The p53 tumor suppressor is a DNA-damage-responsive sequence-specific transcriptional activator. The sustained activation of the p53 response is incompatible with cell growth and viability. To circumvent this issue, a variety of negative feedback loops exist to limit the duration of p53 activation. Very little is known about the effect of transient p53 activation on the long-term expression of p53 target genes. Here the authors used a temperature-sensitive variant of p53 and oligonucleotide microarrays to monitor gene expression during and following reversible p53 activation. It is shown that expression of most p53-induced transcripts was rapidly reversible, consistent with active mRNA decay. Remarkably, short-lived p53 targets were induced faster, reaching maximum transcript levels earlier than the stable p53 targets. Therefore, the evidence indicates that the p53 transcriptional response has evolved with primarily short-lived target mRNAs and that post-transcription processes play a prominent role in the p53 response.
Keywords: cell cycle arrest, gene expression, mRNA decay, microarray, p53 response
Abstract
The p53 tumor suppressor is a DNA-damage-responsive sequence-specific transcriptional activator. The sustained activation of the p53 response is incompatible with cell growth and viability. To circumvent this issue, a variety of negative feedback loops exist to limit the duration of p53 activation. Despite our understanding of p53 regulation, very little is known about the effect of transient p53 activation on the long-term expression of p53 target genes. Here we used a temperature-sensitive variant of p53 and oligonucleotide microarrays to monitor gene expression during and following reversible p53 activation. The expression of most p53-induced transcripts was rapidly reversible, consistent with active mRNA decay. Representative 3′ UTRs derived from short-lived transcripts (i.e., DDB2 and GDF15) conferred instability on a heterologous mRNA, while 3′ UTRs derived from more stable transcripts (i.e., CRYABand TP53I3) did not. The 3′ UTRs derived from unstable p53-induced mRNAs were significantly longer than those derived from stable mRNAs. These 3′ UTRs had high uridine and low cytosine content, leading to a higher density of U-, AU-, and GU-rich sequences. Remarkably, short-lived p53 targets were induced faster, reaching maximum transcript levels earlier than the stable p53 targets. Taken together, the evidence indicates that the p53 transcriptional response has evolved with primarily short-lived target mRNAs and that post-transcription processes play a prominent role in the p53 response.
INTRODUCTION
Mutations in the p53 tumor suppressor are among the most common alterations in cancer (Hollstein et al. 1991). This protein is best known as a transcriptional regulator of messenger RNAs (mRNAs) encoding regulators of apoptosis, senescence, cell cycle arrest, and DNA repair that likely contribute to the anti-neoplastic activity of p53 (Riley et al. 2008). In response to a variety of cellular stresses, including DNA damage, p53 becomes phosphorylated and acetylated, leading to an increase in the half-life of the protein (Ljungman and Lane 2004). Extensive DNA damage can lead to the sustained activation and accumulation of p53 and the consequent induction of apoptosis (Bates et al. 1999; McKay et al. 1999). Moderate levels of DNA damage can lead to transient p53 accumulation associated with short-term cell cycle arrest and DNA repair (Bates et al. 1999; McKay et al. 1999). The expression of p53 itself is negatively regulated by the p53-regulated ubiquitin ligase Mdm2 (Wu et al. 1993; Honda et al. 1997). This protein is able to bind p53, inhibit its transcriptional activity, and target p53 for proteasome-mediated degradation (Momand et al. 1992; Honda et al. 1997). The p53–Mdm2 negative feedback loop is essential to prevent a sustained increase in the expression and activity of p53 (Jones et al. 1995; Montes de Oca Luna et al. 1995). Despite our understanding of this negative feedback mechanism, very little is known about the attenuation of the p53-dependent transcriptional response.
A variety of conditional expression systems have been used to study the onset of the p53 transcriptional response on a genomewide scale (Yu et al. 1999; Kannan et al. 2000, 2001; Zhao et al. 2000; McKay et al. 2004; Smith et al. 2007). A valine-to-alanine substitution at codon 135 of murine p53 (V135A) gives rise to a temperature-sensitive variant of p53 that drives p53-dependent gene expression at the permissive temperature (32°C) (Michalovitz et al. 1990; Gannon and Lane 1991; Yonish-Rouach et al. 1991; Caelles et al. 1994; Merchant et al. 1996; Kannan et al. 2000, 2001; McKay et al. 2000, 2004). Unlike many other conditional p53 expression systems, the activity of the V135A variant of p53 is readily reversible, as evidenced by cell cycle reentry (McKay et al. 2000) and reversal of earlier changes associated with apoptosis (Geske et al. 2001) when cells are returned to the restrictive temperature (38°C). Therefore, this model affords a unique but until now unutilized opportunity to study the reversal of the p53-transcriptional response following transient p53 activation.
Here, gene expression profiling was performed to monitor global changes in gene expression during and following activation of the temperature-sensitive V135A variant of p53. Activation of p53 led to the induction of many known p53 target genes, and most of these p53-responsive transcripts decayed rapidly upon loss of p53 activity. The 3′ UTRs of representative unstable transcripts accelerated the decay of a heterologous reporter mRNA under the control of a tetracycline-regulated promoter, indicating that these 3′ UTRs contribute to accelerated mRNA decay. These unstable 3′ UTRs were uridine-rich at the expense of cytosine, resulting in significantly more U-, AU-, and GU-rich sequences that have been associated with mRNA instability in other systems. In contrast, the 3′ UTRs derived from stable p53-induced transcripts were significantly shorter than expected, exhibited no sequence bias, contained fewer putative mRNA destabilizing elements, and failed to destabilize the heterologous reporter mRNAs. Remarkably, the kinetics of p53-induced gene expression was also dependent on the 3′ UTR of the target gene. We propose that the p53 response has evolved with primarily short-lived target mRNAs to permit the tight temporal regulation of the p53 response under appropriate conditions.
