Intestinal bacteria aid host health and limit bacterial pathogen colonization. However, the influence of bacteria on enteric viruses is largely unknown. We depleted the intestinal microbiota of mice with antibiotics prior to inoculation with poliovirus, an enteric virus. Antibiotic-treated mice were less susceptible to poliovirus disease and supported minimal viral replication in the intestine. Exposure to bacteria or their N-acetylglucosamine-containing surface polysaccharides, including lipopolysaccharide and peptidoglycan, enhanced poliovirus infectivity. We found that poliovirus binds lipopolysaccharide, and exposure of poliovirus to bacteria enhanced host-cell association and infection. The pathogenesis of reovirus, an unrelated enteric virus, also was more severe in the presence of intestinal microbes. These results suggest that antibiotic-mediated microbiota depletion diminishes enteric virus infection and that enteric viruses exploit intestinal microbes for replication and transmission.
Enteric viruses encounter up to 1014 bacteria in the mammalian intestine (1). It is unclear whether commensal microorganisms affect enteric viruses. Poliovirus is an enteric human pathogen transmitted by the fecal-oral route and serves as a model for enteric virus infections (2). Orally acquired poliovirus undergoes a primary replication cycle in the gastrointestinal tract prior to dissemination. Poliovirus occasionally disseminates from the intestine to the central nervous system, resulting in paralytic poliomyelitis days to weeks after initial infection in the gastrointestinal tract. A key question is whether microbiota influence viral replication in the gastrointestinal tract, which aids systemic dissemination.
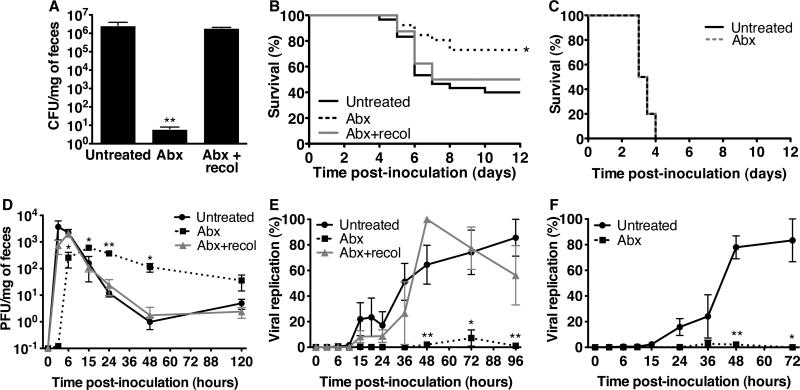
To investigate the effect of intestinal microbiota on poliovirus infection, mice susceptible to poliovirus were treated with antibiotics to deplete microbes, and viral disease was monitored (fig. S1) (3). Murine poliovirus infection requires expression of the human poliovirus receptor, PVR (4-6). PVR-transgenic mice (PVRtg), however, are not susceptible to oral poliovirus infection unless rendered immunodeficient by interferon-α/β receptor gene inactivation (PVRtg-Ifnar1-/-) (7, 8). PVRtg-Ifnar1-/- mice were untreated or treated orally with four antibiotics prior to oral inoculation with poliovirus. Antibiotic treatment reduced culturable intestinal bacteria by a million-fold (Fig. 1A). The mortality of untreated mice was twice that of antibiotic-treated mice (Fig.1B). Reintroduction of fecal bacteria into antibiotic-treated mice enhanced poliovirus disease, suggesting that microbiota promote poliovirus pathogenesis. However, when the intestinal lumen was bypassed by intraperitoneal inoculation of poliovirus, pathogenesis was microbiota-independent (Fig. 1C, fig. S2). Given that orally-inoculated poliovirus enters the intestine and encounters the large number of bacteria that reside there, the microbiota-mediated enhancement of poliovirus pathogenesis in orally inoculated mice is likely initiated in the intestine.
Fig. 1.
Poliovirus pathogenesis, shedding, and replication in microbiota-depleted mice. (A) Bacterial loads in feces. PVRtg-Ifnar1-/- mice (n=4-7) were untreated, antibiotic-treated (Abx) for 10 days, or antibiotic-treated for 8 days and recolonized for 2 days with fecal bacteria (Abx+recol). Feces were plated and grown anaerobically, yielding colony-forming units (CFU) per milligram of feces. (B) Survival of PVRtg-Ifnar1-/- mice orally inoculated with poliovirus (untreated: n=30, Abx: n=26, Abx+recol: n=8). *p=0.012, Log-rank test. (C) Survival of PVRtg-Ifnar1-/- mice intraperitoneally inoculated with poliovirus (n=10 mice each). (D) Poliovirus shedding from PVRtg-Ifnar1-/- mice. Mice were orally inoculated with poliovirus, feces were collected (n=2-26 per interval), and poliovirus was isolated and quantified by plaque assay, yielding plaque-forming units (PFU) per milligram of feces. (E,F) Poliovirus replication in intestinal tracts of PVRtg-Ifnar1-/- (E) or PVRtg (F) mice orally inoculated with light-sensitive poliovirus (n=3-9 mice per interval). Feces were harvested, and virus was quantified +/- light exposure to determine percent replication. Symbols represent mean + SEM, *p<0.05, **p<0.01, Student's t-test. N=2-6 for all experiments.
