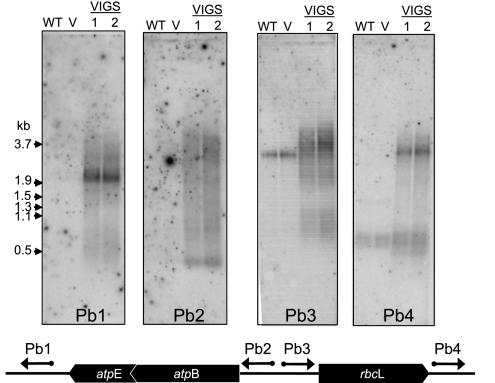
The asRNAs that were detected in Figure 5 were mainly larger than their sense strand counterparts. Their sizes, however, do not permit deduction of their extents with respect to the transcript derived from the opposite strand. To gain more specific information regarding atpBE and rbcL asRNAs, strand-specific probes upstream of and downstream from the genes were used, as shown at the bottom of Figure 6. The hybridizations in Figure 6 show that each probe identified VIGS material-specific asRNAs. In addition, the probe upstream of rbcL (Pb3) identified a band in WT and vector control samples, which was also seen in Figure 5. Somewhat unexpected was the disparity in patterns when the pairs of probes (Pb1/Pb2 and Pb3/Pb4) were compared for each gene. This result suggests that while asRNAs corresponding to upstream, downstream, and within sense strand coding regions are present, no single major species seems to span the entire region. Thus, many different 5′ and 3′ ends likely exist, resulting in complex populations of asRNAs.
FIGURE 6.
The asRNAs cover both the coding regions and UTRs. Total RNA was analyzed as described in the legend to Figure 5. The schematic indicates the positions of the strand-specific probes, which are described in detail in Materials and Methods.
If asRNAs lead to a chlorotic phenotype in VIGS tissue, it would likely be exerted through pairing with sense transcripts. Some natural bacterial asRNAs, for example, can repress translation by sequestering Shine-Dalgarno elements or other regulatory motifs (for review, see Waters and Storz 2009). In chloroplasts, translation can also be impeded when 5′-UTR sequences are engaged in double-stranded structures, which can be relieved through binding of translation activation factors (Rochaix et al. 1989; Prikryl et al. 2010). To investigate the possible presence of dsRNA in RNase J–depleted material, we used mung bean nuclease (MB), which is a single-strand nucleic acid-specific endonuclease. Total RNA purified under nondenaturing conditions was treated with increasing concentrations of MB, then RNAs were examined by gel blots using strand-specific probes. The presence of MB-insensitive transcripts would indicate the presence of dsRNA segments.
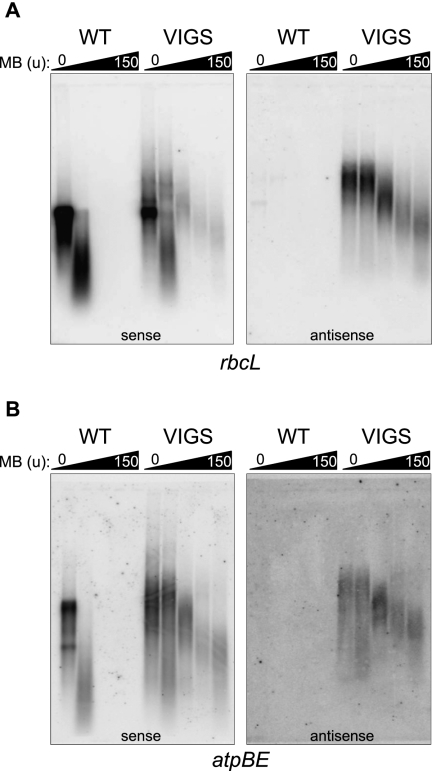
Figure 7, A and B, show results for rbcL and atpBE, respectively. For both genes, the sense strand RNA (left panels) was fully MB-sensitive in the WT, with 1.5 units causing complete degradation. As shown above, with the exception of a weak band for rbcL that was also MB-sensitive, WT does not accumulate asRNAs for these genes (right panels). An opposite result was obtained for VIGS material. For the rbcL sense strand, treatment with 15 units of MB revealed a small proportion that was nuclease-insensitive, and up to 150 units of MB did not fully degrade the sense strand. Instead, a smear of RNAs slightly smaller than the full-length sense strand remained. For the rbcL antisense strand, a smear of the same apparent size was also revealed following treatment with 150 units of MB. This suggests that there is a set of fully paired dsRNAs with heterogeneous ends. We also noted that the rbcL asRNA was completely resistant