Abstract
The study of gene regulation in cells has recently begun to shift from a period dominated by the study of transcription factor-DNA interactions to a new focus on RNA regulation. This was sparked by the still-emerging recognition of the central role for RNA in cellular complexity emanating from the RNA World hypothesis, and has been facilitated by technologic advances, in particular high throughput RNA sequencing and crosslinking methods (RNA-Seq, CLIP, and HITS-CLIP). This article will place these advances in context, and, focusing on CLIP, will explain the method, what it can be used for, and how to approach using it. Examples of the successes, limitations and future of the technique will be discussed.
Crosslinking immunoprecipitation (CLIP), coupled with high throughput sequencing (HITS-CLIP), has caught the attention of the RNA community as a means of achieving a new depth of understanding about how protein-RNA complexes interactions regulate gene expression in living cells1–4. This review will describe the context in which CLIP was developed, and provide an up-to-date review of its uses in developing genome-wide maps of RNA-protein interactions and, more recently, microRNA (miRNA) binding sites. The uses, limitations, and future of CLIP will be discussed.
Keywords: RNA regulation, high throughput sequencing, crosslinking immunoprecipitation, CLIP, HITS-CLIP
A brief historical overview
The study of protein-nucleic acid interactions in cells began with the study of protein-DNA interactions, in longstanding efforts to identify transcription factor binding sites in an unbiased, genome-wide manner. In vitro DNA selection to identify idealized binding sequences for transcription factors5, 6 provided the groundwork for these studies. DNA selection provided binding sites for factors, defining the sequences bound and the affinity with which they were recognized, and provided important information with which to interpret more biologically relevant efforts.
Early attempts were made to co-purify DNA-protein complexes from cells, for example by immunoprecipitation (IP) of transcription factors such as Myc-Max heterodimers bound to DNA elements7. However, co-purification of factors and natively bound DNA elements were generally limited by concerns stemming from the dynamic nature of chromatin, and the transient nature of DNA-protein interactions, as well as the recognition that protein-DNA interactions could rearrange or be lost during purification8. This era ended with the ability to preserve physiologically relevant protein-DNA interactions with improved biochemical purification methods—in particular, protein factor-DNA crosslinking methods9, 10 developed during the 1980s and widely implemented beginning in the late 1990s, for example in the study of histone-DNA interactions8. These efforts have been sufficiently successful to allow machine-learning algorithms to accurately predict transcriptional cooperativity based on disparate arrays of biochemical data (see 11 and references therein12, 13). This success led to new feed-forward loops, both with respect to science and resources, spurring such new initiatives as the ENCODE, modENCODE (www.genome.gov/10005107), and the NIH Roadmap Epigenomics (http://nihroadmap.nig.gov/epigenomics/) projects.
While RNA biochemists are the first to recognize the importance of transcription—they at least acknowledge that RNA is not present in the cell without having been transcribed—they have at the same time recognized limits to DNA complexity, the excitement engendered by the concept of the RNA world and emerging evidence of great RNA complexity (as recently discussed3, 4, 14).
Genome-wide RNA biochemistry
Efforts to determine genome-wide profiles for protein-RNA interactions have paralleled but lagged behind those in the protein-DNA domain. Identification of RNA binding motifs for regulatory proteins began on a strong note, including the demonstration that known U1A-RNA interactions could be rederived from in vitro RNA selection experiments15. In vitro consensus motifs were subsequently enumerated for a large number of RNA binding proteins (RNABPs). However, efforts to apply discrete consensus motifs to a range of in vivo targets were less compelling. This likely relates to the fact that many, if not all, RNA binding proteins bind low-complexity RNA targets—an interesting observation in itself, perhaps relating to greater flexibility in the system and protein-RNA dynamics. Certainly such flexibility may be a greater issue in RNABP:RNA regulation than in the protein-DNA world, as some RNABPs serve multiple functions (e.g. splicing factors or EJC proteins are likely to serve and perhaps coordinate nuclear and cytoplasmic RNA regulation16–19). In any case, the low complexity of RNABP target motifs restricted the usefulness of in vitro RNA selection or related methods such as yeast three-hybrid selection to a subset of proteins that bind very specific sequence motifs (PUF20, Fox1/221, Nova22, 23), and even in these instances, the use of these motifs as a sole means to derive genome-wide insight into target recognition and biology has been limited. Conversely, in vitro selection with other RNABPs has identified binding motifs too complex to identify natural targets, such as the loop-loop pseudoknot or G-quartet motifs characterized as preferred ligands of the FMRP KH and RGG domains, respectively.
RNA IP (“RIP”) strategies again initially paralleled studies in the DNA transcription factor field24, 25. Several studies have used this approach with clear success, as evident in recent analyses of miRNA targets26, 27. However, it is also clear that the signal:noise in such experiments, while discernable, creates difficulties in data interpretation, such that even the most carefully done experiments entail uncertainties that prevents a comprehensive analysis of targets or their direct binding sites28. The problems with RIP were well illustrated in our own early efforts to identify RNA targets of the fragile-X mental retardation protein, FMRP29, 30. In this work, RNA co-immunoprecipitating with FMRP from mouse brain was interrogated on Affymetrix microarrays. Because it was recognized that even under relatively stringent conditions (albeit mild enough to allow RNA-protein interactions to be maintained; in the case of FMRP this was 400mM salt, 0.5% NP40), RNA is sticky and significant background is present in such IPs, a number of controls were done. These included selection of RNAs that were enriched in IPs relative to starting material, elimination of RNAs that co-immunoprecipitated in Fmr1 null brains, and bioinformatic screening of targets for FMRP binding motifs. Yet despite these precautions, only a handful of these targets have subsequently been validated as direct FMRP-regulated transcripts31, 32. This likely relates to a number of factors inherent in such IPs beyond simple noise issues. RNABPs are frequently found in complexes with other RNABPs (a large number are reported to co-IP with FMRP), suggesting that indirect RNA-RNABP interactions may be identified in such co-IPs, with the potential to complicate biologic validation of targets. Moreover, regulatory RNABPs tend to have transient interactions with targets (likely a measure of their off-rates) that may relate to their dynamic nature. It is likely that these points underlie the observation of Mili and Steitz that RNABP-RNA interactions may readily reassociate during co-IPs33. Nonetheless, there remain some scenarios where RIP may be of value, for example in testing if an entire RNA, compared to a processed sequence or degraded fragment, is bound to a protein. Consider for example the differences between CLIP and RIP in analysis of DICER bound to precursor miRNAs.
Crosslinking immunoprecipitation (CLIP)
A breakthrough in mapping RNA-protein interactions in vivo came with the development of crosslinking-immunopurification (CLIP) strategies34–36. In CLIP (Figure 1), whole tissues, organisms or individual cell types are treated with UV-B irradiation. This generates a covalent bond between RNA-protein complexes that are in close contact within the intact cell or tissue. Following formation of this bond, RNABPs can be purified under very stringent conditions. While any purification method can in theory be used, in practice proteins are generally purified by virtue of antibodies to the RNABPs themselves or protein epitope tags. In the course of this purification, RNA is intentionally reduced in size—typically to a modal size of ~50 nt—to facilitate identification of binding sites (e.g. crosslinked RNAs from ~20 nt to 100 nt). Once sufficient purity has been obtained, the protein component of the crosslinked complex is removed with proteinase K and the RNA is purified. Current protocols, still evolving, have used RNA ligase to ligate RNA linkers, followed by cDNA synthesis with an antisense primer and reverse transcriptase (RT), to generate templates for sequencing.
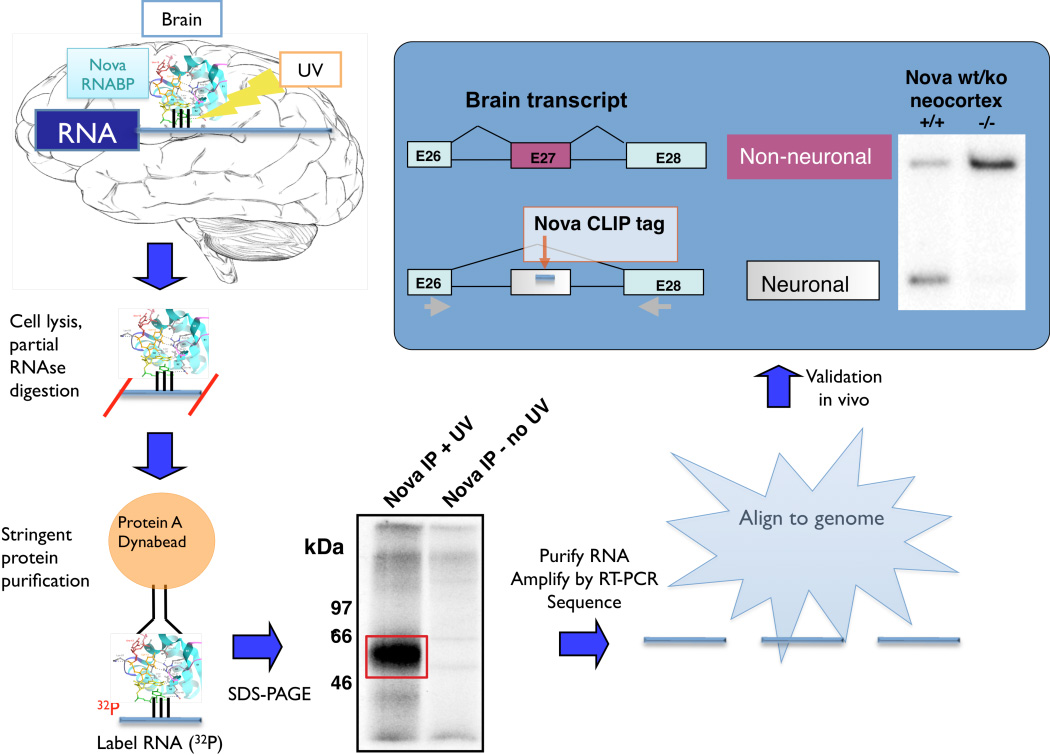
Figure 1. CLIP (crosslinking immunoprecipitation).