To determine whether mice harboring microbiota support more efficient poliovirus replication than mice with depleted microbiota, we quantified viral titers from fecal samples (Fig. 1D, fig. S3A) because poliovirus was undetectable in intestinal tissue (fig. S4) and minimal intestinal pathology was evident (fig. S5). Peak poliovirus titers in feces from antibiotic-treated animals were lower than those from untreated mice, but titers from antibiotic-treated mice were higher at later times. Prolonged shedding from antibiotic-treated mice was due to slower peristalsis, since dye transit also was delayed (fig. S6) (9). We postulated that increased poliovirus titers from antibiotic-treated mice at late times might be due to extended shedding of unreplicated inoculum virus. To differentiate between replicated and inoculum virus, we first quantified fecal shedding of poliovirus from nonpermissive mice lacking PVR and observed elevated late titers in antibiotic-treated mice, suggesting that total viral titers in feces and replication are not linked (fig. S3B). We then quantified viral replication in PVR mice using light-sensitive poliovirus. Poliovirus propagated in the presence of neutral red dye is sensitive to light-induced inactivation by RNA cross-linking but loses light-sensitivity upon replication in the dark inside mice, facilitating assessment of replication (10). We orally inoculated untreated or antibiotic-treated mice with light-sensitive poliovirus and collected feces in the dark. Fecal viruses were light-exposed or unexposed and quantified to determine replication status (fig. S7). PVRtg-Ifnar-1/- and PVRtg mice harboring microbiota supported efficient intestinal poliovirus replication, whereas antibiotic-treated mice did not (Fig. 1E,1F). Therefore, total fecal titers do not reflect viral replication, a fact only revealed by using light-sensitive viruses. Moreover, poliovirus intestinal replication was equivalent in Ifnar1+/+ and Ifnar1-/- mice, suggesting intestinal replication was IFNAR-independent. Because poliovirus infection was lethal for a fraction of antibiotic-treated mice (Fig. 1B), it is possible that either minimal viral replication was sufficient for lethality or inoculum virus breached the epithelium and replicated in extra-intestinal sites, occasionally initiating disease. Collectively, these results indicate that the microbiota enhance gastrointestinal poliovirus replication.
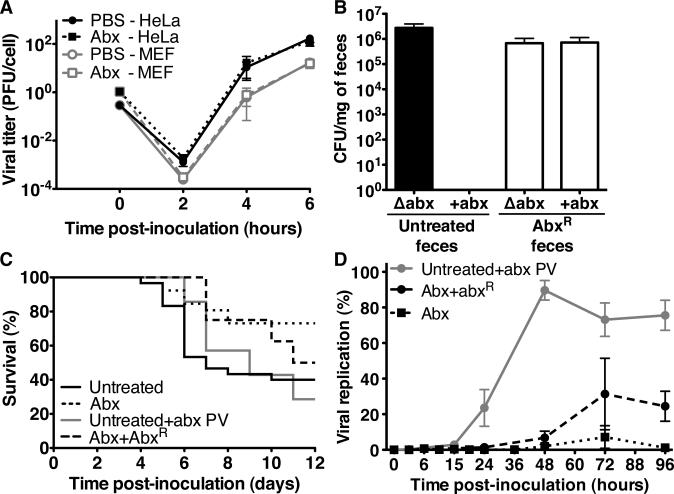
We gathered several lines of evidence suggesting that diminished poliovirus replication and disease in antibiotic-treated mice is due to microbiota depletion rather than direct effects of antibiotic treatment. We found that antibiotics do not directly affect poliovirus since poliovirus replication kinetics were identical in the presence and absence of antibiotics in HeLa cells and PVRtg mouse embryo fibroblasts (MEFs) (Fig. 2A). We next assayed poliovirus replication and pathogenesis in antibiotic-treated mice harboring antibiotic-resistant bacteria. For these experiments, we treated PVRtg-Ifnar1-/- mice with antibiotics to select antibiotic-resistant microbiota (fig. S8). After several weeks, fecal bacteria were insensitive to antibiotics in vitro (Fig. 2B). The multi-antibiotic resistant strain was identified as Ochrobactrum intermedium a Gram-negative aerobe, by 16S rDNA sequencing of fecal-derived subclones (fig. S9). Poliovirus replicated and was pathogenic in antibiotic-treated mice harboring Ochrobactrum intermedium (Fig. 2C,D). Furthermore, poliovirus mixed with antibiotics prior to oral inoculation of mice replicated and was pathogenic (Fig. 2C,D). Therefore, diminished poliovirus replication and pathogenesis in antibiotic-treated mice is not due to direct antiviral effects of antibiotics.