to 0.15 units of MB, the same concentration that degraded nearly all of the sense RNA in the WT. This suggests that the majority of the asRNAs are engaged in dsRNA structures, but these are likely to contain discontinuities such as bulges, which would be susceptible to higher concentrations of MB. For atpBE transcripts in VIGS material, the results paralleled what we observed for rbcL except that a higher proportion of sense strand RNA appeared to be MB-resistant, suggesting that much of the atpBE mRNA in the VIGS material is partly or fully double-stranded. If so, substantial implications for gene expression would be anticipated.
FIGURE 7.
Duplexed antisense and sense RNAs accumulate in the RNase J–deficient leaves. Total RNA isolated from wild-type (WT) or RNase J–deficient material was treated with increasing amounts of mung bean nuclease (0, 0.15, 1.5, 15, or 150 units) for 15 min prior to gel blot analysis. Blots were hybridized with strand-specific rbcL (A) or atpB (B) RNA probes to detect transcripts from the indicated strands.
Antisense RNA sequesters sense RNA in nonpolysomal fractions
If the formation of dsRNA is impeding translation, RNase J deficiency should be correlated with diminished loading of mRNAs onto polysomes. To assess this, total polysomes were sedimented through sucrose gradients, and RNAs from various fractions were analyzed by gel blots, again using strand-specific probes.
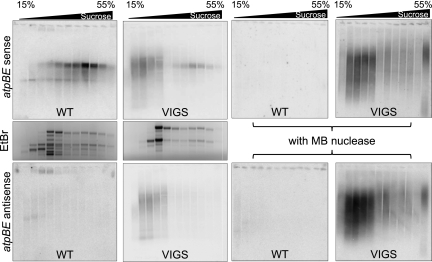
Figure 8 shows results for atpBE. When the sense strand was analyzed from WT material (Fig. 8, middle left panel), as expected, most atpBE transcripts were in the heavier fractions of the gradient, suggesting that they were engaged in translation. When the atpBE sense strand was examined in VIGS material (Fig. 8, middle right panel), most of the hybridization was to RNAs in nonpolysomal fractions closer to the top of the gradient, although some sense RNA was found in the polysomal fractions as well. This implies that unpaired RNA is translatable, whereas most or all RNAs engaged in double-stranded structures fail to be translated or are poorly translated.
FIGURE 8.
Detection of sense and antisense RNAs in polysomal fractions. Crude polysomal pellets were sedimented in 15%–55% sucrose gradients, and RNA was isolated from various fractions, and either treated (right side) or not (left side) with mung bean nuclease. RNA was analyzed by gel blot to detect the atpBE sense strand (top row) or the antisense strand (bottom row). To detect sense transcripts, the blot used to detect asRNA was reprobed. (Center row) Ethidium bromide staining of gels from fractions not treated with MB nuclease.
The lower panels of Figure 8 show results for the antisense strand of atpBE. As shown earlier, asRNA does not accumulate in the WT. For the VIGS lines, results were fully coherent with what was observed for the sense strand, implying that dsRNA was sedimenting in nonpolysomal fractions. We subsequently verified that this was, indeed, dsRNA, by treating RNA from polysomal fractions with MB (Supplemental Fig. S3). This experiment also showed that the small amount of sense RNA that was polysome-loaded in the VIGS material was MB-sensitive, confirming that only ssRNA is translatable.
