Abstract
BioA catalyzes the second step of biotin biosynthesis and this enzyme represents a potential target to develop new antitubercular agents. Herein we report the design, synthesis, and biochemical characterization of a mechanism-based inhibitor (1) featuring a 3,6-dihydropyrid-2-one heterocycle that covalently modifies the pyridoxal 5′-phosphate (PLP) cofactor of BioA through aromatization. The structure of the PLP adduct was confirmed by MS/MS and X-ray crystallography at 1.94 Å resolution. Inactivation of BioA by 1 was time- and concentration-dependent and protected by substrate. We used a conditional knock-down mutant of M. tuberculosis to demonstrate the antitubercular activity of 1 correlated with BioA expression and these results provide support for the designed mechanism of action.
INTRODUCTION
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), has plagued mankind for millennia and is presently the leading cause of bacterial infectious disease mortality.1 Rifampin, the last approved first-line drug for TB chemotherapy was introduced in 1968.2 During the last four decades resistance to all of the first- and second-line antitubercular agents has been reported. Consequently, new drugs that inhibit novel targets in Mtb are needed. Biotin (vitamin H) is an essential cofactor responsible for activation of carbon dioxide in acyl-CoA carboxylases (ACCs) involved in fatty acid metabolism and pyruvate carboxylase (PCC) in gluconeogenesis.3 Biotin starvation of an Mtb biotin auxotroph led to cell death suggesting biotin biosynthesis represents a very attractive pathway for development of new antitubercular agents.4 Significantly, conditional silencing of bioA, which encodes for the second enzyme in the biotin biosynthetic pathway, after establishment of an infection, demonstrated that BioA is essential to maintain a chronic infection in a murine model of TB.5
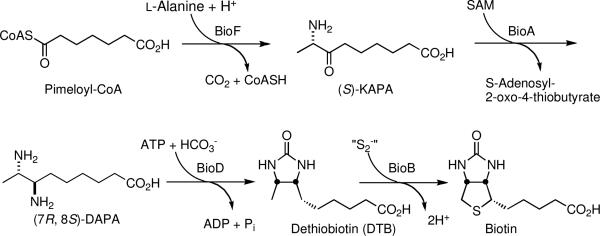
In analogy to the well-studied pathway in E. coli, biotin biosynthesis in Mtb begins from pimeloyl-CoA, which is elaborated to biotin in four steps catalyzed, respectively by BioF, BioA, BioD, and BioB (Scheme 1).6 The first step is carried out by BioF that converts pimeloyl-CoA to 7-keto-8-aminopelargonic acid (KAPA) through decarboxylative condensation with l-alanine.6a In the second step, BioA, a pyridoxal 5′-phosphate (PLP) dependent enzyme, catalyzes the transamination of KAPA into 7,8-diaminopelargonic acid (DAPA) using S-adenosylmethionine (SAM) as the amino donor.6b–d BioD, dethiobiotin synthetase, carboxylates DAPA to form the imidazolidin-2-one ring of dethiobiotin (DTB) in the third step.4a Finally, BioB, an iron-sulfur cluster enzyme, catalyzes the C-H activation and insertion of sulfur in DTB to afford biotin.6e,f Although the source of pimelate in Mtb is unknown, Cronan and co-workers have recently shown that E. coli co-opts the fatty acid synthase type II pathway along with BioC (Mtb ortholog: Rv0089) and BioH (Mtb ortholog: Rv2715) for synthesis of pimeloyl-ACP, which is proposed as the physiologically relevant substrate for BioF.7
Scheme 1.
Biosynthesis of Biotin in Mtb.