RESULTS
HT29-tsp53 cells as a model of reversible p53 activity
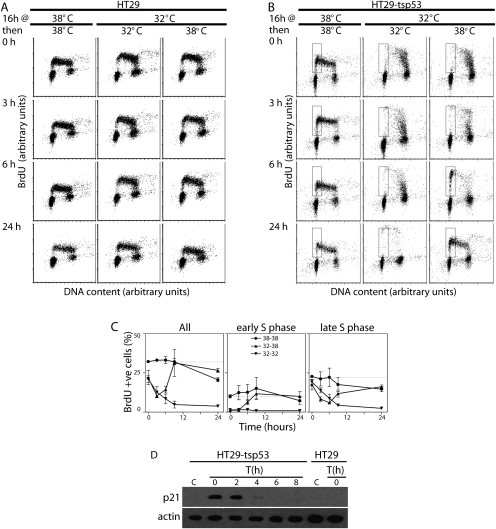
HT29-tsp53 cells express the V135A variant of murine p53 that is temperature-sensitive for nuclear import (Michalovitz et al. 1990; Gannon and Lane 1991; Merchant et al. 1996). Sustained activation of p53 in these cells reportedly leads to prolonged cell cycle arrest with very little apoptosis (Merchant et al. 1996; McKay et al. 2000, 2004). To determine how rapidly the p53 response could be reversed, HT29-tsp53 cells and an isogenic control cell line were subjected to a variety of temperature shifts, incubated in BrdU for 30 min immediately prior to collection, and were then subjected to two parameter flow cytometric analysis to monitor the effect of p53 on cell cycle distribution following transient p53 activation. Cell cycle distribution remained unaltered in control cultures regardless of temperature (Fig. 1A). HT29-stp53 cells were switched to the permissive temperature for up to 40 h (Fig. 1B), and a decrease in the proportion of BrdU-positive HT29-tsp53 cells was observed at the permissive temperature that was consistent with the establishment of a p53-dependent G1 arrest (McKay et al. 2000). The recovery of a prominent early-S-phase population of BrdU-positive cells could be detected within 6 h at the restrictive temperature with the restoration of a typical asynchronous cell cycle distribution within 24 h (Fig. 1B,C). As expected, this reversible G1 arrest was temporally associated with reversible expression of the cyclin-dependent kinase inhibitor p21WAF1 (Fig. 1D). The present results confirm that p53-dependent G1 arrest was reversible and further indicate that these cells represent a powerful and unique model to study reversal of the p53 response.
FIGURE 1.
Reversible cell cycle arrest. HT29 (A) and HT29-tsp53 cells (B) were incubated for 16 h at either 32°C or 38°C, as indicated. Cells incubated at 32°C were then either returned to 38°C or maintained at 32°C for up to 24 h, as indicated. Thirty minutes before collection, BrdU was added to the culture medium to label nascent DNA. Cells were then subjected to two parameter flow cytometric analysis. The early S-phase population in B is indicated by the box. (C) Quantitative analysis of independent experiments like those presented in panel B. Each value represents the mean (±SEM) determined from three independent experiments. (D) HT29 and HT29-tsp53 cells were incubated for 16 h at 32°C and were then returned to 38°C for the indicated time in hours (T). Cell lysates were collected for immunoblot analysis. The control sample (designated C) was collected from cells maintained at the restrictive temperature.
p53-induced transcripts are predominantly short-lived
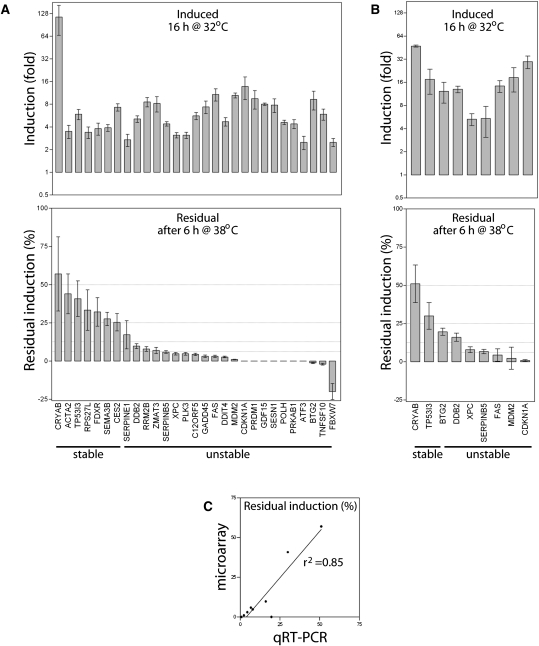
To monitor the attenuation of the p53 response, HT29-tsp53 cells were incubated at the permissive temperature for 16 h to induce p53-dependent gene expression, and then replicate cultures were released at the restrictive temperature for 6 h to remove active p53. Total RNA was collected for oligonucleotide microrarray analysis using Affymetrix HuGene 1.0 ST Arrays. Eighty-four independent transcripts represented by a total of 552 probe sets were induced under these conditions (Supplemental Table 1). The fold-change in expression was determined using all probe sets representing putative p53-responsive transcripts (all probe sets corresponding to the relevant cluster identification number). Transcripts that were induced an average of twofold or greater were compared with a comprehensive list of p53-induced transcripts (Riley et al. 2008), and 29 previously reported direct transcriptional targets of p53 were identified (Supplemental Table 2).
Remarkably, the expression of 21 of these p53-responsive transcripts had decreased at least eightfold within 6 h at the restrictive temperature, indicating that they have half-lives ≤2 h (Fig. 2A). Conversely, seven transcripts decreased less than fourfold during the same period of time (Fig. 2A). Transcripts were grouped by their apparent stability using 25% residual expression as a cutoff (denoted “stable” and “unstable” in Fig. 2A). There was no correlation between the initial fold increase in expression and residual expression following loss of p53 activity (Fig. 2A). Quantitative RT-PCR (qRT-PCR) confirmed the reversible pattern of expression of the p53 targets tested (Fig. 2B). Overall, there was a strong correlation between residual induction determined by microarray and qRT-PCR (Fig. 2C). Taken together, the p53-transcriptional response is readily reversible in these cells, and this implies that most p53-regulated transcripts are short-lived.
FIGURE 2.
Reversible regulation of p53-induced genes. (A) HT29-tsp53 cells were incubated for 16 h at the permissive temperature (32°C) to increase the expression of p53-responsive transcripts. Cells were then returned for 6 h to the restrictive temperature (38°C) to monitor the attenuation of the p53 response. Unstable transcripts were expected to decrease rapidly, whereas stable mRNAs were expected to remain elevated at the restrictive temperature. Total RNA was collected and subjected to oligonucleotide microarray analysis. Twenty-nine known p53-induced transcripts were significantly induced following 16 h at 32°C. Seven transcripts retained at least 25% of their induced level, while the other 22 retained <25% of their induced levels (indicated stable and unstable, respectively). (B) HT29-tsp53 cells were treated as described in panel A. Two of the stable and seven of the unstable transcripts were selected for further study using qRT-PCR. Each value in panels A and B represents the mean (±SEM) response of the indicated transcript determined from a minimum of three independent experiments. (C) Residual expression determined by quantitative RT-PCR is plotted relative to residual expression determined by microarray for the nine transcripts presented in panel B.