Tissue (e.g. brain) is UV-irradiated, covalently crosslinking RNA-protein complexes in situ. After cell lysis and partial RNase digestion to reduce crosslinked RNA to a modal size of ~50 nt, protein-RNA complexes are purified, protein removed with proteinase K, RNA linkers added and RT-PCR products sequenced to identify CLIP “tags” that can be mapped onto the transcriptome, delineating native RNA-protein interaction sites. These point to functional binding sites, which can be validated in the tissue used for CLIP, as illustrated here for a Nova splicing target transcript encoding neogenin (adapted from 36, with permission).
Historically, UV-irradiation37, 38 and formaldehyde treatment39 were initially described in the 1960s as ways to crosslink DNA-protein complexes in vitro. In 1974 Schoemaker and Schimmel showed that UV-irradiation caused a specific crosslink between tRNA and tyrosyl-tRNA synthetase40. It was recognized in the 1980s that these techniques could be applied in vivo9, but they were not used at the time for purification and sequencing of bound nucleic acids. This was due in part to a belief that persisted until recent times that the efficiency of UV-crosslinking was too low to be of value for such strategies 41. In addition, there was concern because crosslinking had been established as a means of blocking reverse transcriptase (RT; for the purposes of mapping RNA-protein binding sites42). Such a blockade was believed to preclude post-crosslinking reverse transcription and/or PCR amplification and sequencing; in fact, when DNA-protein crosslinking was undertaken more recently in Drosophila S2 cells, it was thought to work specifically because the co-purifying, non-crosslinked complimentary strand was available for PCR amplification43. In the meantime, chromatin IP (ChIP) had become widely accepted and utilized, relating in part to the perceived importance of the reversibility of formaldehyde crosslinking8 to allow the analysis of bound DNA-protein complexes by PCR.
Re-evaluation of these issues and technical advances were put together to enable RNA CLIP. An important point was the recognition that the specificity conferred by UV-crosslinking protein-RNA complexes offered an advantage over formaldehyde crosslinking methods. This advantage relates to the mechanism of UV-mediated crosslinking, which, although incompletely understood, is believed to involve absorption of UV light by nucleic acid bases44 to induce ground state electrons to a singlet higher-energy state that enables the electron to partake in a chemical reaction in which a new covalent bond is formed41. The nature of this reaction is such that it occurs only between closely opposed molecules—on the order of Angstroms apart—such that only direct protein-RNA contacts are able to be crosslinked. Notably, UV irradiation does not induce protein-protein crosslinks, although it induces RNA-RNA crosslinks44, 45, which has been used extensively, for example in mapping the structure of the ribosome. The protein-RNA crosslinking reaction occurs on only a minority of contact sites (in our estimates, maximal crosslinking efficiency is on the order of 1–5%, although this may vary with different proteins; see also 41). This specificity of UV-crosslinking to RNA contrasts with formaldehyde crosslinking, in which interactions are generated between large protein-nucleic acid and protein-protein complexes, complicating identification of direct interactions sites.
Moreover, the irreversibility of UV-crosslinking turned out not to be a limitation for further sequence analysis of the RNA. CLIP protocols established conditions in which RT could bypass crosslinking sites. There are several aspects to this—the blockade may be partially efficient, it may be absolute for some but not all crosslink sites, or it may induce errors in copying RNA to DNA at the site of crosslinking—however, in any of these cases, crosslinking proved not to be an absolute barrier to the identification of bound RNA fragments. There may even be advantages to the problems RT encounters at crosslinking sites. Errors at crosslinking sites have been used to map the sites of RNA-protein interaction (see below). Similarly, RT arrest at crosslink sites has recently been taken advantage of as a means of mapping interaction sites in CLIP data46.
One of the main advantages of CLIP is its applicability to biologic systems. The concept of UV-irradiating living cells was first introduced in the 1980’s as a means of analyzing protein-RNA complexes47–49. Dreyfuss and colleagues demonstrated that protein-RNA complexes could be IP’d from irradiated cells50, leading to the generation of antibodies and characterization of hnRNP complexes. These studies set the stage for undertaking CLIP—UV-crosslinking followed by sequencing of the sites at which proteins contact RNA—in living cells. While CLIP was first undertaken in brain, as discussed below, it has since been applied to whole bacteria, fungi, yeast, C. elegans, and a number of mammalian tissue culture cells including human embryonic stem cells (Table 1).
High throughput sequencing (HITS) CLIP
The original CLIP experiments36 entailed sequencing of 340 unique Nova-bound RNAs at a cost of ~$4,000. These studies showed that CLIP was able to identify direct protein-RNA interactions, and that such interactions identified functionally relevant points of RNA-protein interaction (Figure 1). However, the small number of tags precluded drawing any robust generalizations about the nature of the RNA-protein interactions. This limitation was overcome by applying next-generation high throughput sequencing methods to CLIP, termed HITS-CLIP. Analysis of the same RNA-protein interactions—Nova-RNA interactions in mouse brain—with HITS-CLIP yielded over 1000-fold more unique tags for the same cost in 2008. Importantly, tags from multiple different mouse brains could be compared, allowing the biologic reproducibility of the method to be assessed. Given the large (and ever-increasing) amounts of data available using next-generation sequencing technologies, bioinformatic analysis of raw tags is an important aspect of such studies. We discuss below the use of CLIP and HITS-CLIP analysis by several groups to explore biologically relevant protein-RNA interactions.
Examples of successful CLIP studies
A number of proteins have been studied using CLIP (Table 1). These include studies using both low throughput and high throughput (HITS) CLIP. In the following section we review the major CLIP studies published through early 2010.
Binary protein:RNA interactions
RNA maps and insights into coordinated RNA regulation
CLIP was originally developed in an effort to identify RNA targets bound by the neuronal KH-type RNABP Nova in the brain36. The motivation behind this effort has been described in detail51. In brief, Nova is targeted in an autoimmune brain disorder clinically associated with failure of neuronal inhibitory pathways. Biochemical studies, including in vitro RNA selection, of Nova function originally identified two different Nova target RNAs as transcripts encoding subunits of the glycine and GABAA inhibitory neurotransmitter receptors. Although errors in splicing of these transcripts in vitro22, 52 could be correlated with errors in splicing in Nova null brains53, two uncertainties remained. First, it was not possible to know with certainty that Nova acted directly on these transcripts in vivo; even though this seemed likely given an abundance of biochemical evidence, such assays are non-stoichiometric and may not extrapolate to living tissue. Second, the correlation between the clinical syndrome and the RNA targets—both implicating inhibitory synaptic function—were intriguing, but were derived from small assays prone to potential selection bias.
Together, these uncertainties prompted a search for a genome-wide means of identifying direct Nova-RNA interactions within the brain, and hence the motivation to develop CLIP. After several years of development by Kirk Jensen, Jernej Ule applied the system specifically to analysis of Nova targets; together they co-authored the first CLIP paper describing 340 RNA tags crosslinked to Nova (CLIP tags36). These targets were rich in Nova binding sequences (clusters of YCAY elements, as previously defined by in vitro selection experiments22, 23, 54 and X-Ray crystallography55), and included 18 individual tags that flanked alternative exons, of which 7 could be validated as mis-spliced in Nova null mice. This experiment demonstrated that clusters of the known Nova YCAY binding site were highly statistically enriched in the CLIP tags, that these Nova binding site included a subset present in transcripts encoding synaptic proteins, and a subset that could be documented as functional Nova binding sites (mediating Nova-dependent alternative splicing in WT versus Nova KO brain).
Based on this low-throughput CLIP data, and exon junction splicing microarrays56, a series of bioinformatic predictions regarding rules of Nova-dependent splicing were generated, relating the position of Nova binding to the outcome of splicing (exon inclusion or exclusion57). While this prediction was statistically robust and accurately distinguished splicing enhancement from repression, the discrimination of target from non-target had a relatively high false positive rate (50%), underscoring the need for more robust datasets, as well as the need to validate such predictions with functional assays.
Subsequently, we developed HITS-CLIP to identify Nova binding sites on a genome-wide scale58. Perhaps not surprisingly, a wealth of both anticipated and unexpected data ensued. HITS-CLIP allowed a genome-wide biophysical assessment of the bioinformatically generated predictions, confirming and refining the predicted Nova splicing regulatory map. Methodologically, this work established a new approach to CLIP tag analysis enabled (and in fact required) by the large number of tags generated by these experiments. The problem was an embarrassment of riches—to determine which of the 412,686 high throughput sequencing reads were signal and which were noise. These tags were bioinformatically culled to produce a more stringent set for analysis. First, unique tags were identified, sidestepping issues of preferential PCR amplification of tags. This left 168,632 unique CLIP tags to analyze. Second, reasoning that signal would be more biologically reproducible than noise, analysis shifted from individual tags to biologically reproducible tag “clusters” (overlapping tags). 19,156 such tag clusters were identified and found to be highly reproducible (over 90% between littermates).
Biologic complexity (BC) thus established a variable that can be applied to threshold an experiment. Further stringency could be applied by demanding clusters have both a given BC value and number of tags (peak height). Biologically reproducible Nova-bound clusters were found to be highly enriched in YCAY elements, the biochemically defined Nova binding site. Motif analysis by MEME59 revealed a consensus sequence of AUCAUCAUCA in the top 500 clusters (P < 10−8) and YCAY enrichment was evident across all 19,156 clusters (P < 10−227). Moreover, these sites identified CLIP tag clusters in 34 of 39 previously validated Nova-regulated transcripts (identified by analysis of exon junction microarrays in Nova WT versus null mouse brains56 (Figure 2). More generally, the position of 1,085 tags in 71 different Nova-regulated alternative exons mapped to positions that were consistent with the previously predicted Nova bioinformatic map (Figure 3), such that Nova binding within alternate exons or in their upstream introns generally led to exon skipping, while binding to downstream introns led to alternate exon inclusion. Thus, HITS-CLIP data confirmed the hypothesis that the position of Nova binding determines the outcome of splicing regulation by experimentally identifying genome-wide Nova binding footprints.