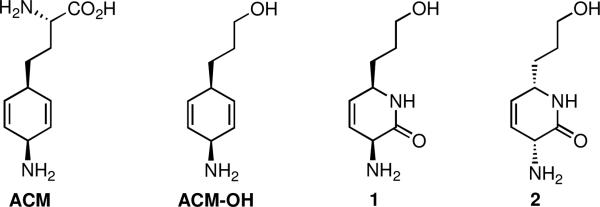
Amiclenomycin (ACM) isolated from Streptomyces lavendulae subsp. amiclenomycini and its simplified amino-alcohol analogue ACM-OH are potent inhibitors of BioA, and we have shown that ACM-OH possesses better whole cell activityw against Mtb (Figure 1).8, 10c Synthetic studies indicated the stereochemistry of the 1,4-cyclohexadiene ring is cis rather than trans as originally assigned.9 Amiclenomycin is a mechanism-based inhibitor through covalent modification of the PLP cofactor of BioA via aromatization.10 Remarkably, ACM exhibits selective antitubercular activity amongst 40 bacterial strains evaluated.8b Chemical complementation of Mtb cultures with DAPA, but not KAPA antagonizes the activity of ACM, thus confirming the mechanism of action.8b Although amiclenomycin has served as an excellent tool compound, it suffers from poor chemical stability due to spontaneous aromatization.10
Figure 1.
Structure of amiclenomycin (ACM), ACM-OH, and dihydropyrid-2-one inhibitors 1 and 2.
Inspired by the unique aromatization mechanism of inhibition by amiclenomycin10 and the extensive previous work by Silverman and co-workers11 on related PLP-dependent mechanism-based inhibitors, we hypothesized that the introduction of a heteroatom into the 1,4-cyclohexadiene ring scaffold would decrease the aromatic stabilization energy and correspondingly would improve the chemical stability of the inhibitor while maintaining the same aromatization mechanism of inhibition (Figure 2). Therefore, a pair of enantiomeric inhibitors based on the simplified ACM-OH structure, containing the 3,6-dihydropyridone heterocycle, was designed to test our hypothesis (Figure 1). Herein, we report the synthesis and kinetic characterization of a mechanism-based inhibitor of BioA with improved chemical stability.
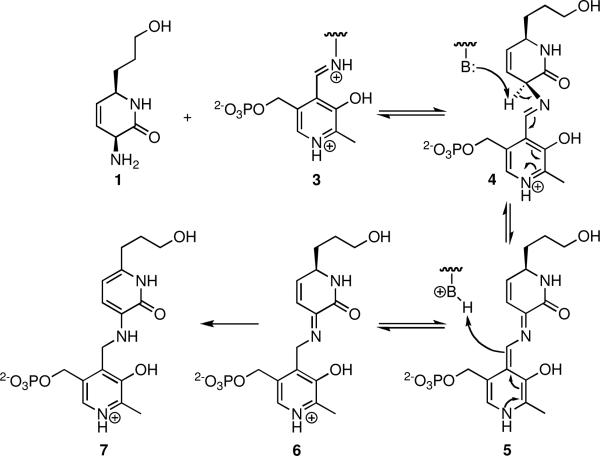
Figure 2.
Proposed aromatization mechanism of 1. Transamination of 1 with enzyme-bound PLP 3 affords the substrate PLP aldimine 4. Subsequent 1,3-prototropic shift via 5 mediated by Lys283 provides ketimine 6. Tautomerization of 6 to 7 is irreversible and leads to formation of a stabile PLP adduct.