Data for rbcL are presented in Supplemental Figure S3. As shown in Figure 7A, the amount of MB-resistant rbcL sense strand RNA was relatively small compared with atpBE. Indeed, in VIGS material, the polysome profile of rbcL was not markedly altered. For rbcL asRNA, transcripts were found in the nonpolysomal fractions, as was the case for atpBE. However, we speculate that rbcL translation is only modestly affected, if at all, in VIGS material, because substantial unpaired sense RNA still accumulates. This implies that depletion of RNase J has variable effects on chloroplast transcripts, depending on the relative proportions of sense and antisense transcripts that accumulate. Given that chloroplast translation is essential for embryo development (Bryant et al. 2010), however, only one ribosomal protein mRNA, for example, would need to be excluded from translation to cause embryo lethality. Given the results here for atpBE, which is a highly transcribed gene, and the fact that we are examining only a partial knockdown of RNase J, it seems axiomatic that translational repression is a major, if not the causal factor of the observation that RNase J mutants are embryo-lethal.
DISCUSSION
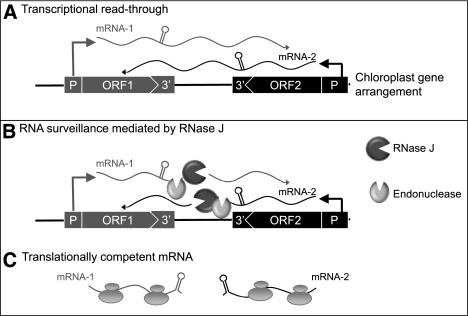
A simple model that takes into account our data, and frames the following discussion, is shown in Figure 9. This model invokes RNase J as an asRNA surveillance enzyme that eliminates the long antisense transcripts and is required for efficient translation of sense strand transcripts. This does not, however, exclude other roles for RNase J, as discussed below.
FIGURE 9.
Model for asRNA surveillance by chloroplast RNase J. (A) 3′ UTRs of chloroplast genes inefficiently terminate transcription, resulting in read-through (mRNA-1 and mRNA-2). Where genes are convergently transcribed, even at a distance, asRNA may be synthesized. (B) These pre-mRNAs are first processed by an endonuclease, which could possibly be RNase J itself or another unidentified endonuclease. This creates substrates for the 5′ → 3′ exonucleolytic activity of RNase J. (C) By removing asRNA, RNase J allows accumulation of single-stranded sense RNA that is translationally competent.
One tenet of the model is that asRNA synthesis is a consequence or read-through transcription (Fig. 9A). It has long been known that transcription termination at the 3′ ends of most genes is inefficient in chloroplasts, necessitating RNA maturation mechanisms to create defined 3′ termini (Stern and Gruissem 1987). A second consequence of read-through transcription, however, is the potential to create antisense transcripts, since chloroplast genomes are compact, and in most cases have an apparently random distribution of genes on one strand versus another (Cui et al. 2006). Because chloroplast genomes are polyploid, polymerase collisions due to symmetric transcription are unlikely to be an impediment to gene expression; however, accumulated antisense transcripts could act in trans to do so, as appears to be the case in RNase J–deficient conditions.
While our examination of the chloroplast genome was not complete, the presence of asRNAs appears to be widespread when RNase J is limited. It might be expected that some regions would not accumulate asRNA due to transcription termination. For example, trnS and perhaps other tRNAs are efficient terminators based on in vitro assays (Stern and Gruissem 1987), and the chloroplast eubacterial enzyme does recognize classical bacterial rho-independent terminators (Chen and Orozco 1988). The long transcripts that accumulate in mutants lacking either the eubacterial polymerase (Legen et al. 2002) or chloroplast RNase E (Walter et al. 2010), as well as indications from pulse labeling that the chloroplast genome is fully transcribed (Legen et al. 2002), however, suggest that post-transcriptional processes are critical in maintaining the population of discrete, functional RNAs.