Fig. 2.
The effects of antibiotic treatment on poliovirus replication and pathogenesis. (A) Poliovirus replication kinetics in MEFs and HeLa cells +/- antibiotics. (B) Fecal bacterial loads from untreated or antibiotic-treated mice harboring antibiotic-resistant (abxR) bacteria. Feces were plated on rich medium +/- four antibiotics. (C) Survival of PVRtg-Ifnar1-/- mice orally inoculated with poliovirus pre-mixed with four antibiotics (Untreated+abx PV, n=9) or poliovirus alone in antibiotic-treated mice harboring AbxR bacteria (Abx+abxR, n=8). (Results from untreated and antibiotic-treated mice are from Fig. 1B.) (D) Replication of light-sensitive poliovirus in untreated mice receiving poliovirus+antibiotics inoculum and antibiotic-treated mice harboring abxR bacteria in comparison to antibiotic-treated mice. (Results from antibiotic-treated mice are from Fig. 1E.) Each symbol represents mean + SEM. A and B, N=2-5 experiments, C and D are from a representative experiment.
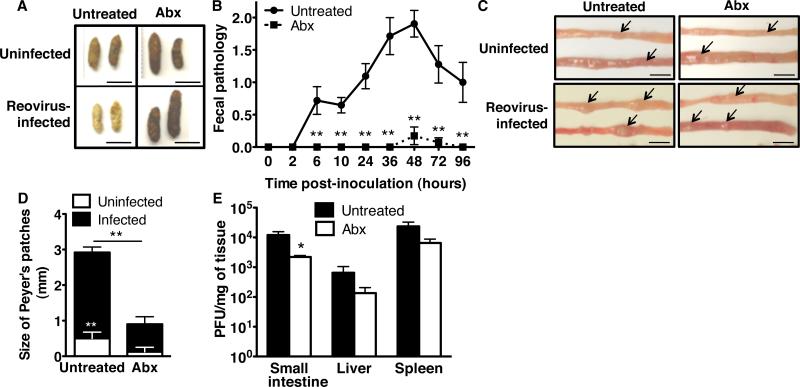
Because all enteric viruses encounter intestinal bacteria within the host, we examined the specificity of the microbiota effects using reovirus, an enteric virus that infects most mammals (11). Although immunocompetent adult mice do not display overt reovirus disease symptoms, immunocompromised adult mice develop nonfatal disease after oral inoculation with reovirus strain T3SA+. We orally inoculated untreated or antibiotic-treated immunocompromised PVRtg-Ifnar1-/- mice with reovirus. Feces from untreated mice were yellow, oily, and hardened, typical of biliary obstruction from T3SA+ reovirus replication and damage (12), whereas feces from antibiotic-treated mice appeared normal (Fig. 3A, 3B). Furthermore, analysis of intestines revealed severe reovirus-induced pathology, with enlarged Peyer's patches in untreated but not antibiotic-treated mice (Fig. 3C, 3D). Reovirus titers in intestines from untreated mice were significantly higher than those from antibiotic-treated mice (Fig. 3E). These results suggest that intestinal microbes promote reovirus disease and, therefore, may promote infection with other enteric viruses.
Fig. 3.
Reovirus pathogenesis in microbiota-depleted mice. (A) PVRtg-Ifnar1-/- mice were either uninfected, untreated (n=5) or antibiotic-treated (n=5), or infected perorally with reovirus, untreated (n=13) or antibiotic-treated (n=15). Feces were collected 24 hours post-inoculation. (B) Fecal pathology (Table S1). (C) Upper (top) and lower (bottom) small intestines were harvested from untreated and antibiotic-treated PVRtg-Ifnar1-/- mice on day 4 post-infection or from uninfected mice. Arrows indicate Peyer's patches. (D) Quantification of Peyer's patch sizes (from C) from uninfected and infected mice. (E) Reovirus titers from day 4 post-infection PVRtg-Ifnar1-/-mouse tissues. Plaque assays were performed using murine L929 cells, yielding PFU per milligram of tissue. For B-E, n=4-9 untreated mice, n=2-9 antibiotic-treated mice. Each symbol or bar denotes the mean + SEM. *p<0.05, **p<0.01, Student's t-test. Scale bars in A and C=5mm. A and C, representative of 3-5 experiments; N=2-4 for B, D, and E.