RESULTS
Synthesis of inhibitor 1 and 2
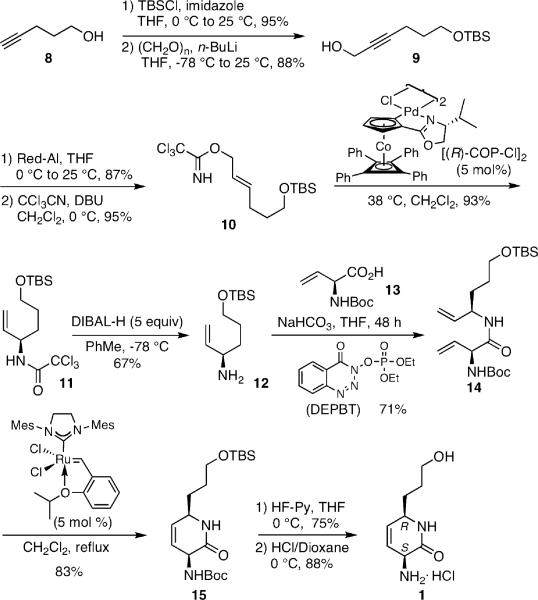
The enantioselective syntheses of 1 and 2 were accomplished in a convergent fashion employing allylic amine 12 and vinylglycine 13 building blocks as depicted in Scheme 2. The allylic amine (R)-12, with high enantiopurity, was efficiently prepared starting with pentynol 8, which was converted to propargylic alcohol 9 by sequential silylation with TBSCl and addition to paraformaldehyde. Red-Al mediated reduction of 9 furnished exclusively the trans-allylic alcohol and treatment with trichloroacetonitrile and DBU afforded trichloroacetimidate 10. Asymmetric Overman rearrangement of 10 employing the cationic Pd(II)-catalyst [(R)-COP-Cl]2 provided the trichloroacetyl protected allylic amine (R)-11 in 93% yield and 96:4 enantiomeric ratio (er).12 Subsequent deprotection with DIBAL-H yielded the desired allylic amine (R)-12. Due to the facile isomerization as well as racemization of vinylglycine derivatives under conventional peptide coupling conditions, we employed DEPBT as the coupling reagent and NaHCO3 as base, a very mild condition proven to minimize racemization,14 for the coupling of vinylglycine (S)-1313a, b with (R)-12 to afford diene 14 in 71% yield. A variety of ring closing metathesis (RCM) catalysts were screened for the RCM reaction of 14, and Hoveda-Grubbs-II catalyst15 was found to be most effective, furnishing (3S, 6R)-dihydropyridone 15 in 83% yield. Finally, the desired (3S, 6R)-dihydropyridone 1 was obtained by sequential deprotection of the TBS and Boc groups with HF·Py and HCl–dioxane, respectively. (3R, 6S)-Dihydropyridone 2 was prepared analogously in 10 steps employing [(S)-COP-Cl]2 and (R)-1313c. Dihydropyridones 1 and 2 showed substantially improved chemical stability compared to ACM, displaying no degradation after 60 days at room temperature under aerobic conditions in DMSO as measured by 1H NMR.
Scheme 2.
Synthesis of 1.
Biochemical evaluation
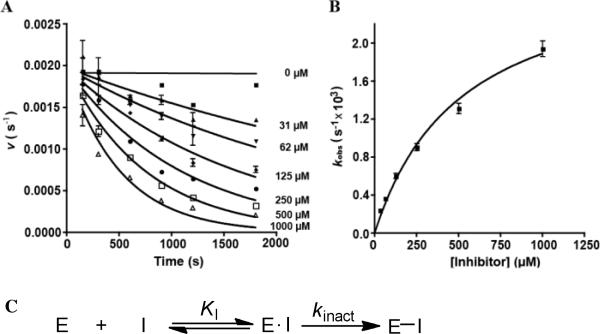
The ability of 1 and 2 to irreversibly inhibit BioA was evaluated by pre-incubation of 0.03–1 mM inhibitor with 2 μM BioA in the absence of substrates (KAPA and SAM). Aliquots were removed at time t and assayed for enzyme activity using a coupled assay employing BioD that measures production of DTB from (S)-KAPA.16 Dihydropyridone 1 exhibited time-dependent inhibition while 2 displayed no activity. Fitting the observed initial velocity as a function of incubation time t for 1 provided the pseudo first-order rate constants of inactivation, kobs at a given inhibitor concentration [I] (Figure 3A and 3B). The hyperbolic relationship between kobs and [I] is consistent with a two-step mechanism of inactivation (Figure 3C). The kinetic parameters kinact, KI (Table 1) were determined from fitting the kobs versus [I] data to equation 2 (see Experimental Section).17 While the kinact of 1 compares favorably with ACM and ACM-OH, the KI is approximately 26-fold higher than ACM-OH. The second-order rate constant for inactivation kinact/KI of 1 is 346 M–1 min–1, which is 80-fold lower than ACM-OH. We also showed that addition of the substrate (S)-KAPA to the initial incubation mixture of BioA and 1 protected against inactivation as expected for a mechanism-based inhibitor. To evaluate potential biochemical selectivity, 1 was also evaluated against two commercially available mammalian PLP-dependent enzymes including alanine aminotransaminase and aspartate aminotransaminase. Inhibitor 1 was inactive against aspartate aminotransaminase, but did show time-dependent inhibition against alanine aminotransaminase. The inhibition was not saturable with up to 2 mM 1; however, the slope of the kobs versus [I] could be fit by linear regression analysis to provide the kinact/KI value of 37 M-1min-1.
