Abstract
Interactions of hematopoietic cells with their microenvironment control blood cell colonization, homing and hematopoiesis. Here, we introduce larval hematopoiesis as the first Drosophila model for hematopoietic colonization and the role of the peripheral nervous system (PNS) as a microenvironment in hematopoiesis. The Drosophila larval hematopoietic system is founded by differentiated hemocytes of the embryo, which colonize segmentally repeated epidermal-muscular pockets and proliferate in these locations. Importantly, we show that these resident hemocytes tightly colocalize with peripheral neurons and we demonstrate that larval hemocytes depend on the PNS as an attractive and trophic microenvironment. atonal (ato) mutant or genetically ablated larvae, which are deficient for subsets of peripheral neurons, show a progressive apoptotic decline in hemocytes and an incomplete resident hemocyte pattern, whereas supernumerary peripheral neurons induced by ectopic expression of the proneural gene scute (sc) misdirect hemocytes to these ectopic locations. This PNS-hematopoietic connection in Drosophila parallels the emerging role of the PNS in hematopoiesis and immune functions in vertebrates, and provides the basis for the systematic genetic dissection of the PNS-hematopoietic axis in the future.
Keywords: Drosophila, Cell proliferation, Cell survival, Hematopoietic colonization, Hematopoiesis, Hemocyte, Larva, Microenvironment, Peripheral nervous system
INTRODUCTION
Drosophila blood cells, or hemocytes, mediate innate immunity, removal of apoptotic cells, wound healing and secretion of extracellular matrix (Evans et al., 2003; Lemaitre and Hoffmann, 2007; Wood and Jacinto, 2007; Brock et al., 2008; Dushay, 2009). Many molecular and cellular aspects of hemocyte development and responses are well conserved between Drosophila and vertebrates (Evans et al., 2003; Hartenstein, 2006; Lemaitre and Hoffmann, 2007; Martinez-Agosto et al., 2007). In Drosophila, undifferentiated prohemocyte progenitors and three differentiated blood cell lineages are being distinguished (Rizki, 1978; Shrestha and Gateff, 1982; Lanot et al., 2001; Evans et al., 2003; Hartenstein, 2006). Macrophages, or plasmatocytes, that express Peroxidasin (Pxn) (Nelson et al., 1994; Tepass et al., 1994; Stramer et al., 2005) and Hemolectin (Hml) (Goto et al., 2001; Sinenko and Mathey-Prevot, 2004), correspond to vertebrate myeloid cells and represent 90-95% of hemocytes at most developmental stages (Tepass et al., 1994; Lanot et al., 2001; Evans et al., 2003). Crystal cells and lamellocytes are invertebrate-specific, minor cell types limited to specific developmental stages and immune challenges (Lanot et al., 2001; Evans et al., 2003; Crozatier et al., 2007). Intermediate committed progenitors of the above lineages have been proposed, yet their molecular markers await further characterization (Rizki, 1978; Fossett and Schulz, 2001; Evans and Banerjee, 2003; Kurucz et al., 2007).
Drosophila hematopoiesis, just like vertebrate hematopoiesis, occurs in several waves (Evans et al., 2003; Hartenstein, 2006; Cumano and Godin, 2007; Martinez-Agosto et al., 2007; Bertrand and Traver, 2009). In the embryo, hemocytes are specified in the procephalic mesoderm by expression of serpent (srp) (Rehorn et al., 1996), and emerge as undifferentiated prohemocytes that undergo four rapid cell divisions during embryonic stages 8-11 (Tepass et al., 1994). Subsequently, these cells stop proliferating and switch to a differentiation program, leading to a fixed number of 600-700 hemocytes, comprising at least 80-90% differentiated plasmatocytes in the late embryo (Tepass et al., 1994). Lymph gland (LG) hematopoiesis is another major wave of blood cell development (Lanot et al., 2001; Jung et al., 2005; Crozatier et al., 2007). However, with the exception of responses to acute parasite infestations, the LG represents a reservoir of hemocytes that become active during metamorphosis and do not play a major role in the larva (Lanot et al., 2001; Krzemien et al., 2007).
Post-embryonic larval hematopoiesis has remained incompletely understood. Previous reports described a population of posterior dorsal-vessel-associated sessile hemocytes as a hematopoietic compartment that is actively involved in larval immunity (Shrestha and Gateff, 1982; Stofanko et al., 2008; Markus et al., 2009), and cell transplantation experiments suggested persistence of embryonic hemocytes (Holz et al., 2003). Because these reports left important open questions regarding the lineage, anatomical locations and inductive microenvironments, we systematically investigated the biology of the Drosophila larval hematopoietic system.
Using genetic and cell biological approaches, we find that the differentiated plasmatocytes of the embryo persist into larval stages, colonize resident hematopoietic sites and expand to form the larval hematopoietic system, exemplifying a rare case of self-renewal of differentiated cells. We demonstrate that the larval hematopoietic compartment is organized in anatomically secluded, segmentally repeated epidermal-muscular pockets, and show that proliferation is proprietary to hemocytes in resident locations. Most importantly, we establish an essential role of the peripheral nervous system (PNS) as an attractive and trophic microenvironment for resident hemocytes. Our findings draw parallels with vertebrate hematopoiesis regarding blood cell colonization and the emerging role of the PNS in the control of hematopoiesis.
MATERIALS AND METHODS
Drosophila strains
W1118 or yw were used as wild type. ato1 (Jarman et al., 1993) homozygotes were identified by absence of TM3 Kr-GAL4, UAS-GFP. UAS/GAL4 system (Brand and Perrimon, 1993) and other lines used were 2-38-GAL4 (Rothenfluh et al., 2006; Corl et al., 2009), elav-GAL4 (Lin and Goodman, 1994), 21-7-GAL4 (Song et al., 2007), repo-GAL4 (Sepp et al., 2001), gliotactin-GAL4 (Sepp and Auld, 1999), en-GAL4 (FlyBase), He-GAL4 (Zettervall et al., 2004), HmlΔ-GAL4 (Sinenko and Mathey-Prevot, 2004), Pxn-GAL4 (Stramer et al., 2005), srpHemo-GAL4 (Brückner et al., 2004), Sal-GAL4 (Thomas et al., 1995), UAS-S/G2/M-Green fucci (Sakaue-Sawano et al., 2008; Nakajima et al., 2010), UAS-EGFP (Halfon et al., 2002), UAS-mCD8 GFP (Lee and Luo, 1999), UAS-Stinger (Barolo et al., 2004), UAS-DN EGFR (A. Michelson, personal communication to FlyBase), UAS-Dt1 (Han et al., 2000), UAS-Rho1.N19 (U. Weber and M. Mlodzik, personal communication to FlyBase), UAS-sc (A. Parks and M. Muskavitch, personal communication to FlyBase), tubP-GAL80ts (McGuire et al., 2003) and svpAE127-lacZ (Mlodzik et al., 1990). GAL4 drivers were recombined with UAS-EGFP to visualize expression patterns. Unless stated otherwise, all genetic crosses were carried out at 25°C.
Generation of transgenic lines
UAS-EOS-FP transgenics were generated by PCR-amplifying EOS-FP (MoBiTec), incorporating EcoRI and XhoI restriction sites to the 5′- and 3′-end, respectively. The Kozak sequence CAAA was introduced in front of the start codon of the EOS-FP sequence. The amplified and digested PCR product was cloned into a digested pUAST vector. Primer sequences for EOS-FP were (restriction site sequences in lower case): Forward, cggaattccaaaATGAGTGCGATTAAGCCAGACATGAAG; Reverse, ccctcgagTTATCGTCTGGCATTGTCAGGCAATC.
HmlΔ-DsRed transgenics were generated by PCR amplifying the truncated HmlΔ promoter sequence (Sinenko and Mathey-Prevot, 2004) from genomic DNA, incorporating Asp718 and BamHI restriction sites to the 5′ and 3′-end, respectively. The amplified and digested PCR product was cloned into a digested pRed H-Pelican vector. Primer sequences for HmlΔ were (restriction site sequences in lower case): Forward, ggggtaccCAAAAGTTATTTCTGTAGGC; Reverse, cgggatccTTTGTTAGGCTAATCGGAAATTG. Two independent insertions on the second chromosome were recombined to gain a bright signal. The same strategy was used to clone HmlΔ into pRed H-Stinger for generating HmlΔ-DsRednls.
Transgenic lines were generated by standard techniques (Ashburner et al., 2004) (Rainbow Transgenic Flies).
EOS-FP tracing, genetic cell ablation and lacZ lineage tracing
Larvae in a 96-well clear bottom plate with a drop of water, or dechorionated embryos in a drop of halocarbon oil, were photoconverted by ultraviolet light (UV) for 8 minutes (larvae) or 5 minutes (embryos) using a 5× objective of a Leica DMI3000 microscope. Limited photoconversion in embryos (to avoid lethality), and some larval experiments, efficiently labeled all hemocytes, yet left a low level of residual green fluorescence. For local photoconversion, larvae were immobilized on a glass slide using double-sided tape. Part of the larva was protected from UV by using black tape, narrowing the field diaphragm and limiting UV exposure to 1-2 minutes. For selective photoswitch of single lateral patches, FRAP Wizard on a Leica SP5 confocal microscope was used, exposing the selected region of interest to 45 bleach cycles (2.6 seconds each, 100% of 405 laser line, 20× objective). Larvae were handled as gently as possibly to avoid mechanical dislodging of resident hemocytes.
For genetic cell ablation experiments, tubP-GAL80ts (McGuire et al., 2003) was used to gain temporal control over 2-38-GAL4. Embryos were collected over 24 hours, aged for 3 days at 18°C, shifted to 29°C for 21 hours to induce toxin expression, and returned back to 18°C for 3-4 days to allow for clearance of apoptotic corpses prior phenotypic analyses.