The production of possibly deleterious asRNA is also likely to occur in plant mitochondria. It has been shown that 3′-end stem–loop structures that flank plant mitochondrial genes are no more efficient in transcription termination than their chloroplast counterparts (Dombrowski et al. 1997). Evidence from plants depleted for mitochondrial PNPase suggests that in mitochondria, this 3′ → 5′ exoribonuclease is responsible for eliminating these products of “relaxed” transcription (Holec et al. 2006). However, while chloroplast PNPase mutants fail to correctly mature 3′ termini, there is no evidence for widespread accumulation of antisense or non-coding transcripts not found in WT plants (Germain et al. 2011). We hypothesize that in chloroplasts RNase J has assumed this surveillance role, whereas mitochondria, lacking RNase J, depend on PNPase for the same function.
RNase J has been studied in several prokaryotes, although it is not present in E. coli (for review, see Condon 2010). It has been implicated in the trimming of 16S rRNA (Britton et al. 2007; Mathy et al. 2007), and the turnover of small RNAs such as the trp and thrS leaders and ΔermC (Even et al. 2005; Deikus et al. 2008; Yao et al. 2009). There is also evidence that RNase J has a broader role in determining mRNA lifetime (Mader et al. 2008; Bugrysheva and Scott 2010). A role in RNA surveillance, i.e., the prevention of the production of aberrant transcripts, however, has not yet been proposed. The previously assigned functions of RNase J have implicated both its endonucleolytic and exonucleolytic activities. While the Arabidopsis enzyme possesses both properties, three recently characterized RNase J proteins of a hyperthermophilic methanogenic archaea are specialized; two were found to be exclusively exonucleases and one displayed only endonucleolytic activity (Levy et al. 2011). The proliferation of RNase J family members in this group of archaea may be related to the likelihood, as suggested by genome sequence examination, that these are the only ribonucleases present in these organisms.
In chloroplasts, RNase J and RNase E have been proposed to be involved in the maturation of polycistronic transcripts that are cleaved into smaller units (Barkan 2011), which would also generate the substrates for RNase J shown in Figure 9B. This hypothesis is based in part on analysis of the PPR10 protein, which defines the 3′ and 5′ ends of two sets of overlapping chloroplast transcripts (Pfalz et al. 2009; Prikryl et al. 2010). PPR10 protects these termini, which are proposed to be generated by stochastic endonucleolytic cleavages outside of the protected region, followed by exonucleolytic trimming. RNase J has two possible functions in this model, cleavage of the primary transcripts, and 5′ → 3′ trimming to generate mature 5′ ends. Given that our recombinant protein possesses a significant endonucleolytic activity, we propose that RNase J may participate in endonuclease cleavages, for example, to remove the 5′ triphosphate, before switching to exonuclease activity.
Because we saw no evidence for accumulation of longer precursors at the expense of mature transcripts, RNase J either does not mediate intercistronic processing or is redundant with another endonuclease. Furthermore, that the asRNA that accumulates in VIGS material apparently does not inhibit endonucleolytic cleavage of polycistronic precursors. On the other hand, our preliminary data suggest that RNase J does trim 5′ ends of maturing sense strand transcripts, as proposed (Barkan 2011). While WT-sized mature transcripts appeared to accumulate on gel blots, 5′ RACE revealed slightly 5′-extended species for atpH, rbcL, and rpl33 (Supplemental Fig. S4). Whether these result from aborted trimming or are directly produced by endonucleolytic cleavages remains to be ascertained.
