Group II introns are mobile genetic elements that self-splice from pre-mRNA transcripts. Some fragmented group II introns found in chloroplastic and mitochondrial genomes are able to assemble and splice in trans. The Ll.LtrB group II intron from the Gram-positive bacterium Lactococcus lactis was shown to splice in trans when fragmented at various locations throughout its structure. Here the authors used Ll.LtrB to assess the contribution of base-pairing interactions between intron fragments during trans-splicing in vivo. By comparing closely located fragmentation sites, the authors show that Ll.LtrB trans-splices more efficiently when base-pairing interactions can occur between the two intron fragments. These findings demonstrate the important contribution of base-pairing interactions for the assembly of group II intron fragments during trans-splicing and rationalize why such interactions were evolutionarily conserved in natural trans-splicing group II introns.
Keywords: Ll.LtrB, Lactococcus lactis, LtrA, conjugation, sex factor
Abstract
Group II introns are mobile genetic elements that self-splice from pre-mRNA transcripts. Some fragmented group II introns found in chloroplastic and mitochondrial genomes are able to assemble and splice in trans. The Ll.LtrB group II intron from the Gram-positive bacterium Lactococcus lactis was shown to splice in trans when fragmented at various locations throughout its structure. Here we used Ll.LtrB to assess the contribution of base-pairing interactions between intron fragments during trans-splicing in vivo. By comparing closely located fragmentation sites, we show that Ll.LtrB trans-splices more efficiently when base-pairing interactions can occur between the two intron fragments. Disruptions and stepwise restorations of specific base-pairing interactions between intron fragments resulted respectively in significant reductions and recoveries of the Ll.LtrB trans-splicing efficiency. Finally, although we confirm that LtrA is an important co-factor for trans-splicing, its overexpression cannot compensate for the reduction in trans-splicing efficiency when the potential base-pairing interactions between intron fragments are disrupted. These findings demonstrate the important contribution of base-pairing interactions for the assembly of group II intron fragments during trans-splicing and rationalizes why such interactions were evolutionarily conserved in natural trans-splicing group II introns.
INTRODUCTION
Group II introns are retromobile elements found in bacteria, bacterial-derived organelles, and archaea (Lambowitz and Zimmerly 2004, 2010). These intervening sequences are RNA enzymes that splice autocatalytically from pre-mRNA transcripts. However, they require the assistance of maturases to fold into their active three-dimensional structure in order to splice in vivo. Despite sharing very little sequence similarity, these large ribozymes fold into a highly conserved RNA secondary structure that consists of six domains (DI to DVI) radiating from a central wheel (e.g., Fig. 1A; Michel et al. 1989; Qin and Pyle 1998; Toor et al. 2001). Each domain contains specific features that contribute to the folding of the intron into its active tertiary conformation. DI, which is the largest domain and the first to be transcribed, provides the scaffold to dock the remaining intron domains. Accordingly, DI takes part in many long-range tertiary interactions with other domains and orchestrates the overall folding of group II introns (Fedorova and Zingler 2007; Pyle et al. 2007). While DII is a small structural domain, DIII functions as a catalytic effector that stimulates splicing (Fedorova et al. 2003). Most group II introns harbor an open reading frame (ORF) in the loop region of DIV that codes for an intron-encoded protein (IEP). IEPs are multifunctional proteins involved in both intron splicing and mobility (Saldanha et al. 1993; Matsuura et al. 2001; Cui et al. 2004). DV is the catalytic domain of group II introns, which positions the catalytic nucleotides and two Mg++ ions at the center of the catalytic core (Pyle 2002). Finally, DVI harbors the branch point nucleotide positioned near the 3′ end of the intron (Lambowitz and Zimmerly 2004, 2010). This highly conserved bulged adenosine is also part of the catalytic center since it initiates the splicing reaction.
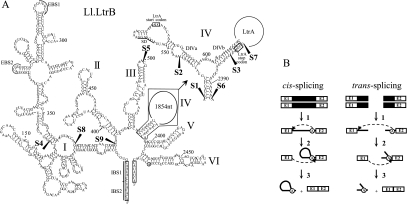
FIGURE 1.
Ll.LtrB secondary structure and splicing pathways. (A) Ll.LtrB secondary structure. The six domains of Ll.LtrB are indicated (I–VI) and the detailed secondary structure of a portion of domain IV is also shown (top right). The ltrA start and stop codons are boxed and the Shine–Dalgarno sequence (SD) is underlined. Exons 1 and 2 at both 5′ and 3′ extremities of the intron are also boxed. Fragmentation sites are mapped with black arrowheads (S1–S9). S4 was not used in this study. EBS1 and EBS2, exon-binding site 1 and 2; IBS1 and IBS2, intron-binding site 1 and 2; branch point, circled A. (B) Group II intron cis- and trans-splicing pathways. Following transcription of the interrupted gene (step 1), the 2′-OH of the bulged adenosine present in domain VI (circled A) performs the initial nucleophilic attack at the exon 1–intron splice junction generating a 2′–5′ linkage (step 2). Then, the 3′-OH of the released exon 1 performs the second nucleophilic attack at the intron–exon 2 splice junction, releasing the intron lariat and ligating the flanking exons (step 3). Fragmented group II introns can also splice in trans using the same pathway (steps 1–3). Because trans-splicing occurs between two independent RNA transcripts, the intron is released as a Y-branched molecule instead of a lariat. Group II intron, black line; exon 1 and 2, E1 and E2; branch point, circled A.