For flipout-lacZ lineage-tracing (Weigmann and Cohen, 1999) of embryonic hemocytes, we crossed tub GAL80ts; UAS-Flp × SrpHemo-GAL4, UAS-srcEGFP; Act>stop>nuc-lacZ. Embryo collections (1 hour) were aged for 2 hours at 25°C and subsequently shifted to 29°C for 5 hours to induce Flp expression. Expression by srpHemoGAL4 was monitored by a co-expressed UAS-srcEGFP, selecting only larvae that showed GFP expression in the embryo, but complete absence of GFP fluorescence in the 1st instar. ‘No heat shock’ controls were continuously maintained at 18°C. X-gal staining was performed as described previously (Ghysen and O’Kane, 1989).
Live imaging, staining and microscopy
Immunostaining was carried out using goat anti-GFP (Rockland), mouse anti-H2 (Kurucz et al., 2007), DyLight 488 anti-HRP (Jackson), rat anti-Elav [Developmental Studies Hybridoma Bank (DSHB)], Alexa Fluor 488-phalloidin (Invitrogen) and secondary antibodies conjugated to Alexa dyes (Invitrogen). Imaging was performed on Leica DMI3000, Leica M205FA, Leica SP5 or Zeiss Axioimager Z1 microscopes.
For cryosections, 3rd instar larvae were embedded using OCT medium in 7×7×5 mm disposable molds (Ted Pella), and snap frozen. Transverse sections of 20 μm thickness were cut using a Leica 3050 S Cryostat at 15-16°C and placed on Superfrost Plus slides (Fisher). Sections were fixed in 4% paraformaldehyde (PFA) for 10 minutes, permeabilized in 0.1% Triton X-100 and 3% bovine serum albumin (BSA) for 3 minutes, followed by staining with phalloidin and DAPI, and mounting in Prolong Gold (Invitrogen).
For larval fillet preps, 3rd instar larvae were pinned down on Sylgard plates and ventrally filleted in a drop of chilled PBS. Gut, fat body and trachea were removed, and the fillet was fixed for 20 minutes in 4% PFA. Fillets were washed in PBS, unpinned, permeabilized and blocked in 0.3% Triton X-100 and 3% BSA, and immunostained overnight with gentle rocking.
For in situ and live fluorescent imaging, a Leica M205FA with motorized focus was used. Larvae were immobilized on a cold metal block. z-stacks were combined into single composite images using the Leica LAS Montage module.
For detailed live imaging, using a Leica SP5 confocal microscope, larvae were immobilized by brief anesthesia with ether (Sigma), positioned on double-sided tape and re-etherized for 10-15 minutes. For 40× imaging, larvae were mounted in halocarbon oil with coverslips that were placed on spacers.
For combined fluorescence and differential interference contrast (DIC) microscopy, whole live larvae and larval fillet preparations of heat-fixed larvae (1 minute at 60°C) were visualized using z-stack functions on a Zeiss AxioImager Z1 motorized microscope.
For cell disturbance experiments, larvae were immobilized on double-sided tape (Scotch Exterior Mounting Tape, as used in VW manipulations) and the hemocyte pattern was manipulated using a paint brush. For 150× time lapse, imaging intervals of 15 seconds for a total duration of 1 hour were used (Leica LAS modules). Final movies were assembled at 10 frames/second using iStopMotion (Boinx).
In all images anterior is left, in lateral images dorsal is up. Images and movies were further processed using Adobe Photoshop, iMovie and Final Cut Express.
Ex-vivo cell analyses
Hemocytes were released by dissecting larvae in a drop of Schneider’s medium (Invitrogen). To bleed circulating hemocytes, larvae were opened at the anterior end and hemolymph was allowed to leak out, applying only gentle pressure. To avoid dislodging of resident hemocytes or contamination with LG hemocytes, larvae were monitored through a fluorescence stereomicroscope. Resident hemocytes were released by opening the remainder of the larva and scraping the body wall with a needle under microscope guidance, avoiding the LG. Cells were allowed to attach to glass slides for 15-30 minutes. Glass slides were pre-treated with 0.5 mg/ml ConcanvalinA (Sigma).
For EdU labeling, larvae were fed 25 mM 5-ethynyl-2′deoxyuridine (EdU) in fly food for 4 hours, or as indicated, at 22°C. Click-iT EdU staining was performed on released cells or dissected larval tissue according to the manufacturer’s instructions (Invitrogen).
For bead phagocytosis followed by EdU incorporation, larvae 96 hours after egg laying (AEL) were capillary injected with 69 nl of 1:200 dilution of red fluorescent latex beads (0.2 μm, Invitrogen) using a Drummond NanojectII injector. Injected larvae were kept on fly food for 3-4 hours to allow phagocytosis of injected beads. Subsequently, larval hemocytes were released, allowed to adhere washed and incubated with 100 μM EdU for 2 hours.
TUNEL labeling was performed on released hemocytes using an In Situ Cell Death Detection Kit (Roche) following the manufacturer’s instructions.
For all ex vivo assays, hemocytes of multiple larvae were used: 3-4 (or 6-8 for backgrounds with reduced hemocyte numbers) larvae for 96 hours AEL, 6-8 for 72 hours AEL, 10-12 for 48 hours AEL. Unless noted otherwise, ex vivo experiments used all larval, i.e. circulating and resident, hemocytes. For in vivo cell counts, 3-5 larvae per condition were assessed. All experiments were confirmed in triplicate. Standard deviations were calculated, and two-tailed two-sample equal variance t-testing was performed using at least two out of three independent experiments.
Larval staging
Larvae were obtained from 1-2 hour embryo collections and aged for time points 24-96 hours AEL at 25°C. Classification as 1st, 2nd or 3rd instar larvae was based on the number of teeth on the mandibular hooks (Demerec, 1994). Age 23-50 hours AEL corresponded to 1st instar, 50-75 hours AEL to 2nd instar, and 75-120 hours AEL to 3rd instar.
RESULTS
Segmental organization of the larval hematopoietic system
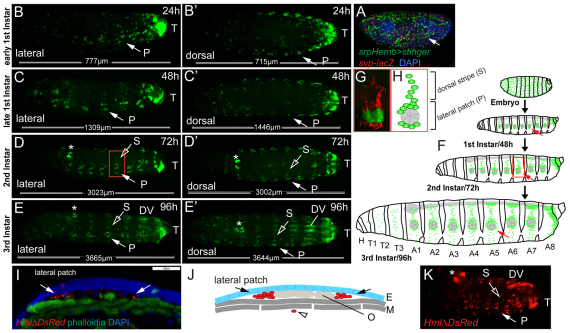
In the embryo, hemocytes are widely dispersed and show no obvious compartmentalization (Fig. 1A). By contrast, studying larval hemocytes marked by GFP under control of the plasmatocyte driver Pxn-GAL4 (Stramer et al., 2005), we found that already in the earliest 1st instar larvae, hemocytes retreat to the terminal segment, and seven doughnut-shaped patches at the lateral midline on each side of abdominal segments A1-A7, to which we will refer as ‘lateral patches’ (Fig. 1B, arrow). Over the 2nd and 3rd larval instars, additional ‘dorsal stripes’ of hemocytes develop, that extend from the lateral patches (Fig. 1D-H). Late during larval life, from 96 hours AEL onwards, hemocytes start to accumulate in segmented clusters along the posterior dorsal vessel (Fig. 1E-F). On the ventral side, stripes of hemocytes appear even later, in wandering stage 3rd instar larvae (115 hours AEL).
Fig. 1.
Organization of larval hemocytes in epidermal-muscular pockets. (A) Stage 17 embryo (srpHemoGAL4, UAS Stinger/+; svpAE127-lacZ/+) showing hemocytes (green), oenocytes (red) and nuclei (DAPI, blue). (B-E′) Time course and hemocyte pattern of Pxn-GAL4, UAS-GFP larvae. Bars indicate actual larval size. (B,B′) Early 1st instar, 24 hours AEL. (C,C′) Late 1st instar, 48 hours AEL. (D,D′) Late 2nd instar, 72 hours AEL. (E,E′) Mid 3rd instar, 96 hours AEL. Arrows indicate lateral patch in segment A5. P, lateral patch; S, dorsal stripe; T, terminal cluster; DV, dorsal-vessel-associated cluster. (F) Schematic (not to scale, lateral view) of larval development (see Hartenstein, 1993). Hemocytes (green), oenocyte clusters (pie-shaped, gray). (G) Lateral patch and dorsal stripe, ∼80 hours AEL, salGAL4,HmlDsRed; UAS-GFP. Oenocytes (green), hemocytes (red). (H) Corresponding area in schematic. (I) Cross-section of an epidermal-muscular pocket, lateral patch, 96 hours AEL. Phalloidin labels muscle layers (green), DAPI and autofluorescence mark nuclei and cuticle (blue). Hemocytes are shown in red (HmlΔDsRednls, arrows). Scale bar: 50 μm. (J) Schematic of panel I showing hemocytes (red, arrows) sandwiched between epidermis (E, blue) and muscle layers (M, dark gray), oenocytes (O, light gray) and a circulating hemocyte (open arrowhead). (K) Bled-out larva, HmlΔDs-Red, 96 hours AEL in which resident hemocytes remain. Asterisk indicates LG.
The distribution of the lateral patches is reminiscent of oenocyte clusters, groups of six or seven large hepatocyte-like cells located along the lateral midline of larval segments A1-7 (Gutierrez et al., 2007). To differentially mark hemocytes and other tissues, we generated hemocyte reporters HmlΔ-DsRed and HmlΔ-DsRednls, which essentially match the expression pattern of the previously described HmlΔ-GAL4 (Sinenko and Mathey-Prevot, 2004) (supplementary material Fig. S1). Combining HmlΔ-DsRed with a driver that marks oenocytes, we found that, indeed, lateral hemocyte patches directly surround oenocyte clusters, and hemocytes are at times interspersed between oenocytes (Fig. 1G,H).
