The in vivo analysis of splicing factors in regulating alternative splicing in animals remains a challenge. Using a microarray-based screen, the authors identified a Caenorhabditis elegans gene, tos-1, that exhibited three of the four major types of alternative splicing: intron retention, exon skipping, and, in the presence of U2AF large subunit mutations, the use of alternative 3′ splice sites. This study provides in vivo evidence that intron retention and exon skipping can be regulated largely by the identities of 3′ splice sites.
Keywords: C. elegans, SFA-1, UAF-1, alternative splicing
Abstract
The in vivo analysis of the roles of splicing factors in regulating alternative splicing in animals remains a challenge. Using a microarray-based screen, we identified a Caenorhabditis elegans gene, tos-1, that exhibited three of the four major types of alternative splicing: intron retention, exon skipping, and, in the presence of U2AF large subunit mutations, the use of alternative 3′ splice sites. Mutations in the splicing factors U2AF large subunit and SF1/BBP altered the splicing of tos-1. 3′ splice sites of the retained intron or before the skipped exon regulate the splicing pattern of tos-1. Our study provides in vivo evidence that intron retention and exon skipping can be regulated largely by the identities of 3′ splice sites.
INTRODUCTION
RNA splicing removes noncoding introns of eukaryotic pre-mRNAs and joins neighboring exons to generate functional coding mRNAs (Reed 2000; Maniatis and Tasic 2002). Alternative splicing generates multiple transcript isoforms from a single pre-mRNA and is believed to be a major molecular mechanism responsible for the generation of the biological complexity of metazoans (Smith and Valcarcel 2000; Graveley 2001; Maniatis and Tasic 2002). In humans, most genes generate alternatively spliced isoforms (Johnson et al. 2003; Pan 2008; Wang et al. 2008). In the nematode Caenorhabditis elegans, ∼20% of the genes are alternatively spliced (Zahler 2005; Ramani et al. 2011).
Four basic types of alternative splicing have been identified: the use of alternative 5′ splice sites, the use of alternative 3′ splice sites, intron retention, and exon skipping (cassette exons) (Nilsen and Graveley 2010). Different genes undergo different numbers or types of alternative splicing events. For example, the splicing of the Drosophila sex-determination genes dsx and transformer involves exon skipping and the use of alternative 3′ splice sites, respectively (Boggs et al. 1987; Burtis and Baker 1989). In C. elegans, egl-15 transcripts contain mutually exclusive exons that are included in different isoforms at distinct developmental stages (Goodman et al. 2003). The human KCNMA1 (SLO) gene pre-mRNA uses alternative 5′ splice sites, alternative 3′ splice sites, and exon skipping to generate >500 splice isoforms (Navaratnam et al. 1997; Rosenblatt et al. 1997). More dramatically, the Drosophila Dscam pre-mRNA has been suggested to generate >30,000 isoforms by combining constitutive exons with different cassette exons (Schmucker et al. 2000).
The 5′ splice site of an intron is recognized by the U1 small nuclear ribonucleoprotein particle (snRNP) (Madhani and Guthrie 1994). SF1/BBP (splicing factor one/branch-point binding protein) and the large and small subunits of U2AF (U2 auxiliary factor) function together to recognize the 3′ splice site of an intron (Krainer and Maniatis 1985; Zamore and Green 1991; Arning et al. 1996; Abovich and Rosbash 1997; Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999a). Alternative RNA splicing is generally achieved by interactions between splicing factors and cis-regulatory sequences in exons and introns. Splicing factors that bind exonic or intronic cis-regulatory nucleotide sequences include arginine-serine-rich RNA-binding SR proteins (Zahler et al. 1992; Fu 1995; Hertel et al. 1997; Long and Caceres 2009) and hnRNP RNA-binding proteins (Smith and Valcarcel 2000). Although >140 splicing factors have been identified in mammals (Zhou et al. 2002), it remains largely unknown how these factors interact to generate alternative splice isoforms of different genes.
By screening for essential C. elegans genes that can affect the rubberband Unc phenotype of unc-93(e1500) animals, we isolated mutations in the C. elegans orthologs of the U2AF large subunit (UAF-1) and SF1/BBP (SFA-1) (Ma and Horvitz 2009). Because these mutations cause conditional lethality—e.g., uaf-1(n4588) and sfa-1(n4562) cause temperature-sensitive lethality and maternal-effect sterility, respectively—they provide novel opportunities to study the in vivo regulation of splicing. To further analyze the functions of these splicing factors, we searched for target genes of UAF-1 by a whole-genome tiling microarray analysis that could detect differential splicing of genes between wild-type and uaf-1(n4588) animals. We identified a gene we named “tos-1” (target of splicing), the splicing of which is altered by mutations affecting both uaf-1 and sfa-1. Our studies provide in vivo evidence that UAF-1 and SFA-1 regulate alternative splicing and that the identities of 3′ splice sites can affect intron retention and exon skipping.
RESULTS
Splicing of K07B1.6 in wild-type animals involves intron retention and exon skipping
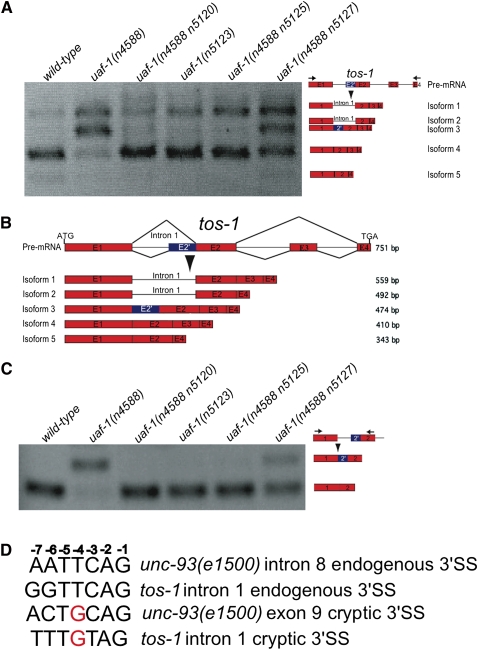
uaf-1(n4588) is a temperature-sensitive mutation in the gene that encodes the C. elegans U2AF large subunit (Ma and Horvitz 2009). To study the in vivo function of uaf-1, we used a whole-genome tiling microarray to seek genes with altered splicing in uaf-1(n4588) animals (see Materials and Methods). We identified 11 candidate genes (Supplemental Table S1). We examined the splicing of all 11 candidate genes by RT-PCR and found that only the splicing of K07B1.6 was dramatically altered in uaf-1(n4588) animals (Fig. 1A) (see below). Because K07B1.6 provides a sensitive readout for studying alternative splicing and the biological function of K07B1.6 remains to be identified (see below), we named K07B1.6 “tos-1” (target of splicing). We subcloned DNA fragments from the RT-PCR products of the tos-1 transcripts in wild-type animals (Fig. 1A, wild-type) and determined the sequences of DNA inserts from 46 independent clones. We identified four major tos-1 splice isoforms (Fig. 1A,B), with isoform 4 being the most abundant (Figs. 1A,B, 2A). Isoforms 1, 2, and 5 were less abundant (Figs. 1A,B, 2A) (for isoform 3, see below). The nature of these isoforms indicates that in wild-type animals, the splicing of tos-1 involves intron 1 retention and exon 3 skipping. The combination of these two types of alternative splicing generates isoforms 1, 2, 4, and 5 (Fig. 1B). Quantification of the RT-PCR products indicated that in wild-type animals isoform 4 constitutes >80% of all transcripts (Fig. 2A).