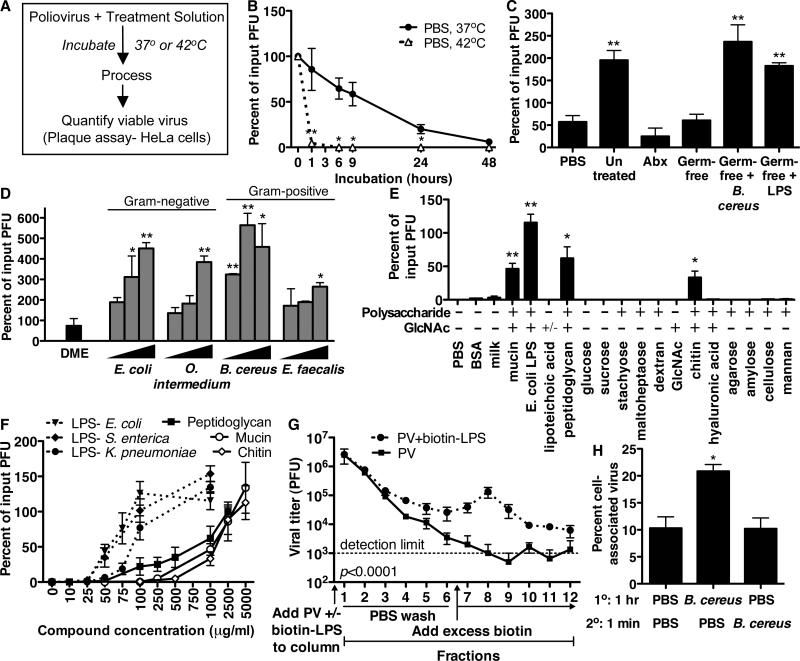
The microbiota-dependent enhancement of poliovirus replication and pathogenesis could be mediated by microbiota-induced host effects, viral effects, or both. To discriminate between these possibilities, we investigated whether intestinal microbes alter poliovirus infectivity. First, we tested whether poliovirus infectivity was altered by exposure to intestinal microbiota in vivo. We orally inoculated untreated, antibiotic-treated, or germ-free mice with poliovirus, harvested lumenal contents from the lower small intestine at two hours post-infection, and quantified infectivity of isolated poliovirus in primary MEFs and HeLa cells. The infectivity in MEFs of poliovirus isolated from untreated mice was twice that of tissue culture-derived virus and antibiotic-treated and germ-free intestinal virus (fig. S10). Second, we developed an ex vivo/in vitro assay to examine poliovirus infectivity (Fig. 4A). Poliovirus was incubated at 37°C or 42°C and viable virus was quantified by plaque assay. Poliovirus incubated in PBS, feces from antibiotic-treated mice, or germ-free feces lost viability (Fig. 4B,4C). However, poliovirus incubated in untreated feces or germ-free feces supplemented with bacteria had significantly increased viability (Fig. 4C). Similarly, poliovirus incubated with Gram-negative (Escherichia coli, Ochrobactrum intermedium) or Gram-positive (Bacillus cereus, Enterococcus faecalis) bacteria had significantly increased viability (Fig. 4D). Exposure to B. cereus increased poliovirus infectivity over 500%. Enhancement of poliovirus infectivity did not require live bacteria (fig. S11). Moreover, poliovirus incubated with certain bacterial surface polysaccharides including lipopolysaccharide (LPS) and peptidoglycan (PG) had significantly enhanced yield over PBS-treated controls (Fig. 4C, E, fig. S12). The enhancement was not due to cellular effects of LPS or PG treatment (fig. S13). We tested a variety of glycans and other compounds, and only N-acetylglucosamine (GlcNAc)-containing polysaccharides demonstrated activity (e.g. chitin, Fig. 4E). Mucin, a host protein modified with GlcNAc-containing polysaccharides, also had activity (13). Of the purified components tested, LPS was the most potent enhancer of poliovirus infectivity, with activity at concentrations >20-fold lower than chitin or mucin (Fig. 4F). Using biotinylated LPS and monomeric avidin columns, we found that poliovirus binds LPS (Fig. 4G). Because B. cereus exposure produced the largest increase in poliovirus yield, we tested whether exposure to B. cereus enhanced radiolabeled poliovirus binding to HeLa cells, aiding infection. Poliovirus incubated with B. cereus displayed two-fold higher HeLa cell adherence compared to controls (Fig. 4H). Overall, poliovirus infectivity was enhanced in the presence of intestinal microbiota in vitro and in vivo, likely contributing to the enhanced replication and pathogenesis in microbiota-harboring mice.