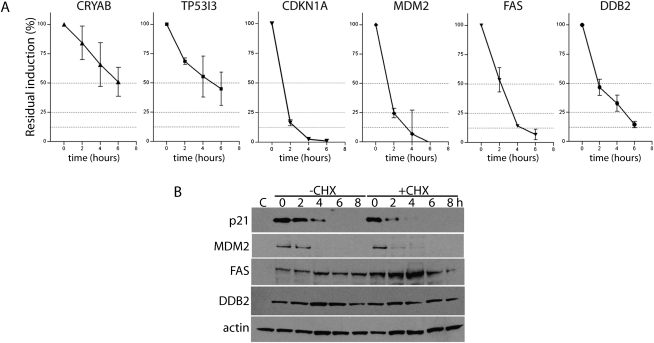
To better monitor the decay of p53-induced transcripts, HT29-tsp53 cells were incubated at the permissive temperature to drive p53-dependent gene expression, and then cells were returned to the restrictive temperature for various lengths of time to allow the decay of p53-induced mRNAs. The expression of several p53-regulated transcripts was monitored as a function of time using qRT-PCR. The expression of all p53-induced transcripts decreased when cells were returned to the restrictive temperature following p53 activation (Fig. 3A). The expression of CRYAB and TP53I3 (also known as PIG3) decreased by ∼50% in 4–6 h (Fig. 3A), indicating that the half-lives of these two p53-induced transcripts are close to the median half-life determined in previous genomewide estimates of mRNA decay (Yang et al. 2003; Friedel et al. 2009; Sharova et al. 2009). Meanwhile, the four apparently unstable transcripts tested (CDKN1A, MDM2, FAS, and DDB2) exhibited half-lives of 2 h or less (Fig. 3) and are thus considered to be short-lived (Yang et al. 2003). Therefore, these groups of transcripts have distinct kinetics of mRNA decay.
FIGURE 3.
The decay of p53-regulated transcripts. (A) HT29-tsp53 cells were incubated at the permissive temperature for 16 h and then switched to the restrictive temperature for between 0 and 6 h. RNA was collected at 2-h intervals and analyzed by qRT-PCR to monitor the decay of p53-induced transcripts. Each value represents the mean (±SEM) determined from a minimum of three independent experiments. (B) HT29-tsp53 cells were incubated at the permissive temperature for 16 h and then switched to the restrictive temperature for between 0 and 8 h in the presence or absence of cycloheximide (CHX). Protein was collected at 2-h intervals and subjected to immunoblot analysis for the indicated proteins. The control sample (C) was collected from cells maintained at the restrictive temperature.
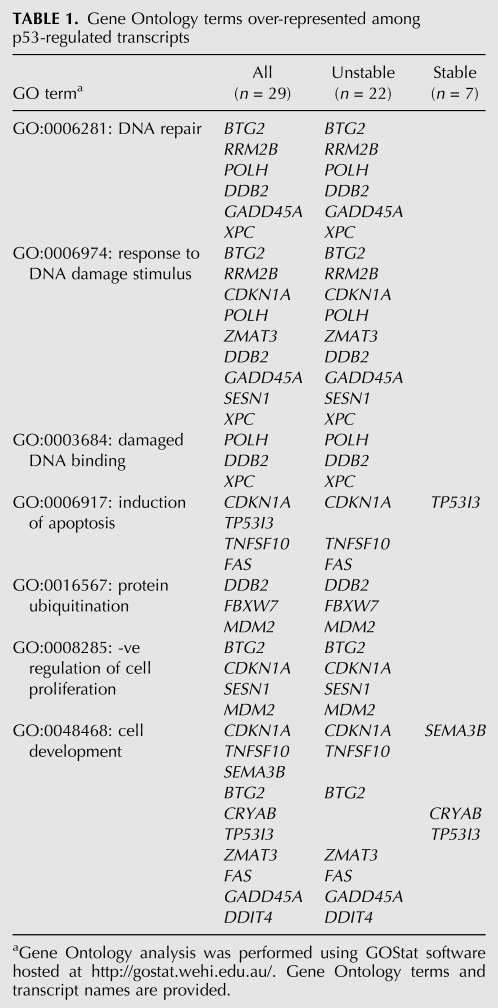
Although all of these p53-induced transcripts are known targets of p53, it was of interest to determine whether these groups comprised different functional classes of transcripts. Therefore, Gene Ontology analysis was performed using GoStat software available online at http://gostat.wehi.edu.au/ (Beissbarth and Speed 2004). Consistent with the p53 response (Riley et al. 2008), statistically over-represented terms in the list of 29 p53-regulated transcripts included “DNA repair” (GO:0006281), “response to DNA damage stimulus” (GO:0006974), “induction of apoptosis” (GO:0006917), and “negative regulation of cell proliferation” (GO:0008285) (Table 1). These terms were statistically over-represented among the unstable (P = 1.1 × 10−8, P = 1.8 × 10−8, P = 4.0 × 10−3, and p = 2.7 × 10−4, respectively), but not the stable group of transcripts. “Cell development” was the only GO term statistically over-represented among stable p53 targets (GO:000048468, P = 0.03), and this term is not typically associated with p53 (Riley et al. 2008). Taken together, the well-characterized p53 response is elicited by the transcriptional induction of primarily short-lived transcripts.
TABLE 1.
Gene Ontology terms over-represented among p53-regulated transcripts
Intriguingly, the reversible expression of some but not all p53-regulated proteins was consistent with rapid mRNA decay. For example, the p21WAF1, Mdm2, Fas, and Ddb2 proteins were strongly induced at the permissive temperature as expected (Fig. 3B). Returning cells to the restrictive temperature led to a rapid decrease in the expression of p21WAF1 and Mdm2, consistent with the rapid decay of their transcripts. In contrast, the expression of Ddb2 and Fas protein remained high at the restrictive temperature. When cells were returned to the restrictive temperature in the presence of the translation inhibitor cycloheximide, p21WAF1 and Mdm2 levels decrease even more rapidly while Fas and Ddb2 levels remained elevated (Fig. 3B). These results indicate that p21WAF1 and Mdm2 are unstable proteins, while Ddb2 and Fas are stable under the present conditions. The discordance between protein and mRNA expression raises the intriguing possibility that reversible mRNA expression may not have evolved exclusively to permit reversible protein expression.
3′ UTRs target p53-responsive transcripts for rapid mRNA decay
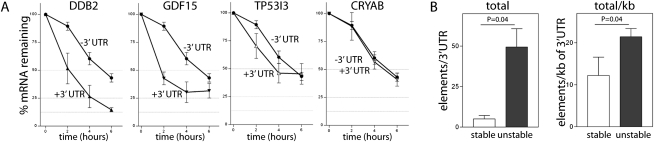
It is well-established that 3′ UTRs are critical determinants of mRNA stability and decay (Grillo et al. 2010). To determine if the 3′ UTRs of p53-induced transcripts affect mRNA decay, heterologous reporter mRNA constructs were generated. The 3′ UTRs of DDB2, GDF15, TP53I3, and CRYAB are 363 nt, 182 nt, 144 nt, and 138 nt, respectively, and they represent the 3′ UTRs of unstable (DDB2 and GDF15) and stable (TP53I3 and CRYAB) mRNAs. These four 3′ UTRs were cloned downstream from a tetracycline-regulated d2EGFP reporter cDNA, and the resulting plasmids were transfected into HeLa Tet-Off cells. Addition of doxycycline to vector control transfected cells (expressing d2EGFP without a heterologous 3′ UTR) led to a reduction in d2EGFP expression with ∼50% of the d2EGFP mRNA remaining 6 h after doxycycline was added (Fig. 4A). The 3′ UTRs of DDB2 and GDF15 led to accelerated loss of d2EGFP expression, while the 3′ UTRs derived of CRYAB and TP53I3 did not significantly affect mRNA decay (Fig. 4A). Similarly, the mean half-lives determined for these reporter transcripts were not equal (P = 0.002 by One-Way ANOVA). The difference could be attributed to decreased half-lives of constructs containing the 3′ UTRs of DDB2 and GDF15 (P ≤ 0.05 and P ≤ 0.01, respectively, by the Tukey Multiple Comparisons Test). Thus, the 3′ UTRs of these unstable p53-induced transcripts are capable of destabilizing a heterologous mRNA, suggesting that cis-acting elements in their 3′ UTRs regulate the decay of p53-responsive transcripts.