FIGURE 1.
uaf-1 mutations alter the splicing pattern of tos-1. (A) RT-PCR experiments examining tos-1 splice isoforms in different genetic backgrounds. Genotypes are labeled on top. Splice isoforms of tos-1 are illustrated on the right. (Arrows) Positions of PCR primers. (B) Major tos-1 splice isoforms identified by subcloning and sequence determination of RT-PCR products. The base-pair length of each isoform is indicated on the right. (Red boxes) Exons. Splicing at the cryptic 3′ splice site in intron 1 caused an altered exon 2′ (blue). (C) RT-PCR experiments examining the recognition of the cryptic 3′ splice site compared to that of the endogenous 3′ splice site of tos-1 intron 1. Splice isoforms are illustrated on the right. (D) Comparison of the endogenous and cryptic 3′ splice sites from tos-1 intron 1 and unc-93(e1500) intron 8 and exon 9. The position of each nucleotide is labeled on top.
FIGURE 2.
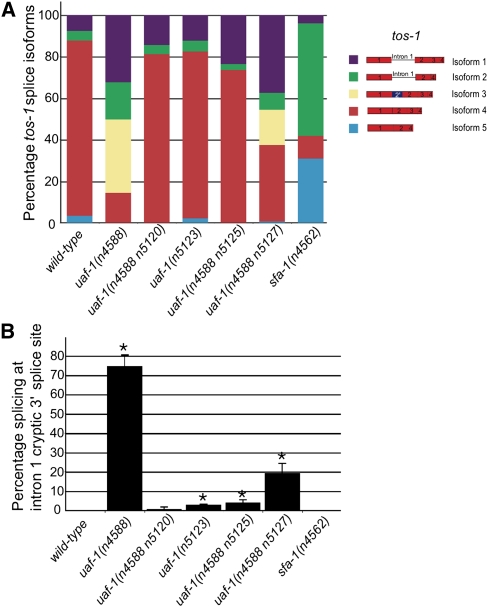
Proportion of each tos-1 splice isoform and the recognition of the tos-1 cryptic 3′ splice site in uaf-1 and sfa-1 mutants. (A) The intensity of each splice isoform was measured using the NIH Image J software from the RT-PCR experiments, and the molar ratio of each isoform is presented as a percentage of all transcripts. (B) Quantitative analysis of the recognition of the 3′ cryptic splice site compared with that of the endogenous 3′ splice site of tos-1 intron 1 by analyzing RT-PCR products separated in agarose gels using the NIH Image J software. (*) p < 0.05.
uaf-1 mutations alter the splicing of tos-1
The uaf-1(n4588) mutation dramatically affected the splicing pattern of tos-1 [Fig. 1A, uaf-1(n4588)]. We analyzed the tos-1 splice isoforms in uaf-1(n4588) animals by subcloning and sequence determination. In uaf-1(n4588) animals, isoform 4 was dramatically reduced, while isoform 1, which was 6% of all transcripts, and a new isoform, 3, which was absent in wild-type animals, were increased to 32% and 33% of all transcripts, respectively (Figs. 1B, 2A). Isoform 3 was generated by the recognition of a cryptic 3′ splice site in intron 1 not recognized in wild-type animals, suggesting that uaf-1(n4588) caused altered recognition between the endogenous 3′ splice site and this cryptic site.
Previously, we isolated four intragenic mutations of uaf-1(n4588)—uaf-1(n4588 n5120), uaf-1(n5123), uaf-1(n4588 n5125), and uaf-1(n4588 n5127)—that partially restored the recognition of the endogenous 3′ splice site of unc-93(e1500) intron 8 in uaf-1(n4588) animals (Ma and Horvitz 2009). We found that these uaf-1 mutants similarly partially restored the splicing of tos-1 to levels intermediate between that of wild-type and uaf-1(n4588) animals (Figs. 1A, 2A).
To examine how different uaf-1 alleles affect the recognition of the cryptic 3′ splice site of tos-1, we performed RT-PCR experiments to specifically visualize the recognition of the normal and cryptic 3′ splice sites of intron 1 (Fig. 1C). Our results indicated that the recognition of the cryptic 3′ splice site occurred in uaf-1(n4588) and uaf-1(n4588 n5127) animals but was not (or was only very weakly) detectable in the wild type and in other uaf-1 mutant animals (Figs. 1C, 2B). We previously showed from a mutagenesis analysis that the n4588 (T180I) mutation caused UAF-1 to favor a G instead of a T at position −4 of 3′ splice sites for the splicing of unc-93(e1500) exon 9 (Ma and Horvitz 2009). A comparison of the normal and cryptic 3′ splice sites from unc-93(e1500) and tos-1 indicates that both normal sites have a T nucleotide at position −4, while both cryptic sites have a G nucleotide at this position (Fig. 1D). Therefore, the altered preference for a G nucleotide at position −4 of 3′ splice sites by UAF-1(T180I) is seen in both unc-93(e1500) and tos-1 animals (see below).
The biological function of tos-1 is unknown
Isoform 4, the major splice isoform of tos-1, encodes a predicted protein of 61 amino acids (Supplemental Fig. S1). Minor splice isoforms 1 and 2 encode proteins of 157 amino acids and 163 amino acids, respectively (Supplemental Fig. S1). Splice isoform 5 encodes a predicted protein of 61 amino acids identical to that encoded by isoform 4 (Supplemental Fig. S1). The new splice isoform 3 in uaf-1(n4588) animals encodes a predicted protein of 127 amino acids (Supplemental Fig. S1). We identified no homologs of TOS-1 in species other than nematodes using the BLAST search algorithm. In the Caenorhabditis elegans genome, the gene D1086.19 encodes a protein of unknown function that is 30% identical to the protein encoded by tos-1 isoform 4.