If RNase J is involved both in maturing functional RNAs and degrading nonfunctional species, it must be able to distinguish between the two. Some functional RNAs are defined by 5′-end RNA-binding proteins that stall 5′ → 3′ exonucleolytic activity (Drager et al. 1998; Nickelsen et al. 1999; Loiselay et al. 2008; Johnson et al. 2010; Prikryl et al. 2010), but other protective mechanisms may also exist. Examples of RNAs probably not protected by PPR-type proteins include natural (i.e., accumulating in the WT) asRNAs (Georg et al. 2010; Hotto et al. 2010; Sharwood et al. 2011), small RNAs involved in splicing or other functions (Goldschmidt-Clermont et al. 1991; Vera and Sugiura 1994), as well as rRNAs, tRNAs, and any mRNAs that do not bind protective 5′-end proteins. Some of these species may escape RNase J degradation by virtue of associating with proteins, such as ribosomal proteins, splicing factors, or translation factors. Others may possess strong 5′-end secondary structures that are refractory to RNase J processivity. Indeed, B. subtilis RNase J is inhibited by an antisense oligonucleotide annealed to the 5′ end (Mathy et al. 2007), although it is unaffected by poly(G), which forms a highly stable tertiary structure that inhibits the cytosolic/nuclear 5′ → 3′ exoribonuclease Xrn1 (Stevens 2001) as well as the chloroplast 5′ → 3′ degradation pathway (Drager et al. 1999). It will be of interest to determine the precise activities of chloroplast RNase J on structured, modified, and protein-bound substrates.
MATERIALS AND METHODS
Plant material
A. thaliana (Col-0) plants used for Agrobacterium infiltration were grown as described in Sharwood et al. (2011). N. benthamiana plants for infiltration were germinated and grown in a greenhouse at 25°C with light provided to ensure a 14-h light/10-h dark cycle.
Cloning, expression, and purification of Arabidopsis RNase J
Arabidopsis RNase J (At5g63420) was PCR-amplified using oligo(dT) cDNA and the primers At RNJ for and At RNJ rev, and cloned into the XhoI and EcoRI restriction sites of the bacterial expression vector pET-28a. In this way, six sequential His residues were added near the N terminus, generating the sequence MGSSHHHHHHSSGLVPRGSHMASMTGGQQMGRGSEF upstream of codon 71 of RNase J. E. coli BL21 Gold cells were transformed with the expression plasmid and grown at 30°C in LB medium with addition of 1 M sorbitol to an O.D.600 of 0.6. Protein expression was induced with 0.1 mM IPTG for 16 h at 18°C. To purify the protein, bacteria were disrupted with a microfluidizer and cleared by centrifugation, the NaCl concentration was brought to 1 M, and the soluble fraction was applied to a Talon-cobalt column, eluted with 150 mM imidazole, dialyzed with buffer containing 50 mM sodium phosphate (pH 7.0), 300 mM NaCl, 5% glycerol, and 0.2 mM DTT. The protein was further purified using a heparin column, passing through Q Sepharose beads and finally dialyzed against a buffer containing 20 mM HEPES (pH 8.0), 100 mM NaCl, 8 mM MgCl2, 5% glycerol, and 0.1 mM DTT. The purified protein was fast-frozen and stored at −80°C. All RNase J preparations were assayed both enzymatically and by immunoblot for contaminating E. coli RNase E, PNPase, and RNase II and R. Only RNase J preparations testing negative for these potential contaminants were used.
Preparation of the RNA substrates for the degradation analysis
The DNA construct used for the transcription of RNA corresponding to the 3′ end of the spinach chloroplast petD gene has been described (Stern and Gruissem 1987). In vitro transcription with T7 RNA polymerase and radioactive labeling were performed as previously described (Portnoy et al. 2008). RNA molecules containing 5′-monophosphate or OH were obtained by the addition of 150 mM GMP and guanosine, respectively, to the transcription reaction mixture (Mathy et al. 2007; Schein et al. 2008). 5′-end labeling was performed by first removing a phosphate from the 5′ end with calf intestinal alkaline phosphatase and subsequently adding [32P] with T4 polynucleotide kinase and [γ-32P]ATP (Schein et al. 2008). A 5′-triphosphorylated transcript that is also [32P]-labeled at this site was produced by including [γ-32P]GTP in the reaction mixture as described (Daou-Chabo and Condon 2009).