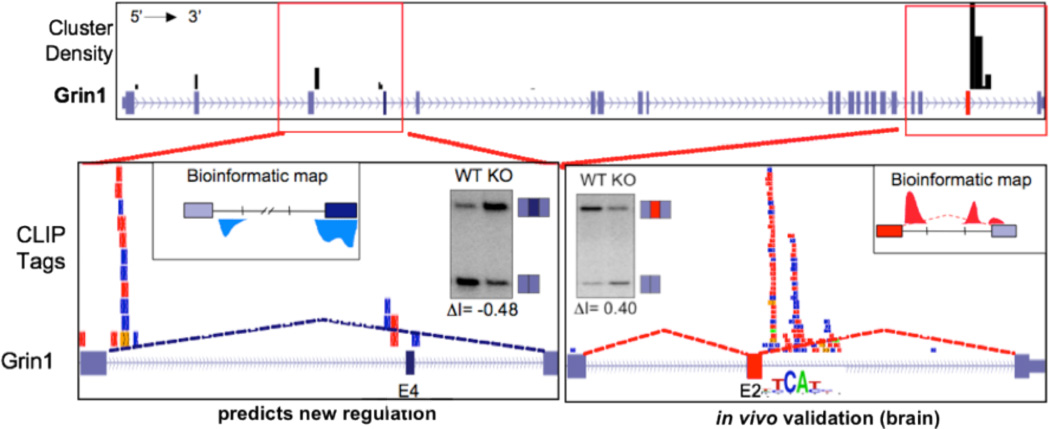
Figure 2. HITS-CLIP tags in the NMDAR1 (Grin1) gene.
Top panel shows locations of Nova CLIP tag clusters (black bars) across the entire Grin1 transcript. Bottom panels show individual tags, with each color representing a different biologic replicate (individual mouse brain). The bottom right panel, depicting a previously validated Nova-dependent exon (E24), shows CLIP tags downstream of the alternate exon, which is included in a Nova-dependent manner (as shown by RT-PCR of WT vs. Nova KO brain). The bottom left panel shows a newly discovered Nova-regulated exon (E4), in which the position of CLIP tags predicts Nova-mediated inhibition of exon inclusion according to a previously generated bioinformatic map, which was again validated by RT-PCR (from 58, with permission).
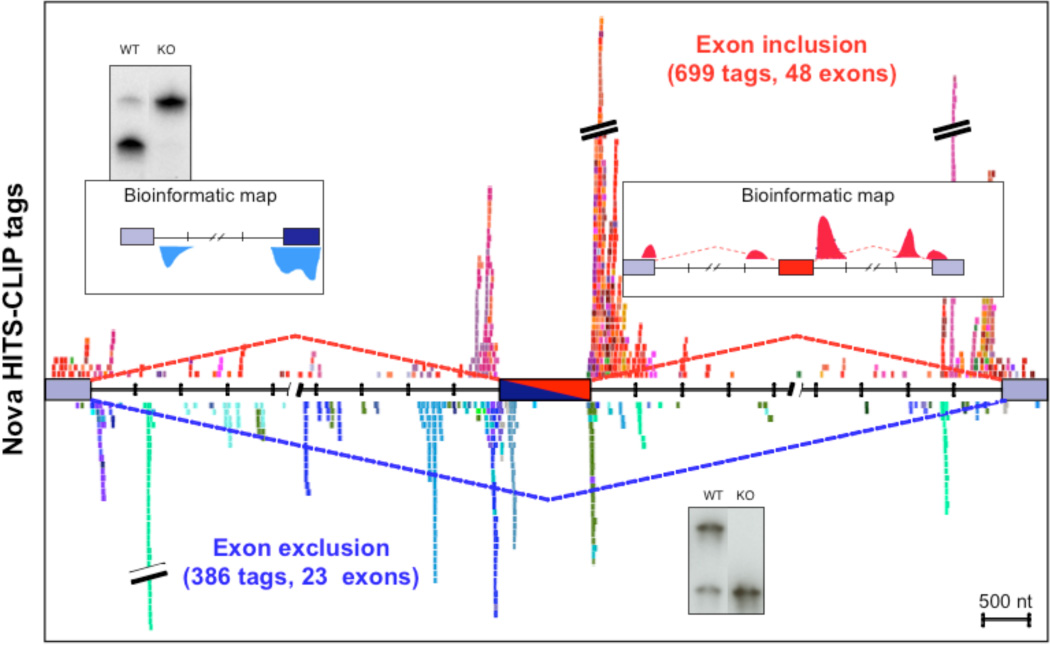
Figure 3. A genome-wide functional Nova interaction map.
CLIP tags are plotted around a composite alternate exon, with each unique color representing tags from an individual Nova-regulated transcript. Tags from transcripts in which Nova inhibits or enhances alternative exon inclusion are represented in cool and warm colors, respectively. Normalization of this data to complexity further refined this map and demonstrated that Nova-RNA interaction sites mapped with HITS-CLIP experimentally confirm predictions from a previously generated bioinformatic map (from 58, with permission).
The approach of applying BC mapping to many HITS-CLIP tags to identify validated sites of exon regulation has recently been extended to another splicing factor, the polypyrimidine tract-binding protein PTB60, by Fu, Zhang and colleagues. PTB has been characterized as a splicing repressor based on careful studies of individual transcripts61. HITS-CLIP analysis of PTB binding sites in HeLa cells identified clusters of CLIP tags that were enriched in CU-containing hexamers, consistent with PTB binding sites defined in vitro. Analysis of PTB tags in the region of a modest subset of PTB-regulated exons (selected from a larger group previously identified by separate studies and by exon junction arrays62) revealed a normalized complexity map showing both similarities and contrasts to the map described for Nova. As for Nova, PTB bound in multiple positions, and those upstream and surrounding the alternate exon appeared to be primary determinants of exon skipping. Unexpectedly, a significant number of exons also showed PTB-dependent exon inclusion, and these appeared to show a disproportionate binding to the downstream intron, particularly proximal to the constitutive 3’ splice site, although the number of analyzed exons was relatively small, and it was not reported how many individual transcripts showed PTB binding to each position. Together with the RNA map derived for Fox2 (see below), these observations suggest that rules governing RNA splicing regulation may be generalized from data on a series of individual splicing factors to reveal general features relating the position of binding to splicing outcome (discussed further in “CLIP promise and caveats”).
Another example in which HITS-CLIP revealed unanticipated new biology came from the finding that a large number of Nova CLIP tag clusters were found in 3’ UTRs, frequently surrounding alternative polyadenylation sites58. Biochemical follow up and analysis of these observations suggested a model paralleling that put forth for Nova-dependent splicing regulation, in which the position of Nova binding around alternative sites determines the outcome of polyadenylation—skipping or utilization of an alternative site—although the number of tags was too small to be conclusive (Figure 4). In fact, the majority of Nova-regulated alternative polyadenylation events culminated in skipping of a poly(A) site such that more distal sites were favored—consistent with data that brain preferentially generates longer 3’ UTRs63, presumably allowing increased potential for regulation (for example by miRNAs64), while dividing cells generate shorter 3’ UTRs65.
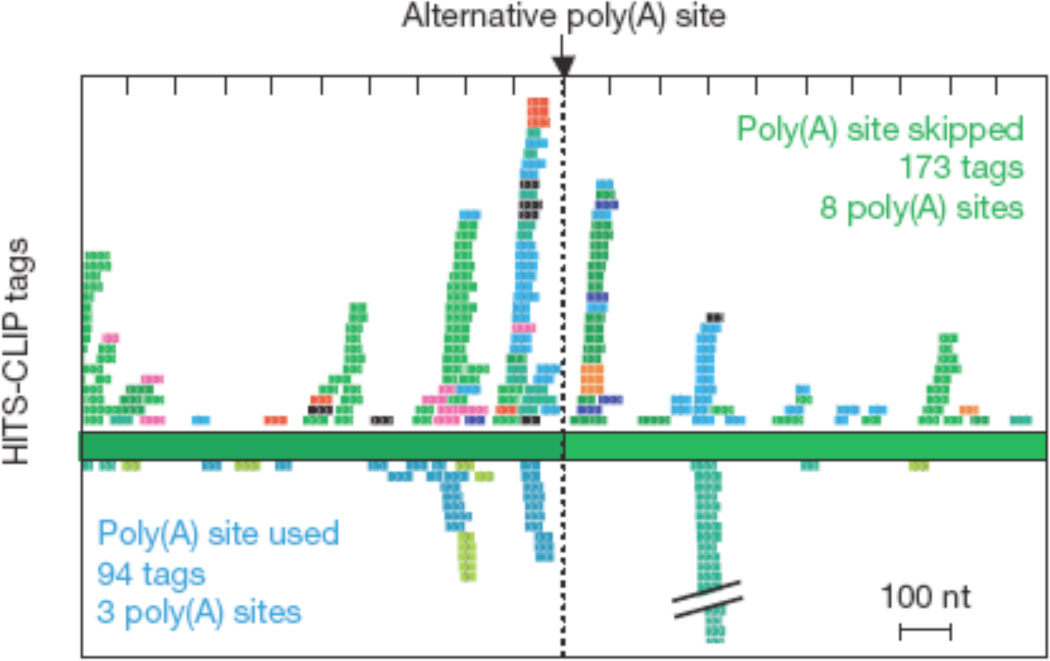
Figure 4. Nova regulation of alternative polyadenylation.
Nova tags surrounding alternative polyadenylation sites are mapped. CLIP tags from transcripts in which Nova is associated with polyadenylation site skipping or inclusion are shown on top and bottom, respectively (from 58, with permission).