Figure 3.
(A) Residual activity of BioA as measured by initial velocity, v, versus preincubation time in the presence of increasing concentrations of 1, showing time dependent inactivation of BioA. (B) Plot of the observed rate of inactivation constants (kobs) versus the concentration of 1, from which the kinetic parameters kinact, KI and the second-order rate constant kinact/KI were determined. (C) Scheme showing two-step mechanism of inactivation of BioA by 1.
Table 1.
Kinetic parameters for inhibition of BioA by ACM and analogues.
| Inhibitor | kinact (min–1) | KI (μM) | kinact/KI (M–1 min–1) |
|---|---|---|---|
| 1 | 0.18 ± 0.01 | 520 ± 72 | (3.5 ± 0.5) × 102 |
| 2 | < 0.01 | > 2000 | < 5 |
| ACMa | 0.35 ± 0.05 | 12 ± 2 | (2.9 ± 0.6) × 104 |
| ACM-OHa | 0.56 ± 0.05 | 20 ± 2 | (2.8 ± 0.4) × 104 |
ref 6c
MS analysis
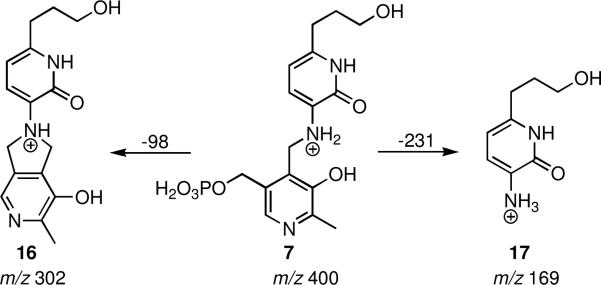
To confirm the formation of the aromatic adduct proposed in Figure 2, MS analysis was performed to identify the formation of the aromatic adduct. We incubated 1 with BioA (360 μM 1, 26 μM BioA, 50 mM Tris pH 8.0, 10 mM 2-mercaptoethanol and 100 μM PLP) at 37 °C for 2 h, which led to complete enzyme inactivation. BioA was subsequently isolated by gel filtration to remove excess 1 and PLP, then analyzed using electrospray ionization tandem mass spectrometry. The PLP-adduct 7 was found to dissociate from the protein complex and the quasimolecular ion was detected at m/z 400, which corresponds to the expected aromatized PLP adduct (Figure 4). The MS/MS spectrum of this ion displayed two fragment ions at m/z 302 [M + H – 98]+ and 169 [M + H – 231]+. The peak at m/z 302 corresponds to the loss of H3O3PO– from the parent molecule suggesting structure 16 while the peak at m/z 169 is consistent with the aromatized inhibitor ion 17 (Figure 4). These results demonstrate formation of the aromatic PLP-adduct and hence provide strong support for the proposed aromatization mechanism.
Figure 4.
Observed fragments by MS/MS of the m/z 400 ion.
Structural characterization
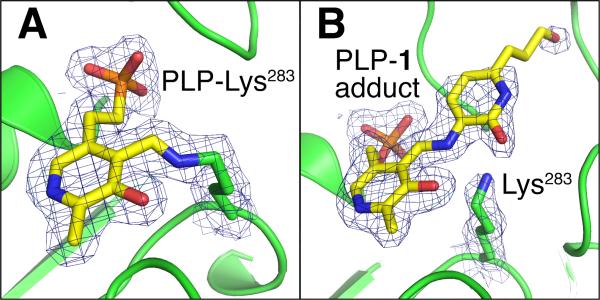
To provide structural insight on adduct formation in the context of the protein active site, we soaked yellow, PLP-loaded crystals of BioA4a with 1 and observed 1 concentrate rapidly to within the crystal affording red crystals before apparently reacting to yield colorless crystals (Figure S2). Crystals soaked with 1 were frozen in liquid nitrogen when visibly red or colorless. Diffraction data to 1.95 Å and 1.94 Å resolution were collected from a red, pre-reaction crystal and a colorless, post-reaction crystal, respectively (Table S1).