Group II introns self-splice from pre-mRNA transcripts by two consecutive trans-esterification reactions (Fig. 1B, cis-splicing; Lambowitz and Zimmerly 2004). Following transcription of the interrupted gene (step 1), the 2′-OH of the bulged adenosine residue in DVI (circled A) initiates the first nucleophilic attack at the exon 1–intron splice junction (step 2) which generates a 2′–5′ linkage. Then, the 3′-OH of the released exon 1 performs the second nucleophilic attack at the intron–exon 2 splice junction (step 3), releasing the intron lariat and ligating the flanking exons (Lehmann and Schmidt 2003). The intron-free mature mRNA can then be translated and the encoded protein synthesized.
Some group II introns fragmented in two or three pieces have been found in chloroplasts of algae as well as in chloroplasts and mitochondria of higher plants (Glanz and Kuck 2009; and references within). These fragmented introns splice in trans using the same pathway as cis-splicing introns (Fig. 1B, trans-splicing). Following transcription of the separate gene pieces (step 1), the intron fragments presumably assemble and fold into the active tertiary structure allowing the two trans-esterification reactions to occur (steps 2 and 3). Because trans-splicing takes place between two RNA transcripts, the intron is released as a Y-branched molecule instead of a lariat.
The Ll.LtrB group II intron from the Gram-positive bacterium Lactococcus lactis is 2.5 kb in length and encodes for LtrA, a 599-amino acid multifunctional protein with reverse transcriptase, maturase, DNA binding, and endonuclease activities (Mills et al. 1996; Lambowitz and Zimmerly 2004, 2010). LtrA is a very important splicing co-factor for both the cis- (Mills et al. 1996; Ichiyanagi et al. 2002; Belhocine et al. 2007) and trans-splicing (Belhocine et al. 2007, 2008) of Ll.LtrB in L. lactis. It was shown to bind the Ll.LtrB intron RNA at a high affinity-binding site located in DIVa (Wank et al. 1999; Singh et al. 2002) and to multiple secondary sites located in DI, DII, and DVI (Singh et al. 2002; Dai et al. 2008); overall LtrA binds with a 2:1 stoichiometry, suggesting that it may function as a dimer (Saldanha et al. 1999; Rambo and Doudna 2004). Ll.LtrB interrupts the ltrB gene, which codes for a relaxase enzyme in three L. lactis conjugative elements: the pRS01 and pAH90 plasmids and an integrative and conjugative element called the sex factor (SF) (Mills et al. 1996). The LtrB relaxase is a single-strand endonuclease that nicks the origin of transfer of conjugative elements and initiates their transfer between L. lactis strains. Because this enzyme is essential for the initiation of conjugative transfer, splicing of Ll.LtrB is absolutely required for the intercellular transfer of its host elements (Shearman et al. 1996; Klein et al. 2004). Taking advantage of the relationship between the conjugative transfer of the SF and splicing of Ll.LtrB from the relaxase transcript, we developed a highly sensitive trans-splicing/conjugation assay in L. lactis. Using this assay we demonstrated that Ll.LtrB is able to splice in trans when fragmented at natural group II intron fragmentation sites (Belhocine et al. 2007). We also showed that Ll.LtrB trans-splices efficiently when fragmented at various other locations throughout its structure, therefore demonstrating the versatility of Ll.LtrB compared to trans-splicing group II introns found in nature (Belhocine et al. 2008).
In this study we use the Ll.LtrB group II intron as a model system to assess the contribution of base-pairing interactions between intron fragments during trans-splicing in vivo. Using the trans-splicing/conjugation assay that we developed, we observe that Ll.LtrB variants with potential base-pairing interactions between the two intron fragments trans-splice more efficiently than variants that are fragmented at close proximity but do not have any potential interactions between the two intron fragments. We also show that abolishing specific base-pairing interactions between intron fragments of Ll.LtrB variants fragmented in DI, DIII, DIVa, or DIVb resulted in important reductions in trans-splicing efficiency. Moreover, we demonstrate that stepwise restorations of base-pairing interactions between intron fragments by either the re-establishment of the wild-type sequence or sequence complementarity resulted in significant recovery of the trans-splicing efficiency. Finally, although we confirm that LtrA is an important Ll.LtrB trans-splicing co-factor, overexpression of LtrA cannot compensate for the reduction in Ll.LtrB trans-splicing efficiency when the potential base-pairing interactions between intron fragments are disrupted. Altogether these findings demonstrate the important contribution of base-pairing for the assembly of intron fragments during trans-splicing and rationalize why such interactions were evolutionarily conserved between nucleotides located on either side of fragmentation sites in natural trans-splicing group II introns.
MATERIALS AND METHODS
Strains and plasmids
The L. lactis strains NZ9800ΔltrB::tet (Klein et al. 2004) and LM0231 (Shearman et al. 1996) were grown at 30°C without shaking in M17 media (Oxoid) supplemented with 0.5% glucose (GM17). The Escherichia coli DH10β strain was used for cloning and was grown at 37°C in LB broth (Wisent) with shaking. When necessary, antibiotics were added at the following concentrations: 3 μg/mL for tetracycline (Tet), 300 μg/mL for spectinomycin (Spc), 25 μg/mL for fusidic acid (Fus), and 10 μg/mL for chloramphenicol (Cam).
Plasmids pDL-P232, pDL-P232-ltrB, pDL-P232-S1-WT, pDL-P232-S2-WT, pDL-P232-S3-WT, and pDL-P232-S5-WT were previously engineered (Belhocine et al. 2007). The other fragmented variants of Ll.LtrB (pDL-P232-S6-WT to pDL-P232-S9-WT) were generated by PCR amplification using the interrupted ltrB gene as the template. Each construct was created using two pairs of primers (Supplemental Table S1). The first primer pairs (NotI) were used to amplify the 5′ end of the ltrB gene, from the beginning of the 5′ exon up to the respective fragmentation points within the intron. The second primer pairs (BssHII) were used to amplify the region from the fragmentation sites within the intron up to the end of the 3′ exon (Belhocine et al. 2007). Plasmids pDL-P232-S1-ΔORF to pDL-P232-S9-ΔORF were similarly created but using as the PCR template the ltrB gene interrupted by the ΔORF version of Ll.LtrB (Supplemental Table S1).