FIGURE 4.
Heterologous reporter constructs. (A) The pTRE-d2EGFP and a series of derivative pTRE-d2EGFP-3′UTR reporter plasmids were transfected into HeLa Tet-Off cells. Twenty-four hours following transfection, doxycycline was added to block transcription of the reporter mRNAs. Residual d2EGFP transcript levels were monitored as a function of time using qRT-PCR. Each point represents the mean (±SEM) determined from three independent experiments. The mean half-lives of the heterologous transcripts were 5.6 ± 0.5, 2.0 ± 0.9, 1.5 ± 0.3, 4.8 ± 0.7, and 5.5 ± 0.8 for control, DDB2, GDF15, TP53I3, and CRYAB, respectively. (B) Sequence analysis of 3′ UTRs led to the identification of known destabilizing elements. The mean number of U-, AU-, and GU-rich other miscellaneous elements (see Supplemental Table 4) per 3′ UTR is presented for the seven stable and the 22 unstable p53-induced transcripts. The mean number of these elements per 3′ UTR and the mean number of elements per kilobase of 3′ UTR is presented. Each point represents the mean (±SEM) determined for seven stable and 22 unstable transcripts. The indicated P-values were determined by a Student's t-test.
The 3′ UTRs of p53-induced transcripts tend to be uridine-rich
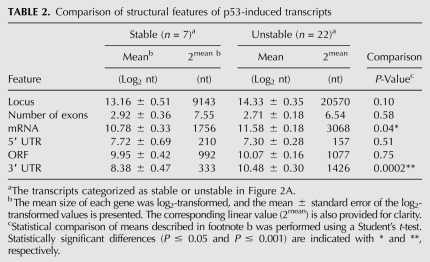
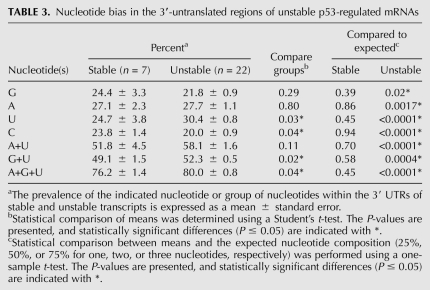
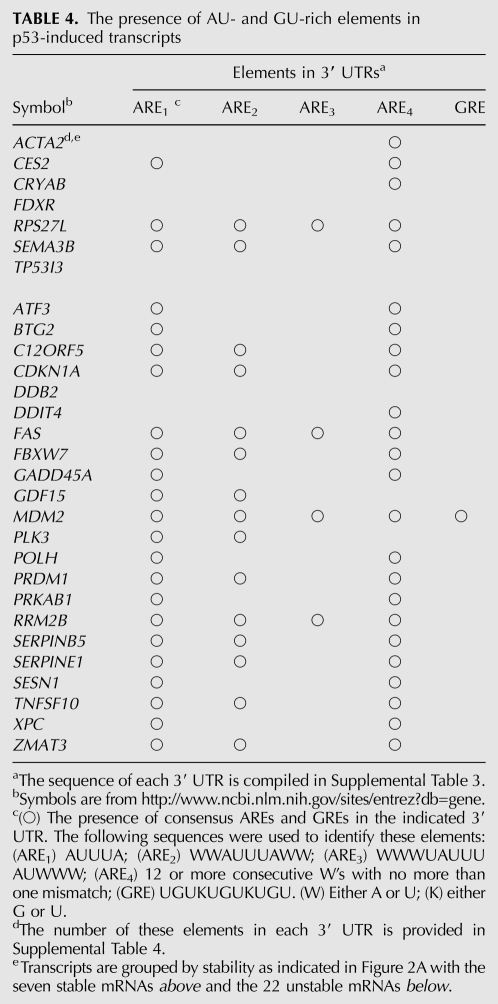
Genomewide analysis of mRNA turnover has identified several structural features that affect the stability of mRNAs. This includes the number of exon junctions per open reading frame (ORF) and a variety cis-acting determinants typically located in 3′ UTRs (Barreau et al. 2005; Sharova et al. 2009; Lee et al. 2010). Structural features of all 29 p53-induced transcripts were examined in more detail. We found no significant difference in the number of exons, the mean length of loci, the mean length of 5′ UTRs, or the size of open-reading frames (ORFs) between the stable and unstable groups of transcripts. However, the stable mRNAs were significantly shorter on average than those from the unstable group, and this was attributed to significantly shorter 3′ UTRs than expected based on genomewide estimates (Table 2; Mignone et al. 2002; Grillo et al. 2010). Sequence comparison indicated that the 3′ UTRs of the longer unstable transcripts had significantly higher uridine content (30%) at the expense of cytosine (20%), and this was not observed among the shorter, more stable p53-regulated transcripts (Table 3). With this sequence disequilibrium, there was a tendency for the unstable 3′ UTRs to be AU-rich and GU-rich, but this only reached significance for GU-rich sequences. Surprisingly, the presence of consensus AU-rich elements (AREs) and GU-rich elements (GREs) (Barreau et al. 2005; Vlasova et al. 2008; Lee et al. 2010; Rattenbacher et al. 2010) did not clearly distinguish stable and unstable groups of transcripts (Table 4). Therefore, the instability of p53-regulated transcripts was not easily predicted from the presence or absence of these well-characterized destabilizing elements.
TABLE 2.
Comparison of structural features of p53-induced transcripts
TABLE 3.
Nucleotide bias in the 3′-untranslated regions of unstable p53-regulated mRNAs
TABLE 4.
The presence of AU- and GU-rich elements in p53-induced transcripts
Recently, a systems biology approach was used to identify short sequences that are over-represented in highly unstable transcripts (Lee et al. 2010). The most significantly over-represented hexamers were poly(U) sequences, followed by GU-rich and then AU-rich sequences, but most of these did not precisely match published consensus ARE and GRE sequences (Barreau et al. 2005; Lee et al. 2010; Rattenbacher et al. 2010). We found that these U-, AU-, and GU-rich sequences were significantly enriched in the 3′ UTRs of the unstable p53-regulated mRNAs compared with the more stable 3′ UTRs (Fig. 4B). Furthermore, the total number of these hexameric sequences was significantly elevated even when the number of elements was normalized to the length of the 3′ UTRs (Fig. 4B). Taken together, most p53-regulated transcripts decayed rapidly following transient activation of p53, representative 3′ UTRs from these unstable transcripts conferred accelerated decay in a heterologous reporter system, and that is consistent with a high density of AU- and GU-rich sequences.