Figure 4.
Effects of bacteria and polysaccharides on poliovirus. (A) Strategy for in vitro poliovirus infectivity experiments. (B) Poliovirus recovered after incubation in PBS. (C) Poliovirus infectivity following exposure to PBS, feces, or feces supplemented with Bacillus cereus or lipopolysaccharide (LPS) (6 hours/37°C). (D) Poliovirus infectivity after exposure to medium (DME) or bacterial strains (107, 108, or 109 CFU) (6 hours/37°C). (E) Poliovirus infectivity after incubation with compounds (1 mg/ml) (6 hours/42°C). (F) Poliovirus infectivity after incubation with various concentrations of compounds (6 hours/42°C). (G) Poliovirus binding to LPS. Poliovirus was incubated +/- biotinylated LPS for 1 hour at 37°C. A monomeric avidin column was loaded with samples and washed with PBS to collect fractions 1-6. Excess biotin was added to elute (fractions 7-12). Poliovirus was quantified yielding PFU per fraction, p<0.0001, 2-way ANOVA. (H) Binding of radiolabeled poliovirus to HeLa cells. 35S-labeled poliovirus was incubated with PBS or 108 CFU B. cereus for 1 hour at 37°C. An equal volume of PBS or B. cereus was added followed by immediate incubation with HeLa cells. After washing, cell-associated radioactivity was quantified. For all experiments, N=2-8 and bars and symbols denote mean + SEM, *p<0.05, **p<0.01, Student's t-test.
Despite the well-known beneficial effects of intestinal microbes, we discovered that they augment enteric virus pathogenesis by enhancing viral replication. Intestinal microbes also induce egg hatching of an intestinal nematode in mice (14), suggesting that diverse pathogens exploit intestinal microbes for propagation. Our work implies that antibiotic-mediated microbiota depletion can have antiviral effects, although we do not advocate the use of antibiotics to prevent viral disease. However, understanding how microbiota promote enteric virus infections may reveal new antiviral strategies. Our results suggest that poliovirus binds specific microbe-associated surface polysaccharides, enhancing viral thermostability and attachment to host cells. Contrary to the known benefits of intestinal microbiota to the host (1), enteric viruses may have evolved to use intestinal microbes as a trigger for replication at a site optimal for transmission.
Supplementary Material
Supplemental Figure Legends
Supp. Figure 1. Treatment strategy used for microbiota depletion and viral infection.
Treatment timelines for untreated, antibiotic-treated (abx), and antibiotic-treated/recolonized (abx + recol) mice. Fecal bacterial loads were determined (◇) prior to and after treatments. White bars denote extent of antibiotic treatment, and gray bars represent feces collection times post-infection.
Supp. Figure 2. Poliovirus pathogenesis in intraperitoneally inoculated PVRtg mice.
Survival of untreated and antibiotic-treated (Abx) immune competent PVRtg mice inoculated intraperitoneally with 1 × 108 PFU of poliovirus (untreated: n=14, Abx: n=10). N=2-3 experiments.
Supp. Figure 3. Poliovirus fecal shedding kinetics. (A) Poliovirus shedding from immune competent PVRtg mice. After oral inoculation with poliovirus, feces were collected from untreated and antibiotic-treated (Abx) mice (n=2-18 at each interval). Poliovirus was isolated from feces and quantified by plaque assay, yielding PFU per milligram of feces. N=2-6 experiments. (B) Poliovirus shedding in feces from non-PVR C57BL/6 mice (n=5 per treatment group) orally inoculated with poliovirus. Mice were untreated, antibiotic-treated, antibiotic-treated/recolonized (Abx + recol), or germ-free (GF, n=6). Germ-free mice became colonized after 48 hours of housing outside of gnotobiotic chambers. Therefore, only data from the first 48 hours are shown. Data are from a representative experiment (non-PVRtg C57BL/6 mice, symbols indicate the mean) or are means from two experiments (GF mice, symbols represent mean + SEM). *p<0.05, **p<0.01 compared to untreated, Student's t-test.