We isolated a tos-1 deletion mutation, n5384Δ, which removed part of exon 1, all of intron 1, and part of exon 2 (Supplemental Fig. S2). n5384Δ is predicted to delete 64 amino acids after amino acid 25 for isoforms 1 and 2 and would cause a frameshift after amino acid 25 if the truncated transcript is expressed, suggesting that n5384Δ is likely a null allele of tos-1. Previously, we proposed that an unknown gene with splicing regulated by uaf-1 and sfa-1 might be required for the expression of the rubberband Unc phenotype of unc-93(e1500) animals (Ma and Horvitz 2009). The n5384Δ mutation did not cause an obviously abnormal phenotype and failed to suppress the rubberband Unc phenotype of unc-93(e1500) animals (L Ma and HR Horvitz, unpubl.). Similarly, reducing the expression of tos-1 by RNAi did not suppress the rubberband Unc phenotype of unc-93(e1500) animals (L Ma and HR Horvitz, unpubl.). Furthermore, uaf-1(n4588) unc-93(e1500); tos-1(n5384Δ) animals were suppressed for the Unc phenotype as well as uaf-1(n4588) unc-93(e1500) animals (Ma and Horvitz 2009), suggesting that the function of tos-1 is not required for the suppression of unc-93(e1500) by uaf-1(n4588).
sfa-1(n4562) dramatically alters the splicing of tos-1
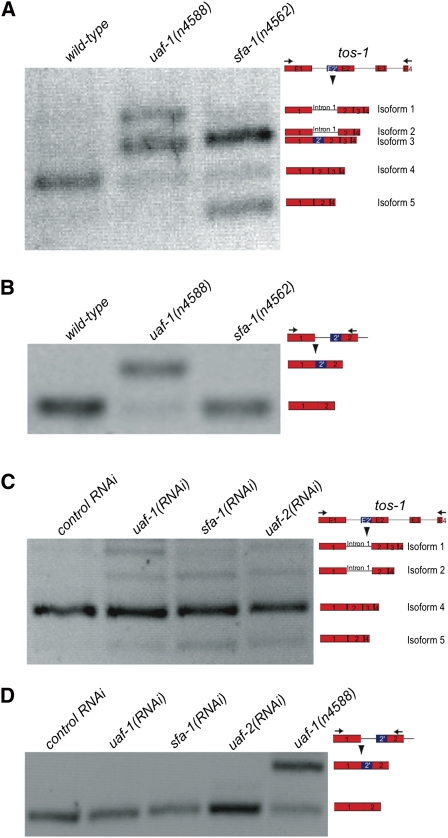
The mutation n4562 causes a nonsense mutation in and is a loss-of-function allele of the sfa-1 gene (Ma and Horvitz 2009). sfa-1 encodes the C. elegans ortholog of the splicing factor SF1/BBP (Mazroui et al. 1999; Ma and Horvitz 2009). sfa-1(n4562) does not alter recognition between the endogenous 3′ splice site in unc-93(e1500) intron 8 and the cryptic 3′ splice site in unc-93(e1500) exon 9 (Ma and Horvitz 2009). We examined whether sfa-1(n4562) altered the splicing of tos-1. As shown in Figures 2A and 3A, sfa-1(n4562) dramatically increased isoforms 2 and 5 and reduced isoform 4 of tos-1. Subcloning and sequence determination of the tos-1 RT-PCR fragments did not identify isoform 3, suggesting that sfa-1(n4562) did not cause recognition of the cryptic 3′ splice site in intron 1. This result was confirmed by RT-PCR analysis to specifically visualize the recognition of the cryptic 3′ splice site and the endogenous 3′ splice site of intron 1 (Fig. 3B).
FIGURE 3.
sfa-1(n4562) alters the splicing pattern of tos-1. (A) RT-PCR experiments (similar to Fig. 1A) showing the effect of sfa-1(n4562) on tos-1 alternative splicing. (B) RT-PCR experiments (similar to Fig. 1C) showing the effect of sfa-1(n4562) on the recognition of the cryptic 3′ splice site of tos-1 intron 1. (C) RT-PCR experiments showing the effect of reducing the expression of uaf-1, sfa-1, or uaf-2 by RNAi feeding on tos-1 alternative splicing. (D) RT-PCR experiments showing the effect of reducing the expression of uaf-1, sfa-1, or uaf-2 by RNAi feeding on the recognition of the cryptic 3′ splice site of tos-1 intron 1.
We also examined the splicing of tos-1 in animals with the expression of uaf-1, sfa-1, or uaf-2 reduced by RNAi feeding. uaf-2 encodes the U2AF small subunit, which interacts with UAF-1 and SFA-1 to regulate 3′ splice site recognition (Zorio and Blumenthal 1999b). As shown in Figure 3, C and D, reducing expression of uaf-1, sfa-1, or uaf-2 by RNAi all caused a similarly weak increase of isoforms 2 and 5 and did not cause recognition of the cryptic 3′ splice site of intron 1, suggesting that uaf-1, sfa-1, and uaf-2 similarly regulate tos-1 alternative splicing in vivo. Compared with the major tos-1 isoform, isoform 4, isoform 2 results from intron 1 retention and exon 3 skipping, and isoform 5 results from exon 3 skipping, suggesting that reducing expression of uaf-1, sfa-1, or uaf-2 increases intron 1 retention and exon 3 skipping.