RNA degradation assays
In vitro RNA degradation assays were performed using the recombinant protein and either [32P]UTP-uniformly labeled or 5′ [32P]-labeled RNA substrates as described above. In a common reaction, a protein (0.2 mM) was incubated at 25°C with 0.06 μM RNA for the times indicated in the figure legends. Following incubation, the RNA was either purified and analyzed by denaturing PAGE or directly applied to a TLC plate (Portnoy et al. 2008). Nucleoside mono, di-, and triphosphates (5 μg of each) were separated on the same TLC plate and visualized by fluorescence quenching.
Virus-Induced Gene Silencing of RNase J
The Arabidopsis RNJ locus was targeted for VIGS as previously described (Sharwood et al. 2011). The partial N. bethanamiana RNJ gene (U431192) was identified in the Solanaceae Genome database (Mueller et al. 2005; http://www.solgenomics.net). Primer pairs RNJ T1 for/rev and RNJ T2 for/rev were used to amplify unique regions of NbRNJ. The amplicons were sequenced and inserted into PYL170 as previously described (Sharwood et al. 2011). VIGS constructs were electroporated into Agrobacterium tumefaciens GV3101 and grown on plates containing 100 μg/mL rifampicin and 50 μg/mL kanamycin. Cultures for infiltration were prepared as described by Sharwood et al. (2011) except the O.D.600 for infiltration in tobacco leaves was adjusted to 0.4. N. benthamiana plants at the 4–5 leaf stage were used for infiltration.
RNA isolation and gel blot analysis
RNA was isolated from Arabidopsis rosette and tobacco leaves using Tri-Reagent according to the manufacturer's instructions. For polysomal RNA analysis, 100 mg of WT and N. benthamiana RNase J VIGS tissue was used for extraction, and polysomes were fractionated through 15%–55% sucrose density gradients as previously described (Barkan 1998). RNA was separated in 1.2% formaldehyde-agarose gels buffered by sodium phosphate (pH 6.8) and transferred to GeneScreen N using transfer solution (1 M ammonium acetate and 5 mM NaOH). RNA blots to be hybridized with PCR-generated DNA probes were prehybridized in Church and Gilbert buffer (Church and Gilbert 1984). Blots to be hybridized with synthetic RNA probes (see below) were prehybridized in 50% formamide buffer (5× SSC, 2% [w/v] BSA, 0.6% [w/v] SDS, 200 μg/mL salmon sperm DNA) for >6 h. Hybridization was performed overnight at 65°C. Signal intensity for each blot was measured on a Storm Scanner PhosphorImager.
Construction of DNA templates for DNA and strand-specific RNA probe synthesis
DNA probes were amplified for specified plastid genes using the primers listed in Supplemental Table 1. Amplicons were used as templates for random hexamer-primed labeling of DNA probes with [32P]CTP (3000 Ci/mmol) using Klenow according to the manufacturer's instructions. The atpB and rbcL DNA probe templates for in vitro RNA synthesis were amplified from N. benthamiana plastid DNA using the primer pairs Nb atpB sense T7/Nb atpB Rev, Nb atpB antisense T7/Nb atpB for and Tob rbcL sense T7/Tob rbcL rev, and Tob rbcL antisense T7/Tob rbcL.