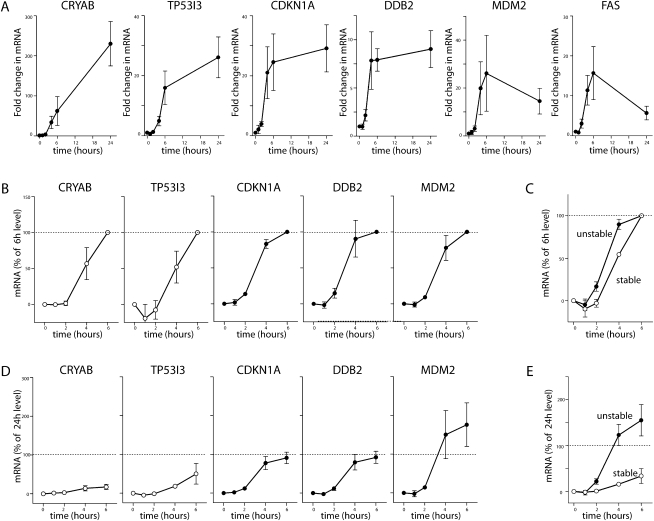
Time course for p53-induced gene expression
Recent evidence suggests that mRNA stability affects the kinetics of gene induction (Lam et al. 2001; Elkon et al. 2010). Therefore, RNA was collected following incubation for various times at the permissive temperature, and the expression of eight unstable and two stable p53-responsive transcripts was assessed by qRT-PCR. A significant increase in the expression of all p53-responsive transcripts was detected (Fig. 5A). There was an initial lag in the induction of all 10 transcripts tested, but this was greater for the two stable transcripts. Fold increase in expression was heterogeneous, and the duration of this increase in expression varied considerably as well (Fig. 5A). With the heterogeneity in expression levels, it was difficult to interpret the rate of mRNA accumulation. As an alternative way to visualize the induction of transcripts over defined periods of time, expression data were normalized to 6 h (Fig. 5B) and 24 h (Fig. 5D). There was a small but statistically significant (P = 0.01, Two-Way ANOVA) delay in the expression of the stable transcripts relative to the unstable mRNAs (Fig. 5A–C). More striking, the expression of the unstable transcripts reached a plateau by 4–6 h, while the expression of stable transcripts continued to increase (Fig. 5A,D). There was a statistically significant difference (P = 0.007, Two-Way ANOVA) in the mean response of stable and unstable transcripts (Fig. 5E). The present data suggest that unstable transcripts reach equilibrium between synthesis and decay, while the stable mRNAs continued to increase with sustained p53 activity.
FIGURE 5.
p53-induced gene expression. (A) HT29-tsp53 cells were incubated at the permissive temperature for the indicated time (between 0 and 24 h), and RNA was collected for qRT-PCR to monitor the induction of 10 separate p53-induced transcripts (CRYAB, TP53I3, BTG2, CDKN1A, DDB2, FAS, GDF15, MDM2, SERPINB5, and XPC). Expression of the indicated representative mRNA is expressed as fold-change compared with samples maintained at the restrictive temperature. (B) Data in A were normalized to the expression level determined following 6 h at the permissive temperature. (C) The mean response of two stable and eight unstable p53-induced transcripts, as normalized in B. (D) Data in A were normalized to the expression level determined following 24 h at the permissive temperature. (E) The mean response of two stable and eight unstable p53-induced transcripts, as normalized in D. Each value in A, B, and D represents the mean (±SEM) determined from a minimum of three independent experiments.
DISCUSSION
The 3′ UTRs of p53-induced transcripts
Using a temperature-sensitive variant of p53 that permits the rapid and reversible control of p53 activity (Merchant et al. 1996), we were able to monitor the decay of p53-induced transcripts following transient p53 activation. Remarkably, the expression of most p53-induced transcripts returned to baseline levels within 2–6 h with apparent half-lives of 2 h or less. Therefore, most p53-induced transcripts should be considered short-lived. The 3′ UTRs from representative members of the stable and unstable groups of p53-induced transcripts were cloned downstream from a d2EGFP cDNA to create heterologous reporter mRNAs under control of a tetracycline-repressible promoter. In this model system, the presence of the 3′ UTRs of DDB2 and GDF15 (unstable transcripts) resulted in accelerated mRNA decay, and the rate of decay was remarkably similar to that of the endogenous mRNA following transient p53 activation. Conversely, the 3′ UTRs of CRYAB and TP53I3 (stable transcripts) had very little effect on the decay of their respective heterologous reporter mRNAs. Therefore, the 3′ UTRs of p53-induced transcripts appear to target their mRNAs for rapid decay.
It is well-established that 3′ UTRs are critical determinants of mRNA stability and decay (Grillo et al. 2010). Based on sequences compiled in the UTR database (http://utrdb.ba.itb.cnr.it/), the length of human 3′ UTRs ranges from ∼20 to >8500 bases with an average length of 1030 nt (Mignone et al. 2002; Grillo et al. 2010). Here, the stable mRNAs were significantly shorter than the unstable transcripts, and this was attributed to the presence of short 3′ UTRs (median lengths of 312 nt and 1317 nt for stable and unstable transcripts, respectively). The 3′ UTRs of the stable p53-regulated mRNAs were shorter than the average length of human 3′ UTRs (P = 0.0089), while unstable transcripts were not. Recent evidence links the length of 3′ UTRs to accelerated decay through the nonsense-mediated decay pathway (NMD) (Hogg and Goff 2010; Nicholson et al. 2010). While NMD is generally considered a quality-control measure that eliminates transcripts with premature termination codons, between 1% and 10% of mRNAs may be subject to regulation by NMD machinery, independent of premature termination codons (Mendell et al. 2004; Rehwinkel et al. 2005, 2006). Other targets of NMD include mRNAs with introns downstream from termination codons, transcripts with upstream ORFs, and transcripts with long 3′ UTRs (Muhlrad and Parker 1999; Rebbapragada and Lykke-Andersen 2009; Hogg and Goff 2010; Hogg 2011). It is possible that some of the longer 3′ UTRs of p53-regulated transcripts are targeted for degradation through NMD.