Supp. Figure 4. Quantification of poliovirus in tissues. PVRtg mice were orally inoculated with poliovirus, and tissues were harvested at 10, 24, or 48 hours post-inoculation (hpi) (n=3 each). Virus was extracted from tissues and quantified by plaque assay. MLN = mesenteric lymph node. Data are representative of two experiments.
Supp. Figure 5. Intestine pathology. Intestinal architecture and cellular changes were analyzed following hematoxylin and eosin staining of tissue sections from uninfected PVRtg mice (untreated or antibiotic-treated) and poliovirus-infected PVRtg mice (untreated or antibiotic-treated; 48 hours post-infection by the oral route) (n=3 each). Scale bars = 50µm. Data are representative of two experiments.
Supp. Figure 6. Intestinal transit time in mice. Untreated or antibiotic-treated (Abx) PVRtg-Ifnar1-/- mice were orally administered Evan's blue dye, and feces were collected at the times shown post-inoculation. Feces were suspended in PBS and the amount of dye excreted was scored. Symbols represent the mean + SEM. *p<0.05, **p<0.01, Student's t-test. N=3 experiments.
Supp. Figure 7. Poliovirus fecal titer data following oral inoculation with lightsensitive virus. (A,B,C) Poliovirus titer data for unexposed (total) and light-exposed (light-insensitive) fecal samples from untreated (A), antibiotic-treated (Abx) (B), and antibiotic-treated/recolonized (Abx+recol) (C) PVRtg-Ifnar1-/- mice. These data were used to calculate the percent replication results shown in Fig. 1E. Bars denote the mean + SEM. N=2-6 experiments.
Supp. Figure 8. Strategy for isolating and identifying antibiotic resistant (abxR) bacteria.
Supp. Figure 9. Identification of antibiotic-resistant (AbxR) bacteria in antibiotic treated mice. Feces were harvested from antibiotic-treated (Abx) mice harboring AbxR bacteria, genomic DNA was isolated, and the 16S rDNA region was PCR amplified, cloned into plasmids, and sequenced. One representative sequence is shown aligned to Ochrobactrum intermedium with 96-100% sequence identity for all seven clones derived from fecal PCR products and both clones derived from AbxR colony PCR products.
Supp. Figure 10. Poliovirus infectivity ex vivo. Infectivity of tissue culture-derived (TC) or mouse intestine lumenal content-derived poliovirus. Intestine lumenal contents were collected from the lower small intestine of untreated (n=5) or antibiotic-treated (Abx) (n=4) PVRtg mice or germ-free (GF) non-PVR C57BL/6 (n=3) mice two hours following oral inoculation with poliovirus. Since intestinal cells are minimally susceptible to poliovirus, and differences in viral infectivity may be more apparent in minimally susceptible cells, poliovirus infectivity was compared using minimally susceptible, freshly harvested PVRtg mouse embryonic fibroblasts (MEFs) and highly susceptible HeLa cells. Seven infectious center assays were performed, and MEF titers are presented as a percentage of HeLa cell titers to reflect relative infectivity. Bars denote the mean + SEM. *p<0.05, **p<0.01, Student's t-test. N=7 experiments.
Supp. Figure 11. Poliovirus recovery after exposure to inactivated bacteria.
Poliovirus was incubated with UV-inactivated bacteria (107 CFU per strain) or minimal medium (DME) for 6 hours at 37°C, followed by plaque assay on HeLa cells. Bars denote the mean + SEM. N=2 experiments. Similar results were obtained with heat-killed bacterial strains (data not shown).
Supp. Figure 12. Poliovirus infectivity in the presence or absence of LPS.
Poliovirus was exposed to 1000 μg/ml E. coli LPS at 37°C or 42°C for the times shown, and viable virus was quantified by plaque assay using HeLa cells. Symbols indicate the mean + SEM. N=2-4 experiments.
Supp. Figure 13. Effect of LPS or peptidoglycan (PG) treatment on HeLa cells. (A) Pretreatment of HeLa cells with E. coli LPS or B. subtilis PG. To control for potential cell changes induced by LPS or PG exposure, HeLa cells were treated for 30 minutes with LPS or PG at the same concentration plated in the experiments shown in Fig. 4E after thousand-fold virus dilution (1 μg/ml final concentration). Cells were then washed, poliovirus was added, and virus was quantified by a plaque assay. (B) Poliovirus was treated with 1000 μg/ml LPS or PG, followed by immediate thousand-fold dilution and plating on HeLa cells for a plaque assay. Data are displayed as the percentage of control (PBS)- treated plaque forming units. Bars denote the mean + SEM. N=2 experiments.
Supp. Table 1. Scoring strategy for reovirus-infected mouse feces. Scores were based on color and consistency. See Fig. 3B for graphical representation.