In the pre-reaction complex, the electron density describes a well-defined PLP covalently bound to Lys283 in a manner identical to previous reports4 (Figure 5A). New positive difference density accumulated adjacent to PLP, presumably due to the binding of 1 during soaking (Figure S3). In the post-reaction complex, Lys283 is very clearly no longer covalently bound to PLP and the planar electron density is consistent with the aromatized adduct 7 (Figure 5B, Figure S4-B). The observed adduct possesses small differences in conformation between the two crystallographically unique molecules in the BioA dimer. In the second molecule in the asymmetric unit, electron density for the aromatized adduct is less well defined and positioned on the opposite side of a flipped Trp64 (Figure S4). Nevertheless, degradation of the internal aldimine appears complete in both molecules in the post-reaction structure.
Figure 5.
Final 2Fo-Fc electron density contoured at 1.0 for the pre-reaction complex (A) and the post-reaction complex (B).
In vivo antitubercular activity
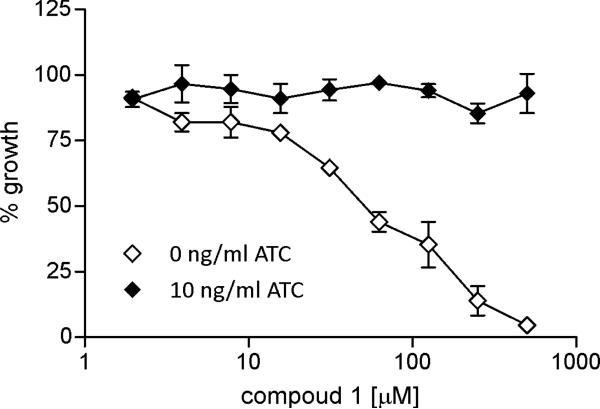
Lastly, we evaluated the antitubercular activity of 1 in biotin-free medium against a mutant strain of M. tuberculosis H37Rv termed Mtb bioA-TetON-1 that enables conditional expression of BioA.5 In the presence of 10 ng/mL anhydrotetracycline (ATC), this strain overexpresses BioA, whereas in the absence of ATC, this strain expresses approximately 7% BioA compared to wt Mtb. Overexpression of BioA conferred resistance to 1 while underexpression of BioA restored sensitivity (Figure 6). The correlation between antitubercular activity of 1 and BioA expression level provides support for the designed mechanism of action. Additionally, the active enantiomer 1 was evaluated against two mammalian cell lines (Vero and CHO-K1) to provide an initial assessment of potential toxicity and 1 showed no inhibition of cell viability up to 1 mM, the maximum concentration employed.
Figure 6.
Anti-TB activity of 1. Mtb bioA-TetON-1 was grown in biotin-free Sauton's medium containing 0 ng/ml or 10 ng/ml ATC and the indicated concentration of compound 1. Growth was measured as optical density at 580 nm and normalized to growth without 1. Error bars represent the standard error of the mean of triplicate measurements.
DISCUSSION
Rationale and design of 1
The 1,4-cyclohexadiene warhead of amiclenomycin (ACM) is both thermodynamically and kinetically unstabile due to the large aromatic stabilization energy of approximately 36 kcal/mol. Replacement of one of the olefins in ACM with an amide isostere provides a 3,6-dihydropyridone heterocycle, which has an estimated aromatic stabilization of only 25 kcal/mol28, and hence is predicted to be substantially more stabile. Mann and Ploux reported on the synthesis of the simplified ACM analog that we term ACM-OH wherein the polar amino acid moiety on the C-4 side-chain was replaced with a simple neutral hydroxy group.6c We independently synthesized ACM-OH and demonstrated that this structural simplification did not adversely impact antitubercular activity.10c Consequently, we elected to use this modification in our designed 3,6-dihydropyridone analogs. Although the X-ray structure of BioA from Mtb was reported,4a simple molecular docking studies were unable to reveal whether 1 or 2 would be more active, thus both enantiomers were prepared.