A variety of cis-acting determinants of mRNA decay have been described, including AREs and GREs (Yang et al. 2003; Barreau et al. 2005; Bakheet et al. 2006; Vlasova et al. 2008; Sharova et al. 2009; Lee et al. 2010; Rattenbacher et al. 2010). Sequence comparison of the 29 3′ UTRs identified through our microarray analysis indicated that unstable UTRs contained more uridine residues than expected, while cytosine was statistically under-represented (30% uridine and 20% cytosine). The 3′ UTRs of stable transcripts exhibited no such sequence disequilibrium (25% uridine and 24% cytosine). Therefore, there is a clear statistical bias in the sequence composition of the 3′ UTRs of the unstable p53-regulated mRNAs. These unstable U-rich 3′ UTRs contain a significantly higher density of U-, AU-, and GU-rich sequences that are thought to be associated with accelerated mRNA decay (Yang et al. 2003; Hau et al. 2007; Sharova et al. 2009; Lee et al. 2010). Taken together, most p53-induced transcripts contain 3′ UTRs with sequences predictive of their short half-lives. We suggest that the transcriptional induction of short-lived mRNAs encoding short-lived proteins like p21WAF1 and Mdm2 is important to the p53 response in a variety of ways.
p21WAF1 expression and reversible G1 arrest
HT29-tsp53 cells underwent a prolonged G1 cell cycle arrest with very little apoptosis when these cells were maintained at the permissive temperature (McKay et al. 2000, 2004). Remarkably, S-phase entry could be detected here within 6 h following the return to the restrictive temperature. The p53-dependent G1 arrest is mediated largely through the transcriptional induction of the cyclin-dependent kinase inhibitor p21WAF1 (el-Deiry et al. 1993, 1994). In our experiments, the half-life of the CDKN1A transcript, encoding p21WAF1, was <1 h following transient activation of the p53 response. This estimate is consistent with previous measurements obtained under other experimental conditions (Gorospe et al. 1998; Wang et al. 2000). Remarkably, the expression of p21WAF1 protein decreased significantly within 2 h and had returned to basal levels within 4–6 h at the restrictive temperature (Figs. 1D, 3B). Therefore, the timing of cell cycle reentry coincided temporally with reversible p21WAF1 expression. The reversible expression of p21WAF1 is likely the consequence of the short half-life of the CDKN1A mRNA coupled with proteasome-mediated degradation of the p21WAF1 protein (Blagosklonny et al. 1996). We propose that the rapid decay of the CDKN1A mRNA is absolutely required for reversible p53-dependent G1 arrest.
Role for mRNA decay in the oscillatory behavior of the p53–Mdm2 feedback loop
Under normal conditions, p53 expression and its ability to transactivate target genes are inhibited largely through its interaction with the E3 ubiquitin ligase, Mdm2, itself a p53-regulated protein (Momand et al. 1992; Honda et al. 1997). Recent single-cell analysis of the p53 response has indicated that activation of p53 leads to repeated oscillatory cycles of p53-dependent induction of Mdm2 followed by proteasome-mediated degradation of p53 (Geva-Zatorsky et al. 2006; Lahav 2008; Proctor and Gray 2008; Batchelor et al. 2009; Zhang et al. 2009). Importantly, oscillatory circuits cannot function with long-lived components. Here we show that the half-life of the MDM2 mRNA was <1 h following transient activation of the p53 response, and the corresponding protein returned to baseline levels within 4 h at the restrictive temperature (Fig. 3B). We propose that the extremely short half-life of the MDM2 transcript is integral to the oscillatory behavior of p53. Importantly, the number of p53 oscillations following the activation of p53 by DNA double-strand breaks is proportional to the extent of DNA damage, and sustained p53–Mdm2 oscillations are thought to contribute to the decision to undergo apoptosis in response to p53 activation (Lahav 2004, 2008; Geva-Zatorsky et al. 2006; Proctor and Gray 2008; Batchelor et al. 2009; Zhang et al. 2009). Therefore, the short half-lives of MDM2 mRNA and protein are likely integral to p53-dependent cell fate decisions.
The induction of short-lived RNAs
Several groups have assessed mRNA decay on a genomewide scale using oligonucleotide microarrays (Raghavan and Bohjanen 2004; Mata et al. 2005; Sharova et al. 2009). Gene Ontology analysis of these data sets indicates that specific functional classes of transcripts are far more likely to be subject to negative regulation by mRNA decay (Lam et al. 2001; Yang et al. 2003; Raghavan and Bohjanen 2004; Mata et al. 2005; Sharova et al. 2009). Salient to the p53 response, regulators of apoptosis and cell cycle are over-represented among short-lived transcripts, while transcripts encoding metabolic enzymes and structural proteins tend to have longer half-lives (Yang et al. 2003; Raghavan and Bohjanen 2004; Mata et al. 2005; Sharova et al. 2009). It is intuitively obvious that spurious activation of apoptosis or cell cycle checkpoints could pose a challenge for development and cell survival, so mRNA decay could play a protective role by limiting the expression of these classes of transcripts. The rapid turnover of some p53-responsive mRNAs is consistent with this concept; however, it is more difficult to reconcile the evolutionary advantage of targeting transcripts encoding other classes of p53 targets such as DNA repair proteins. In fact, the most highly over-represented GO terms among the unstable p53-induced transcripts were “response to DNA damage stimulus” and “DNA repair,” while none of the stable transcripts encode proteins associated with these terms (Table 1). There is no obvious survival advantage conferred by limiting the p53-dependent expression of DNA repair genes. It is noteworthy that some p53-induced proteins (i.e., DDB2 and FAS) appear to be stable, giving rise to prolonged induction of the proteins despite depletion of the mRNA. These issues raise the possibility that mRNA decay is not simply required to shut off the p53-dependent transcriptional response.
There is accumulating evidence that mRNA stability affects the kinetics of gene induction (Lam et al. 2001; Elkon et al. 2010). For example, 30% of ionomycin- and lipopolysaccharide-induced transcripts have half-lives of 2 h or less (Lam et al. 2001). Furthermore, transcripts induced within the first 2 h following exposure to bacterial lipopolysaccharide, tumor necrosis factor, and γ-interferon were short-lived compared with transcripts induced at later times (Elkon et al. 2010). Here, we found a striking bias toward short-lived transcripts among 29 p53-induced mRNAs with >70% of these p53-responsive mRNAs exhibiting half-lives of 2 h or less. In time-course experiments, we noted a small yet statistically significant difference in the kinetics of induction such that short-lived targets were induced slightly faster than the longer-lived mRNAs. So our data are consistent with the emerging concept that unstable mRNAs are poised for rapid transcriptional regulation (Elkon et al. 2010).
More striking than this small difference was the fact that the expression of unstable targets reached a plateau within 4–6 h, while the expression of stable transcripts continued to increase. It appears that the unstable p53-responsive mRNAs reach equilibrium between production and decay. Intriguingly, the expression of several unstable transcripts, including MDM2 and FAS, decreased between 6 and 24 h at the permissive temperature. It is conceivable that p53 increases the expression of RNA binding proteins or microRNAs that accelerate decay of specific p53-responsive transcripts. We also recognize that p53-induced transcripts may be subject to post-transcription stabilization under specific conditions. It will be important to determine to what extent the decay of specific p53-regulated transcripts is regulated under relevant conditions.