Plasmids containing different intron variants with disruptions and restorations of base-pairing interactions in DI, DIII, DIVa, and DIVb were engineered either by site-directed mutagenesis or by PCR using the Ll.LtrB ΔORF intron as the template (Supplemental Table S1). Disruptions of the base-pairing interactions were done by replacing G↔C and T↔A. The unpaired nucleotides located between stems were not modified.
The pLE-Pnis-ltrA plasmid was constructed by first cloning the nisin-inducible promoter (Pnis) into the unique BamHI site of the Gram-positive/Gram-negative shuttle plasmid pLE1. The PCR-amplified ltrA gene (NotI) was then introduced at the engineered NotI site directly downstream from Pnis (Supplemental Table S1). Integrity of all plasmids was confirmed by sequencing (Génome Québec).
Conjugation assay
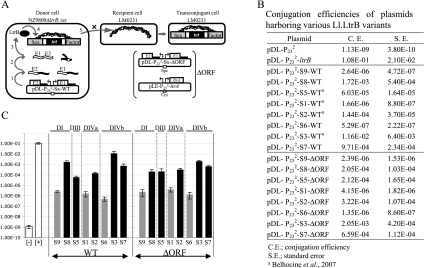
Mating was done on 5% non-fat dried milk plates (Carnation milk) containing 1% glucose and 1.5% agar. L. lactis NZ9800ΔltrB::tet (TetR) was used as the donor strain while L. lactis LM0231 (FusR) was used as the recipient strain. Both strains were diluted from saturated overnight cultures (0.4 mL into 10 mL) and grown for 7 h at 30°C. Cells were collected by centrifugation, mixed, spread on milk plates, incubated at 30°C for 16 h, and recovered with PBS 1×. Serial dilutions were plated on GM17 plates containing the specific antibiotic to select for donor, recipient or transconjugant cells. Conjugation efficiency of the chromosomal SF was assessed with two different assays (Fig. 2A). The first assay measured the SF conjugation efficiency when LtrA is encoded within the Ll.LtrB intron (pDL-P232-Sx-WT series). The second assay determined the SF transfer efficiency when LtrA is provided in trans from pLE-P232-ltrA (pDL-P232-Sx-ΔORF series). The conjugation efficiencies were calculated as the ratio of transconjugant (FusR/TetR) to donor cells (Spc for LtrA provided in cis, Spc/Cam for LtrA provided in trans) for three independent assays.
FIGURE 2.
Ll.LtrB trans-splicing/conjugation assay and SF conjugation rates. (A) The trans-splicing efficiency of Ll.LtrB is monitored by the conjugation rate of the SF between two L. lactis strains. The donor cell harbors the chromosomal SF with a defective relaxase gene (NZ9800ΔltrB::tet) while the recipient cell lacks the SF (LM0231). The Ll.LtrB intron, along with portions of its exons, was replaced in the chromosome of NZ9800 by a tetracycline resistance marker (tet), which prevents expression of the relaxase (LtrB). The LtrB deficient strain is complemented by providing the interrupted ltrB gene from a plasmid. Following transcription of the gene pieces (step 1), intron fragments assemble, Ll.LtrB trans-splices, and the flanking exons are ligated (step 2). Mature mRNA is translated and leads to expression of the relaxase enzyme (LtrB) (step 3), which recognizes the SF origin of transfer (oriT) (step 4) and initiates its transfer from a donor to a recipient cell by conjugation (step 5). Transfer efficiency of conjugative elements between L. lactis strains was previously shown to be directly proportional to Ll.LtrB splicing from the relaxase transcript (Klein et al. 2004; Belhocine et al. 2007). In a second version of the assay, the pLE-LtrA plasmid was introduced in various L. lactis strains (NZ9800ΔltrB::tet/pDL-P232-Sx-ΔORF) to provide LtrA in trans (between brackets). Ll.LtrB group II intron, black line; exon 1 and 2, E1 and E2; P23 promoter, bent arrows; SF, gray; intron RNA fragments, black wavy lines; RNA exons, white wavy lines; LtrB, black oval; spectinomycin resistance marker, Spc; chloramphenicol resistance marker, Cm; L. lactis chromosome, scribble. (B,C) SF conjugation efficiency in the presence of the ltrB gene interrupted by various Ll.LtrB fragmented introns (S1–S9 WT and ΔORF). Gray bars, bipartite introns with no potential base-pairing interactions; black bars, bipartite introns with potential base-pairing interactions; white bars, negative ([−], pDL-P232, empty vector) and positive ([+], pDL-P232-ltrB, full length Ll.LtrB) controls.