Coordinate regulation of biologically coherent sets of transcripts
Data suggesting that RNABPs regulate transcripts encoding related sets of proteins that mediate coherent biologic programs has long been suggested, but has been difficult to demonstrate, particularly on a genome-wide basis (discussed in 4). The study of Nova targets, identified by analysis of alternative splicing in WT vs. Nova null brain, had suggested that Nova regulated transcripts encoded a biologic subset of functions56, 66. However, as with similar analyses of other RNABPs, these conclusions suffered from the caveat that it was not possible to determine which transcripts were directly regulated (i.e. bound) by Nova, and which might be indirectly regulated through an action on an intermediary splicing factor. Nova HITS-CLIP allowed the first assessment of a set of regulated RNAs that were also directly bound by the RNABP, confirming that Nova does indeed directly regulate a specific biologic subset of brain transcripts encoding synaptic proteins58.
Since these studies, HITS-CLIP has been used by several labs to address genome-wide RNA target identification for other RNABPs. The first of these was a genome-wide study of the binding sites of Fox2 (Rbm9) by Gene Yeo, Fred Gage and colleagues67. A point of nomenclature—these authors renamed HITS-CLIP as “CLIP-seq”, a reference to RNA-Seq, a shotgun method of high throughput RNA sequencing. We find this nomenclature to be unclear, as sequencing per se is intrinsic to the original (non-high throughput) CLIP method. Yeo and colleagues applied CLIP to study Fox2 binding sites in human embryonic stem cells (hESCs) and generated a binding map by identifying over 6,000 tag clusters. Motif analysis demonstrated that the most enriched hexamer in these clusters corresponded to the Fox2 binding site UGCAUG (p < 10−70), although only 22% of FOX2 CLIP clusters have the UGCAUG motif, compared to 11% expected by chance. The reasons for the signal:noise issue here is not clear, but appears to relate to this particular experiment, as opposed to a more general problem with CLIP. For example, by comparison, the Nova-binding YCAY tetramer is ~5–6 fold enriched in Nova CLIP clusters68, although the Nova motif is 64-fold more degenerate. Even after combining FOX CLIP data with the presence of the motif, there is still a substantial fraction of false positives (6/23 or 26%), while FOX bioinformatic predictions alone were 55–60% accurate69. In contrast, combining Nova CLIP data with the presence of the less well defined motif has an accuracy of ~75% for the top 70 targets identified by microarray analysis68. One general issue relating to background problems in CLIP experiments relates to issues of experimental stringency (see “Signal:noise issues in CLIP”). Another factor that may have contributed to the background in this study (also relevant to the PTB study by this group) is that clusters were defined in a bioinformatic rather than purely biochemical manner. All raw FOX2 tags mapping to unique sites in the genome were analyzed, and each read was computationally extended to an average size of 100 nt, which were then corrected for computationally to derive a defined set of clusters.
Regardless of these issues, Fox2 binding sites could be found to be significantly enriched in introns 50–100 nt downstream of 5’ splice sites flanking alternative exons. Interestingly, analysis of 23 such targets selected for validation suggested a similar positional map to that seen for Nova, in which Fox2 binding to upstream introns inhibited alternate exon inclusion, while binding downstream led to enhancement of exon inclusion, consistent with the map derived from a combination of comparative genomic and microarray data analysis69. Gene ontology analysis of Fox2 targets indicated that the protein has clusters in a non-random set of transcripts, encoding RNA binding proteins and serine/threonine kinases, suggesting a coherent biologic role for Fox2 in regulating RNA metabolism and signaling pathways in hESCs.
A very recent and focused analysis of RNABP function comes from a HITS-CLIP study of the yeast Khd1 protein, an RNABP hypothesized by Fink and colleagues to play a role in the development of filamentous growth in diploid yeast70. HITS-CLIP demonstrated that the major target of Khd1 (2 million out of 16 million raw CLIP tags!) is the FLO11 transcript, which encodes a cell wall protein required for filamentous growth. Khd1 bound to the coding region of FLO11 and 54 other transcripts encoding cell surface proteins, suggesting a means by which Khd1 coordinately regulates assembly of the cell wall to permit filamentous growth.
Unexpected activities and disease connections revealed by CLIP
Caceres and colleagues were the first to undertake CLIP on a mammalian RNABP other than Nova, in a 2007 paper analyzing RNA binding sites of hnRNPA1, with what have proven to be astounding results. Two hundred hnRNP A1 CLIP tags were sequenced, one of which was, unexpectedly, a miRNA, miR-18a71. The authors went on to show that hnRNP A1 bound directly to the stem-loop of the miR-18a precursor (pri-miRNA), specifically facilitating generation of miR-18a but not other miRNAs present in the pri-miRNA. A follow-up study refined the hnRNP A1 binding site to the terminal and internal loops of pri-miR-18a, elegantly demonstrating that this binding has a functional consequence, to relax the stem and facilitate Drosha binding and hence miR-18a generation72. In the context of CLIP, this set of studies provided a second clear demonstration that the method is able to identify sites of functional RNA-protein interactions, and to discover previously unanticipated new biology.
CLIP was used to explore what was an unexpected action of DJ-1 (PARK7) as an RNABP, a protein whose gene is mutated in recessively inherited Parkinson’s-like movement disorder. The protein had previously been co-purified with an incompletely characterized RNA binding protein activity in tissue culture cells, prompting Cookson and colleagues to ask whether DJ-1 itself might have RNA binding activity73. They used CLIP as an assay to demonstrate crosslinking to RNA and to clone a small number of tags. Although the biology of such “non-professional” RNA binding proteins remains to be clarified, this work illustrates the possibility of using CLIP as a reliable assay for RNA binding activity and further analysis.
A number of human neurologic disorders have been associated with triplet repeat expansions. In some cases, including myotonic dystrophy, FXTAS and spinocerebellar ataxias, these repeats are believed to act at the RNA level to sequester RNA binding proteins74. These actions are complex, as evidenced by RNA-Seq studies of myotonic dystrophy, which suggest a variety of effects from sequestration of different CUG-repeat binding proteins, including MBNL75 and CUGBP176, proteins involved in regulating alternative splicing. Recently Swanson, Ranum and colleagues have begun to use CLIP as a means of addressing the gain and loss of function of RNABPs in these disorders, focusing on spinocerebellar ataxia type 8 (SCA877). They analyzed 315 CUGBP1-associated RNA tags identified by CLIP, representing 206 genes with 53 having more than one tag. As seen with the splicing regulator Nova, most tags were intronic (64%), but 25% were positioned within 3′ untranslated regions (UTRs). CUGBP1 CLIP tags were enriched in UG repeats, consistent with three-hybrid and SELEX studies identifying (UG)N and (UGUU)N repeats as CUGBP1 binding motifs. One CUGBP1 CLIP tag (overlapping exon 7 of the GABA transporter 4 (Gabt4, Slc6a11) transcript) was validated in detail; increased levels of transcript including exon 7 were identified in SCA8 brain, as well as increased protein levels. Since the Gabt4 exon 7-minus isoform would introduce a premature stop codon, it was hypothesized that this isoform, normally downregulated during the transition from fetal to adult life, would be subjected to NMD-mediated decay in adults. Thus this CLIP tag pointed to a mechanism for the high levels of Gabt4 in SCA8 patients and in early development. These experiments are the first to demonstrate the potential of CLIP in the study of triplet repeat/RNA-sequestration disorders.
RNA regulation in subcellular compartments: SFRS1 (SF2/ASF), Nova and Rrm4
Sanford and colleagues used CLIP to study RNA targets of the splicing factor SF2/ASF (now termed SFRS178). Like many splicing factors, SFRS1 shuttles between the nucleus and cytoplasm, prompting these investigators to fractionate cells after crosslinking to separately examine SFRS1 RNA CLIP tags in the nuclear, cytoplasmic and polysome fractions of HEK293T cells. Although a relatively small number of unique tags were analyzed (326), several were identified in multiple fractions, suggesting binding of the same RNAs in multiple subcellular compartments.
Interestingly, while a consensus binding site for SFRS1 could not be clearly determined from these initial studies, one became evident after SFRS1 HITS-CLIP experiments79. These studies identified a stringent set of 135,318 unique CLIP tags and further culled them to 681 clusters of biologic complexity 3 (those found in 3 out of 4 experiments). MEME analysis revealed a GAAGAA consensus binding site, which was similar to prior in vitro selection motifs. Interestingly, this motif also matched a known orphan splicing enhancer—an element identified computationally as an exonic splicing enhancer, but for which no corresponding RNABP regulator had been clearly identified80. The SFRS1 binding sites identified by CLIP may be relevant to human disease, as 181 of the 21,700 single-nucleotide substitutions present in the Human Gene Mutation Database (HGMD; www.hgmd.org) are present in potential SFRS1 binding sites, some of which are known to cause aberrant splicing in patient transcripts.
Recent studies of nuclear and cytoplasmic Nova CLIP tags also suggest that CLIP may point to transcripts that are coordinately regulated at the level of splicing within the nucleus and subcellular localization within the neuronal cytoplasm16. Nova binds both to intronic sequences of the inhibitory glycine receptor GlyRα2 transcript (Glra2) (originally identified bioinformatically and confirmed by HITS-CLIP) resulting in alternative exon inclusion, as well as 3’ UTR elements, identified by HITS-CLIP, to affect mRNA localization in primary neuronal cultures and transfected cell lines. Further support for a link between CLIP data, splicing data and subcellular localization came from immunoelectron microscopy and immunogold in situ hybridization studies of spinal cord motor neurons, demonstrating co-localization of Nova protein and GlyRα2 mRNA in the neuronal dendrite. Therefore, CLIP can be an effective means of addressing issues of RNA regulation within discrete cellular compartments.