Antitubercular activity
Park and co-workers recently developed Mtb conditionally-regulated knockdown strains of BioA.5 Significantly, it was shown that BioA activity must be reduced by approximately 99% to prevent bacterial growth.5 It should be noted that these were among the first experiments to address the extent of inactivation, or vulnerability, required to halt replication against any target in Mtb. Based on these results, BioA is not particularly vulnerable to inhibition since nearly complete inactivation is required. Consequently, irreversible inhibitors or tight-binding inhibitors with slow dissociation rates are likely to be more effective than simple rapidly reversible inhibitors.29
The Mtb strains that enable conditional BioA expression as shown herein are also extremely useful to assess on-target activity of compounds, since these permit the correlation of antitubercular activity with enzyme expression levels. In the present case, overexpression of BioA in strain Mtb bioA-TetON-1 renders Mtb resistant to 1, presumably by stoichiometric titration of the inhibitor. This result is readily reconciled, since the amount of BioA produced in this conditional strain is approximately 100-fold greater in the presence of the small molecule anhydrotetracycline (ATC), versus in its absence.
Biochemical Activity and Mechanism
The kinact values for 1 is only 3-fold lower than ACM-OH showing that replacement of the 1,4-cyclohexadiene warhead with the dihydropyridone has little impact on the rate of inactivation. Other described mechanism-based inhibitors of PLP dependent enzymes are typically on the order of ~1 min-1.11 The slow rate of inactivation is commensurate with the relatively slow turnover rate of PLP-dependent enzyme due to slow transamination between the internal and external aldimines.30
Mechanism-based inhibitors of PLP-dependent enzymes that operate through an aromatization mechanism are fascinating and have examples beyond amiclenomycin and 3,6-dihydropyridone 1, including the natural product gabaculine, isogabaculine, 4-amino-4,5-dihydro-2-furancarboxylic acid, 4-amino-4,5-dihydro-2-thiophenecarboxylic acid, d-cycloserine, and gostatin.11,34 Interestingly, d-cycloserine is a second-line antitubercular agent, whose mechanism of action is due to irreversible inactivation of the mycobacterial PLP-dependent enzyme d-alanine racemase.2d-Cycloserine has also been shown to inhibit host PLP-dependent enzymes including γ-aminobutyric acid aminotransferase (GABA-AT)11a and serine palmitoyltransferase.32 To investigate the potential selectivity of 1 toward other PLP-dependent enzymes we screened it against porcine alanine aminotransaminase and aspartate aminotransaminase. 3,6-Dihydropyridone 1 was inactive against aspartate aminotransaminase, but showed weak time-dependent, yet nonsaturable inhibition against alanine aminotransaminase with a kinact/KI value of 37 M1min-1 demonstrating a promising level of biochemical selectivity for BioA.
Several mechanisms of BioA inactivation are plausible including an aromatization mechanism, an enamine mechanism and a Michael addition mechanism.31 However, the isolation of the PMP adduct 7 by mass spectrometry and X-ray analysis provides unequivocal support of the aromatization mechanism of inhibition. Interestingly, inactivation of GABA-AT by 4-amino-4,5-dihydro-2-furancarboxylic acid does not operate through an aromatization mechanism, but rather a Michael-addition mechanism. The resulting adduct between GABA-AT and 4-amino-4,5-dihydro-2-furancarboxylic acid is not stabile and undergoes slow hydrolysis resulting in restoration of enzyme activity. By contrast, the corresponding thiophene derivative 4-amino-4,5-dihydro-2-thiophenecarboxylic acid inactivates GABA-AT via an aromatization mechanism and the resulting adduct is stabile. The divergence in the mechanisms of these two structurally related inhibitors has been reconciled by consideration of the lower aromatic stabilization energy of furan (16 kca/mol) relative to thiophene (29 kcal/mol).33 As noted earlier, the aromatic stabilization of 1 is 25 kcal/mol,28 which appears to be a perfect balance ensuring improved chemically stability of the inhibitor, yet adequate to favor reaction via an aromatization pathway.