To determine the position of hemocytes and oenocytes relative to the layers of the larval body wall, we examined cryosections of larvae with HmlΔ-DsRednls-marked hemocytes, and studied larvae using fluorescence and DIC microscopy (Fig. 1I; supplementary material Movie 1). Resident hemocyte clusters are secluded from the open hemocoel, being sandwiched between the epidermal and muscle layers of the larva. We will refer to these locations as ‘epidermal-muscular pockets’ (Fig. 1J). Consistently, when we released hemocytes from larvae by gentle bleeding, resident hemocytes were retained with the larval tissues (Fig. 1K). The ability to release hemocytes by bleeding correlated with larval age, i.e. 1st and early 2nd instar bleeds yielded barely any Pxn-positive hemocytes, suggesting lower fractions of circulating hemocytes.
The larval hemocyte pattern was confirmed using additional hemocyte markers. Comparing HmlΔ-DsRed with Pxn-GAL4,UAS-GFP and He-GAL4,UAS-GFP, a GAL4 driver expressed in 80% of all larval hemocytes (Kurucz et al., 2003; Zettervall et al., 2004), all markers labeled largely overlapping sets of hemocytes in 3rd instar larvae (supplementary material Fig. S1). To determine whether young larvae contain a significant population of undifferentiated prohemocytes, we studied 1st instar larvae co-expressing HmlΔDsRed and srpHemo-GAL4, an early hemocyte driver (Brückner et al., 2004). Only a minimal fraction of cells were single positive for srpHemoGAL4 (supplementary material Fig. S1H,I), suggesting absence of substantial numbers of undifferentiated prohemocytes. Likewise, comparing HmlΔDsRed expression with the pan-circulating hemocyte marker H2 (Kurucz et al., 2007), showed expression in almost identical hemocyte populations (supplementary material Fig. S1G,J). Co-expression with the posterior compartment marker engrailed-GAL4 (en-GAL4) demonstrated localization of resident hemocytes to the anterior compartment of the larval segments (supplementary material Fig. S1D).
In summary, larval hemocytes retreat to anatomically secluded resident locations in epidermal-muscular pockets of the larval body wall, segregating resident and circulating hemocyte populations. The larval hematopoietic system shows no indication of large prohemocyte reservoirs, and larval hemocytes display mostly overlapping patterns of several known plasmatocyte markers.
Dynamics of resident hemocyte clusters
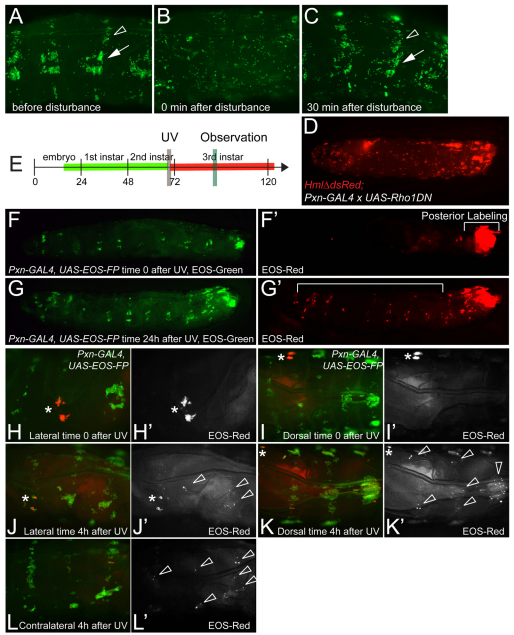
Previous studies investigated the dynamics of circulating-, dorsal-vessel-associated-, and terminal cluster hemocytes of the larva (Babcock et al., 2008; Welman et al., 2010). Focusing on epidermal-muscular pockets, we found that physical manipulation can perturb the pattern of resident hemocytes, which spontaneously re-formed within 30-60 minutes, independently of new hemocyte formation (Fig. 2A-C; supplementary material Movie 2; data not shown). Time-lapse microscopy demonstrated that, after disturbance, floating hemocytes re-attached and subsequently moved laterally, progressively consolidating into lateral patches and dorsal stripes over time (supplementary material Movie 2), consistent with ‘homing’ of hemocytes to their resident locations. Expression of dominant-negative Rho1 in larval hemocytes suppressed formation of the pattern (Fig. 2D), suggesting involvement of the actin cytoskeleton.
Fig. 2.
Dynamics of resident hemocyte clusters. (A-C) Re-formation of the resident hemocyte pattern after disturbance. Lateral view of larva HmlΔ-GAL4, UAS-GFP, 96 hours AEL. (A) Before disturbance. (B) Time 0 after disturbance. (C) Same segments, 30 minutes after disturbance. Arrow, lateral patch; arrowhead, dorsal stripe. (D) Expression of dominant-negative Rho1 in hemocytes. 96 hours AEL, HmlΔdsRed;Pxn-GAL4 × UAS-Rho1DN. (E-L′) EOS-FP tracing of local hemocyte populations, Pxn-GAL4, UAS-EOS-FP. (E) Timeline of posterior EOS-FP tracing (corresponding to F-G′). Vertical bars mark UV induction and observation. (F,F′) Time 0 after UV. (G,G′) Same larva 24 hours after UV. Red hemocytes can be found in areas anterior to the original photoconversion area (bracket). (H-I′) EOS-FP tracing of one individual lateral patch (asterisk), segment A4, 96 hours AEL. (H-I′) Time 0 after UV; (H,I) combined channels; (H′,I′) red channel only. (J-L′) Same larva at 4 hours after UV. Labeled hemocytes have moved to various lateral patches and dorsal stripes on the same and contralateral sides, and to dorsal-vessel-associated clusters (arrowheads). (J,K,L) combined channels; (J′,K′,L′) red channel only.
To determine whether hemocytes would exchange between resident locations, we used EOS-FP tracing in live larvae. EOS-FP is a color-switch fluorescent protein that fluoresces green in its native state, and by exposure to UV is photoconverted to a red fluorescent form, which is very stable and well detected even 24 hours after the color-switch (Wiedenmann et al., 2004). We generated the transgenic line UAS-EOS-FP and expressed it under control of Pxn-GAL4. Larvae were asymmetrically labeled, UV photoconverting only the most posterior part (Fig. 2E-F′). After 24 hours, we re-examined the localization of EOS-red cells and found that tail-derived hemocytes had reached complementary areas, i.e. lateral patches and dorsal stripes of the anterior half of the larva, and dorsal-vessel-associated clusters (Fig. 2G,G′). Using a laser to precisely photoswitch hemocytes of a single lateral patch without affecting circulating hemocytes (Fig. 2H-I′), we found that, already at 4 hours post labeling, hemocytes had moved to various lateral patches and dorsal stripes of the same and the contralateral side, and dorsal-vessel-associated clusters, preferably reaching areas between the originating segment and the posterior end of the larva (Fig. 2J-L′). This dynamic exchange was repeatedly observed at various larval instars. Interestingly, hemocyte dynamics seemed further elevated at stages when the majority of hemocytes is resident (e.g. 48 hours AEL, not shown).
Altogether, establishment of the larval resident hemocyte pattern involves hemocyte attachment, some form of lateral movement of hemocytes and possibly active cytoskeletal processes. At least a fraction of the hemocytes in resident locations are in a dynamic steady state, leaving their locations and joining hemocyte clusters at distant places.
Larval hemocytes expand by self-renewal in the differentiated state
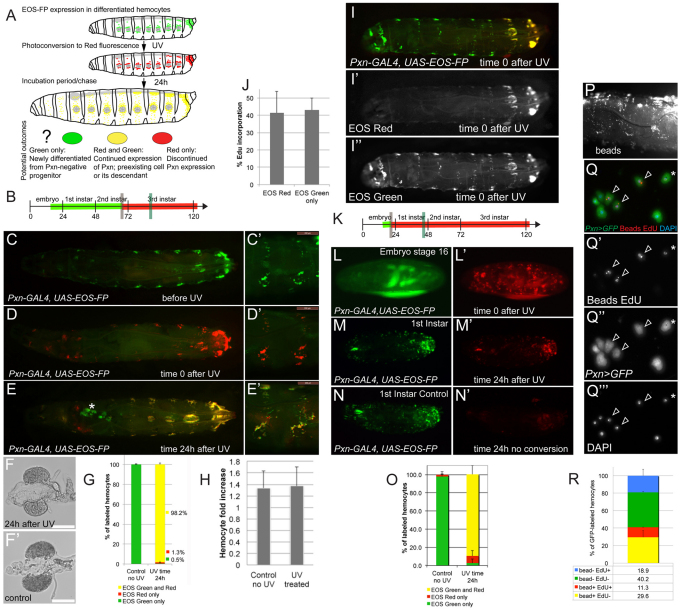
Next, we investigated the lineage, origin and proliferative capacity of larval hemocytes. Performing flipout-lacZ lineage tracing (Weigmann and Cohen, 1999), we confirmed that larval hemocytes derive from embryonic hemocytes (supplementary material Fig. S2A-D). Assessing hemocyte counts over the course of larval development (supplementary material Fig. S2E) illustrated hemocyte increase from <400 in the early 1st instar to >6000 in wandering 3rd instar larvae. Consistently, in vivo labeling by the thymidine nucleoside analog EdU showed high incorporation rates, suggesting hemocyte proliferation throughout larval life (supplementary material Fig. S2F).