SUMMARY
This study indicates that the p53 response is dominated by the induction of short-lived transcripts. This engenders the p53 response with several important features. First, the rapid turnover of p53-responsive transcripts protects cells from the spurious p53 activation because sustained activation of p53 is incompatible with life (Jones et al. 1995; Montes de Oca Luna et al. 1995). Second, the rapid turnover of p53-regulated mRNAs permits reversal of the p53 response following transient activation of p53. Third, the rapid synthesis of unstable mRNAs likely affords the opportunity for post-transcriptional regulation under specific conditions, providing plasticity to the p53 transcriptional response. These novel concepts have broad implications for the tumor suppressor activity of p53.
MATERIALS AND METHODS
Cell culture
All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; Thermo Fisher Scientific) in a humidified atmosphere with 5% CO2. HT29 cells are human colon cancer cells homozygous for an arginine-to-histine mutation at position 273 (R273H) of p53 that renders it transcriptionally inactive (Rodrigues et al. 1990). HT29-tsp53 cells were derived from this parental line following transfection of cDNA encoding a temperature-sensitive variant of murine p53 that has a substitution of valine-to-alanine at position 135 (V135A) (Merchant et al. 1996). At the restrictive temperature (38°C), this variant of p53 is in a mutant conformation and remains cytoplasmic (Michalovitz et al. 1990; Gannon and Lane 1991; McKay et al. 2000, 2004). At the permissive temperature (32°C), p53 rapidly switches to wild-type conformation and localizes to the nucleus, where it acts as a transcriptional activator (Michalovitz et al. 1990; Gannon and Lane 1991; McKay et al. 2000, 2004). Where indicated, these cells were incubated for 16 h at the permissive temperature (32°C) to strongly induce functional p53 and were then returned to the restrictive temperature for the indicated time to allow cells to recover from p53 activation. HeLa Tet-Off cells were purchased from Clontech Laboratories.
Flow cytometry
The incorporation of 5-bromo-2′-deoxyuridine (BrdU) was used to identify actively replicating S-phase cells by two parameter flow cytometry. BrdU (30 μM; Sigma-Aldrich Canada Ltd.) was added to cultures 30 min before cell collection to label nascent DNA. Labeled cells were collected and fixed at −20°C in 70% ethanol for a minimum of 1 h. Detection of BrdU incorporation in single-stranded DNA using a primary anti-BrdU antibody (1:100; PharMingen) and secondary anti-mouse FITC-conjugated antibody (1:15; Sigma-Aldrich Canada Ltd.) was performed, as described previously (Stubbert et al. 2007). Propidium iodide (30 μM in PBS) was added, and then samples were analyzed by fluorescence-activated cell sorting using a Beckman Coulter FACS station and FCS Express 3 software (DeNovo Software).
Microarray analysis
HT29-tsp53 cells were switched from the restrictive temperature (38°C) to the permissive temperature (32°C) for 16 h, and they were then returned for 6 h to 38°C. Samples were collected following p53 activation or following attenuation of the p53 response (t = 0 or 6 h at restrictive temperature, respectively) and compared with a control sample maintained at the restrictive temperature. Total RNA was isolated using the RNeasy RNA isolation kit (QIAGEN) according to the manufacturer's specifications and submitted for analysis at the Ottawa Genomics Innovation Center Affymetrix Gene-Chip Facility (Ottawa, ON) using the Human Gene 1.0 ST oligonucleotide arrays (Affymetrix). Probe sets in these arrays represent individual exons, while cluster identification numbers represent loci, which, in turn, are made up of multiple exons and are thus represented by multiple probe sets. FlexArray 1.4.1 software with the Affymetrix Power Tools software package was used for statistical comparison of probe sets using an empirical Bayesian algorithm (Wright and Simon 2003). A threshold of P ≤ 0.001 was used to identify differentially expressed probe sets. The fold-change in expression of each mRNA was determined using expression data for all linked probe sets represented by the relevant cluster identification number. The residual expression of all of these statistically induced transcripts was similarly determined 6 h following return to 38°C. The term “residual induction” refers to the fold increase in expression of p53-induced transcripts after being retuned to the restrictive temperature for 6 h, expressed as a percentage of the fold increase observed at 0 h, as defined above. A value of 0% indicates that the expression of the indicated transcript had returned to the baseline level within 6 h at 38°C, while 100% indicates that there was no measurable decrease in expression within 6 h.
Quantitative reverse transcriptase polymerase chain reaction
Cells were trypsinized, harvested, and washed with PBS. Total RNA was isolated using the RNeasy RNA isolation kit (QIAGEN) according to the manufacturer's specifications. Five micrograms of total RNA was reverse-transcribed by using the first-strand cDNA synthesis kit (MBI Fermentas). Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed using the Sybr green fluorescent DNA stain (Invitrogen) and the LightCycler 2 quantitative PCR machine with Light-Cycler software version 3 (Roche Diagnostics).
The following primers were used to measure the relative expression of DDB2 (CCACCTTCATCAAAGGGATTGG and CTCGGATCTCGCTCTTCTGGTC), CDKN1A (CTCAAATCGTCCAGCGACCTT and CATTGTGGGAGGAGCTGTGAA), FAS (CCCAGAATACCAAGTGCAGATG and TCCTTTCTGTGCTTTCTGCATG), MDM2 (AGGTGATTGGTTGGATCAGG and GAAGCCAATTCTCACGAAGG), TP53I3 (TCTCTATGGTCTGATGGG and TTGCCTATGTTCTTGTTG), BTG2 (GTGAGCGAGCAGAGGCTTAAGG and TGCGGTAGGACACCTCATAGGG), SERPINB2 (CTTTTCTGTGGATGCCGATT and CCTGCCAGGGCTTAACATAA), CRYAB (AGCCGCCTCTTTGACCAGTTCTT and GCGGTGACAGCAGGCTTCTCTTC), and XPC (AAGTTCACTCGCCTCGGTTGC and TTCTTTCCTGATTTTAGCCTTTTT). Expression of each transcript was normalized to the expression of ACTB (GGGCATGGGTCAGAAGGAT and GTGGCCATCTCTTGCTCGA) and/or GAPDH (AACAGCGACACCCACTTCTC and GGAGGGGAGATTCAGTGTGG).