Acknowledgments
We thank B. Duerkop, C. Behrendt-Boyd, J. Charles, J. Richardson, and S. Gore for assistance, B. Levine and V. Sperandio for manuscript comments, and S. Koike for PVRtg mice. This research was supported by Public Health Service awards T32 AI007520 (S.K.K.), F32 NS071986 (A.J.P.), T32 AI07611 (J.M.F.), R37 AI38296, P30 CA68485, and P60 DK20593 (T.S.D.), R01 AI74668 (J.K.P.), the Elizabeth B. Lamb Center for Pediatric Research (T.S.D.), and a Pew Scholar award (J.K.P.). The data reported in the paper are tabulated in the main manuscript and in the supporting online materials. The sequence data are available in GenBank (accession #: BankIt1475845 Seq10 JN613288).
Footnotes
Publisher's Disclaimer: “This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Summary Intestinal microbiota enhance poliovirus and reovirus pathogenesis in mice.
References and Notes
- 1.Garrett WS, Gordon JI, Glimcher LH. Cell. 2010;140:859. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pallansch MA, Roos RP. In: Virology. Fields BN, et al., editors. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 723–775. [Google Scholar]
- 3.Materials and methods are available as supporting material on Science online.
- 4.Ren RB, et al. Cell. 1990;63:353. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- 5.Koike S, et al. Proc Natl Acad Sci U S A. 1991;88:951. doi: 10.1073/pnas.88.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ida-Hosonuma M, et al. Arch Virol. 2003;148:29. doi: 10.1007/s00705-002-0910-7. [DOI] [PubMed] [Google Scholar]
- 7.Ida-Hosonuma M, et al. J Virol. 2005;79:4460. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohka S, et al. J Virol. 2007;81:7902. doi: 10.1128/JVI.02675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrams GD, Bishop JE. Proc Soc Exp Biol Med. 1967;126:301. doi: 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- 10.Kuss SK, Etheredge CA, Pfeiffer JK. PLoS Pathogens. 2008;4 doi: 10.1371/journal.ppat.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyler KL. In: Fields virology. Knipe DM, Howley PM, editors. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 1729–1945. [Google Scholar]
- 12.Barton ES, et al. J Clin Invest. 2003;111:1823. doi: 10.1172/JCI16303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podolsky DK. J Biol Chem. 1985;260:8262. [PubMed] [Google Scholar]
- 14.Hayes KS, et al. Science. 2010;328:1391. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer JK, Kirkegaard K. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virgin HW, et al. J Virol. 1988;62:4594. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash HL, et al. Science. 2006;313:1126. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, et al. Cell. 118:229. 204. [Google Scholar]
- 19.Brandenburg B, et al. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure Legends
Supp. Figure 1. Treatment strategy used for microbiota depletion and viral infection.
Treatment timelines for untreated, antibiotic-treated (abx), and antibiotic-treated/recolonized (abx + recol) mice. Fecal bacterial loads were determined (◇) prior to and after treatments. White bars denote extent of antibiotic treatment, and gray bars represent feces collection times post-infection.
Supp. Figure 2. Poliovirus pathogenesis in intraperitoneally inoculated PVRtg mice.
Survival of untreated and antibiotic-treated (Abx) immune competent PVRtg mice inoculated intraperitoneally with 1 × 108 PFU of poliovirus (untreated: n=14, Abx: n=10). N=2-3 experiments.
Supp. Figure 3. Poliovirus fecal shedding kinetics. (A) Poliovirus shedding from immune competent PVRtg mice. After oral inoculation with poliovirus, feces were collected from untreated and antibiotic-treated (Abx) mice (n=2-18 at each interval). Poliovirus was isolated from feces and quantified by plaque assay, yielding PFU per milligram of feces. N=2-6 experiments. (B) Poliovirus shedding in feces from non-PVR C57BL/6 mice (n=5 per treatment group) orally inoculated with poliovirus. Mice were untreated, antibiotic-treated, antibiotic-treated/recolonized (Abx + recol), or germ-free (GF, n=6). Germ-free mice became colonized after 48 hours of housing outside of gnotobiotic chambers. Therefore, only data from the first 48 hours are shown. Data are from a representative experiment (non-PVRtg C57BL/6 mice, symbols indicate the mean) or are means from two experiments (GF mice, symbols represent mean + SEM). *p<0.05, **p<0.01 compared to untreated, Student's t-test.
Supp. Figure 4. Quantification of poliovirus in tissues. PVRtg mice were orally inoculated with poliovirus, and tissues were harvested at 10, 24, or 48 hours post-inoculation (hpi) (n=3 each). Virus was extracted from tissues and quantified by plaque assay. MLN = mesenteric lymph node. Data are representative of two experiments.