To address which blood cell lineage provides the founders of the larval hematopoietic system, i.e. whether larval hemocytes differentiate from a prohemocyte or derive from another source, we again utilized EOS-FP-tracing. Here, we took advantage of the fact that expressing EOS-FP exclusively in differentiated plasmatocytes under the control of Pxn-GAL4 makes it possible to distinguish between descendants of these differentiated hemocytes (marked by EOS-red), or newly differentiated hemocytes deriving from an undifferentiated, i.e. Pxn-negative, prohemocyte (marked by absence of EOS-red). We photoconverted larvae or embryos at time 0, and examined their hemocytes 24 hours later, quantifying the fractions of green and red cells ex vivo (Fig. 3A-O). Labeling late 2nd instars and analyzing early 3rd instars, we found that, surprisingly, all hemocytes that were EOS-green also carried the red label (Fig. 3B-E,G). To determine whether these hemocytes were daughters of the originally labeled hemocytes, rather than the same cells that did not divide over time, we assessed the expansion of EOS-labeled hemocytes after photoconversion. Comparing EOS-red hemocytes in UV-treated larvae, with EOS-green hemocytes in non-UV controls, we found increases in hemocyte numbers of comparable magnitude (Fig. 3H), indicating that the experimental conditions did not affect the ability of hemocytes to divide. In the LG, we found ‘EOS-green-only’ hemocytes in a number of larvae (Fig. 3E). In these larvae, LG hemocytes were not fully differentiated and, consequently, did not show Pxn-GAL4 driven EOS-FP expression at the time of photoconversion (Fig. 3C), yet LG hemocyte differentiation proceeded within the 24 hour incubation, resulting in green-only hemocytes (Fig. 3E). When extracting larval hemocytes from these larvae (omitting the LG proper), we did not find any ‘green-only’ hemocytes, indicating that LG hemocytes did not contribute to the pool of larval hemocytes assessed in this assay (Fig. 3G). Consistently, LGs of larvae exposed to UV and incubated for 24 hours did not show any signs of premature disintegration (Fig. 3F,F′). In a ‘partial switch’ experiment in which only the posterior end of the larva was UV treated, thereby avoiding labeling of LG hemocytes, we found that EOS-red labeled hemocytes and non-UV switched ‘EOS-green-only’ hemocytes had comparable abilities to incorporate EdU, once more ruling out the possibility that the observed hemocyte expansion relates to a contribution of LG hemocytes (Fig. 3I-J). Altogether, this suggests that larval hemocytes derive from the Pxn-positive, pre-existing population of differentiated larval hemocytes, which expand by proliferation.
Fig. 3.
Larval hemocytes expand in the differentiated state. (A) Schematic of EOS-FP tracing of differentiated hemocytes, illustrating possible outcomes. (B) Timeline of EOS-FP labeling in the larva, corresponding to panels C-E′, marking UV switch (gray vertical bar) and observation (green vertical bar) times. (C-E′) Pxn-GAL4, UAS-EOS-FP. Dorsal views of a whole larva (C-E) and lateral close up of two segments (C′-E′) over time. (C,C′) Before UV. (D,D′) Time 0 after photoconversion. (E,E′) 24 hours after photoconversion, asterisk marks LG. There is incomplete overlap of channels due to minor movement of the larva. Anterior red areas in E reflect nonspecific expression in gastric cecae and proventriculus. (F) LG of UV-switched larva 24 hours after UV. (F′) LG of non-UV control. (G) Ex vivo quantification of EOS fluorescence in hemocytes of larvae similar to those shown in C-E′. (H) Fold increase in larval hemocyte numbers in EOS-red photoconverted hemocytes and EOS-green hemocytes from non-UV controls over the period of a 24-hour experiment. (I-J) Local UV photoconversion of the posterior end of a 3rd instar larva, followed by EdU feeding for 4 hours. (I-I′) Photoconverted larva. (J) EdU incorporation rates in EOS-red (i.e. UV-treated) and EOS-green-only (i.e. non-UV treated) hemocytes. (K) Timeline of EOS-FP labeling of stage 16 embryos. (L,L′) Time 0 after UV. Residual EOS-green and some autofluorescence in green channel. (M,M′) 1st instar larva ∼46 hours AEL from embryo in L. (N,N′) Non-UV control larva processed in parallel, 46 hours AEL. (O) Quantification of EOS-FP-green and -red in released hemocytes of experiment and non-UV controls shown in M-N′. (P-R) Consecutive phagocytosis and EdU incorporation assay. (P) HmlΔ-GAL4, UAS-GFP 3rd instar larvae injected with fluorescent latex beads, lateral view of live larva. (Q-Q′′) Released hemocytes from larva shown in P, after ex vivo EdU incorporation showing hemocytes (GFP, green), EdU (red, nuclear stain), fluorescent beads (red, cytoplasmic inclusions) and nuclear DAPI (blue). (R) Quantification of EdU incorporation in bead-positive and bead-negative hemocytes. Error bars represent s.d.
To determine whether the same was true for embryonic hemocytes transitioning into the larval stages, we repeated the experiment, photoconverting stage 16 embryos and observing 1st instar larvae the next day (Fig. 3K-O). Again, we found that the vast majority of EOS-green hemocytes was also positive for the red label, indicating that the majority of larval hemocytes derive directly from the Pxn-positive, differentiated population of embryonic hemocytes, rather than from undifferentiated prohemocytes.
To confirm that the proliferating Pxn-positive hemocytes of the larva are indeed fully differentiated plasmatocytes, we tested the capacity of Pxn-positive hemocytes to phagocytose and subsequently incorporate EdU (Fig. 3P-R). In this experiment, we first injected fluorescent latex beads into larvae, followed by an ex vivo EdU incorporation assay. Indeed, a large proportion of hemocytes were double-positive for phagocytosed beads and the EdU label, and EdU ratios were similar between the groups of bead-positive and bead-negative hemocytes (Fig. 3Q-R).
In summary, we conclude that late embryonic plasmatocytes, positive for the differentiation marker Pxn, constitute the majority of hemocytes of the 1st larval instar, thereby founding the larval hematopoietic system. Over the course of larval development, hemocytes proliferate while being committed to the plasmatocyte fate and showing signs of full differentiation, such as their readiness to phagocytose.
Larval hemocytes proliferate in resident clusters
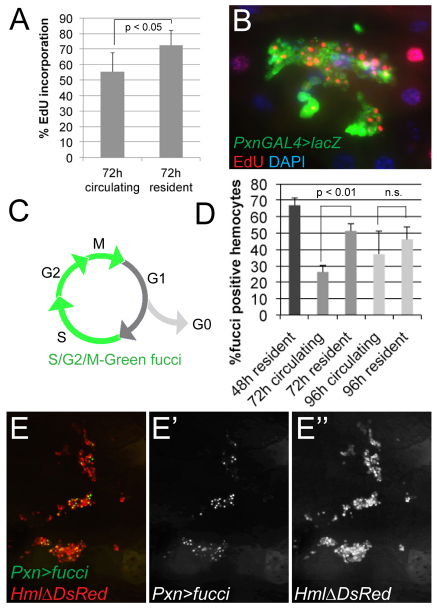
Considering the anatomical compartmentalization and expansion of larval hemocytes, we determined whether hemocyte proliferation might correlate with certain microenvironments, such as the epidermal-muscular pockets. Indeed, when assessing in vivo EdU incorporation of resident versus circulating hemocytes (Fig. 4A,B), we found that resident hemocytes incorporated EdU at a higher rate than circulating hemocytes, suggesting elevated levels of proliferation (Fig. 4A). Examining resident hemocytes in situ, we observed a large proportion of EdU-positive hemocytes in the lateral patches and terminal segment (Fig. 4B; data not shown).
Fig. 4.
Hemocytes proliferate in resident sites. (A) In vivo EdU incorporation rates of circulating and resident hemocytes marked by HmlΔ-GAL4, UAS-GFP, omitting the developing LG. Larvae 72 hour AEL fed with EdU for 4 hours. (B) In vivo EdU incorporation in a lateral patch of an early 3rd instar, EdU-fed for 2 hours. EdU (red), Pxn-GAL4, UAS-GFP/UAS-lacZ (stained for β-Gal, green), DAPI (blue). (C-E′) In vivo labeling of proliferating hemocytes using UAS-S/G2/M-Green fucci. (C) S/G2/M-Green emits green fluorescence in the SG2-M phases of the cell cycle; quiescent cells in G1 are negative for fucci fluorescence. (D) HmlΔ-DsRed/CyO; Pxn-GAL4, UAS-S/G2/M-Green. Fucci-positive hemocytes among HmlΔ-DsRed positive circulating and resident hemocyte populations. At 48 hours AEL, all hemocytes are resident correlating with high fucci-positive rates. At 96 hours AEL, differences between resident and circulating hemocytes become insignificant, possibly owing to increased mobility and exchange between the hemocyte populations. (E-E′) Lateral hemocyte patches of a late 2nd instar larva. Pxn-GAL4, UAS-S/G2/M-Green (green), HmlΔ-DsRed (red). Error bars represent s.d.
To complement these experiments, we used S/G2/M-Green fucci (fluorescent ubiquitination-based cell cycle indicator) (Sakaue-Sawano et al., 2008) to quantify hemocyte proliferation in live larvae. UAS-S/G2/M-Green (Nakajima et al., 2010) shows fluorescence during the S/G2/M phases of the cell cycle, and absence of fluorescence during G1, corresponding to cell cycle quiescence (Fig. 4C). Separating the hemocyte populations, we observed significantly higher fucci positive rates in resident versus circulating hemocytes (Fig. 4D-E′).
Taken together, this indicates that hemocytes located in epidermal-muscular pockets are actively proliferating, whereas proliferation rates are reduced in circulating hemocytes. It further suggests that the microenvironment surrounding resident hemocytes provides inductive factors that support hemocyte expansion.