Effects of uaf-1 mutations and sfa-1(n4562) on tos-1 intron 1 retention and exon 3 skipping
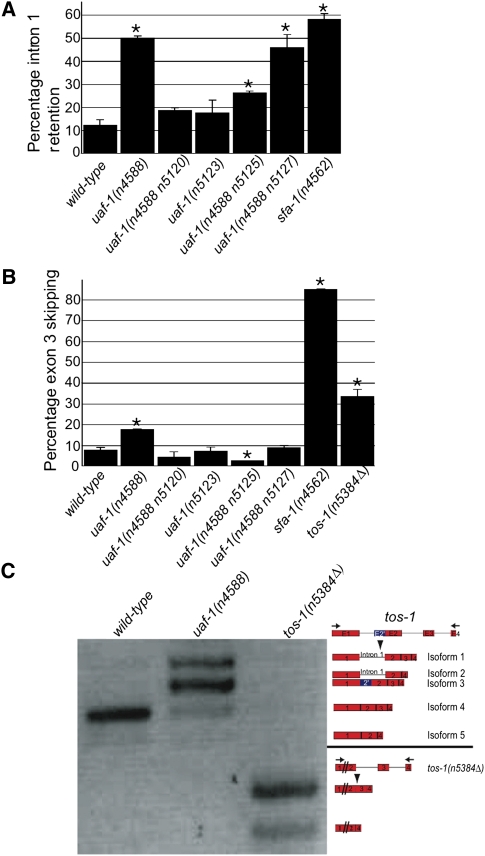
Because the splicing of tos-1 involves intron 1 retention, exon 3 skipping, and, in the case of uaf-1 mutations, alternative 3′ splice site selections, an analysis of these different splicing events might reveal how each type of alternative splicing is regulated by uaf-1 and sfa-1. Figure 2B presents a quantitative analysis of the recognition of the cryptic 3′ splice site. We similarly examined intron 1 retention and exon 3 skipping. As shown in Figure 4A, ∼12% of tos-1 transcripts have intron 1 retention in wild-type animals. uaf-1(n4588) and uaf-1(n4588 n5127) increased intron 1 retention to 50% and 46% of all transcripts, respectively. A weaker uaf-1 mutation, uaf-1(n4588 n5125), caused a twofold increase of intron 1 retention to 26%, while other weak uaf-1 mutations, such as uaf-1(n4588 n5120) and uaf-1(n5123), did not cause an apparent increase of intron 1 retention. sfa-1(n4562) caused the most significant increase of intron 1 retention, to almost 60%, which was about a fivefold increase compared with the wild type.
FIGURE 4.
Quantitative analysis of tos-1 intron 1 retention and exon 3 skipping and RT-PCR analysis of exon 3 skipping in tos-1(n5384Δ) animals. (A) The molar ratio of all tos-1 splice isoforms with intron 1 retention, presented as a percentage of all isoforms combined. Error bars: standard deviation. (*) p < 0.05. (B) The molar ratio of all tos-1 splice isoforms with exon 3 skipping, presented as a percentage of all isoforms combined. Error bars: standard deviation. (*) p < 0.05. (C) RT-PCR analysis of exon 3 skipping in tos-1(n5384Δ) animals. Splice isoforms are indicated on the right. (Arrows) PCR primers.
We also analyzed the proportions of exon 3 skipping (Fig. 4B). In wild-type animals, exon 3 skipping was found in 8% of transcripts. uaf-1 mutations have differing effects on exon 3 skipping. uaf-1(n4588) caused a more than twofold increase of exon 3 skipping (to 18% of all transcripts), while uaf-1(n4588 n5120), uaf-1(n5123), and uaf-1(n4588 n5127) did not cause apparent changes in exon 3 skipping. Interestingly, uaf-1(n4588 n5125) appeared to reduce exon 3 skipping to 3% of all transcripts. sfa-1(n4562) dramatically increased exon 3 skipping to 85% of all transcripts, which was more than a 10-fold increase over the frequency seen in the wild type (Fig. 4B).
It has been reported that with a reporter transgene a reduced splicing of the first intron often results in a reduced splicing of the second intron (Zhang and Blumenthal 1996). With the availability of the tos-1(n5384Δ) allele, we tested whether the deletion of intron 1 (n5384Δ) (Supplemental Fig. S1) would affect exon 3 skipping. As shown in Figure 4, B and C, exon 3 skipping was increased to ∼32% of all transcripts in tos-1(n5384Δ) animals, which was a fourfold increase compared with wild-type animals. This result suggests that the sequences deleted by tos-1(n5384Δ) are important for reducing exon 3 skipping. However, whether cis-elements other than those required for the splicing of intron 1 are involved remains to be investigated (see below).
3′ splice sites regulate the alternative splicing of tos-1
To investigate the roles of 3′ splice sites in regulating tos-1 alternative splicing, we used site-directed mutagenesis to modify 3′ splice sites and analyzed these modified 3′ splice sites in transgenic animals. In C. elegans, TTTTCAG is the major consensus 3′ splice site and is strongly recognized in both in vitro and in vivo experiments (Hollins et al. 2005; Ma and Horvitz 2009). 3′ splice sites of lower frequency in the genome are likely to be recognized less efficiently. In tos-1, both intron 1 and intron 2 contain 3′ splice sites (Fig. 5A) that are less frequent in the genome (Kent and Zahler 2000a,b), while intron 3 contains the consensus TTTTCAG (Fig. 5A). Interestingly, the 3′ splice site of intron 3 was similarly strongly recognized in all uaf-1 and sfa-1 mutants tested (see above).
FIGURE 5.
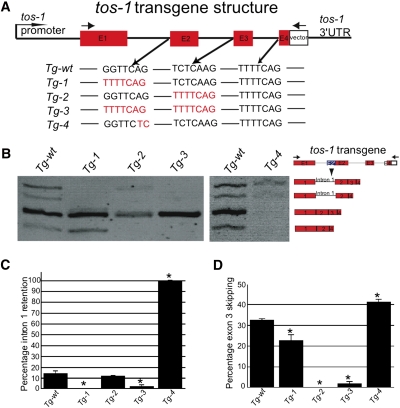
3′ splice site substitution and transgenic experiments identified 3′ splice sites being important for regulating intron 1 retention and exon 3 skipping. (A) Illustration of the transgene structure and the substitutions of 3′ splice sites. (Arrows) PCR primers for detecting transgene-specific transcripts. (B) RT-PCR experiments to detect splice patterns of each indicated transgene (top of micrograph) in wild-type background. Splice isoforms are indicated on the right. (C) Quantitative analysis of intron 1 retention of each transgene using the NIH Image J software. (D) Quantitative analysis of exon 3 skipping of each transgene using the NIH image J software. (*) p < 0.05.
We replaced the 3′ splice sites of intron 1 (GGTTCAG) and intron 2 (TCTCAAG) with the consensus site TTTTCAG in a tos-1 reporter gene (Fig. 5A) and generated transgenic animals in a wild-type background. Substituting the 3′ splice site of intron 1 with TTTTCAG caused the apparent disappearance of isoforms 1 and 2 and hence a failure of intron 1 retention (Fig. 5B,C, Tg-1). Similarly, substituting the 3′ splice site of intron 2 with TTTTCAG led to the disappearance of isoforms 2 and 5 and exon 3 skipping (Fig. 5B,D, Tg-2). As expected, when the 3′ splice sites in both intron 1 and intron 2 were substituted with TTTTCAG, isoforms 1, 2, and 5 were barely detectable, and both intron 1 retention and exon 3 skipping were greatly reduced (Fig. 5B–D, Tg-3).