RT-PCR
To qualitatively determine the residual level of RNJ expression within Arabiodpsis, RT-PCR was performed with cDNA that was generated by Supercscript III from 1.0 μg of RQ1 DNase-treated total RNA using the At-RNJ, At-RNR1, and UBQ10 reverse primers and At RNJ VIGS Rev, RNR1 3′ and UBQ2 forward primers, respectively. RNJ- and RNR1-containing cDNA was then amplified using 25 PCR cycles using the primer pairs At RNJ VIGS 5′ RT/At RNJ VIGS Rev and RNR1 5′/RNR1 3′. UBQ10 cDNA was amplified as a control (28 cycles) using the primers UBQ1 and UBQ2. For N. benthamiana VIGS material, RT-PCR was performed with cDNA generated as described above using the reverse primers Tob RNJ Sq2 rev and Tob Actin Rev, respectively. RNJ and actin cDNAs were amplified using 28 cycles and the primer pairs RNJ SQ2 for/RNJ SQ2 rev and Tob Actin for/Tob Actin Rev. The method for 5′ RACE is described in the legend to Supplemental Figure S4 (see Steglich et al. 2008).
In vivo analysis of RNA duplexes using mung bean nuclease treatment
Total RNA was extracted from N. benthamiana VIGS material and the vector control (V) as described above. Reactions (20 μL) consisting of 1.2 μg of RNA were treated with 1.5–150 U of mung bean nuclease for 15 min at 37°C. For polysome duplex RNA analysis, 10 μL of each fractionated RNA sample was treated with 15 U of mung bean nuclease. Assays were stopped by adding RNA loading buffer and heated for 15 min to 65°C before separation in 1.2% formaldehyde-agarose gels as described above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Chloe Marchive for performing the YFP targeting experiment shown in Supplemental Figure S1 and Erica Tiberia for her contributions in developing the Arabidopsis VIGS approach. This work was supported by awards from the Israel–US Binational Science Foundation (#2009253) and the Binational Agricultural Research and Development Fund (#IS-4152-08) to G.S. and D.B.S., and award DE-FG02-90ER20015 to D.B.S. from the Department of Energy's Energy Bioscience Program.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028043.111.
REFERENCES
- Barkan A 1993. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A 1998. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Barkan A 2011. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D 2005. RNR1, a 3′–5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB 2009. The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 [DOI] [PubMed] [Google Scholar]
- Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol 63: 127–138 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D 2010. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis thaliana. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Scott JR 2010. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol Microbiol 75: 731–743 [DOI] [PubMed] [Google Scholar]
- Chen LJ, Orozco EMJ 1988. Recognition of prokaryotic transcription terminators by chloroplast RNA polymerase. Nucleic Acids Res 16: 8411–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G, Gilbert W 1984. Genomic sequencing. Proc Natl Acad Sci 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet-d'Orval B, Rinaldi D, Quentin Y, Carpousis AJ 2010. Euryarchaeal β-CASP proteins with homology to bacterial RNase J have 5′- to 3′-exoribonuclease activity. J Biol Chem 285: 17574–17583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C 2010. What is the role of RNase J in mRNA turnover? RNA Biol 7: 316–321 [DOI] [PubMed] [Google Scholar]
- Cui L, Leebens-Mack J, Wang LS, Tang J, Rymarquis L, Stern DB, dePamphilis CW 2006. Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evol Biol 6: 13 doi: 10.1186/1471-2148-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou-Chabo R, Condon C 2009. RNase J1 endonuclease activity as a probe of RNA secondary structure. RNA 15: 1417–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikus G, Condon C, Bechhofer DH 2008. Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J Biol Chem 283: 17158–17167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Sierra-Gallay IL, Zig L, Jamalli A, Putzer H 2008. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol 15: 206–212 [DOI] [PubMed] [Google Scholar]
- Dombrowski S, Brennicke A, Binder S 1997. 3′-Inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J 16: 5069–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager RG, Girard-Bascou J, Choquet Y, Kindle KL, Stern DB 1998. In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J 13: 85–96 [DOI] [PubMed] [Google Scholar]
- Drager RG, Higgs DC, Kindle KL, Stern DB 1999. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J 19: 521–532 [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H 2005. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res 33: 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Honsel A, Voss B, Rennenberg H, Hess WR 2010. A long antisense RNA in plant chloroplasts. New Phytol 186: 615–622 [DOI] [PubMed] [Google Scholar]
- Germain A, Herlich S, Larom S, Kim SH, Schuster G, Stern DB 2011. Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J 67: 381–394 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Choquet Y, Girard-Bascou J, Michel F, Schirmer-Rahire M, Rochaix JD 1991. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65: 135–144 [DOI] [PubMed] [Google Scholar]