Larval hemocytes depend on the PNS
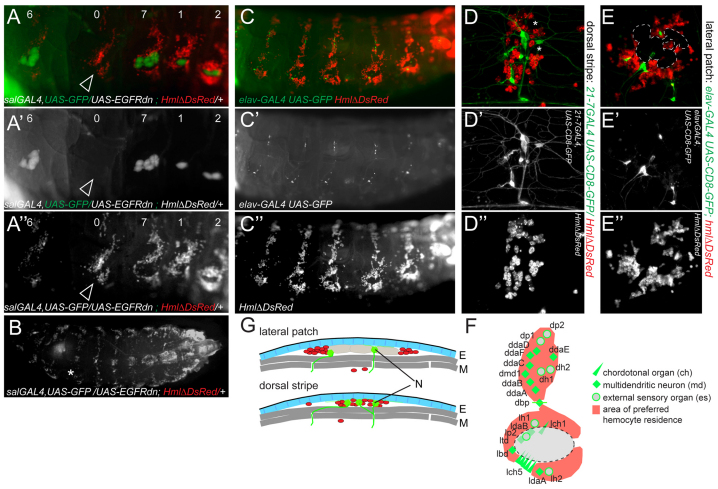
Next, we investigated which tissue provides a microenvironment for resident hemocytes in epidermal-muscular pockets. First, we considered whether oenocytes might play a role. Suppressing the specification of oenocyte fate by inhibition of EGFR (epidermal growth factor receptor) signaling in embryonic progenitors, we obtained larvae with an occasional complete loss of oenocytes in certain segments (Fig. 5A-B). Despite the lack of oenocytes, hemocytes still resided as segmental lateral patches and dorsal stripes (Fig. 5A-A′). Lateral patches changed their outline from the typical doughnut shape to a collapsed sigmoid shape, suggesting that oenocytes usually prevent hemocytes from accessing an area of preferred residence or elevated attraction. Interestingly, the collapsed sigmoid shape resembled the pattern of lateral peripheral neuron clusters, which show a stereotyped organization in abdominal segments A1-A7 and are known to overlap in location with oenocyte clusters (Fig. 5F) (Bodmer et al., 1989).
Fig. 5.
Larval hemocytes colocalize with the PNS. (A-B) Oenocytes are dispensable for hemocyte attraction. Suppression of oenocyte specification by UAS-EGFRdn under control of sal-GAL4, visualization of remaining oenocytes by UAS-GFP. Numbers indicate the number of remaining oenocytes per segment. Note one segment that completely lacks oenocytes, yet hemocytes still accumulate (arrowhead). (A-A′) Lateral views. (B) Dorsal view, asterisk marks ‘0’ oenocyte segment. (C-G) Larval hemocytes co-localize with the PNS. (C-C′) Co-labeling of neurons (elav-GAL4, UAS-CD8-GFP in green) and hemocytes (HmlΔ-DsRed in red) in live larvae. (D-D′) Dorsal stripe, 21-7-GAL4, UAS-CD8-GFP/HmlΔDs-Red. Hemocytes (red), md neurons (green). Asterisks mark approximate positions of unlabeled es neurons (see also F). (E-E′) Lateral patch. elav-GAL4, UAS-CD8-GFP; HmlΔ-DsRed. Hemocytes localize next to lateral chordotonal organs; oenocytes marked by dashed outline. (F) Model of lateral and dorsal PNS neuron clusters and areas of hemocyte attraction (red) in abdominal segments A1-A7. External sense organs (es) and chordotonal organs (ch) are depicted by just one symbol each, representing neurons plus their accompanying support cells. (G) Model of hemocyte-PNS colocalization in the lateral patch and dorsal stripe areas. Hemocytes (red), neurons (N, green), epidermis (E, blue), oenocytes (light gray), muscle layers (M, dark gray).
We therefore investigated whether the PNS might interact with hemocytes. Examining live larvae double-labeled for neurons and hemocytes, we found colocalization of Elav-positive neurons with lateral hemocyte patches, dorsal hemocyte stripes and the terminal hemocyte cluster (Fig. 5C-E′; data not shown). Confocal microscopy of resident hemocytes revealed striking association of resident hemocytes with neuronal cell bodies, or somata, in particular of multiple dendritic (md) neurons in the dorsal stripe area, and lateral chordotonal organs (lch) (Fig. 5D-E′). In some areas, we observed layering of neuronal extensions with hemocytes (not shown). Neurons in the lateral patch colocalized with hemocytes only partially (Fig. 5E-E′), presumably owing to displacement of hemocytes by the bulky oenocyte clusters (see above and Fig. 5A-A′,F,G). Combined fluorescence and DIC microscopy confirmed the spatial colocalization of neurons and hemocytes in commonly shared epidermal-muscular pockets (Fig. 5G; data not shown).
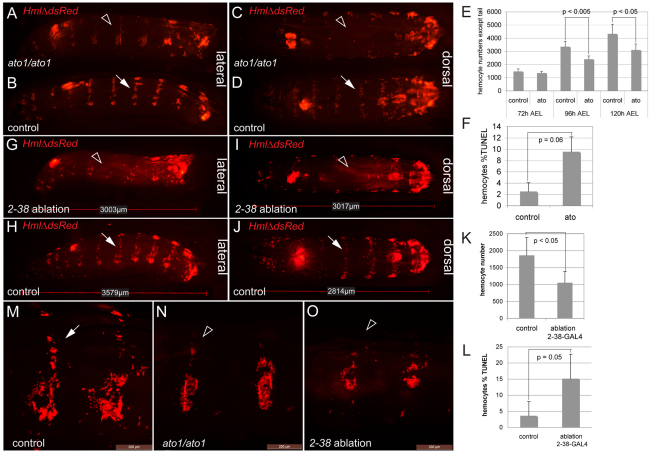
Next, we addressed functional dependence of resident hemocytes on the PNS. We examined hemocytes in atonal (ato) mutants, which lack all chordotonal organs, a few multiple dendritic neurons (v’pda and v’td2), and oenocytes, and are viable to adulthood (Jarman et al., 1993; Jarman et al., 1995). At 96 hours AEL, all ato1 homozygotes showed a defective hemocyte pattern that lacked dorsal stripes (Fig. 6A-D,N). In the lateral patches, the hemocyte pattern reflected absence of oenocytes in ato1 mutants and, in addition, appeared somewhat less organized than in oenocyte suppression experiments (Fig. 5A-A′). Hemocyte numbers progressively declined in ato1 mutants (Fig. 6E) and showed increased rates of TUNEL labeling (Fig. 6F), suggesting that hemocytes of ato1 mutants die apoptotically owing to a lack of survival-promoting factors. In vivo EdU incorporation showed a mild but statistically not significant reduction in hemocytes of ato1 mutants (not shown). Rescue of ato1 mutants by expression UAS-ato under control of Pxn-Gal4 did not improve the hemocyte phenotype (not shown), supporting the hypothesis that hemocyte defects in ato1 mutants are caused by an insufficiency in the hemocyte microenvironment, rather than hemocyte-autonomous functions of ato.
Fig. 6.
The PNS as an attractive and trophic microenvironment for larval hemocytes. (A-D) ato1/ato1 homozygotes and heterozygous controls (early 3rd instar) imaged side-by-side. Dorsal stripes are almost completely absent in ato1/ato1 mutants (A,C, arrowheads) compared with controls (B,D, arrows). Genotypes are HmlΔ-DsRed/+; ato1 / ato1 for ato homozygous mutants and HmlΔ-DsRed/+; ato1 / TM3, Kr-GAL4, UAS-GFP for controls. (E) In vivo hemocyte counts in ato1 homozygotes and yw controls. (F) TUNEL rates of hemocytes from ato1 homozygotes and controls, 96 hours AEL. (G-L) Genetic ablation of peripheral neurons. Genotypes are 2-38-GAL4, UASCD8GFP, HmlΔ-DsRed/tub-GAL80ts; UAS-Dt1/+ for ablation and 2-38-GAL4, UASCD8GFP, HmlΔ-DsRed/+; tub-GAL80ts/+ for controls. (G-J) Lateral and dorsal views of early 3rd instar larvae, 2-38-GAL4 ablation (G,I) and control (H,J). Note absence of dorsal stripes (arrowhead). Dorsal stripe in control (arrow). (K) In vivo hemocyte counts in 2-38-GAL4 ablation and controls, early 3rd instar. (L) TUNEL rates of hemocytes from 2-38-GAL4 ablation and control larvae. (M-O) Lateral areas of control, ato1 and 2-38-GAL4 ablation larvae. Genotypes as above, 3rd instar larvae. Arrow, dorsal stripe in control larva; arrowheads, dorsal stripe in ato1 and 2-38-GAL4 ablation larvae. Scale bars: 200 μm. Error bars represent s.d.
To gain independent evidence for a functional role of the PNS in larval hematopoiesis, we pursued a genetic cell-ablation approach. We induced PNS neuron ablation by expressing UAS-diphteria toxin (Dt1) (Han et al., 2000) under control of 2-38-GAL4 (Rothenfluh et al., 2006; Corl et al., 2009), which is active in chordotonal organs and other neurons of the PNS and CNS (supplementary material Fig. S3). To circumvent embryonic lethality, expression of Dt1 was temporally restricted using tub-GAL80ts (McGuire et al., 2003). Ablation using 2-38-GAL4 led to defects in the pattern, number and survival of hemocytes (Fig. 6G-L,O), phenocopying and exceeding the phenotype observed in ato1 mutants. Hemocyte EdU incorporation rates appeared to be reduced in ablation larvae, but were not statistically significant (not shown). Altogether, these findings suggest functional dependence of the larval hematopoietic system on the PNS microenvironment with respect to hemocyte localization and trophic survival.