We also mutated the conserved AG nucleotides at positions −1 and −2 of the 3′ splice site of intron 1 to TC, which should eliminate splicing at this site (Fig. 5A), and examined the splicing of the transgene. As shown in Figure 5B (Tg-4), isoforms 4 and 5 disappeared, while isoforms 1 and 2 were still detectable. We observed 100% intron 1 retention (Fig. 5C) and, interestingly, increased exon 3 skipping (Fig. 5D). This result suggests that lack of intron 1 splicing increases exon 3 skipping, which is consistent with the conclusion obtained from analyzing the tos-1(n5384Δ) mutant (Fig. 4C). In agreement with this notion, animals carrying transgene Tg-1 (which substituted the endogenous 3′ splice site of intron 1 with TTTTCAG) showed a much more efficient splicing of intron 1 (Fig. 5B,C), which resulted in a reduced exon 3 skipping (Fig. 5B,D). Interestingly, these effects were not as strong as those that resulted from substituting the 3′ splice site preceding exon 3 with the consensus sequence TTTTCAG (Fig. 5A,B,D, Tg2), which caused a complete disappearance of exon 3 skipping. Therefore, splicing at a strong 3′ splice site, such as TTTTCAG, might be less affected by the splicing of preceding introns. That the splicing at the consensus 3′ splice site TTTTCAG is not affected by the splicing of other introns is consistent with our previous study of unc-93(e1500) splicing, in which a substitution with the consensus sequence TTTTCAG of the weak 3′ splice sites at either intron 8 or exon 9 completely abolished the effects of surrounding sequences on splicing at these sites (Ma and Horvitz 2009).
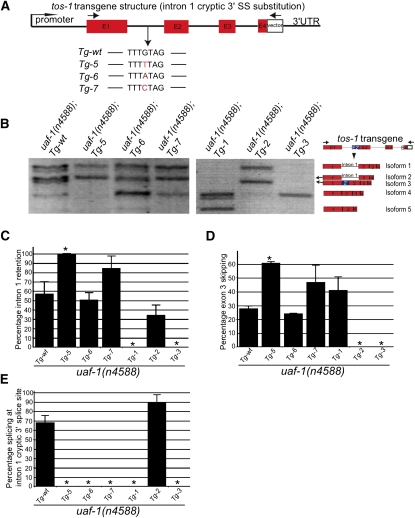
To examine whether the G nucleotide at position −4 of the cryptic 3′ splice site in intron 1 is important for the recognition of this site by UAF-1(T180I) (see above), we replaced this nucleotide with a T, A, or C and tested the effects of these substitutions on the splicing of tos-1 transgenes in uaf-1(n4588) animals (Fig. 6A). Substituting the G with a T, A, or C all caused altered splicing of the tos-1 transgenes in uaf-1(n4588) background (Fig. 6B). A G-to-T substitution (Tg-5) increased intron 1 retention (Fig. 6C) and exon 3 skipping (Fig. 6D) and abolished the recognition of the cryptic 3′ splice site (Fig. 6E). A G-to-A (Tg-6) or a G-to-C (Tg-7) substitution similarly abolished the recognition of the cryptic 3′ splice site but did not affect intron 1 retention and exon 3 skipping (Fig. 6C–E). These results confirmed and extended our previous analysis (Fig. 1D; Ma and Horvitz 2009) that a G at position −4 of the cryptic 3′ splice site is important for the recognition by UAF-1(T180I). It is interesting to note that the G-to-T substitution appeared to cause an decrease of isoform 4, while the G-to-A substitution caused an increase of isoform 4 (Fig. 6B), implying the presence of a potential regulatory sequence for intron 1 splicing where these nucleotide reside.
FIGURE 6.
3′ splice site substitution and transgenic experiments identified the G nucleotide at position −4 of the intron 1 cryptic 3′ splice site as important for recognition by UAF-1(T180I). (A) Illustration of transgene structure and substitutions at the cryptic 3′ splice site. (Arrows) PCR primers for detecting transgene-specific transcripts. (B) Splice patterns of the transgenes indicated in a uaf-1(n4588) background as detected by RT-PCR. Splice isoforms are indicated on the right. (C) Quantitative analysis of intron 1 retention of each transgene obtained using the NIH Image J software. (D) Quantitative analysis of exon 3 skipping of each transgene obtained using the NIH image J software. (E) Percentage of the recognition of the intron 1 cryptic 3′ splice site of each transgene calculated as the percentage of isoform 3 in the total of isoforms 3, 4, and 5. (*) p < 0.05.
We also tested the splicing of Tg-1, Tg-2, and Tg-3 in uaf-1(n4588) mutants (Fig. 6B–E). As in wild-type animals, when the weak 3′ splice site of intron 1 was substituted with the consensus TTTTCAG in Tg-1, intron 1 retention was abolished (Fig. 6B,C). When the weak 3′ splice site of intron 2 was substituted with the consensus TTTTCAG in Tg-2, exon 3 skipping was abolished (Fig. 6B,D). Both intron 1 retention and exon 3 skipping were abolished when both weak 3′ splice sites in introns 1 and 2 were substituted with TTTTCAG in Tg-3 (Fig. 6B–D). No recognition of the cryptic 3′ splice site was detected in Tg-1 (Fig. 6B,E), suggesting that the presence of the consensus TTTTCAG at the endogenous 3′ splice site of intron 1 suppressed the recognition of this cryptic site. This result is consistent with our mutagenesis analysis of the unc-93(e1500) transcript (Ma and Horvitz 2009), in which substituting either the weak endogenous 3′ splicing site in intron 8 or the cryptic 3′ splice site in exon 9 with the consensus TTTTCAG abolished splicing at the neighboring weak site and caused splicing exclusively at the consensus site.