RNA isolation and qRT-PCR
L. lactis strains containing pLE-Pnis-ltrA and one of the pDL-P232-Sx-ΔORF variant were grown to logarithmic phase (OD600 = 0.5), aliquoted, and induced for 3 h with serial dilutions of nisin (Sigma) (0 ng/mL, 0.14 ng/mL, 0.28 ng/mL, 0.42 ng/mL, 0.56 ng/mL, 0.70 ng/mL, 0.84 ng/mL, 0.98 ng/mL). Cell pellets were mixed with 500 mL of TRIzol (Invitrogen life technologies) and 250 mg of acid-washed glass beads (Sigma). The mixture was vortexed for 3 min and incubated at 55°C for 5 min; this treatment was repeated a total of three times. The remainder of the RNA extraction was performed according to the manufacturer's protocol. Total RNA concentration and quality was determined using a NanoDrop 1000 instrument (Thermo Scientific). Total RNA (10 μg) was treated with 2 U of DNAseI (New England Biolabs) for 30 min at 37°C in a final volume of 50 μL and heat-inactivated for 15 min at 75°C in the presence of 5 mM EDTA. Quantitative RT-PCR was performed with ∼0.5 μg of treated total RNA using the EXPRESS One-Step SYBR GreenER kit (Invitrogen) and the Rotor Gene RG-3000 software from the Thermo LightCycler instrument (Corbetz Research) (Supplemental Table S1). Amplification conditions were: (95°C, 20 sec; 60°C, 1 min) × 45. Slope noise correction and dynamic tube normalization was applied to the raw fluorescence data prior to analysis. The Ct values recorded during amplification, which correspond to the point at which the level of fluorescence rises significantly above the background, were analyzed. In order to obtain the ΔCt of each sample, we subtracted the level of fluorescence recorded in our negative control to the fluorescence detected for each sample. Inter-run variations were adjusted using as a calibrator the pDL-P232-S1 variant grown in the absence of nisin. The final fold differences were obtained with the comparative quantification algorithm: 2−ΔΔCt, where ΔΔCt = (CtltrB – CtldhB)sample − (CtltrB – CtldhB)calibrator. The values were normalized using the ldhB housekeeping gene as a reference (Supplemental Table S1). All assays were performed in triplicate to confirm reproducibility and data presented are the average and standard errors. Specificity of the PCR reaction was analyzed on 2% agarose gel and the PCR product was confirmed by DNA sequencing (Genome Québec).
RESULTS
Ll.LtrB trans-splicing efficiency is significantly different between introns fragmented in close proximity in DI, DIII, DIVa, or DIVb
In order to assess the contribution of base-pairing interactions during trans-splicing, Ll.LtrB was split in two fragments (Fig. 1A, S1 to S9). Then, trans-splicing efficiency was compared between bipartite introns with close fragmentation sites that allowed for or lacked potential base-pairing interactions between the two fragments. Ll.LtrB trans-splicing was monitored by measuring the conjugation efficiency of the chromosomal SF between L. lactis strains (Fig. 2A; Belhocine et al. 2007). Following transcription of the two gene pieces (step 1), Ll.LtrB trans-splices and ligates its flanking exons (step 2). This leads to expression of the relaxase enzyme (LtrB) (step 3), which recognizes the origin of transfer of the SF (step 4) and initiates SF transfer from a donor to a recipient cell by conjugation (step 5). Transfer efficiency of the SF between L. lactis strains is directly proportional to Ll.LtrB splicing from the relaxase transcript (Klein et al. 2004; Belhocine et al. 2007).
Fragmentation of Ll.LtrB at position S8 in DI provides a potential interaction of 16 bp between the two intron RNA fragments while fragmentation at position S9, only 18 nt downstream, does not. Ll.LtrB fragmented at S8 supports conjugative transfer of the SF more efficiently than S9 (Fig. 2B). This is similar to what was previously observed when comparing the trans-splicing efficiency of Ll.LtrB fragmented at positions S1 and S2 in DIVa, which are only 19 nt apart. Ll.LtrB fragmented at S2, which allows for a potential interaction of 16 bp between the two intron fragments, trans-splices more efficiently than S1, which in contrast does not allow for any potential base-pairing (Fig. 2B; Belhocine et al. 2007). Next, we compared close fragmentation sites in other domains of Ll.LtrB. In accordance with our previous observations in DI and DIVa, Ll.LtrB fragmented at the tip of DIII at position S5 trans-splices more efficiently than if the fragmentation site is located 26 nt downstream at position S1 (Fig. 2B). This suggests that the potential interaction of 17 bp between the two RNA fragments contributes to intron assembly during trans-splicing in L. lactis. Next, we compared the trans-splicing efficiency of Ll.LtrB fragmented at position S6, located at the bottom of the stem in DIV, with two intron variants fragmented in DIVb at positions S3 and S7 allowing for potential interactions of 15 and 22 bp, respectively, between the two intron fragments. The Ll.LtrB variants fragmented at positions S3 and S7 were significantly more efficient than S6 (Fig. 2B).
Fragmentation sites located in both DIVa (S1, S2) and DIVb (S3, S6, S7) are adjacent to the extremities of the ltrA gene and may affect LtrA expression levels (Fig. 1A). Since LtrA was shown to be a very important splicing co-factor (Mills et al. 1996; Ichiyanagi et al. 2002; Belhocine et al. 2007, 2008), it is crucial to keep its expression level constant while assessing the contribution of base-pairing interactions during trans-splicing. In order to ensure comparable levels of LtrA expression for all fragmented intron variants, we modified our trans-splicing/conjugation assay by removing most of the LtrA coding region (ΔORF) from the fragmented introns and providing LtrA in trans from a second plasmid (pLE-P232-ltrA) (Fig. 2A, ΔORF; Belhocine et al. 2007). The trans-splicing efficiencies observed for the ΔORF fragmented introns show the same trend as their wild-type counterparts; that is, the bipartite introns allowing for potential base-pairing interactions between the intron fragments trans-splice more efficiently than the variants that do not allow for any interactions (Fig. 2B,C, cf. WT and ΔORF).
Base-pairing interactions between intron fragments contribute to Ll.LtrB trans-splicing in L. lactis
To further investigate the contribution of base-pairing interactions between intron fragments during trans-splicing, we disrupted potential interactions near various engineered fragmentation sites in different domains of Ll.LtrB ΔORF, as well as in the non-fragmented ΔORF cis-splicing intron. Next, we performed stepwise restoration of base-pairing interactions by either wild-type sequence restoration or sequence complementarity.