CLIP has been effectively applied to study subcellular RNA-protein interactions in non-mammalian systems. CLIP was used to demonstrate that a TAP-tagged RNABP termed Rrm4 could be purified after crosslinking to RNA in vivo, and therefore functioned as an RNA binding protein in the filamentous fungus Ustilago maydis81. This underscores a flexible point about CLIP, which is that any protein purification scheme can be employed to purify RNA-protein complexes, although issues of stoichiometry need to be considered (see below) if not using endogenous proteins. In this regard the term CLIP is perhaps a technical misnomer, as TAP-tag purification (see also below) need not use antibody for protein purification. Subsequent sequence analysis of 78 CLIP tags revealed that Rrm4 binds to CA-rich motifs, and study of target transcripts identified by CLIP were used to demonstrate that the Rrm-RNPs are colocalized by FISH and are transported on microtubules within filaments82.
CLIP to abundant RNAs; protein-rRNA analyses
The study of RNA-protein interactions with very abundant RNAs has special considerations with respect to signal:noise and validation that has been successfully negotiated by several groups. The issue is that the molar concentration of any individual high complexity RNA (e.g. mRNA) may be vastly lower than that of even a moderately low complexity RNA (such as snoRNA or rRNA). As a rough approximation, an mRNA present at 10 copies per cell, in a cell with 105 mRNA molecules, will be present at a ~106 lower molar abundance than rRNA, all of which is the same sequence and which constitutes ~98% of total cellular RNA. Hence, even if the efficiency of CLIP purification is very high, background issues for abundant RNAs of low complexity may present an issue in data analysis.
This issue was first addressed by Tollervey and colleagues in the context of their studies of snoRNA and rRNA biogenesis, using yeast as a model organism83. They took advantage of the versatility of yeast genetics to create a series of tagged proteins that could be readily purified in an antibody-independent “CLIP” method they termed CRAC (to emphasize the affinity purification rather than immunopurification strategy; see discussion below). Using tagged constructs of Rrp9, a known U3 snoRNP, the authors were able to estimate that ~3% of U3 snoRNA was UV-crosslinked to Rrp9, consistent with estimates of crosslinking efficiency initially made with Nova. UV-irradiation was effective in intact yeast cells, and HITS-CLIP was then performed with tagged versions of other snRNP proteins, including Nop1, Nop56 and Nop58. ~ 70–90% of sequenced RNAs accurately mapped to discrete positions in box C/D snoRNAs, and analysis of short reads (15–18 nt long) allowed precise mapping of protein-RNA binding footprints in U3 snoRNA.
Tollervey and colleagues took a particularly interesting approach to independently corroborate their HITS-CLIP results for Rrp9, as well as those in a second HITS-CLIP study mapping rRNA binding sites for the helicase Prp4384. They noticed that sequencing of CLIP tags revealed a disproportionate number of mutations and deletions at sites of crosslinking; for example, 48% of U3 tags crosslinked to Nop58 contained substitutions at a G residue (G323) two nucleotides upstream of box D, but these same mutations were rare in CLIP tags seen with other snoRNPs. Similar observations were made for the interaction of Prp43 and functional sites within helix 44 of 18S rRNA and snoRNAs (snR51, snR60 and snR72). The authors concluded that such sites of disproportionate mutagenesis corresponded to sites of protein-RNA contact that were crosslinked. Similar mutations were seen with Nova35, and presumably reflect sites of residual amino-acid-RNA adducts left after proteinase K digestion in CLIP, and the subsequent difficulty of RT traversing such crosslink sites. These observations both point to the utility of this observation in mapping potential crosslinking sites and in distinguishing background and crosslinked tags derived from abundant RNAs.
In addition to studies in yeast and filamentous fungus discussed above, CLIP has been applied to several other simple organisms. Wurtmann and Wolin were the first to apply CLIP to the study of eubacteria as an extension of a longstanding interest in the biology of the Ro autoantigen85. Ro is an RNA binding protein that is believed to function during stress to control the quality of non-coding RNAs, although there has not been a comprehensive understanding of its RNA substrates. CLIP studies of the radiation-resistant eubacterium Deinococcus radiodurnas were used to identify RNA targets of the eubacterial Ro homologue Rsr following cellular stress, revealing an interaction with rRNA. Interestingly, this interaction was again validated bioinformatically by observing a disproportionate number of point mutations (23%) in Rsr-rRNA crosslinks that were not present in controls. These results, coupled with prior studies that Rsr interacts with the exonuclease polynucleotide phosphorylase and additional functional analysis, led to a model in which the protein plays a biologically important role to bring this exonuclease to rRNA during stress to allow degradation of ribosomal subunits, conferring a selective advantage to the organism.
CLIP detection of higher order RNP structures
Ule and colleagues undertook HITS-CLIP analysis of hnRNP C binding sites in HeLa cells, incorporating a new strategy termed iCLIP to help precisely map protein-interaction sites by detection of sites of RT arrest46. These studies revealed hnRNP C to be a major RNABP, in that a majority (55%) of annotated transcripts showed protein binding to U-rich elements, the binding motif predicted from in vitro binding studies. Remarkably, secondary peaks of binding were found at distances of 165 and 300 nt from the primary U-rich binding site, and these secondary peaks were also found to be U-rich. This suggested a higher-order structure in which hnRNP might form a nucleosome-like hnRNP particle wrapping up target RNAs. Moreover, these regions were considered in the context of an RNA splicing map, determined by overlaying HITS-CLIP data with analysis of hnRNP C dependent exon usage in knock-down HeLa cells. Interestingly, silenced exons and their proximal introns were preferentially those predicted to be incorporated into hnRNP particles, suggesting a mechanism by which hnRNP C wraps up alternate exons to insulate them from splicing and ensure their exclusion.
Ternary protein:small-RNA:RNA interactions
Ago-mRNA-miRNA CLIP
MiRNAs are small non-coding RNAs that are believed to regulate mRNA expression by directly binding transcripts and inhibiting translation, promoting deadenylation and/or decreasing mRNA stability. This action is most commonly through interactions in the 3’ UTR, and requires a “seed” match of only 6–8 nucleotides between the miRNA and mRNA. However, systematic means of determining the sites of miRNA-mRNA interaction has been hindered by the inherent difficulty in identifying binding sites of complexity (¼)6 (~1/4,000 nt). Despite intense efforts, including a focus on elements that are conserved across species, predictive bioinformatic algorithms still have ~50–70% false positive rates86–88.
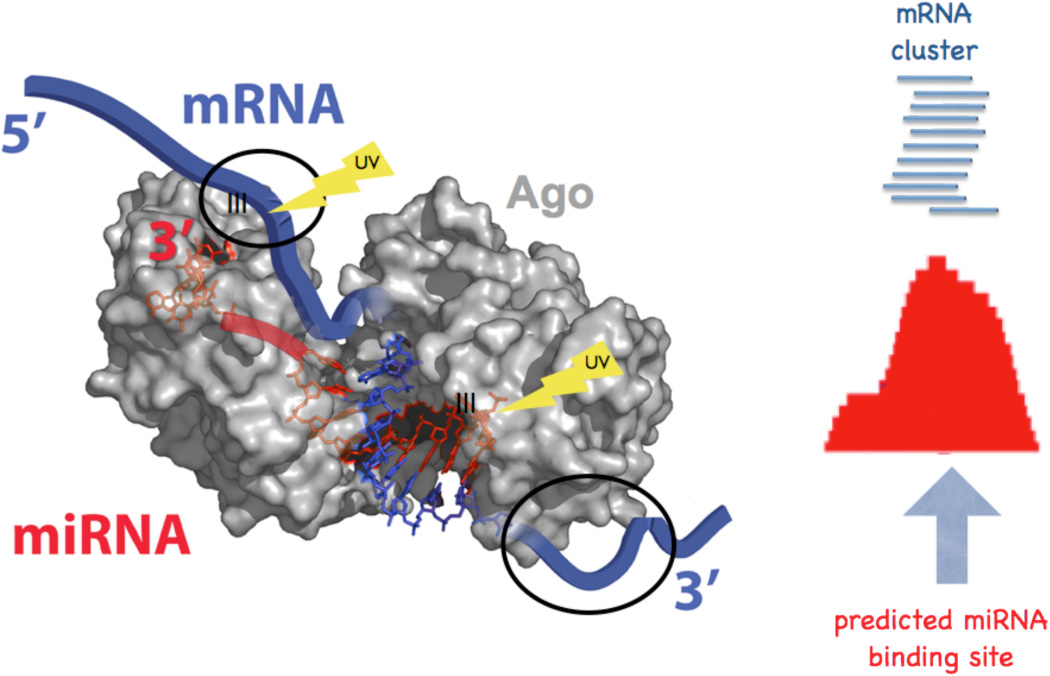
Significant progress toward resolving this difficulty was provided by the development of Ago HITS-CLIP in 200989 (Figure 5). Ago HITS-CLIP analysis of mouse brain tissue identified Ago-miRNA interactions, but also, somewhat unexpectedly, revealed a footprint of Ago-mRNA binding sites. These two datasets were overlaid to identify specific miRNA seed sequences present within the Ago-mRNA footprints, effectively decoding which miRNAs bound to specific sites within individual mRNAs. When performed on a genome-wide scale, these studies were able to decode a map identifying both the mRNAs and the sites within those mRNAs to which ~90% of brain miRNAs bound89.
Figure 5. Schematic of Ago HITS-CLIP.
UV crosslinking of intact tissues creates covalent bonds between Ago-miRNA and, separately, Ago in contact with neighboring mRNA sequences (structure on left is adapted from 117). Alignment of the latter produces an Ago-mRNA footprint; analysis of this restricted sequence space allows prediction and validation of miRNA binding sites. (Figure adapted from 89).
Searching Ago-mRNA footprints for binding sites for the best-validated mammalian miRNA, miR-124, confirmed a robust set of previously validated targets27 in 16 (or 21) of 22 cases (depending on stringency of analysis). Moreover, after transfecting miR-124 (a brain-specific miRNA) into HeLa cells, 17 new clusters appeared at miR-124 sites in these 22 cases. These and additional analyses demonstrated that Ago HITS-CLIP identified sites of functional miRNA regulation, and went on to identify additional predicted sites of regulation in brain mRNAs, mediated not only by miR-124, but by each of the 20 most abundant brain miRNAs. Such Ago HITS-CLIP maps serve as a means of decoding maps of miRNA-mediated RNA regulation, and offer the possibility of specifically targeting their actions.