DISCUSSION
tos-1 splicing could be used to analyze the in vivo regulation of alternative splicing
Genetic analysis of C. elegans unc-52 mutants identified the splicing factors SMU-1, SMU-2, and MEC-8 as regulators of unc-52 exon 17 skipping (Lundquist et al. 1996; Spike et al. 2002; Spartz et al. 2004). The mutually exclusive splicing of exons of the C. elegans genes egl-15 and let-2 at different developmental stages has been used to identify regulatory splicing factors (Kuroyanagi et al. 2006; Ohno et al. 2008). We have shown that altered splicing of unc-93(e1500) exon 9 can serve as a reporter for studying the in vivo recognition of alternative 3′ splice sites (Ma and Horvitz 2009). Splicing of tos-1 involves three of the four basic classes of alternative splicing events that can be easily detected and are affected by mutations of uaf-1 and sfa-1. We suggest that tos-1 could serve as a sensitive and efficient in vivo reporter for studying splicing regulation.
uaf-1 can affect multiple aspects of alternative splicing
Numerous splicing factors and intronic and exonic cis-regulatory elements act in alternative splicing (Maniatis and Tasic 2002; Zhou et al. 2002; Wang and Burge 2008; Barash et al. 2010). However, the in vivo function of the U2AF large subunit in regulating alternative splicing in animals remains to be fully understood. In this study, we analyzed the effects of a series of uaf-1 mutations on the pattern of tos-1 alternative splicing. We found that uaf-1(n4588) caused increased intron 1 retention, increased exon 3 skipping, and recognition of the cryptic 3′ splice site (Supplemental Table S2). Intragenic suppressors of uaf-1(n4588) exhibited variable effects on these splicing events. For example, uaf-1(n4588 n5127) caused increased intron 1 retention and recognition of the cryptic 3′ splice site but did not cause increased exon 3 skipping, while uaf-1(n4588 n5125) caused increased intron 1 retention, decreased exon 3 skipping, and displayed a weak recognition of the cryptic 3′ splice site (Supplemental Table S2). Two other uaf-1(n4588) intragenic suppressors, uaf-1(n4588 n5120) and uaf-1(n5123), did not obviously alter the recognition of the tos-1 endogenous 3′ splice sites (Supplemental Table S2) or tos-1 splicing (see above). The strength of each uaf-1 mutation in recognizing the cryptic 3′ splice site of tos-1 correlated with its strength in altering the splicing of unc-93(e1500) exon 9 (Ma and Horvitz 2009). These results suggest that in animals, UAF-1 might play important roles in regulating intron retention and exon skipping and possibly in the choice of alternative 3′ splice sites.
Although we identified 11 candidate genes from our microarray study, our RT-PCR analysis found that only the splicing of tos-1 was dramatically affected by uaf-1(n4588) (Fig. 1). Several factors could contribute to this low validation rate. First, microarray experiments can be noisy and inaccurate (Tan et al. 2003). Second, we performed the screen with only one set of arrays (see Materials and Methods), so that noise or false-positive signals could not have been eliminated by replicate microarrays. Third, the uaf-1(n4588) mutation does not affect the recognition of the consensus site TTTTCAG and probably of other strong 3′ splice sites based on our nucleotide substitution analyses and examination of the splicing of several other genes (Ma and Horvitz 2009); also, the altered recognition of certain weak 3′ splice sites by UAF-1(T180I) is context-dependent (Ma and Horvitz 2009; this study). Hence, splicing events affected by uaf-1(n4588) might be limited in number and/or have effects too weak to be detected by our microarray study.
n4588 is likely both a loss-of-function and an altered-function mutation of uaf-1
We previously postulated that n4588 might cause a loss of function or an altered function of UAF-1, or both (Ma and Horvitz 2009). In this study, we found that uaf-1(n4588) caused increased intron 1 retention and exon 3 skipping, which is similar to the effect of reducing uaf-1 expression by RNAi (Fig. 3C) (see above). However, uaf-1(n4588) also caused recognition of the intron 1 cryptic 3′ splice site, which was not detectable in either sfa-1(n4562) mutants or in animals treated with RNAi targeting uaf-1, sfa-1, or uaf-2. This result suggests that the recognition of the cryptic 3′ splice site by uaf-1(n4588) per se might be caused by an altered function of UAF-1. We propose that n4588 is a mutation that causes both a loss of function and an altered function of UAF-1.
sfa-1 might regulate alternative splicing of a subset of genes in C. elegans
We previously failed to detect an effect of sfa-1(n4562) on the splicing of the unc-93(e1500) transcript. Our findings are consistent with the conclusions of Guth and Valcarcel (2000) and Tanackovic and Kramer (2005), who suggested that SF1/BBP might be required for the splicing of a subset of transcripts. In this study, we found that splicing of tos-1 is dramatically altered by sfa-1(n4562), providing in vivo evidence that SFA-1 can affect alternative splicing in C. elegans. In addition, sfa-1(n4562) did not cause recognition of the cryptic 3′ splice site in tos-1 intron 1 (Fig. 3B; Supplemental Table S2), which is consistent with our previous finding that sfa-1(n4562) did not cause recognition of a cryptic 3′ splice site in exon 9 of unc-93(e1500) (Ma and Horvitz 2009).
Reducing the expression of uaf-1, sfa-1, or uaf-2 by RNAi caused a slight increase of isoforms 2 and 5, similar to that caused by the sfa-1(n4562) mutation. The similarity of sfa-1(n4562) and sfa-1(RNAi) in affecting tos-1 alternative splicing is consistent with our previous conclusion that n4562 is a loss-of-function allele of sfa-1 (Ma and Horvitz 2009). In addition, the similarity of uaf-1(RNAi), sfa-1(RNAi), and uaf-2(RNAi) in affecting tos-1 splicing is consistent with the notion that UAF-1, SFA-1, and UAF-2 can interact to regulate the recognition of 3′ splice sites. However, it remains to be investigated whether UAF-1, SFA-1, and UAF-2 always function together to regulate splicing of the same set of genes in vivo.
Intron retention and exon skipping could be regulated by 3′ splice sites in C. elegans
The 3′ splice sites of intron 1 (GGTTCAG) and intron 2 (TCTCAAG) are relatively uncommon in C. elegans introns (Kent and Zahler 2000a,b) and are likely weakly recognized by the U2AF splice factors compared with the consensus site TTTTCAG, based on in vitro binding assays (Hollins et al. 2005). Because intron 1 retention and exon 3 skipping are caused by altered recognition at these two sites and splicing at the 3′ splice site of intron 3 (a consensus sequence TTTTCAG) was not altered by uaf-1 and sfa-1 mutations (Supplemental Table S2), we tested whether the identities of 3′ splice sites (Fig. 5) play important roles in regulating intron retention and exon skipping. We found that substituting the less frequent endogenous 3′ splice sites in intron 1 and intron 2 with the consensus sequence TTTTCAG caused a persistent recognition of the substituted sites, the abolishment of intron 1 retention and exon 3 skipping, and a dramatic change of the splicing patterns of the tos-1 transgenes. Our nucleotide substitution analysis indicates that the G nucleotide at position −4 of the intron 1 cryptic 3′ splice site is important for the recognition of this site by UAF-1(T180I) (Fig. 6), confirming and extending our previous nucleotide substitution analysis of the unc-93(e1500) cryptic 3′ splice site (Ma and Horvitz 2009).