Reporter constructs with heterologous 3′ UTRs
HT29-tsp53 cells were incubated at the permissive temperature for 16 h to induce p53 target genes, then RNA was collected and reverse-transcribed to generate cDNA, as described above. Specific primers containing either EcoRI or XbaI linker sequences were used to PCR-amplify the 3′ UTRs of DDB2 (GATCGGAATTCGAGACACTAAAGAAGGTGTG and AAGCATCTAGACATATCAAAAGAGCACAAATC), GDF15 (GATCGGAATTCGCAGTCCTGGTCCTTCCAC and AAGCATCTAGAAGACCAGCCCCCGAGTCC), TP53I3 (GATCGGAATTCAGGAGGATGGGGCAGGACA and AAGCATCTAGACCAGTTCACTCTTTATTTC), and CRYAB (GATCGGAATTCCCCCACCAGTGAATGAAAGTCTTGT and AAGCATCTAGAAGCTTGATAATTTGGGCCTGCCCT) using a GeneAmp PCR system 9700 (Applied Biosystems). The PCR products were cloned directly into a pCR2.1 TOPO vector (Invitrogen Canada). Individual 3′ UTRs of 363 nt, 182 nt, 144 nt, and 76 nt corresponding to the 3′ UTRs of DDB2, GDF15, TP53I3, and CRYAB, respectively, were then subcloned into the pTRE-d2EGFP reporter plasmid (Clontech Laboratories) using EcoRI and XbaI restriction sites. The identity of all constructs was confirmed by sequencing. The resulting plasmids were transiently transfected into HeLa Tet-Off cells (Clontech Laboratories), and the expression of the d2EGFP mRNA was determined by quantitative RT-PCR using the following primers: CGACGGCAACTACAAGACC and CCATCATCCTGCTCCTCCAC.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We acknowledge the assistance of the Microarray and DNA Sequencing Core Facilities of StemCore Laboratories in the Sprott Center for Stem Cell Research at the Ottawa Hospital Research Institute. This work was supported by the Canadian Institutes of Health Research to B.C.M. We also thank the Ottawa Regional Cancer Foundation for seed funding. B.D.M. held an Ontario Graduate Scholarship in Science and Technology. B.C.M. was supported with a Research Scientist Award from the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030122.111.
REFERENCES
- Bakheet T, Williams BR, Khabar KS 2006. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res 34: D111–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Lahav G 2009. The ups and downs of p53: Understanding protein dynamics in single cells. Nat Rev Cancer 9: 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Hickman ES, Vousden KH 1999. Reversal of p53-induced cell-cycle arrest. Mol Carcinog 24: 7–14 [DOI] [PubMed] [Google Scholar]
- Beißbarth T, Speed TP 2004. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465 [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Wu GS, Omura S, el-Deiry WS 1996. Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem Biophys Res Commun 227: 564–569 [DOI] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M 1994. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370: 220–223 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. 1994. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res 54: 1169–1174 [PubMed] [Google Scholar]
- Elkon R, Zlotorynski E, Zeller KI, Agami R 2010. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11: 259 doi: 10.1186/1471-2164-11-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, Dölken L, Ruzsics Z, Koszinowski UH, Zimmer R 2009. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 37: e115 doi: 10.1093/nar/gkp542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon JV, Lane DP 1991. Protein synthesis required to anchor a mutant p53 protein which is temperature-sensitive for nuclear transport. Nature 349: 802–806 [DOI] [PubMed] [Google Scholar]
- Geske FJ, Lieberman R, Strange R, Gerschenson LE 2001. Early stages of p53-induced apoptosis are reversible. Cell Death Differ 8: 182–191 [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, Liron Y, Polak P, Lahav G, et al. 2006. Oscillations and variability in the p53 system. Mol Syst Biol 2: 2006.0033 doi: 10.1038/msb4100068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, Wang X, Holbrook NJ 1998. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol 18: 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. 2010. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38: D75–D80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR 2007. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem 100: 1477–1492 [DOI] [PubMed] [Google Scholar]
- Hogg JR 2011. This message was inspected by Upf1: 3′UTR length sensing in mRNA quality control. Cell Cycle 10: 372–373 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC 1991. p53 mutations in human cancers. Science 253: 49–53 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Kannan K, Amariglio N, Rechavi G, Givol D 2000. Profile of gene expression regulated by induced p53: connection to the TGF-β family. FEBS Lett 470: 77–82 [DOI] [PubMed] [Google Scholar]
- Kannan K, Amariglio N, Rechavi G, Jakob-Hirsch J, Kela I, Kaminski N, Getz G, Domany E, Givol D 2001. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20: 2225–2234 [DOI] [PubMed] [Google Scholar]
- Lahav G 2004. The strength of indecisiveness: Oscillatory behavior for better cell fate determination. Sci STKE 2004: pe55 doi: 10.1126/stke.2642004pe55 [DOI] [PubMed] [Google Scholar]
- Lahav G 2008. Oscillations by the p53–Mdm2 feedback loop. Adv Exp Med Biol 641: 28–38 [DOI] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2: research0041–research0041.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ 2010. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS ONE 5: e11201 doi: 10.1371/journal.pone.0011201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M, Lane DP 2004. Transcription—guarding the genome by sensing DNA damage. Nat Rev Cancer 4: 727–737 [DOI] [PubMed] [Google Scholar]
- Mata J, Marguerat S, Bähler J 2005. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci 30: 506–514 [DOI] [PubMed] [Google Scholar]
- McKay BC, Ljungman M, Rainbow AJ 1999. Potential roles for p53 in nucleotide excision repair. Carcinogenesis 20: 1389–1396 [DOI] [PubMed] [Google Scholar]
- McKay BC, Chen F, Perumalswami CR, Zhang F, Ljungman M 2000. The tumor suppressor p53 can both stimulate and inhibit ultraviolet light-induced apoptosis. Mol Biol Cell 11: 2543–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC 2004. Regulation of ultraviolet light-induced gene expression by gene size. Proc Natl Acad Sci 101: 6582–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Merchant AK, Loney TL, Maybaum J 1996. Expression of wild-type p53 stimulates an increase in both Bax and Bcl-xL protein content in HT29 cells. Oncogene 13: 2631–2637 [PubMed] [Google Scholar]
- Michalovitz D, Halevy O, Oren M 1990. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell 62: 671–680 [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G 2002. Untranslated regions of mRNAs. Genome Biol 3: reviews0004–reviews0004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor CJ, Gray DA 2008. Explaining oscillations and variability in the p53–Mdm2 system. BMC Syst Biol 2: 75 doi: 10.1186/1752-0509-2-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan A, Bohjanen PR 2004. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief Funct Genomic Proteomic 3: 112–124 [DOI] [PubMed] [Google Scholar]
- Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St. Louis-Vlasova IA, Bohjanen PR 2010. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol 30: 3970–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol 21: 394–402 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E 2005. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E 2006. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, Lane DP 1990. p53 mutations in colorectal cancer. Proc Natl Acad Sci 87: 7555–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS 2009. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Stubbert LJ, Hamill JD, McKay BC 2007. The contribution of transactivation subdomains 1 and 2 to p53-induced gene expression is heterogeneous but not subdomain-specific. Neoplasia 9: 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbert LJ, Hamill JD, Spronck JC, Smith JM, Becerril C, McKay BC 2007. DDB2-independent role for p53 in the recovery from ultraviolet light-induced replication arrest. Cell Cycle 6: 1730–1740 [DOI] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, SternJohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. 2008. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell 29: 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M 2000. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GW, Simon RM 2003. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19: 2448–2455 [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ 1993. The p53–mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr 2003. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M 1991. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352: 345–347 [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B 1999. Identification and classification of p53-regulated genes. Proc Natl Acad Sci 96: 14517–14522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Liu F, Cheng Z, Wang W 2009. Cell fate decision mediated by p53 pulses. Proc Natl Acad Sci 106: 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 14: 981–993 [PMC free article] [PubMed] [Google Scholar]