Structural Characterization
Consistent with the mass spectrometry results, the crystal structure clearly shows the formation of an adduct of 1 with PLP that is consistent with the proposed structure 7 (Figure 5). Lys283 is positioned to mediate the 1,3-prototropic shift (Figure 2). This adduct is conformationally flexible and possesses few hydrogen bonds to the surrounding protein, which likely explains the discontinuous electron density for the length of the adduct (Figure 5B, Supplemental Figure 3B). The adduct forms in a time- and concentration-dependent manner with diffraction data collected from intermediate time points showing a mixture of pre- and post-reaction states (data not shown). The initial red color observed upon soaking crystals of BioA with 1 likely corresponds to a minor, transient build-up of intermediate 5 (Figure 2).
CONCLUSION
In conclusion, we have designed, synthesized and kinetically and structurally characterized a mechanism-based inhibitor of BioA. The dihydropyrid-2-one heterocycle of 1 is an isostere of the 1,4-cyclohexadiene warhead of amiclenomycin (ACM) and we demonstrated that it irreversibly inactivates the PLP cofactor of BioA via an aromatization mechanism. The process is highly selective as the enantiomeric inhibitor 2 is completely inactive. We expect the greater chemical stability of 1 compensates for the attenuated inhibitor efficiency relative to ACM as measured by the second order rate constant kinact/KI. The inverse relationship between antitubercular activity of 1 and BioA protein expression provides support for the designed mechanism of action. In view of the importance of drug-target residence time to efficacy of new drugs, selective mechanism-based inhibitors such as these may have a higher likelihood of translating into useful therapeutics.29 The post-reaction complex structure will be used to guide the generation of derivative compounds that will possess higher specificity and activity.
EXPERIMENTAL SECTION
General methods
All chemical reactions were performed under an inert atmosphere of dry Ar or N2 in oven-dried (150 °C) glassware. 1H and 13C NMR spectra were recorded on a Varian 600 MHz spectrometer. Proton chemical shifts are reported in ppm from an internal standard of residual chloroform (7.26 ppm) or methanol (3.31 ppm), and carbon chemical shifts are reported using an internal standard of residual chloroform (77.0 ppm) or methanol (49.1 ppm). Proton chemical data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, p = pentet, m = multiplet, br = broad), integration, coupling constant. Melting points were measured on a Büchi 535 melting point apparatus and uncorrected. High resolution mass spectra were obtained on an Agilent TOF II TOF/MS instrument equipped with either an ESI or APCI interface. TLC analyses were performed on TLC silica gel 60F254 from EMD Chemical Inc., and were visualized with UV light, iodine chamber, 10% sulfuric acid or 10% PMA solution. Optical rotations values were obtained on a Rudoldf Autopol III Polarimeter. Purifications were performed by flash chromatography on silica gel (Dynamic Adsorbents, 60A). Enzymatic activity, kinetic parameters, and inhibition assays were performed on a molecular devices (Sunnyvale, CA, USA) M5e multi-mode plate reader. ESI MS/MS experiments were performed on a 2000 QTRAP LC/MS/MS spectrometer (Applied Biosystems).