- Hasenohrl D, Konrat R, Blasi U 2011. Identification of an RNase J ortholog in Sulfolobus solfataricus: Implications for 5′-to-3′ directional decay and 5′-end protection of mRNA in Crenarchaeota. RNA 17: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks A, Drager RG, Higgs DC, Stern DB 2002. An mRNA 3′ processing site targets downstream sequences for rapid degradation in Chlamydomonas chloroplasts. J Biol Chem 277: 3325–3333 [DOI] [PubMed] [Google Scholar]
- Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D 2006. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol 26: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Huston ZE, Stern DB 2010. Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol 10: 213 doi: 10.1186/1471-2229-10-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O 2010. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishine M, Takabayashi A, Munekage Y, Shikanai T, Endo T, Sato F 2004. Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis. Plant Mol Biol 55: 595–606 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S 2004. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Lam E 1995. Domain analysis of the plant DNA-binding protein GT1a: Requirement of four putative α-helices for DNA binding and identification of a novel oligomerization region. Mol Cell Biol 15: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM 2002. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Levy S, Portnoy P, Admon J, Schuster G 2011. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol doi: 10.4161/rna.8.6. [DOI] [PubMed] [Google Scholar]
- Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y 2008. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol 28: 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Hüttenhofer A 2006. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res 34: 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader U, Zig L, Kretschmer J, Homuth G, Putzer H 2008. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol 70: 183–196 [DOI] [PubMed] [Google Scholar]
- Madhugiri R, Evguenieva-Hackenberg E 2009. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett 583: 2339–2342 [DOI] [PubMed] [Google Scholar]
- Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, Wu H, Fernie AR, Fait A, Stern DB 2009. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol 151: 905–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C 2007. 5′-to-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129: 681–692 [DOI] [PubMed] [Google Scholar]
- Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung WI, Day A 2008. A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J Exp Bot 59: 2597–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. 2005. The SOL Genomics Network: A comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J, Fleischmann M, Boudreau E, Rahire M, Rochaix J-D 1999. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A 2009. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy V, Palnizky G, Yehudai-Resheff S, Glaser F, Schuster G 2008. Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. RNA 14: 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A 2010. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J-D, Kuchka M, Mayfield S, Schirmer-Rahire M, Girard-Bascou J, Bennoun P 1989. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J 8: 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein A, Sheffy-Levin S, Glaser F, Schuster G 2008. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I 2008. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schweer J, Loschelder H, Link G 2006. A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett 580: 6617–6622 [DOI] [PubMed] [Google Scholar]
- Sharwood RE, Hotto AM, Bollenbach TJ, Stern DB 2011. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 17: 230–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR 2008. The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet 4: e1000173 doi: 10.1371/journal.pgen.1000173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Gruissem W 1987. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51: 1145–1157 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR 2010. Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stevens A 2001. 5′-Exoribonuclease 1: Xrn1. Methods Enzymol 342: 251–259 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. 2004. Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Sugiura M 1994. A novel RNA gene in the tobacco plastid genome: Its possible role in the maturation of 16S rRNA. EMBO J 13: 2211–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Kilian J, Kudla J 2002. PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J 21: 6905–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R 2010. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB 1996. CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Sharp JS, Bechhofer DH 2009. Bacillus subtilis RNase J1 endonuclease and 5′ exonuclease activities in the turnover of ΔermC mRNA. RNA 15: 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G 2003. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell 15: 2003–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S 2008. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]