Supp. Figure 5. Intestine pathology. Intestinal architecture and cellular changes were analyzed following hematoxylin and eosin staining of tissue sections from uninfected PVRtg mice (untreated or antibiotic-treated) and poliovirus-infected PVRtg mice (untreated or antibiotic-treated; 48 hours post-infection by the oral route) (n=3 each). Scale bars = 50µm. Data are representative of two experiments.
Supp. Figure 6. Intestinal transit time in mice. Untreated or antibiotic-treated (Abx) PVRtg-Ifnar1-/- mice were orally administered Evan's blue dye, and feces were collected at the times shown post-inoculation. Feces were suspended in PBS and the amount of dye excreted was scored. Symbols represent the mean + SEM. *p<0.05, **p<0.01, Student's t-test. N=3 experiments.
Supp. Figure 7. Poliovirus fecal titer data following oral inoculation with lightsensitive virus. (A,B,C) Poliovirus titer data for unexposed (total) and light-exposed (light-insensitive) fecal samples from untreated (A), antibiotic-treated (Abx) (B), and antibiotic-treated/recolonized (Abx+recol) (C) PVRtg-Ifnar1-/- mice. These data were used to calculate the percent replication results shown in Fig. 1E. Bars denote the mean + SEM. N=2-6 experiments.
Supp. Figure 8. Strategy for isolating and identifying antibiotic resistant (abxR) bacteria.
Supp. Figure 9. Identification of antibiotic-resistant (AbxR) bacteria in antibiotic treated mice. Feces were harvested from antibiotic-treated (Abx) mice harboring AbxR bacteria, genomic DNA was isolated, and the 16S rDNA region was PCR amplified, cloned into plasmids, and sequenced. One representative sequence is shown aligned to Ochrobactrum intermedium with 96-100% sequence identity for all seven clones derived from fecal PCR products and both clones derived from AbxR colony PCR products.
Supp. Figure 10. Poliovirus infectivity ex vivo. Infectivity of tissue culture-derived (TC) or mouse intestine lumenal content-derived poliovirus. Intestine lumenal contents were collected from the lower small intestine of untreated (n=5) or antibiotic-treated (Abx) (n=4) PVRtg mice or germ-free (GF) non-PVR C57BL/6 (n=3) mice two hours following oral inoculation with poliovirus. Since intestinal cells are minimally susceptible to poliovirus, and differences in viral infectivity may be more apparent in minimally susceptible cells, poliovirus infectivity was compared using minimally susceptible, freshly harvested PVRtg mouse embryonic fibroblasts (MEFs) and highly susceptible HeLa cells. Seven infectious center assays were performed, and MEF titers are presented as a percentage of HeLa cell titers to reflect relative infectivity. Bars denote the mean + SEM. *p<0.05, **p<0.01, Student's t-test. N=7 experiments.
Supp. Figure 11. Poliovirus recovery after exposure to inactivated bacteria.
Poliovirus was incubated with UV-inactivated bacteria (107 CFU per strain) or minimal medium (DME) for 6 hours at 37°C, followed by plaque assay on HeLa cells. Bars denote the mean + SEM. N=2 experiments. Similar results were obtained with heat-killed bacterial strains (data not shown).
Supp. Figure 12. Poliovirus infectivity in the presence or absence of LPS.
Poliovirus was exposed to 1000 μg/ml E. coli LPS at 37°C or 42°C for the times shown, and viable virus was quantified by plaque assay using HeLa cells. Symbols indicate the mean + SEM. N=2-4 experiments.
Supp. Figure 13. Effect of LPS or peptidoglycan (PG) treatment on HeLa cells. (A) Pretreatment of HeLa cells with E. coli LPS or B. subtilis PG. To control for potential cell changes induced by LPS or PG exposure, HeLa cells were treated for 30 minutes with LPS or PG at the same concentration plated in the experiments shown in Fig. 4E after thousand-fold virus dilution (1 μg/ml final concentration). Cells were then washed, poliovirus was added, and virus was quantified by a plaque assay. (B) Poliovirus was treated with 1000 μg/ml LPS or PG, followed by immediate thousand-fold dilution and plating on HeLa cells for a plaque assay. Data are displayed as the percentage of control (PBS)- treated plaque forming units. Bars denote the mean + SEM. N=2 experiments.
Supp. Table 1. Scoring strategy for reovirus-infected mouse feces. Scores were based on color and consistency. See Fig. 3B for graphical representation.