Recently Ago-miRNA-mRNA ternary HITS-CLIP studies have been replicated in C. elegans by Pasquinelli, Yeo and colleagues90. This approach offers an advantage in that only a single Ago protein, ALG-1, mediates miRNA regulation in worms, and ALG-1 mutants exist that provide good negative controls (see below). 4806 clusters with a biologic complexity of 2 (2 out of 3 replicate experiments) were identified from wildtype but not alg-1 mutant samples. However, a significant number of raw clusters (over 800) were identified in control CLIP experiments (done in alg-1 mutants) that needed to be removed from analysis, suggesting that the biochemical stringency could be further optimized. For example, nuclease digestion was done after IP, which may have impacted the signal:noise, and the definition of clusters was less stringent than that used in prior Ago HITS-CLIP studies89. A number of validation studies were done, including the observation that 9 of 13 well studied miRNA-regulated transcripts had ALG-1 clusters over cognate miRNA sites. Interestingly, analysis of sequences surrounding ALG-1 clusters revealed a number of features—sequence conservation, sequence accessibility (single strandedness), and, most unexpectedly, 3’ UTR clusters contained and were flanked by CU regions, consistent with the possibility of accessory factors associating with Ago-miRNA target recognition.
As with mammalian Ago, binding sites were found to be distributed in transcripts in proportions similar to that seen for mammalian Ago-HITS-CLIP clusters89, with a large set in 3’ UTRs, very few in 5’ UTRs, and surprisingly high numbers in introns and coding exons. These observations re-emphasize the likelihood of undiscovered Ago biology, underscored also by the finding that only in Ago 3’ UTR clusters, but not coding sequence clusters, was binding correlated with effects on steady-state mRNA levels. Analysis of genes bound and presumably regulated by ALG-1 revealed evidence for autoregulatory feedback loops, such that there was a preponderance of ALG-1 clusters in transcripts, such as the alg-1 3’ UTR itself, that encode proteins implicated in the miRNA pathway. In summary, this paper confirms and extends insights into miRNA regulation made through the use of Ago HITS-CLIP.
CLIP to piRNA and other small RNAs; MSY2 CLIP
MSY2 is a germ cell-specific DNA/RNA binding protein that was studied by CLIP. Although the properties of MSY2 as an mRNA binding protein have previously been studied, recent work suggested that it might also bind small RNAs. Therefore a “directed-CLIP” experiment was done in which small RNA-protein complexes were isolated on the basis of elecrophoretic mobility91. 231 CLIP tags were sequenced and found to be small RNAs expressed in testes ranging in length from 18 to 36 nt. This included a small subset that matched known piRNAs, and a larger set that was distinct from both miRNAs and piRNAs, and whose presence was unaltered in Miwi-null mice. These experiments nicely demonstrate this dimension of CLIP, seen also in Ago HITS-CLIP experiments, which is the ability to use size-selection of crosslinked complexes to specifically interrogate a subset of protein-RNA complexes.
CLIP promise and caveats
CLIP: Descriptive biology versus mechanism
There is no doubt that much of the power of CLIP comes from its ability to delineate massive numbers of protein-nucleic acid binding sites with high resolution and fidelity. There may be a temptation to consider CLIP studies as descriptive or unable to deliver mechanistic results. These observations are in part true, but may be more pessimistic than necessary. Certainly in the DNA transcription field, such criticisms are not leveled at ChIP-seq experiments, and the many reasons for this acceptance should find some resonance in the study of RNA-protein interactions. These techniques certainly expand mechanistic studies performed on individual transcripts by allowing those studies, which are always limited in their generality, to be, well, generalized.
HITS-CLIP derived RNA-protein maps can reveal rules (with implied mechanisms) that were not otherwise obvious. For example, Nova regulation of alternative splicing was initially observed in traditional single gene studies, but was shown to operate according to rules such that the position of binding determines the outcome of splicing (exon inclusion/exclusion), observations made possible by a combination of genome-wide splicing studies56, bioinformatic predictions57, and HITS-CLIP58, as recently discussed4. Moreover, the ability to “zoom out” and derive such genome-wide rules governing RNA-protein regulation does not preclude the ability to “zoom in” in a robust manner; since CLIP works equally well with tissue culture or living tissues (yeast, worms, mammalian brains), biologically relevant targets can be chosen for mechanistic studies. Again, using Nova as an example, individual alternative exons proven to be directly regulated by Nova in the brain were also studied mechanistically with detailed boundary mapping, mutagenesis52, 92, in vitro splicing, psoralen crosslinking and analysis of RNA intermediates66 to determine that Nova binding at the exon/intron junction competes with U1 snRNP to block utilization of the proximal, but not upstream intron to inhibit exon inclusion66. This potential of CLIP should not be overlooked as an adjunct to more traditional approaches to studying mechanisms of RNA regulation60, 93.
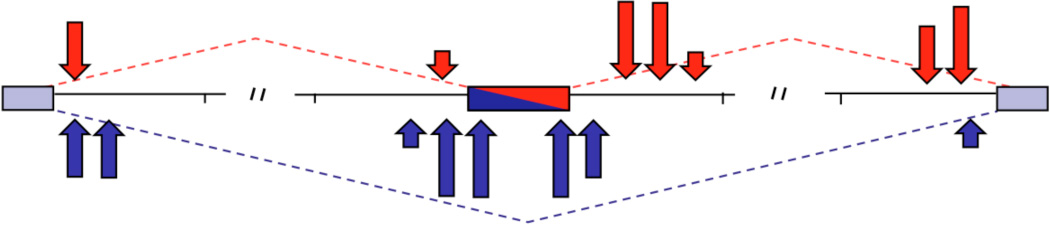
Others have remarked on the similarities and differences in HITS-CLIP derived RNA regulatory maps for different RNABPs1. These contrasts illustrate an interesting future direction for CLIP, which is the overlay of multiple RNA maps to derive both rules of regulation and to understand combinatorial regulation of RNA metabolism. Careful analysis of the three currently available RNA regulatory maps for splicing factors suggests that common features may be emerging. Specifically, the data suggest that binding very close to (or within) alternative exon splice junctions (or within the exon) inhibit exon inclusion, while binding further downstream of the exon, and/or binding at or near constitutive exon donor/acceptors, lead to enhancement of alternate exon inclusion (Figure 6). It is likely that several mechanisms are involved in mediating this regulation57, 93. One point that awaits clarification, given so many different binding sites on these composite maps, is how combinatorial binding to different positions, in cis on a given transcript, contributes to splicing regulation. An important point to keep in mind regarding HITS-CLIP data is that it delivers population averages, rather than information about binding on a single molecule. One future approach to these problems will be to apply machine learning techniques used successfully in the analysis of transcriptional control11, 12. A recent study reports the first integration of HITS-CLIP data in a Bayesian network to study Nova regulation of alternative splicing, producing completely unexpected data that were not identified from either HITS-CLIP or other analyses in isolation68.
Figure 6. Position-dependent binding of RNABPs determines the outcome of splicing regulation.
The map is a schematic based on HITS-CLIP studies of Nova, PTB, hnRNP C and Fox2; see Figure 3 for comparison. The most consistent effects seen (tallest arrows) reveal that binding proximal to or within alternate exons (middle exon) inhibits exon inclusion (blue arrows); binding downstream enhances exon inclusion (red arrows).
The unbiased nature of CLIP can offer the discovery of new binding sites not previously anticipated, and thereby go beyond descriptive work to generate new hypotheses regarding RNA-protein regulation. The Caceres study exemplifies this idea—prior to CLIP there had been no hint that hnRNP A1 might regulate pre-miRNA processing71, an observation subsequently pursued mechanistically72, and one which may have more general implications94. Similarly, HITS-CLIP revealed an unexpected role for Nova in the regulation of alternative polyadenylation58. Finally, a plethora of new Ago footprints, outside of the 3’ UTR, suggests additional roles for Ago-miRNA regulation of RNA transcripts89, 90. Hence while it is certainly true that CLIP itself is a descriptive tool, it offers a clear opportunity to divulge new insights, including mechanistic ones, into RNA-protein regulation.
CLIP validation
We have previously discussed the synergy that can emerge from combining biophysical descriptions of RNA-protein interactions afforded by CLIP, with the description of RNA variants, afforded for example by RNA-Seq analysis of different tissues or genetic backgrounds4. Such genome-wide analyses can be validated by independent RNA analyses35, 58, 67, 77, sequence analysis84, 89, 90, 95, functional assays of RNA-protein interactions89, physiology96, 97, or even cellular studies of RNA localization16 or cell migration98.
Another parameter of validation is reproducing CLIP results with independent methods. This has been accomplished in a number of current CLIP studies. For example, genome-wide Nova HITS-CLIP studies have been confirmed both in our laboratory and independently, through the use of microarrays35, 58 and bioinformatic analyses68, 99. Conversely, predictions of a position-dependent code for Nova-dependent splicing66 were independently validated by HITS-CLIP in our lab58, and these studies have very recently been validated bioinformatically by an independent group99. Similar cross-validation using comparison of different platforms from different labs to confirm CLIP data include studies of Fox2 (67; with data independently confirmed bioinformatically69) and PTB (60; with data independently confirmed biochemically62, 100 and bioinformatically101. Finally, it should be noted that three groups have published HITS-CLIP studies of Ago89, 90, 95; while done in quite different systems, the results are largely supportive of each other. Taken together, these observations reflect to the robust nature of CLIP methodology.