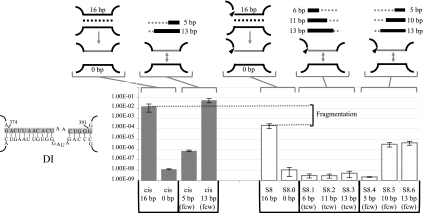
The first fragmentation site investigated was S8 in DI. Site-directed mutagenesis was used to replace the 16 pairing nucleotides at the 5′ end of the second intron fragment with the complementary sequence, abolishing base-pairing interactions between the two fragments (Fig. 3, S8.0). As a result, trans-splicing efficiency dropped by ∼19,000-fold. Then, we used two different approaches to sequentially restore the wild-type base-pairing interactions. Stepwise restorations of 6, 11, or 13 bp, directed toward the central wheel (tcw), did not lead to recovery of trans-splicing efficiency (Fig. 3, S8.1 to S8.3). In contrast, stepwise restorations starting from the central wheel (fcw) led to partial but significant recovery of the Ll.LtrB trans-splicing activity (Fig. 3, S8.4 to S8.6). While a 5-bp restoration was not sufficient to increase trans-splicing efficiency, 10- and 13-bp restorations stimulated Ll.LtrB trans-splicing by ∼300-fold. However, partial restoration of the same stem (fcw) in the cis-splicing intron led to complete splicing recovery, indicating that a 13-bp restoration is sufficient to support wild-type splicing efficiency (Fig. 3, cis 13 bp). The decreased rescue of splicing in the fragmented intron indicates that even if base-pairing at S8 significantly contributes to Ll.LtrB assembly and trans-splicing, fragmentation of the intron at this position also considerably affects the tertiary structure of the ribozyme.
FIGURE 3.
Sex factor conjugation efficiency of the Ll.LtrB S8ΔORF variants. Nucleotides modified to abolish base-pairing interactions in S8 and in the cis-splicing intron are shown on the left (gray box). Schematics of the different cis- and trans-splicing intron variants are illustrated at the top showing the modified RNA strand (gray line). The sequential base pair restorations toward the central wheel (tcw) or from the central wheel (fcw) are identified as gray and black lines while dotted lines depict unpaired residues. The S8 fragmentation site is represented by an arrowhead. Conjugation efficiencies are from three independent assays (Supplemental Table S2).
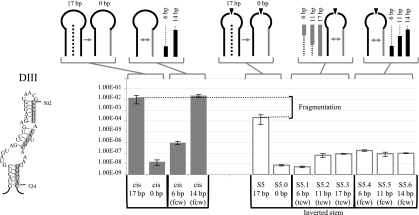
Next, we assessed the contribution of base-pairing interactions during trans-splicing of Ll.LtrB fragmented in DIII at position S5. We abolished the 17-bp interaction between the two intron fragments by substituting the pairing residues at the beginning of the second intron fragment for their complementary nucleotides (Fig. 4, S5.0); this led to ∼28,000-fold reduction in trans-splicing efficiency. We approached restoration of base-pairing in two ways. First, we introduced sequence complementarity at the 3′ end of the first intron fragment, resulting in an inverted stem. Base-pairing interactions were recovered in three consecutive steps moving toward the central wheel (tcw). The Ll.LtrB variant allowing for a 6-bp interaction between fragments did not show any recovery in trans-splicing activity (Fig. 4, S5.1), while variants with 11- and 17-bp interactions showed a slight increase in trans-splicing efficiency (Fig. 4, S5.2 and S5.3). We also restored the wild-type base pair in three steps from the central wheel (fcw). These variants, allowing for 6-, 11-, and 14-bp interactions, showed a slight increase in trans-splicing activity similar to S5.2 and S5.3 (Fig. 4, S5.4 to S5.6). On the other hand, partial restoration of the base-pairing interactions in the cis intron led to complete splicing recovery (Fig. 4, cis 14 bp). This indicates that base-pairing at S5 significantly contributes to Ll.LtrB assembly during trans-splicing. However, similar to what was observed in DI, fragmentation of Ll.LtrB at the tip of DIII may affect the tertiary structure of the ribozyme.
FIGURE 4.
Sex factor conjugation efficiency of the Ll.LtrB S5ΔORF variants. Nucleotides modified to abolish base-pairing interactions in S5 and in the cis-splicing intron are shown on the left (gray box). Schematics of the different cis- and trans-splicing intron variants are illustrated at the top showing the modified RNA strand (gray line). The sequential base pair restorations toward the central wheel (tcw) or from the central wheel (fcw) are identified as gray and black lines while dotted lines depict unpaired residues. The S5 fragmentation site is represented by an arrowhead. Conjugation efficiencies are from three independent assays (Supplemental Table S2).
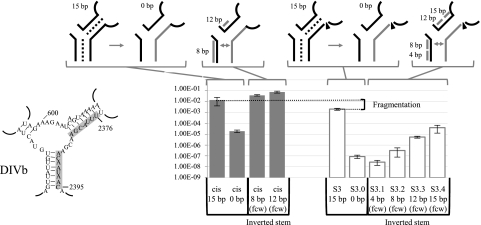
The contribution of base-pairing between intron fragments during Ll.LtrB trans-splicing was also analyzed in DIV. We first abolished the potential interactions of 15 bp in DIVb at position S3 by replacing the pairing residues at the beginning of the second intron fragment with their complementary nucleotides (Fig. 5, S3.0). Removing base-pairing between the two intron RNA fragments reduced the trans-splicing efficiency of Ll.LtrB by ∼2,300-fold. Subsequent stepwise restoration of base-pairing interactions from the central wheel (fcw) was done in four consecutive steps, by sequence complementation of residues within the first intron fragment (Fig. 5, S3.1 to S3.4); this resulted in an inverted stem. While a 4-bp interaction between the two intron fragments was not sufficient to recover trans-splicing activity, the restoration of 8-, 12-, and 15-bp interactions gradually increased Ll.LtrB trans-splicing efficiency by ∼3.6-fold, ∼63-fold, and ∼456-fold, respectively. As observed in DI and DIII, cis-splicing is completely recovered when the stem is partially restored by complementation (Fig. 5, cis 8 and 12 bp), indicating that even if base-pairing significantly contributes to Ll.LtrB assembly and trans-splicing, fragmentation of the intron at S3 seems to also affect the tertiary structure of the ribozyme.