Attraction of larval hemocytes by PNS neurons
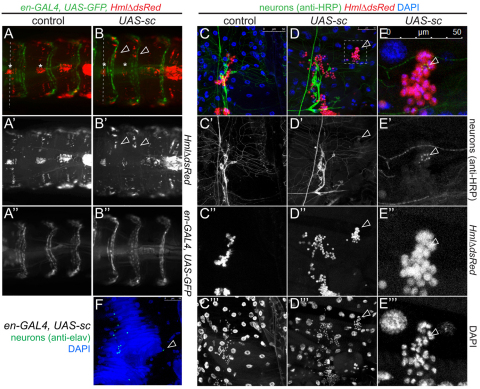
In order to investigate whether hemocytes would be directly attracted by the PNS, we induced supernumerary peripheral neurons by en-GAL4-driven expression of the proneural gene scute (sc) (Chien et al., 1996). en-GAL4 demarcates an area typically devoid of PNS neurons and hemocytes (supplementary material Fig. S1D). Indeed, upon sc overexpression, hemocytes of the dorsal stripe shifted into the en-domain, which was particularly evident in late 3rd instar larvae (Fig. 7A-E′′). Subtle effects were also seen in the lateral hemocyte pattern (not shown). Dorsal-vessel-associated hemocyte clusters were not affected by overexpression of sc, suggesting an alternative mechanism of hemocyte accumulation. To confirm that hemocytes were attracted to ectopic neurons, and not PNS-unrelated cell types in the en-domain, we examined filleted sc-overexpressing larvae using immunohistochemistry (Fig. 7E-E′′). Indeed, sc overexpressors showed small numbers of ectopic neurons positive for the neuronal markers HRP (Jan and Jan, 1982) and Elav (O’Neill et al., 1994) in the dorsal area of the en-domain (Fig. 7D-F), and hemocytes were tightly gathered around these ectopic neurons (Fig. 7D-E′′). This suggests that the PNS, i.e. peripheral neurons or their recruited glia or support cells, provide an attractive microenvironment for larval hemocytes, driving the colonization of epidermal-muscular pockets, and directly or indirectly promoting the trophic survival of hemocytes.
Fig. 7.
Ectopic peripheral neurons attract larval hemocytes. (A-F) Overexpression of scute generates ectopic neurons that re-direct larval hemocytes. (A-A′,C-C′′) control. (B-B′,D-E′′) sc overexpression. Genotypes are en-GAL4, UAS-GFP, HmlΔDsRed/UAS-sc and en-GAL4, UAS-GFP, HmlΔDsRed/+ as control in A,B; en-GAL4, HmlΔDsRed/UAS-sc and HmlΔDsRed/+ as control in D,E. (A) Control; dorsal hemocyte stripes are in line (dashed line) with dorsal-vessel-associated hemocyte clusters (asterisks). (B) sc expression. Hemocytes accumulate in the posterior compartment of each segment (arrowheads relative to asterisks; dashed line marks wt position of hemocytes). (C-E′′) Double-labeling of PNS neurons (anti-HRP, green), hemocytes (red) and nuclei (DAPI, blue) in fillets of sc misexpressing and control larvae. (D-D′′) Ectopic neuron surrounded by cluster of hemocytes (arrowhead). (E-E′′) Close-up of region marked in D. (F) Ectopic neuron in sc misexpression larva positive for anti-Elav (green, arrowhead). DAPI (blue); en-GAL4/UAS-sc. Owing to extensive staining, hemocytes were lost from specimen.
DISCUSSION
Hematopoietic expansion by self-renewal of differentiated cells
Previous reports suggested that embryonic hemocytes persist into postembryonic stages (Holz et al., 2003), and that larval hemocyte numbers increase over time (Lanot et al., 2001; Holz et al., 2003; Stofanko et al., 2008). However, the identity of the founders of the larval hematopoietic system, and their lineage during expansion, remained unclear. We now demonstrate that it is the differentiated plasmatocytes of the embryo that persist into larval stages and proliferate to constitute the population of larval hemocytes. Embryonic plasmatocytes comprise 80-90% of a population of 600-700 hemocytes that are BrdU-negative in the late embryo and that do not expand in number, even upon experimental stimulation of their phagocytic function (Tepass et al., 1994), suggesting their exit from the cell cycle. Thus, proliferation of these hemocytes in the larva implies re-entry into (or progression in) the cell cycle, and expansion by self-renewal in the differentiated state. This finding contrasts with the common mechanism of cell expansion (Buttitta and Edgar, 2007), in which undifferentiated prohemocytes expand by proliferation, which ceases once cell differentiation ensues (Tepass et al., 1994; Evans et al., 2003; Martinez-Agosto et al., 2007). In Drosophila, another case of self-renewing differentiated cells has been described in the developing adult tracheal system (Sato et al., 2008; Weaver and Krasnow, 2008), and expression of oncogenes such as RasV12 triggers expansion of differentiated larval hemocytes (Asha et al., 2003; Zettervall et al., 2004). In vertebrates, differentiated cell populations that self-renew and expand are known for hematopoietic and solid, ‘self-duplicating’ or ‘static’, tissues (Dor and Melton, 2004; Duncan et al., 2009; Geissmann et al., 2010), and neoplasias such as leukemias can develop from differentiated cells that re-gain the ability to expand (Krivtsov et al., 2006; Guibal et al., 2009). Controlling the proliferation of differentiated cells is pivotal in regenerative medicine and cancer biology, and Drosophila larval hemocytes may be an attractive system to study this phenomenon in the future.
The Drosophila resident hematopoietic pocket
Previous publications reported dorsal-vessel-associated hemocyte clusters as a ‘larval posterior hematopoietic organ’ that plays a role in larval immunity (Shrestha and Gateff, 1982; Stofanko et al., 2008; Markus et al., 2009). We now reveal that the earliest compartmentalization of the larval hematopoietic system is based on epidermal-muscular pockets that persist throughout larval development. The retreat of larval hemocytes to secluded hematopoietic environments parallels the vertebrate seeding of hematopoietic sites by hematopoietic stem cells (HSCs) or committed progenitors, which occur at multiple times during development (Lambolez et al., 2006; Cumano and Godin, 2007; Dzierzak and Speck, 2008; Laird et al., 2008; Bertrand and Traver, 2009).
Correlation of hemocyte residency with elevated proliferation levels and anti-apoptotic cell survival are consistent with the idea that inductive and trophic local microenvironments support hemocytes in epidermal-muscular pockets. Using gain- and loss-of-function analyses, we identify the PNS as such a functional hematopoietic microenvironment. Correspondingly, in vertebrates, HSCs or committed progenitors typically require an appropriate microenvironment, or niche, that provides signals to ensure the survival, maintenance and controlled proliferation and differentiation of these cells. Examples include the bone marrow niche (Ehninger and Trumpp, 2011), known to require PNS activity (Katayama et al., 2006; Lucas et al., 2008; Mendez-Ferrer et al., 2008; Mendez-Ferrer et al., 2010), and inducible peripheral niches in tissue repair, revascularization and tumorigenesis (Adams and Scadden, 2006; Kaplan et al., 2007).
Larval resident hemocytes are in a dynamic equilibrium, showing at least partial exchange between various resident locations. Based on our real-time and time-lapse studies, and consistent with the previously reported adhesion-based recruitment of circulating hemocytes to wound sites (Babcock et al., 2008), and hemocyte dynamics in the terminal cluster (Welman et al., 2010), some of this exchange may be attributed to the detachment, circulation and subsequent re-attachment of hemocytes to resident sites. However, lateral movement of hemocytes during re-formation of the resident pattern suggests that hemocytes can also travel continuously, presumably within the epidermal-muscular layer. This idea is further supported by the elevated hemocyte exchange in young larvae, in which most of the hemocytes reside in epidermal-muscular pockets. We define the (re-)colonization of resident sites as hemocyte ‘homing’, which might be based on active processes such as cell migration, and/or passive processes that might involve circulation of the hemolymph or undulation. Negative effects of dominant-negative Rho1 on the resident hemocyte pattern suggest a role for active cytoskeletal processes. These findings show intriguing parallels with vertebrates, in which hematopoietic stem and progenitor cells cycle between defined microenvironments and the peripheral blood (Adams and Scadden, 2006; Kaplan et al., 2007; Mendez-Ferrer et al., 2009; Ehninger and Trumpp, 2011).
The PNS as a microenvironment for hematopoiesis
We identified the PNS as a microenvironment that supports hemocyte attraction and trophic survival. Resident hemocytes colocalize with lateral ch and other lateral and dorsal PNS neurons such as md, and loss of ch neurons in ato1 mutants results in distinct hemocyte pattern and number defects. Likewise, genetic ablation of ch and other peripheral neurons strongly affects larval hemocytes regarding their resident pattern and trophic survival. Overexpression of the proneural gene sc induces supernumerary ectopic neurons that effectively attract hemocytes in 3rd instar larvae, providing evidence for a direct role of peripheral neurons or their recruited and closely associated glia or support cells (Banerjee and Bhat, 2007) in hemocyte attraction. This, together with the direct or indirect trophic dependence of hemocytes on the PNS, clearly distinguishes our findings from the previously reported role of hemocytes in dendrite and axon pruning, which typically is initiated at the onset of metamorphosis (Watts et al., 2003; Han et al., 2011). A functional connection of the PNS with the hematopoietic system might be of fundamental importance across species: in vertebrates, PNS activity governs regulation of HSC egress from the bone marrow and proliferation (Mauer, 1965; Katayama et al., 2006; Tsinkalovsky et al., 2007; Lucas et al., 2008; Mendez-Ferrer et al., 2008; Mendez-Ferrer et al., 2010), and immune responses in lymphocytes and myeloid cells (Mignini et al., 2003; Shepherd et al., 2005). Indeed, all hematopoietic tissues, such as bone marrow, thymus, spleen and lymph nodes, are highly innervated by the sympathetic and, in some cases in addition, the sensory nervous system (Shepherd et al., 2005; Nance and Sanders, 2007). However, since in Drosophila the PNS largely comprises sensory neurons (Brewster and Bodmer, 1996) rather than autonomic neurons (LaJeunesse et al., 2010), future studies will determine mechanistic parallels in the use of these distinct subsets of the PNS with respect to hematopoiesis in different phyla. As direct sensory innervation is present in the mammalian bone marrow and lymph nodes (Shepherd et al., 2005; Nance and Sanders, 2007), our work in Drosophila provides important precedence for a role of the sensory nervous system in hematopoiesis.