Signal:noise issues in CLIP
Not all CLIP is equal. Different signal:noise in CLIP results is proportional to the biochemical effort initially put in to maximize stringency of conditions, to the specificity of reagents (especially antibodies) used for protein purification, and to the bioinformatic analysis of data—the degree of subtraction of background tags from biologically robust tags. In general, because of the plethora of tags and data available from HITS-CLIP, we have erred in the direction of using very stringent biochemical purifications and discarding large numbers of tags to analyze more stringent datasets, leading to highly predictive results. Biologic reproducibility is an important adjunct to this analysis, and cannot be replaced by deeper sequencing (which may provide a measure of technical reproducibility); this is particularly important in biologically noisy systems (e.g. tissues such as brain). Since different numbers of replicates are done between experiments and among investigators, this should be reported as a fraction (not a decimal; i.e. the significance of defined clusters with a biologic complexity of 2 differs if this is from a total of 2 or 5 experiments; thus the term is more usefully identified as BC 2/3 or 2/5). In sum, analysis of CLIP data using BC as a measure of reproducibility is a key concept.
Ideally, stringency can be tuned in part by establishing negative controls—most informatively RNABP nulls—that can be used to optimize signal:noise throughout CLIP. Note however that such controls are most important in optimizing the early biochemical steps of CLIP, as we have found that low stringency in early steps may be difficult to overcome even with the best downstream efforts, and conversely that in stringent CLIP experiments only extremely small numbers of negative control tags can be detected46, 58.
Along these lines, it should be noted that good signal:noise in Western blots does not necessarily predict good signal:noise in radioactive co-IP’s. We have found that different antibodies to the same protein may immunoprecipitate variable amounts of cross-linked 32P-labeled RNA, presumably due to cross-reacting RNABPs that are not evident by Western but that show interfering bands by autoradiography. This issue is discussed in more detail in several methods reviews34, 35, 102.
What CLIP is not
Some shortcuts invoking CLIP have been reported in which investigators undertake the UV-crosslinking step, and then IP protein and analyze RNA by RT-PCR. However, such studies do not have the stringency of CLIP experiments. Such co-precipitated RNA has all of the problems surrounding the analysis of standard RNA IPs discussed earlier, including the presence of background RNAs in the IP, reassociation artifacts, and co-precipitating RNA binding proteins bringing down additional RNAs. In other words, crosslinking is only of value if it is accompanied by stringent purifying steps to remove irrelevant RNABP:RNA complexes and non-crosslinked RNA—stringent IPs, boiling in SDS prior to size separation on denaturing gels, and transfer to nitrocellulose membranes—that are embedded in the CLIP protocols. Thus it is probably best to reserve the term CLIP as defined here, and those studies using UV-crosslinking (or formaldehyde crosslinking) without such stringent purification of RNA-protein complexes47, 103, 104 need to consider validating their protein-RNA interactions by independent means.
CLIP concerns
Along with efforts to develop improvements to CLIP, some concerns about the original method have been raised 95. One is that the efficiency of crosslinking by UV irradiation is low. While this is accurate (in pilot experiments with purified protein and radiolabeled RNA, we have estimated efficiency to be on the order of 1–5%), this would be a major concern only if the complexity of tags that could be generated from a single CLIP experiment were rate limiting in data analysis. In general, this does not seem to be a major concern, as HITS-CLIP studies reported here have typically generated on the order of 105 up to >4 × 106 unique tags per experiment.
A second concern raised has been regarding DNA and RNA damage responses that are induced after UV irradiation. Several studies have documented cellular RNA responses to UV irradiation105–108, but these invariably require cells to be cultured at physiologic temperature after radiation, typically for 30 minutes or longer, presumably because these radiation responses are the result of well-studied biologic cascades that need to emerge in living cells. Since CLIP is typically done on acutely prepared cells kept at 4 °C during the irradiation, and are then immediately lysed or frozen, these processes are not likely to impinge on most CLIP experiments. However, the issue is worth bearing in mind if CLIP were to be attempted on tissues that were kept living after irradiation.
CLIP limitations
The specificity of crosslinking remains an area that is incompletely understood at the biophysical level. While there is some reported preference for UV to crosslink certain amino acids and nucleotides, this is not well established, in part because of the lack of consensus regarding either the means of assessing protein-nucleic acid crosslinking or conclusions regarding mechanism41. Studies of RNA-protein interaction have demonstrated that UV crosslinking can induce covalent bond formation between a large variety of amino acids with both purines and pyrimidines109. As a general point, CLIP has been used to identify binding targets for RNABPs that recognize both specific elements with a wide variety of sequence variation (YCAY, CU, U-rich, GA, elements) and RNABPs that do not appear to have any particular sequence bias (Ago crosslinking to mRNAs surrounding a variety of miRNA binding sites). In fact, Ago was able to form footprints in mRNA sequences surrounding 21 of 22 previously validated miR-124 sites; each such binding site was determined by the miR-124 seed, and appeared to be largely independent of any surrounding mRNA sequence bias, as were sequences surrounding 11,118 genome-wide Ago-mRNA CLIP clusters89. Moreover, in several cases (Nova, PTB, Ago-miR-124-mRNA footprints), binding sites identified by CLIP match very closely to predicted sites identified by independent means (e.g. exon junction array and bioinformatics). Thus in practical terms, if there is some degree sequence bias introduced by UV-crosslinking, it is likely relatively subtle, and does not appear to have had a major impact on studies reported in this review. Moreover, we have been able to crosslink all RNABPs attempted (8/8) to date in our laboratory, with efficiencies that vary at most by a factor of less than one order of magnitude as assessed by the number of unique tags detected.
Another situation where CLIP might miss specific interactions could arise if antibody-epitope interactions preclude analysis (i.e. crosslinking obscures an epitope); for this reason we have found comparison of CLIP results with different purification strategies (different antibodies) to be of value89. In addition, it should be recognized that some RNAs are post-transcriptionally modified—tRNA or rRNA harboring modified (pseudo-U) nucleotides, for example—which may not allow efficient reverse transcription and sequencing. Stretches of low complexity may crosslink efficiently, but yield results that are difficult interpret; for example, consider interpreting the results of CLIP with poly(A) binding protein.
CLIP was designed as an in vivo method to analyze mouse brain tissue. RNA-protein interactions studied in vitro may give only a snapshot of the extent of endogenous interactions, and these may represent a biased snapshot, reflecting stabilities and stoichiometries in vitro that are not necessarily reflective of those present within cells. In fact, Tollervey and colleagues noted significant differences in the sites of crosslinking in vivo and in vitro between Prp43 and rRNA84, likely reflecting such differences.
HITS-CLIP is increasingly dependent on bioinformatic analyses. As discussed for studies of FOX HITS-CLIP, bioinformatic strategies may have significant impact on data interpretability. Currently, the promise of HITS-CLIP as a quantitative tool has not been entirely realized. For example, the number of tags per transcript clearly relates to transcript abundance, but at least in our own Nova HITS-CLIP studies, we have not been able to normalize tags appropriately. The reason for this is that Nova binds both to intronic and exonic sequences, and normalization of intronic binding sites to transcript levels measured only on Affymetrix arrays, which are sensitive to exon but not intron levels, is not possible. In theory, normalization could be achieved through RNA-Seq, but this will need to await greater read depth in order to quantitate both intron and exon levels.
Future CLIP
CLIP improvements
An attractive advance in CLIP has been to use tagged proteins as a solution to the problem of lack of high quality antibodies or other means of protein purification. As discussed, several tags have successfully been used to purify RNABPs for CLIP studies83. One advantage to this approach is that they provide built-in negative controls—untransfected (tag-) cells, or even better, cells harboring constructs expressing tag alone (e.g. for large tags such as GFP; we have found that such tags can have background RNA binding that the investigator would want to be aware of). However, such tags also come at a price. Most notably, they invoke a need to attend to stoichiometry. Certainly blind overexpression of a tagged protein (e.g. from strong promoters) is likely to lead to identification of adventitious RNA-protein interactions.
A number of efforts have been made to improve on biases inherent in the original CLIP protocols. Direct RNA sequencing is an emerging technology110 that if applied to CLIP could eliminate the need for PCR amplification with its inherent biases and artifacts. Biases are introduced into the original CLIP protocol by the use of RNase A, which cleaves only at the 3’ end of unpaired pyrimidine residues. Yeo and colleagues addressed this by introducing the use of micrococcal nuclease, a relatively non-specific single stranded endo/exonuclease that can be inactivated by EGTA, although it hydrolyzes 5’ of A or U residues much faster than G or C residues111. Alkaline hydrolysis would offer an unbiased means of reducing RNA size, but has not yet been tested.
Recently, in CLIP studies of the RNABP TIA-1, Ule and colleagues have introduced several additional improvements, including the use of RNase I (which has no sequence specificity), an improved ligation protocol that used PEG 6000 to increase linker ligation efficiency, and primers that were optimized for paired-end Illumina high-throughput sequencing protocols102.
Another strategy has been considered to improve crosslinking efficiency, which relies on using substituted nucleotide analogs in tissue culture cells. Mourelatos and colleagues demonstrated that cells transfected with in vitro transcribed mRNAs and treated with 4-thio-Uridine and UV-irradiation could be immunoprecipitated with an antibody to Ago to interrogate protein-RNA interactions112. A very recent methods paper by Tuschl and colleagues describes using this strategy to undertake CLIP of tagged transfected RNABPs95. This strategy, termed PAR-CLIP, offers several opportunities, including high crosslinking efficiency, which may be helpful if native proteins show difficulty in efficient crosslinking through standard CLIP methods. In addition, the mutagenesis conferred by using nucleic acid analogs was capitalized on to allow mapping of direct crosslinking sites. While similar analyses proved valuable even in the absence of mutagens (e.g. as discussed above, Tollervey and colleagues mapped crosslinking sites by analyzing UV-damage induced mutations in CLIP tags84, and Ule and colleagues have developed iCLIP to map UV-generated crosslink sites46), the generality of using UV-CLIP to monitor crosslinked sites remains to be determined. Some concerns regarding PAR-CLIP are its use of a uridine analogue, which currently restricts its usefulness to tissue culture studies and confers cellular toxicity; applications in vivo would face a similar toxicity concerns.