In C. elegans and other animals, alternative splicing is regulated by exonic and intronic cis-regulatory elements and trans-splicing factors that bind these cis-elements (Maniatis and Tasic 2002; Zahler 2005; Kuroyanagi et al. 2006). The contribution of 3′ splice sites in intron retention and exon skipping remains to be fully understood. Our study suggests that in addition to the activities of the U2AF factors and SF1/BBP, the sequences of 3′ splice sites play important roles in intron retention and exon skipping in C. elegans. By changing 3′ splice sites or the activities of the U2AF factors and SF1/BBP, we could dramatically alter the splicing pattern of tos-1. Our study also provides an in vivo system, which includes a sensitive endogenous reporter, tos-1, and mutations affecting UAF-1 and SFA-1 that can be used for analyzing other trans-splicing factors in the regulation of alternative splicing in C. elegans.
MATERIALS AND METHODS
Strains
C. elegans strains were grown at 20°C as described (Brenner 1974), except where otherwise specified. N2 (Bristol) was the wild-type strain. Mutations used in this study include LGIII: uaf-1(n4588, n5120, n5123, n5125, n5127) (Ma and Horvitz 2009), unc-93(e1500) (Greenwald and Horvitz 1980); LGIV: sfa-1(n4562) (Ma and Horvitz 2009); and LGV: tos-1(n5384Δ) (this study). The translocation nT1 IV;V with the dominant gfp marker qIs51 (Belfiore et al. 2002) was used to balance the sfa-1 locus.
Whole-genome tiling microarray-based screen and gene expression analysis
Total RNA was prepared from synchronized L1 wild-type and uaf-1(n4588) animals using TRIzol as described by the manufacturer (Invitrogen). Double-stranded cDNA was prepared using a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and was labeled and hybridized to a C. elegans whole-genome tiling microarray according to the manufacturer's instructions (Affymetrix). Each double-stranded cDNA sample was analyzed using one microarray chip each for wild-type and uaf-1(n4588) animals. Raw data were processed using the Tiling Analysis Software (TAS v1.1.02; Affymetrix) using quantile normalization, with both samples normalized together. Probe intensity data were then overlaid with gene annotations based on genomic coordinates (C. elegans assembly WS120/ce2, 2004). Genes with at least three probes exhibiting an expression difference of fourfold or more between uaf-1(n4588) and wild-type animals were examined using the Integrated Genome Browser (http://www.bioviz.org) with TAS output and annotated public domain exon structure data. We focused on genes with annotated splice isoforms that were altered in uaf-1(n4588) animals. We identified 11 such genes (Supplemental Table S1) as candidates and examined their mRNA expression in both wild-type and uaf-1(n4588) animals using RT-PCR experiments (see below). Only one gene (tos-1) was confirmed to exhibit apparent altered splicing in uaf-1(n4588) animals.
RT-PCR and quantitative analysis of splice isoforms
Total RNA was prepared using TRIzol according to the manufacturer's instructions (Invitrogen), treated with RNase-Free DNase I (New England Biolabs), and incubated for 10 min at 75°C to inactivate DNase I. First-strand cDNA was synthesized with random hexamer primers (New England Biolabs) using the SuperScript III First-Strand Synthesis Kit (Invitrogen). RT-PCR was performed using an Eppendorf cycler with the following conditions: 30 sec at 94°C, 30 sec at 52°C, and 1 min at 72°C, 30 cycles. PCR primers for full-length tos-1 cDNA were 5′-ATGATCTACGGATTCGAGTCGTCACCATC-3′ and 5′-TCAAGCGCTATCCTCCAGTGACTTC-3′. PCR primers for analyzing the recognition of the intron 1 endogenous and cryptic 3′ splice sites were 5′-ATCTACGGATTCGAGTCGTCACCATC-3′ and 5′- GAAGAAATCTTCCAGTCCGAAGGG-3′. PCR primers for analyzing transgene-specific transcripts were 5′-ATGATCTACGGATTCGAGTCGTCACC-3′ and 5′- GCGAATTCACTAGTGATTACCGGTAC-3′. Two or three independent samples for each genotype of animals at mixed stages were analyzed, and the proportions of each splice isoform were quantified from each biological replicate using the NIH ImageJ software to measure the intensity of each isoform separated by agarose gels. We confirmed that our PCR assay could quantitatively amplify each tos-1 splice isoform by using ∼106 copies of purified cDNA for each isoform as a template and obtaining similar molar amounts of each isoform in the final product (Supplemental Fig. S3). For transgene analysis in a wild-type background, one stable line was analyzed for each of the transgenes Tg-1, Tg-2, and Tg-4 (Fig. 5), and two stable lines were analyzed for each of the transgenes Tg-wt and Tg-3 (Fig. 5). For transgene analysis in a uaf-1(n4588) background, one stable line for Tg-3 and two stable lines for each of the trangenes Tg-wt, Tg-1, Tg-2, Tg-5, Tg-6, and Tg-7 were analyzed (Fig. 6). Proportions of intron 1 retention or exon 3 skipping were calculated as the combined ratios of all transcripts with intron 1 retention or exon 3 skipping, respectively.
Identification of splice isoforms
RT-PCR samples were resolved with 2.5% agarose gels, and all visible PCR bands were isolated using the QIAquick Gel Extraction Kit (QIAGEN) as a mixture. The PCR product was subcloned to a pGEM-T Easy vector (Promega), and the sequences of each individual insert of 40 or more clones were determined. The nucleotide sequences of each splice isoform and the amino acid sequences of their encoded protein products are listed in Supplemental Figure S1.
Isolation of deletion alleles
Genomic DNA pools from EMS-mutagenized animals were screened for deletions using PCR as described (Jansen et al. 1997; Liu et al. 1999). Deletion mutant animals were isolated from frozen stocks and backcrossed to the wild type at least three times. tos-1(n5384Δ) removes nucleotides 16,301–16,640 of cosmid K07B1.
RNA interference
Young adult animals [wild-type or unc-93(e1500)] were fed HT115(DE3) bacteria containing plasmids directing the expression of dsRNAs targeting uaf-1, sfa-1, uaf-2, or tos-1 on NGM plates with 1 mM IPTG and 0.1 μg/mL ampicillin (Timmons et al. 2001). F1 progeny of wild-type or unc-93(e1500) animals were examined for obviously abnormal phenotypes or suppression of locomotion defects, respectively. We generated the DNA construct expressing dsRNA targeting tos-1 (see below). RNAi feeding for uaf-1 and sfa-1 was performed as described (Ma and Horvitz 2009). The uaf-2 RNAi clone was derived from an RNAi library (Kamath et al. 2003), and the sequence of the insert was determined to confirm the presence of the uaf-2 coding region.