FIGURE 5.
Sex factor conjugation efficiency of the Ll.LtrB S3ΔORF variants. Nucleotides modified to abolish base-pairing interactions in S3 and in the cis-splicing intron are shown on the left (gray box). Schematics of the different cis- and trans-splicing intron variants are illustrated at the top showing the modified RNA strand (gray line). The sequential base pair restorations from the central wheel (fcw) are identified as gray lines. The S3 fragmentation site is represented by an arrowhead. Conjugation efficiencies are from three independent assays (Supplemental Table S2).
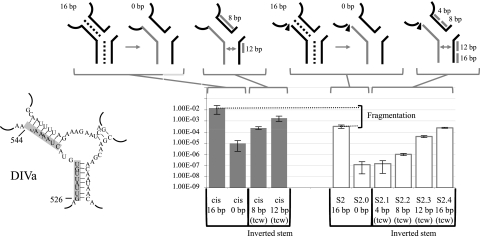
In order to disrupt the base-pairing interaction between the two intron RNAs of Ll.LtrB fragmented at position S2 in DIVa, we replaced the pairing residues at the end of the first intron fragment with their complementary nucleotides (Fig. 6, S2.0). Abolishment of the potential base-pairing interaction between intron fragments again resulted in a significant drop in trans-splicing efficiency of ∼2,700-fold. This potential interaction between the two intron fragments was then restored by stepwise complementation going toward the central wheel (tcw), resulting in an inverted stem. Residues from the second intron fragment were modified 4 nt at a time (Fig. 6, S2.1 to S2.4). Restoration of 4 bp did not improve Ll.LtrB trans-splicing efficiency (Fig. 6, S2.1). However, restoration of 8-, 12-, and 16-bp interactions between the two intron fragments led to a gradual recovery of the Ll.LtrB trans-splicing activity of ∼8.5-fold, ∼340-fold, and ∼2050-fold, respectively (Fig. 6, S2.2 to S2.4). Strikingly, restoration of the 16-bp interactions by complementation resulted in complete recovery of the trans-splicing activity (Fig. 6, cf. S2 and S2.4) while 12 bp were not sufficient to completely restore the cis-splicing activity (Fig. 6, cis 12 bp). This indicates that base-pairing at S2 contributes significantly to Ll.LtrB assembly and trans-splicing, and that unlike what was observed at S3, S5, and S8, fragmentation of the intron at this position does not strongly affect the tertiary structure of the intron.
FIGURE 6.
Sex factor conjugation efficiency of the Ll.LtrB S2ΔORF variants. Nucleotides modified to abolish base-pairing interactions in S2 and in the cis-splicing intron are shown on the left (gray box). Schematics of the different cis- and trans-splicing intron variants are illustrated at the top showing the modified RNA strand (gray line). The sequential base pair restorations toward the central wheel (tcw) are identified as gray lines. The S2 fragmentation site is represented by an arrowhead. Conjugation efficiencies are from three independent assays (Supplemental Table S2).
Influence of the LtrA protein during Ll.LtrB trans-splicing
LtrA is an important splicing co-factor (Mills et al. 1996; Ichiyanagi et al. 2002; Belhocine et al. 2007) that binds to Ll.LtrB at a high affinity-binding site in DIVa (Wank et al. 1999; Singh et al. 2002) and to multiple secondary sites located in DI, DII, and DVI (Singh et al. 2002; Dai et al. 2008). We thus wanted to address the role of this protein in the assembly and folding of Ll.LtrB intron fragments in vivo and its relative importance compared to base-pairing interactions during trans-splicing.
In the trans-splicing/conjugation assay used above (Fig. 2A, ΔORF), LtrA was expressed at a constant level from the P23 constitutive promoter (pLE-P232-ltrA), independently from the two intron fragments. In order to control the expression level of LtrA, the ltrA gene was cloned in pLE downstream from the nisin inducible promoter (pLE-Pnis-ltrA). Western blots were used to identify the nisin concentration at which LtrA expression levels plateaued in L. lactis. The levels of LtrA linearly increased between 0 and 1 ng/mL of nisin and stayed constant at higher nisin concentrations (Supplemental Fig. S1).
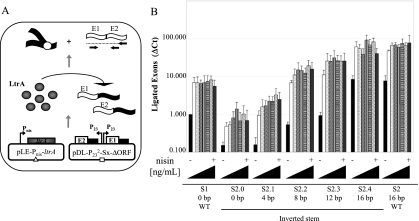
To evaluate the importance of LtrA during Ll.LtrB trans-splicing, we co-transformed the pLE-Pnis-ltrA plasmid in L. lactis with plasmids containing various bipartite introns fragmented at position S2 and harboring different potential base-pairing interactions between the two intron fragments (Fig. 7, S2.0 to S2.4 and S2). Ll.LtrB trans-splicing efficiency was evaluated by monitoring the presence of ligated exons by qRT-PCR at 10 different nisin concentrations ranging from 0 to 0.98 ng/mL (Fig. 7B).
FIGURE 7.