Developmental control of PNS-hematopoietic regulation
In Drosophila larva, hemocyte attraction to specific PNS locations is developmentally regulated: although the abdominal PNS clusters are maintained from embryonic stages onward (Bodmer et al., 1989; Banerjee and Bhat, 2008), they do not associate with hemocytes in the embryo. In the larva, attraction of resident hemocytes to PNS clusters proceeds in several steps, starting with the lateral PNS cluster (lateral patch) and posterior sensory organs (terminal cluster), and expanding at ∼72 hours AEL to the dorsal PNS cluster (dorsal stripe). Only late during larval development, from ∼110 hours AEL, can hemocytes be found in ventral locations. This suggests differential upregulation of certain factors that attract hemocytes in otherwise similar classes of neurons or their associated cells, and/or changes in the responsiveness of hemocytes over time.
In all backgrounds examined, PNS-dependent hemocyte phenotypes become most apparent from mid-larval development onwards, coincident with the developmental emergence of dorsal hemocyte stripes. We hypothesize an increasing limitation of trophic factors or a developmental loss of redundancy in directional and/or trophic support. The observed phenotypes might be direct or indirect, e.g. involving glia or other closely associated cells (zur Lage et al., 1997; Sepp and Auld, 2003; Parker and Auld, 2006). Likewise, sc misexpression experiments show potent attraction of hemocytes by ectopic neurons predominantly in late 3rd instar larvae, suggesting the need for some level of anatomical or molecular differentiation or maturation. All PNS manipulations showed only mild effects on lateral hemocyte patches, suggesting redundant signals of a larger group of neurons or glia, which we have not been able to manipulate in aggregate without inducing embryonic lethality (K.M. and K.B., unpublished). Also, resident hemocyte homing and induction might involve complex combinations of attractive and/or repulsive signals, similar to the cues operating in axon guidance and directed cell migrations in Drosophila and vertebrates (Dickson, 2002). Alternatively, attraction of hemocytes to the lateral patches might rely on additional, yet to be identified, microenvironments. Dorsal-vessel-associated hemocyte clusters (Shrestha and Gateff, 1982; Stofanko et al., 2008; Markus et al., 2009) do not colocalize with peripheral neurons and are not affected by manipulations of the PNS. As these clusters build up quickly after resident hemocyte disturbance, we speculate that their formation might relate to the accumulation of circulating hemocytes, consistent with previous observations (Babcock et al., 2008).
Outlook
In vertebrates, efforts to characterize at a molecular level the emerging connection between the PNS and the hematopoietic system are ongoing. Both indirect effects, via PNS signals to stromal cells of the bone marrow niche that engage in SDF-1/CXCR4 signaling (Yamazaki and Allen, 1990; Katayama et al., 2006; Mendez-Ferrer et al., 2010), and direct effects through stimulation of HSCs with neurotransmitters (Spiegel et al., 2007) have been reported. Drosophila larval hematopoiesis will allow the systematic dissection of the cellular and molecular factors that govern PNS-hematopoietic regulation. Future studies will reveal molecular evolutionary parallels and inform our understanding of PNS-controlled hematopoiesis in vertebrates. Furthermore, the system will allow investigation of the mechanisms of self-renewal of differentiated cells in a simple, genetically tractable model organism.
Supplementary Material
Acknowledgments
We thank M. Frasch, M. Galko, C. Han, U. Heberlein, D. Hultmark, Y. N. Jan, L. Kockel, L. Luo, B. Mathey-Prevot, J. Schulte, K. Sepp, S. Sinenko, S. Younger, the Bloomington Stock Center and the Drosophila Genomics Resource Center for stocks, plasmids or sequence information. We especially thank A. Miyawaki and K. Sugimura for UAS-S/G2/M-Green transgenic flies. We thank Lutz Kockel, Rik Derynck, Katharine Sepp, Yuh Nung Jan, Chun Han, Sijun Zhu, David Casso, Prashanth Rao, Todd Nystul and members of the Brückner laboratory for discussion and comments on the manuscript. K.M. thanks Prashanth Rao for valuable suggestions.
Footnotes
Funding
This work was supported by a Human Frontier Science Program postdoctoral fellowship [K.M.], Sandler Foundation Startup, Broad Research Incubator, Hellman Fellows and Program for Breakthrough Biomedical Research awards [K.B.]. This investigation was in part conducted in a facility constructed with support from the Research Facilities Improvement Program [C06-RR16490] from the National Center for Research resources (National Institutes of Health). Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.067322/-/DC1
References
- Adams G. B., Scadden D. T. (2006). The hematopoietic stem cell in its place. Nat. Immunol. 7, 333–337 [DOI] [PubMed] [Google Scholar]
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I., Dearolf C. R. (2003). Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K. M., Hawley R. S. (2004). Drosophila: A Laboratory Handbook. New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Babcock D. T., Brock A. R., Fish G. S., Wang Y., Perrin L., Krasnow M. A., Galko M. J. (2008). Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proc. Natl. Acad. Sci. USA 105, 10017–10022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Bhat M. A. (2007). Neuron-glial interactions in blood-brain barrier formation. Annu. Rev. Neurosci. 30, 235–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Bhat M. A. (2008). Glial ensheathment of peripheral axons in Drosophila. J. Neurosci. Res. 86, 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., Castro B., Posakony J. W. (2004). New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques 36, 436–440, p442 [DOI] [PubMed] [Google Scholar]
- Bertrand J. Y., Traver D. (2009). Hematopoietic cell development in the zebrafish embryo. Curr. Opin. Hematol. 16, 243–248 [DOI] [PubMed] [Google Scholar]
- Bodmer R., Carretto R., Jan Y. N. (1989). Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron 3, 21–32 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- Brewster R., Bodmer R. (1996). Cell lineage analysis of the Drosophila peripheral nervous system. Dev. Genet. 18, 50–63 [DOI] [PubMed] [Google Scholar]
- Brock A. R., Babcock D. T., Galko M. J. (2008). Active cop, passive cop: developmental stage-specific modes of wound-induced blood cell recruitment in Drosophila. Fly (Austin) 2, 303–305 [DOI] [PubMed] [Google Scholar]
- Brückner K., Kockel L., Duchek P., Luque C. M., Rørth P., Perrimon N. (2004). The PDGF/VEGF Receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73–84 [DOI] [PubMed] [Google Scholar]
- Buttitta L. A., Edgar B. A. (2007). Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. T., Hsiao C. D., Jan L. Y., Jan Y. N. (1996). Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl. Acad. Sci. USA 93, 13239–13244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corl A. B., Berger K. H., Ophir-Shohat G., Gesch J., Simms J. A., Bartlett S. E., Heberlein U. (2009). Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell 137, 949–960 [DOI] [PubMed] [Google Scholar]
- Crozatier M., Krzemien J., Vincent A. (2007). The hematopoietic niche: a Drosophila model, at last. Cell Cycle 6, 1443–1444 [PubMed] [Google Scholar]
- Cumano A., Godin I. (2007). Ontogeny of the hematopoietic system. Annu. Rev. Immunol. 25, 745–785 [DOI] [PubMed] [Google Scholar]
- Demerec M. (1994). Biology of Drosophila. Plainview: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Dickson B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964 [DOI] [PubMed] [Google Scholar]
- Dor Y., Melton D. A. (2004). How important are adult stem cells for tissue maintenance? Cell Cycle 3, 1104–1106 [PubMed] [Google Scholar]
- Duncan A. W., Dorrell C., Grompe M. (2009). Stem cells and liver regeneration. Gastroenterology 137, 466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushay M. S. (2009). Insect hemolymph clotting. Cell. Mol. Life Sci. 66, 2643–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Speck N. A. (2008). Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger A., Trumpp A. (2011). The bone marrow stem cell niche grows up: mesenchymal stem cells and macrophages move in. J. Exp. Med. 208, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. J., Banerjee U. (2003). Transcriptional regulation of hematopoiesis in Drosophila. Blood Cells Mol. Dis. 30, 223–228 [DOI] [PubMed] [Google Scholar]
- Evans C. J., Hartenstein V., Banerjee U. (2003). Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 5, 673–690 [DOI] [PubMed] [Google Scholar]
- Fossett N., Schulz R. A. (2001). Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation 69, 83–90 [DOI] [PubMed] [Google Scholar]
- Geissmann F., Manz M. G., Jung S., Sieweke M. H., Merad M., Ley K. (2010). Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., O’Kane C. (1989). Neural enhancer-like elements as specific cell markers in Drosophila. Development 105, 35–52 [DOI] [PubMed] [Google Scholar]
- Goto A., Kumagai T., Kumagai C., Hirose J., Narita H., Mori H., Kadowaki T., Beck K., Kitagawa Y. (2001). A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem. J. 359, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibal F. C., Alberich-Jorda M., Hirai H., Ebralidze A., Levantini E., Di Ruscio A., Zhang P., Santana-Lemos B. A., Neuberg D., Wagers A. J., et al. (2009). Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood 114, 5415–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E., Wiggins D., Fielding B., Gould A. P. (2007). Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445, 275–280 [DOI] [PubMed] [Google Scholar]
- Halfon M. S., Gisselbrecht S., Lu J., Estrada B., Keshishian H., Michelson A. M. (2002). New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis 34, 135–138 [DOI] [PubMed] [Google Scholar]
- Han C., Jan L. Y., Jan Y. N. (2011). Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. USA 108, 9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. D., Stein D., Stevens L. M. (2000). Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development 127, 573–583 [DOI] [PubMed] [Google Scholar]
- Hartenstein V. (1993). Atlas of Drosophila Development. New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Hartenstein V. (2006). Blood cells and blood cell development in the animal kingdom. Annu. Rev. Cell Dev. Biol. 22, 677–712 [DOI] [PubMed] [Google Scholar]
- Holz A., Bossinger B., Strasser T., Janning W., Klapper R. (2003). The two origins of hemocytes in Drosophila. Development 130, 4955–4962 [DOI] [PubMed] [Google Scholar]