Plasmids
We cloned a full-length tos-1 genomic DNA (from ATG to TGA) into a pGEM-TA easy vector (Promega) and subcloned a PstI/ApaI fragment (670 bp) from this construct into pPD129.36 using PstI and ApaI sites. This construct contains sequences from the first exon to a portion of the third intron of tos-1 and was used for the RNAi feeding experiment. To examine the effects of different 3′ splice sites on tos-1 intron 1 retention and exon 3 skipping, a genomic sequence of tos-1 containing a 1.3-kb promoter region, the whole coding sequence without the stop codon, and a 0.6-kb 3′ region was subcloned into a pGEM-TA easy vector (Promega). A short multiple cloning sequence of the pGEM-TA vector was inserted between the coding region and the 3′ region, which was used for detecting transgene-specific transcripts. Mutations in transgenes for splice site substitution analysis were introduced as described with primers containing corresponding mutations (Chiu et al. 2004). PCR was performed using Eppendorf Cyclers, and DNA products were resolved using agarose gels. DNA sequence determination was performed with an ABI Prism 3100 Genetic Analyzer and an ABI 3730XL DNA Analyzer.
Transgene experiments
Germline transgene experiments were performed as described (Mello et al. 1991). Transgene mixtures generally contained 50 μg/mL 1-kb DNA plus ladder (Invitrogen), 20 μg/mL Arabidopsis genomic DNA, 5 μg/mL pPD93.97 as a coinjection marker, and 5 μg/mL the transgene of interest. pPD93.97 drives the expression of GFP in body-wall muscles.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Horvitz laboratory, especially Daniel Denning, Nicolas Paquin, Takashi Hirose, and Dengke Ma, for critical suggestions. We thank Na An and Yaoyu Zhong for strain maintenance and Rita Droste, Fei Wang, and Liange Huang for technical support. We thank Andy Fire for providing the pPD plasmids we used to construct our RNAi plasmids. Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. Some genomic sequences and polymorphisms were identified by the WUSTL Genome Center. H.R.H. is the David H. Koch Professor of Biology at MIT and an Investigator of the Howard Hughes Medical Institute. This study was supported by a Natural Science Foundation of China grant (30971639) and a National Basic Research Program of China grant to L.M. (2011CB510005) and NIH Grant GM24663 to H.R.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027458.111.
REFERENCES
- Abovich N, Rosbash M 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412 [DOI] [PubMed] [Google Scholar]
- Arning S, Gruter P, Bilbe G, Kramer A 1996. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA 2: 794–810 [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Belfiore M, Mathies LD, Pugnale P, Moulder G, Barstead R, Kimble J, Puoti A 2002. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3′ untranslated region in Caenorhabditis elegans. RNA 8: 725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs RT, Gregor P, Idriss S, Belote JM, McKeown M 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50: 739–747 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis KC, Baker BS 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010 [DOI] [PubMed] [Google Scholar]
- Chiu J, March P, Lee R, Tillett D 2004. Site-directed, Ligase-Independent Mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res 32: e174 doi: 10.1093/nar/gnh172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680 [PMC free article] [PubMed] [Google Scholar]
- Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ 2003. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development 130: 3757–3766 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Horvitz HR 1980. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics 96: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth S, Valcarcel J 2000. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J Biol Chem 275: 38059–38066 [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Maniatis T 1997. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol 9: 350–357 [DOI] [PubMed] [Google Scholar]
- Hollins C, Zorio DA, MacMorris M, Blumenthal T 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11: 248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet 17: 119–121 [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM 2000a. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res 10: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM 2000b. The intronerator: Exploring introns and alternative splicing in Caenorhabditis elegans. Nucleic Acids Res 28: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T 1985. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell 42: 725–736 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M 2006. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods 3: 909–915 [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, et al. 1999. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res 9: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, Shaw JE 1996. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122: 1601–1610 [DOI] [PubMed] [Google Scholar]
- Ma L, Horvitz HR 2009. Mutations in the Caenorhabditis elegans U2AF large subunit UAF-1 alter the choice of a 3′ splice site in vivo. PLoS Genet 5: e1000708 doi: 10.1371/journal.pgen.1000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1994. Dynamic RNA–RNA interactions in the spliceosome. Annu Rev Genet 28: 1–26 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236–243 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Puoti A, Kramer A 1999. Splicing factor SF1 from Drosophila and Caenorhabditis: Presence of an N-terminal RS domain and requirement for viability. RNA 5: 1615–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V 1991. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841 [DOI] [PubMed] [Google Scholar]
- Navaratnam DS, Bell TJ, Tu TD, Cohen EL, Oberholtzer JC 1997. Differential distribution of Ca2+-activated K+ channel splice variants among hair cells along the tonotopic axis of the chick cochlea. Neuron 19: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G, Hagiwara M, Kuroyanagi H 2008. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev 22: 360–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol 12: 340–345 [DOI] [PubMed] [Google Scholar]
- Rosenblatt KP, Sun ZP, Heller S, Hudspeth AJ 1997. Distribution of Ca2+-activated K+ channel isoforms along the tonotopic gradient of the chicken's cochlea. Neuron 19: 1061–1075 [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684 [DOI] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem Sci 25: 381–388 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Herman RK, Shaw JE 2004. SMU-2 and SMU-1, Caenorhabditis elegans homologs of mammalian spliceosome-associated proteins RED and fSAP57, work together to affect splice site choice. Mol Cell Biol 24: 6811–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Davies AG, Shaw JE, Herman RK 2002. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129: 4999–5008 [DOI] [PubMed] [Google Scholar]
- Tan PK, Downey TJ, Spitznagel ELJ, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC 2003. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res 31: 5676–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Kramer A 2005. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell 16: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Wang ZW, Burge C 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835 [DOI] [PubMed] [Google Scholar]
- Zahler AM 2005. Alternative splicing in C. elegans. WormBook 2005: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB 1992. SR proteins: A conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR 1991. Biochemical characterization of U2 snRNP auxiliary factor: An essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J 10: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Blumenthal T 1996. Functional analysis of an intron 3′ splice site in Caenorhabditis elegans. RNA 2: 380–388 [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T 1999a. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T 1999b. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]