Influence of LtrA levels on Ll.LtrB trans-splicing in L. lactis. (A) Ll.LtrB trans-splicing efficiency was assessed in the presence of various levels of LtrA by monitoring the level of ligated exons by quantitative RT-PCR. LtrA expression was under the control of the Pnis promoter (pLE-Pnis-ltrA). (B) Levels of ligated exon were compared amongst introns having different number of base pairs between the two intron fragments (S2.0ΔORF to S2.4ΔORF, S2ΔORF, S1ΔORF). Ll.LtrB trans-splicing efficiency was assessed in the presence of different levels of LtrA expressed from the nisin inducible promoter: black, 0 ng/mL nisin; white, 0.14 ng/mL; dotted, 0.28 ng/mL; upward diagonals, 0.42 ng/mL; vertical lines, 0.56 ng/mL; horizontal lines, 0.70 ng/mL; downward diagonals, 0.84 ng/mL; gray, 0.98 ng/mL. Exon 1 and exon 2, E1 and E2; Ll.LtrB variants, black lines; ltrA gene, gray box; P23 and Pnis promoters, bent arrows; Ll.LtrB RNA intron fragments, black wavy lines; RNA exons, white wavy lines; LtrA, gray circles; primers used for the qRT-PCR, horizontal arrows.
Despite the gradual increase in LtrA levels, no significant differences in trans-splicing efficiency is observed above 0.14 ng/mL of nisin, regardless of the number of base pairs between the two intron fragments (Fig. 7B). Further, the maximum level of trans-splicing obtained for each variant was proportional to the number of base pairs between the two intron fragments. All constructs showed a residual trans-splicing activity in the absence of nisin, suggesting that the nisin promoter is leaky and that small amounts of LtrA are produced under non-induced conditions (Fig. 7, black bars). Residual trans-splicing activity under non-induced conditions also increased proportionally with the number of base pairs between the two intron fragments.
DISCUSSION
In this study, we used the Ll.LtrB group II intron as a model to assess the contribution of base-pairing interactions between intron fragments during trans-splicing in vivo. Using a trans-splicing/conjugation assay that provides LtrA either in cis (WT) or in trans (ΔORF), we found that bipartite Ll.LtrB variants with the potential for base-pairing between the two intron fragments trans-splice more efficiently than variants fragmented at close proximity, but lack the potential to base-pair. Interestingly, we observed this trend across four different locations throughout the intron structure: DI (S8 vs. S9), DIII (S5 vs. S1), DIVa (S2 vs. S1), and DIVb (S3 vs. S6 and S3 vs. S7) (Fig. 2, WT and ΔORF). This suggests that base-pairing interactions between intron fragments are involved during intron assembly and trans-splicing in vivo. However, the trans-splicing introns lacking base-pairing interactions are fragmented in the central wheel (Fig. 1A, S1, S6, and S9). Since the positions of these fragmentation sites are between residues that lie close to the catalytic center and that are most likely involved in multiple tertiary interactions (Dai et al. 2008; Toor et al. 2010), structural defects probably also contributed to the observed decrease in trans-splicing efficiency.
To further investigate the potential contribution and importance of base-pairing interactions between intron RNA fragments during trans-splicing, we introduced mutations to disrupt base-pairing interactions at various fragmentation sites in different domains of Ll.LtrB. The abolishment of specific base-pairing interactions between intron fragments of Ll.LtrB variants fragmented in DI (Fig. 3, S8), DIII (Fig. 4, S5), DIVb (Fig. 5, S3), or DIVa (Fig. 6, S2) led to significant reductions in trans-splicing efficiency. In all tested domains, disruption of base-pairing was more deleterious than intron fragmentation at positions nearby within the central wheel that do not allow for base-pairing interactions. This further suggests a significant contribution of base-pairing interactions between intron fragments during trans-splicing.
Next, we demonstrated that stepwise restorations of base-pairing interactions between intron fragments by either wild-type sequence re-establishment at DI and DIII or sequence complementarity at DIII, DIVa, and DIVb led to significant trans-splicing recovery. Complementation in DIVa and DIVb led to complete or nearly complete recovery of trans-splicing efficiencies, while base pair restoration in DI and DIII led to partial but significant trans-splicing recovery. This suggests that base-pairing interactions are contributing to intron assembly but that other factors have been affected by the engineered fragmentations and nucleotide substitutions. Indeed, some of the nucleotides substituted in DI and DIII are known to be involved in long-range tertiary interactions (Singh et al. 2002; Dai et al. 2008). In vitro studies suggest that G374 interacts with G37, helping the intron to fold into its native structure by positioning DIc1 and DIa along a helical turn of DI(ii) (Dai et al. 2008). Similarly in DIII, long-range interactions involving residues G494, C500, G501, and A523 were also shown to be involved in the three-dimensional folding of the intron (Dai et al. 2008). Disruption of these tertiary interactions most likely affected the final ribozyme structure and may explain why trans-splicing was only partially recovered in DI and DIII. In contrast, trans-splicing gradually increased as base-pairing interactions were progressively restored by complementation in both DIVa and DIVb. While base pair restorations by complementation in DIVb led to high trans-splicing efficiency, base pair restoration by complementation in DIVa resulted in complete trans-splicing recovery. The important recovery of Ll.LtrB trans-splicing activity by sequence complementarity in DIVa toward the central wheel and DIVb from the central wheel indicates that base-pairing interactions are contributing significantly, in a sequence independent manner, to intron fragment assembly. Accordingly, the modified residues in DIVa and DIVb do not appear to be involved in any long-range tertiary interactions (Singh et al. 2002; Dai et al. 2008).