In theory, DNA CLIP43 may provide a means of crosslinking/sequencing sites of DNA-protein interaction that would have higher resolution than current ChIP-Seq methods. However, given the success of ChIP-Seq in resolving binding sites bioinformatically113, potential advantages of DNA CLIP remain to be demonstrated.
As CLIP is applied to biologic systems, there is an increasing need to consider more refined CLIP studies. For example, a number of RNABPs are believed to be associated with cell-specific disorders114, 115; cell-type specific CLIP would offer an improved signal:noise in comparing RNA-protein interactions specifically in healthy and diseased cells. Combining tagged proteins with cell-specific promoters in model systems may provide a means of generating cell-specific CLIP from a complex tissue.
For such refined CLIP studies to be feasible, the sensitivity of the method will need to be improved. Optimization of RNA sequencing methods will be a major focus. Already improved linker ligation protocols are a step in this direction102; other approaches may be to eliminate linker ligation completely, using tailing methods as described for ribosome footprinting116, or direct RNA sequencing, eliminating PCR amplification (and hence maintaining complexity of starting populations).
Another parameter to be considered is time. Low intensity lasers with rapid pulse time can crosslinking RNA-protein complexes relatively efficiently, and if there were an indication for such fine resolution, these methods could be developed together with the CLIP methods reviewed here. For example, a single 50–100 mJ pulse of less than 10 nanosecond duration delivers a sufficient number of photons to induce measurable crosslinking41, suggesting that in the future CLIP may be able to be used to detect rapid (nanosecond scale) biological processes.
Finally, as HITS-CLIP data is acquired in combination with other high throughput data, such as RNA-Seq for the analysis of RNA variants, bioinformatics will take a greater role in data interpretation 4. For such data to be of the greatest power, it is essential that it be deposited in common databases such as GEO. The power of pooling many datasets has recently been underscored, through analysis of microarray studies, motifs and other features, in the breakthrough efforts to develop a complex RNA splicing code99. Going further, combining diverse data sets in Bayesian networks has demonstrated the predictive power of bioinformatics combined with CLIP-based biochemical in vivo footprints to yield new biologic insights into RNA splicing regulation68.
Conclusion
CLIP and HITS-CLIP are methods that can be used in conjunction with the analysis of RNA regulation by such means as RNA Seq profiling, and with bioinformatic tools, to produce genome-wide maps of sites of RNA regulation. The versatility of the method is evident in the wide variety of publications reviewed here, nearly all published since 2008, that include application of CLIP to a range of living organisms and tissues, subcellular compartments, and RNA-protein interactions. In the future refinement on all of these fronts should allow greater understanding of the role of RNA regulation in normal and disease biology.
Supplementary Material
Footnotes
Tag cluster: An array of two or more tags that overlap, or, in some cases, are predicted to overlap (Yeo and colleagues have estimated overlap given that tag length is often greater than Illumina read length, by counting tags within 50–100 nt as overlapping).
Biologic complexity (BC): The number of independent biologic replicates showing tags at a given location (cluster).
References
- 1.Corrionero A, Valcarcel J. RNA processing: Redrawing the map of charted territory. Mol Cell. 2009;36:918–919. doi: 10.1016/j.molcel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliphant AR, Brandl CJ, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollock R, Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990;18:6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 9.Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A. 1984;81:4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Zhang XS, Xia Y. Predicting eukaryotic transcriptional cooperativity by Bayesian network integration of genome-wide data. Nucleic Acids Res. 2009;37:5943–5958. doi: 10.1093/nar/gkp625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cech TR. Crawling out of the RNA world. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Tsai DE, Harper DS, Keene JD. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991;19:4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Racca C, et al. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh YY, et al. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 25.Steitz J. Immunoprecipitation of ribonucleoproteins using autoantibodies. Meth Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- 26.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000238. e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X Syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 30.Darnell JC, et al. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 31.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 33.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 37.Alexander P, Moroson H. Cross-linking of deoxyribonucleic acid to protein following ultra-violet irradiation different cells. Nature. 1962;194:882–883. doi: 10.1038/194882a0. [DOI] [PubMed] [Google Scholar]
- 38.Smith KC. Dose dependent decrease in extractability of DNA from bacteria following irradiation with ultraviolet light or with visible light plus dye. Biochem Biophys Res Commun. 1962;8:157–163. doi: 10.1016/0006-291x(62)90255-3. [DOI] [PubMed] [Google Scholar]
- 39.Ilyin YV, Georgiev GP. Heterogeneity of deoxynucleoprotein particles as evidencec by ultracentrifugation of cesium chloride density gradient. J Mol Biol. 1969;41:299–303. doi: 10.1016/0022-2836(69)90395-7. [DOI] [PubMed] [Google Scholar]
- 40.Schoemaker HJ, Schimmel PR. Photo-induced joining of a transfer RNA with its cognate aminoacyl-transfer RNA synthetase. J Mol Biol. 1974;84:503–513. doi: 10.1016/0022-2836(74)90112-0. [DOI] [PubMed] [Google Scholar]
- 41.Fecko CJ, et al. Comparison of femtosecond laser and continuous wave UV sources for protein-nucleic acid crosslinking. Photochem Photobiol. 2007;83:1394–1404. doi: 10.1111/j.1751-1097.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- 42.Urlaub H, Hartmuth K, Luhrmann R. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 2002;26:170–181. doi: 10.1016/S1046-2023(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 43.Law A, Hirayoshi K, O'Brien T, Lis JT. Direct cloning of DNA that interacts in vivo with a specific protein: application to RNA polymerase II and sites of pausing in Drosophila. Nucleic Acids Res. 1998;26:919–924. doi: 10.1093/nar/26.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brimacombe R, Stiege W, Kyriatsoulis A, Maly P. Intra-RNA and RNA-protein cross-linking techniques in Escherichia coli ribosomes. Methods Enzymol. 1988;164:287–309. doi: 10.1016/s0076-6879(88)64050-x. [DOI] [PubMed] [Google Scholar]
- 45.Zwieb C, Ross A, Rinke J, Meinke M, Brimacombe R. Evidence for RNA-RNA cross-link formation in Escherichia coli ribosomes. Nucleic Acids Res. 1978;5:2705–2720. doi: 10.1093/nar/5.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1838. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, et al. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 48.Mayrand S, Setyono B, Greenberg JR, Pederson T. Structure of nuclear ribonucleoprotein: identification of proteins in contact with poly(A)+ heterogeneous nuclear RNA in living HeLa cells. J Cell Biol. 1981;90:380–384. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayrand S, Pederson T. Nuclear ribonucleoprotein particles probed in living cells. Proc Natl Acad Sci U S A. 1981;78:2208–2212. doi: 10.1073/pnas.78.4.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreyfuss G, Choi YD, Adam SA. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4:1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darnell RB. Developing global insight into RNA regulation. Cold Spring Harb Symp Quant Biol. 2006;71:321–327. doi: 10.1101/sqb.2006.71.002. [DOI] [PubMed] [Google Scholar]
- 52.Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Molecular & Cellular Biology. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen KB, et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 54.Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci U S A. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 56.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 57.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 58.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 62.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayr C, Bartel DP. Widespread shortening of 3' UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Frias MA, Mele A, Licatalosi D, Darnell RB. Integrative modeling defines a comprehensive splicing-regulatory network and its combinatorial controls. Science. 2010 doi: 10.1126/science.1191150. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf J, Fink G. Feed-forward regulation of a cell fate determinant by an RNA-binding protein in yeast. Genetics. 2010 doi: 10.1534/genetics.110.113944. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 72.Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Brug MP, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000600. e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanford JR, et al. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS ONE. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 81.Becht P, Konig J, Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci. 2006;119:4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- 82.Konig J, et al. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 2009;28:1855–1866. doi: 10.1038/emboj.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wurtmann EJ, Wolin SL. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 88.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu M, Medvedev S, Yang J, Hecht NB. MIWI-independent small RNAs (MSY-RNAs) bind to the RNA-binding protein, MSY2, in male germ cells. Proc Natl Acad Sci U S A. 2009;106:12371–12376. doi: 10.1073/pnas.0903944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hafne RM, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang CS, et al. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 97.Ruggiu M, et al. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci U S A. 2009;106:3513–3518. doi: 10.1073/pnas.0813112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010 doi: 10.1016/j.neuron.2010.05.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 100.Xing Y, et al. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 103.Poon MM, Chen L. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A. 2008;105:20303–20308. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burd CJ, Kinyamu HK, Miller FW, Archer TK. UV radiation regulates Mi-2 through protein translation and stability. J Biol Chem. 2008;283:34976–34982. doi: 10.1074/jbc.M805383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leverkus M, Yaar M, Eller MS, Tang EH, Gilchrest BA. Post-transcriptional regulation of UV induced TNF-alpha expression. J Invest Dermatol. 1998;110:353–357. doi: 10.1046/j.1523-1747.1998.00154.x. [DOI] [PubMed] [Google Scholar]
- 107.Li B, Si J, DeWille JW. Ultraviolet radiation (UVR) activates p38 MAP kinase and induces post-transcriptional stabilization of the C/EBPdelta mRNA in G0 growth arrested mammary epithelial cells. J Cell Biochem. 2008;103:1657–1669. doi: 10.1002/jcb.21554. [DOI] [PubMed] [Google Scholar]
- 108.Munoz MJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 109.Havron A, Sperling J. Specificity of photochemical cross-linking in protein-nucleic acid complexes: identification of the interacting residues in RNase- pyrimidine nucleotide complex. Biochemistry. 1977;16:5631–5635. doi: 10.1021/bi00644a038. [DOI] [PubMed] [Google Scholar]
- 110.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 111.Krupp G, Gross HJ. Rapid RNA sequencing: nucleases from Staphylococcus aureus and Neurospora crassa discriminate between uridine and cytidine. Nucleic Acids Res. 1979;6:3481–3490. doi: 10.1093/nar/6.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirino Y, Mourelatos Z. Site-specific crosslinking of human microRNPs to RNA targets. RNA. 2008;14:2254–2259. doi: 10.1261/rna.1133808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 114.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 116.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.