- Jan L. Y., Jan Y. N. (1982). Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA 79, 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P., Grau Y., Jan L. Y., Jan Y. N. (1993). atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73, 1307–1321 [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Sun Y., Jan L. Y., Jan Y. N. (1995). Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121, 2019–2030 [DOI] [PubMed] [Google Scholar]
- Jung S. H., Evans C. J., Uemura C., Banerjee U. (2005). The Drosophila lymph gland as a developmental model of hematopoiesis. Development 132, 2521–2533 [DOI] [PubMed] [Google Scholar]
- Kaplan R. N., Psaila B., Lyden D. (2007). Niche-to-niche migration of bone-marrow-derived cells. Trends Mol. Med. 13, 72–81 [DOI] [PubMed] [Google Scholar]
- Katayama Y., Battista M., Kao W. M., Hidalgo A., Peired A. J., Thomas S. A., Frenette P. S. (2006). Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421 [DOI] [PubMed] [Google Scholar]
- Krivtsov A. V., Twomey D., Feng Z., Stubbs M. C., Wang Y., Faber J., Levine J. E., Wang J., Hahn W. C., Gilliland D. G., et al. (2006). Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 [DOI] [PubMed] [Google Scholar]
- Krzemien J., Dubois L., Makki R., Meister M., Vincent A., Crozatier M. (2007). Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 446, 325–328 [DOI] [PubMed] [Google Scholar]
- Kurucz E., Zettervall C. J., Sinka R., Vilmos P., Pivarcsi A., Ekengren S., Hegedus Z., Ando I., Hultmark D. (2003). Hemese, a hemocyte-specific transmembrane protein, affects the cellular immune response in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2622–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E., Vaczi B., Markus R., Laurinyecz B., Vilmos P., Zsamboki J., Csorba K., Gateff E., Hultmark D., Ando I. (2007). Definition of Drosophila hemocyte subsets by cell-type specific antigens. Acta. Biol. Hung. 58 Suppl., 95–111 [DOI] [PubMed] [Google Scholar]
- Laird D. J., von Andrian U. H., Wagers A. J. (2008). Stem cell trafficking in tissue development, growth, and disease. Cell 132, 612–630 [DOI] [PubMed] [Google Scholar]
- LaJeunesse D. R., Johnson B., Presnell J. S., Catignas K. K., Zapotoczny G. (2010). Peristalsis in the junction region of the Drosophila larval midgut is modulated by DH31 expressing enteroendocrine cells. BMC Physiol. 10, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambolez F., Arcangeli M. L., Joret A. M., Pasqualetto V., Cordier C., Di Santo J. P., Rocha B., Ezine S. (2006). The thymus exports long-lived fully committed T cell precursors that can colonize primary lymphoid organs. Nat. Immunol. 7, 76–82 [DOI] [PubMed] [Google Scholar]
- Lanot R., Zachary D., Holder F., Meister M. (2001). Postembryonic hematopoiesis in Drosophila. Dev. Biol. 230, 243–257 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- Lin D. M., Goodman C. S. (1994). Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron 13, 507–523 [DOI] [PubMed] [Google Scholar]
- Lucas D., Battista M., Shi P. A., Isola L., Frenette P. S. (2008). Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell 3, 364–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus R., Laurinyecz B., Kurucz E., Honti V., Bajusz I., Sipos B., Somogyi K., Kronhamn J., Hultmark D., Ando I. (2009). Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 106, 4805–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Agosto J. A., Mikkola H. K., Hartenstein V., Banerjee U. (2007). The hematopoietic stem cell and its niche: a comparative view. Genes Dev. 21, 3044–3060 [DOI] [PubMed] [Google Scholar]
- Mauer A. M. (1965). Diurnal variation of proliferative activity in the human bone marrow. Blood 26, 1–7 [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Lucas D., Battista M., Frenette P. S. (2008). Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Chow A., Merad M., Frenette P. S. (2009). Circadian rhythms influence hematopoietic stem cells. Curr. Opin. Hematol. 16, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Michurina T. V., Ferraro F., Mazloom A. R., Macarthur B. D., Lira S. A., Scadden D. T., Ma’ayan A., Enikolopov G. N., Frenette P. S. (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignini F., Streccioni V., Amenta F. (2003). Autonomic innervation of immune organs and neuroimmune modulation. Auton. Autacoid Pharmacol. 23, 1–25 [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Hiromi Y., Weber U., Goodman C. S., Rubin G. M. (1990). The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell 60, 211–224 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Kuranaga E., Sugimura K., Miyawaki A., Miura M. (2010). Nonautonomous apoptosis is triggered by local cell cycle progression during epithelial replacement in Drosophila. Mol. Cell. Biol. 31, 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance D. M., Sanders V. M. (2007). Autonomic innervation and regulation of the immune system (1987-2007). Brain Behav. Immun. 21, 736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. E., Fessler L. I., Takagi Y., Blumberg B., Keene D. R., Olson P. F., Parker C. G., Fessler J. H. (1994). Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 13, 3438–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E. M., Rebay I., Tjian R., Rubin G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137–147 [DOI] [PubMed] [Google Scholar]
- Parker R. J., Auld V. J. (2006). Roles of glia in the Drosophila nervous system. Semin. Cell Dev. Biol. 17, 66–77 [DOI] [PubMed] [Google Scholar]
- Rehorn K. P., Thelen H., Michelson A. M., Reuter R. (1996). A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development 122, 4023–4031 [DOI] [PubMed] [Google Scholar]
- Rizki T. M. (1978). The circulatory system and associated cells and tissues. In The Genetics and Biology of Drosophila (ed. Ashburner M., Wright T. R. F.), pp 397–452 New York: Academic Press; [Google Scholar]
- Rothenfluh A., Threlkeld R. J., Bainton R. J., Tsai L. T., Lasek A. W., Heberlein U. (2006). Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell 127, 199–211 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 [DOI] [PubMed] [Google Scholar]
- Sato M., Kitada Y., Tabata T. (2008). Larval cells become imaginal cells under the control of homothorax prior to metamorphosis in the Drosophila tracheal system. Dev. Biol. 318, 247–257 [DOI] [PubMed] [Google Scholar]
- Sepp K. J., Auld V. J. (1999). Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics 151, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp K. J., Auld V. J. (2003). Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J. Neurosci. 23, 8221–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp K. J., Schulte J., Auld V. J. (2001). Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev. Biol. 238, 47–63 [DOI] [PubMed] [Google Scholar]
- Shepherd A. J., Downing J. E., Miyan J. A. (2005). Without nerves, immunology remains incomplete - in vivo veritas. Immunology 116, 145–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R., Gateff E. (1982). Ultrastructure and cytochemistry of the cell types in the larval hematopoietic organs and hemolymph of Drosophila melanogaster. Dev. Growth Differ. 24, 65–82 [DOI] [PubMed] [Google Scholar]
- Sinenko S. A., Mathey-Prevot B. (2004). Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene 23, 9120–9128 [DOI] [PubMed] [Google Scholar]
- Song W., Onishi M., Jan L. Y., Jan Y. N. (2007). Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc. Natl. Acad. Sci. USA 104, 5199–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel A., Shivtiel S., Kalinkovich A., Ludin A., Netzer N., Goichberg P., Azaria Y., Resnick I., Hardan I., Ben-Hur H., et al. (2007). Catecholaminergic neurotransmitters regulate migration and repopulation of immature human CD34+ cells through Wnt signaling. Nat. Immunol. 8, 1123–1131 [DOI] [PubMed] [Google Scholar]
- Stofanko M., Kwon S. Y., Badenhorst P. (2008). A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics 180, 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M. J., Redd M. J., Jacinto A., Parkhurst S. M., Martin P. (2005). Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168, 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Fessler L. I., Aziz A., Hartenstein V. (1994). Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120, 1829–1837 [DOI] [PubMed] [Google Scholar]
- Thomas U., Jonsson F., Speicher S. A., Knust E. (1995). Phenotypic and molecular characterization of SerD, a dominant allele of the Drosophila gene Serrate. Genetics 139, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinkalovsky O., Smaaland R., Rosenlund B., Sothern R. B., Hirt A., Steine S., Badiee A., Abrahamsen J. F., Eiken H. G., Laerum O. D. (2007). Circadian variations in clock gene expression of human bone marrow CD34+ cells. J. Biol. Rhythms 22, 140–150 [DOI] [PubMed] [Google Scholar]
- Watts R. J., Hoopfer E. D., Luo L. (2003). Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871–885 [DOI] [PubMed] [Google Scholar]
- Weaver M., Krasnow M. A. (2008). Dual origin of tissue-specific progenitor cells in Drosophila tracheal remodeling. Science 321, 1496–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigmann K., Cohen S. M. (1999). Lineage-tracing cells born in different domains along the PD axis of the developing Drosophila leg. Development 126, 3823–3830 [DOI] [PubMed] [Google Scholar]
- Welman A., Serrels A., Brunton V. G., Ditzel M., Frame M. C. (2010). Two-color photoactivatable probe for selective tracking of proteins and cells. J. Biol. Chem. 285, 11607–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J., Ivanchenko S., Oswald F., Schmitt F., Rocker C., Salih A., Spindler K. D., Nienhaus G. U. (2004). EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 101, 15905–15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., Jacinto A. (2007). Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat. Rev. Mol. Cell Biol. 8, 542–551 [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Allen T. D. (1990). Ultrastructural morphometric study of efferent nerve terminals on murine bone marrow stromal cells, and the recognition of a novel anatomical unit: the “neuro-reticular complex”. Am. J. Anat. 187, 261–276 [DOI] [PubMed] [Google Scholar]
- Zettervall C. J., Anderl I., Williams M. J., Palmer R., Kurucz E., Ando I., Hultmark D. (2004). A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. USA 101, 14192–14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage P., Jan Y. N., Jarman A. P. (1997). Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr. Biol. 7, 166–175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.