The IEP LtrA is a very important Ll.LtrB splicing co-factor that binds to multiple sites located throughout the intron (Mills et al. 1996; Ichiyanagi et al. 2002; Belhocine et al. 2007, 2008). We thus wanted to address the role of LtrA in the assembly of Ll.LtrB intron fragments and its relative importance compared to base-pairing interactions during trans-splicing. The trans-splicing efficiency of Ll.LtrB variants fragmented at position S2 was assessed by qRT-PCR in the presence of increasing amounts of LtrA (Fig. 7). The trans-splicing efficiency of Ll.LtrB plateaued rapidly at low concentrations of LtrA regardless of the number of base pairs between the two intron fragments. Interestingly, the results obtained by qRT-PCR are consistent with our conjugation data. First, abolishment of the potential base-pairing interactions between the two intron fragments significantly decreased trans-splicing efficiency (Figs. 6, 7, cf. S2 and S2.0). Second, trans-splicing efficiency of the Ll.LtrB variant fragmented at position S1 (0 bp) is intermediate between S2 (16 bp) and S2.0 (0 bp) (Figs. 7, 2, cf. S1 and S2 and S2.0). Finally, stepwise restoration of the potential base-pairing interactions resulted in a gradual increase of the Ll.LtrB trans-splicing efficiency, ultimately reaching complete recovery (Figs. 6, 7, cf. S2 and S2.0 to S2.4). We thus confirmed that LtrA is an important Ll.LtrB trans-splicing co-factor in vivo, which allows each Ll.LtrB variant to reach its maximal trans-splicing potential. However, we also showed that overexpression of LtrA cannot compensate for the trans-splicing deficiencies induced by altering the potential base-pairing interactions between the two intron fragments.
In nature, some chloroplastic and mitochondrial genomes harbor trans-splicing group II introns fragmented into two or three pieces (Lambowitz and Zimmerly 2004; Glanz and Kuck 2009). Regardless of the position of the fragmentation site(s), these introns can still be folded into the conserved secondary structure (e.g., Fig. 1A). Analysis of the secondary structures of six chloroplastic (psaA-i1, psaA-i2, petD-i1, psaC-i1, rbcL-i1, rbcL-i2) and six mitochondrial (nad1-i1, nad2-i2, nad5-i3, nad1-i4, nad3-i1, nad3-i2) fragmented group II introns showed that even if the secondary structures were not always perfectly conserved in the vicinity of the fragmentation sites, these fragmented introns have a series of five to 20 potential base-pairing interactions between nucleotides located on either side of the fragmentation sites (Supplemental Table S3). The conservation of these potential base-pairing interactions between intron RNA fragments suggests that they were maintained because they contribute to intron fragment assembly during trans-splicing in vivo.
Taken together our data show the important contribution of base-pairing interactions for the assembly of intron fragments during trans-splicing in vivo even though other factors, such as long-range tertiary interactions, seem to also play a significant role. Our work also rationalizes why base-pairing interactions were evolutionarily conserved between nucleotides located on either side of fragmentation sites in natural trans-splicing group II introns.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Lauren Narcross and Dr. Karen K. Yam for providing comments on the manuscript. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (#227826 to B.C.). B.C. is a McGill University William Dawson Scholar and a Chercheur-Boursier Senoir from Fonds de la Recherche en Santé du Québec (FRSQ). C.Q. was the recipient of an external post-doctoral fellowship from Consejo Nacional de Investigaciones Cientfícas y Técnicas from Argentina (CONICET). A.F. was the recipient of an Undergraduate Summer Research Award from FRSQ.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028886.111.
REFERENCES
- Belhocine K, Mak AB, Cousineau B 2007. Trans-splicing of the Ll.LtrB group II intron in Lactococcus lactis. Nucleic Acids Res 35: 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhocine K, Mak AB, Cousineau B 2008. Trans-splicing versatility of the Ll.LtrB group II intron. RNA 14: 1782–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM 2004. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol 340: 211–231 [DOI] [PubMed] [Google Scholar]
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, Lambowitz AM, Zimmerly S 2008. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 30: 472–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Zingler N 2007. Group II introns: Structure, folding and splicing mechanism. Biol Chem 388: 665–678 [DOI] [PubMed] [Google Scholar]
- Fedorova O, Mitros T, Pyle AM 2003. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol 330: 197–209 [DOI] [PubMed] [Google Scholar]
- Glanz S, Kuck U 2009. Trans-splicing of organelle introns—a detour to continuous RNAs. Bioessays 31: 921–934 [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M 2002. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol Microbiol 46: 1259–1272 [DOI] [PubMed] [Google Scholar]
- Klein JR, Chen Y, Manias DA, Zhuo J, Zhou L, Peebles CL, Dunny GM 2004. A conjugation-based system for genetic analysis of group II intron splicing in Lactococcus lactis. J Bacteriol 186: 1991–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2010. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K, Schmidt U 2003. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 38: 249–303 [DOI] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Mills DA, McKay LL, Dunny GM 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol 178: 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM 2002. Metal ions in the structure and function of RNA. J Biol Inorg Chem 7: 679–690 [DOI] [PubMed] [Google Scholar]
- Pyle AM, Fedorova O, Waldsich C 2007. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci 32: 138–145 [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol 8: 301–308 [DOI] [PubMed] [Google Scholar]
- Rambo RP, Doudna JA 2004. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43: 6486–6497 [DOI] [PubMed] [Google Scholar]
- Saldanha R, Mohr G, Belfort M, Lambowitz AM 1993. Group I and group II introns. FASEB J 7: 15–24 [DOI] [PubMed] [Google Scholar]
- Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083 [DOI] [PubMed] [Google Scholar]
- Shearman C, Godon JJ, Gasson M 1996. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol Microbiol 21: 45–53 [DOI] [PubMed] [Google Scholar]
- Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM 2002. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol 318: 287–303 [DOI] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7: 1142–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM 2010. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM 1999. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell 4: 239–250 [DOI] [PubMed] [Google Scholar]