Materials
Chemicals and solvents were purchased from Sigma-Aldrich Company or Acros Organic Fischer Company, and were used as received. An anhydrous solvent dispensing system (J. C. Meyer) using 2 packed columns of neutral alumina was used for drying THF, Et2O, and CH2Cl2 while 2 packed columns of molecular sieves were used to dry DMF and the solvents were dispensed under argon. (S)-N-Boc-vinylglycine 1313a,b, (R)-N-Boc-vinylglycine enan-1313c, (8S)-7-keto-8-aminopelargonic acid hydrochloride salt (KAPA·HCl)16 and fluorescein-tagged dethiobiotin (Fl-DTB)16 were synthesized according to the reported procedures. BioA and BioD were cloned, overexpressed, and purified following the reported protocols.16 For crystallization, the published purification protocols were altered to replace Tris and DTT with HEPES and TCEP, respectively.4a
Chemistry experimental procedures for inhibitor 1
tert-Butyldimethyl(pent-4-ynyloxy)silane (S1)
To a solution of pent-4-yn-1-ol 8 (8.4 g, 100 mmol, 1.0 equiv) and imidazole (10.2 g, 150 mmol, 1.5 equiv) in THF (200 mL) at 0 °C, was added TBSCl (16.6 g, 110 mmol, 1.1 equiv) portionwise. The reaction was gradually warmed from 0 °C to 25 °C over 0.5 h and stirred for 2 h at 25 °C. The reaction was quenched by addition of saturated aqueous NH4Cl (100 mL), and the aqueous layer was separated and extracted with Et2O (3 × 50 mL). The combined organic layers were washed with saturated aqueous NaCl (50 mL), dried (MgSO4), and concentrated to afford the crude product as colorless oil. Distillation under reduced pressure afforded the title compound (18.8 g, 95%) as a colorless oil: bp 63–65 °C (P = 10 mbar); 1H NMR (600 MHz, CDCl3) δ 0.01 (s, 6H), 0.85 (s, 9H), 1.63 (p, J = 7.0 Hz, 2H), 1.81 (t, J = 2.7 Hz, 1H), 2.22 (td, J = 7.0, 2.7 Hz, 2H), 3.64 (t, J = 6.0 Hz, 2H); 13C NMR (150 MHz, CDCl3) δ –5.4, 15.2, 18.5, 26.2, 30.9, 62.2, 68.3, 84.0. All data are consistent with reported values.18
6-(tert-Butyldimethylsilyloxy)hex-2-yn-1-ol (9)
To a solution of S1 (1.98 g, 10 mmol, 1.0 equiv) in THF (50 mL) at –78 °C was added n-BuLi (4.8 mL, 2.5 M in hexanes, 12 mmol, 1.2 equiv) dropwise over 15 min. The solution was stirred at –78 °C for 30 min then paraformaldehyde (1.5 g, 50 mmol, 5.0 equiv) was added portionwise over 1 h. The mixture was gradually warmed up to 25 °C and stirred 16 h. The reaction was quenched by addition of saturated aqueous NH4Cl solution (50 mL) and the aqueous layer was separated and extracted with Et2O (3 × 25 mL). The combined organic layers were washed with saturated aqueous NaCl (50 mL), dried (MgSO4) and concentrated. Purification by flash chromatograph afforded the title compound (2.01 g, 88%) as colorless liquid: Rf 0.30 (15% EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 0.03 (s, 6H), 0.87 (s, 9H), 1.68 (p, J = 6.0 Hz, 2H), 2.12 (br t, J = 6.0 Hz, 1H), 2.27 (tt, J = 6.0, 2.4 Hz, 2H), 3.66 (t, J = 6.0 Hz, 2H), 4.21 (t, J = 2.4 Hz, 2H); 13C NMR (150 MHz, CDCl3) δ –5.1, 15.4, 18.5, 26.1, 31.8, 51.4, 61.8, 78.7, 86.1. All data are consistent with reported values.18
(E)-5-(tert-Butyldimethylsilyloxy)pent-2-en-1-ol (S2)
To a solution of 9 (1.14 g, 5 mmol, 1.0 equiv) in Et2O (50 mL) at 0 °C was added Red-Al (3.75 mL, 65% wt% in toluene, 12.5 mmol, 2.5 equiv) dropwise over 20 min. The solution was warmed to 23 °C over 1 h, then stirred 6 h at 23 °C. The reaction was cooled down to 0 °C and quenched by the dropwise addition of MeOH (0.5 mL). The reaction mixture was then diluted with Rochelle's salt solution (50 mL) and stirred for 30 min at 23 °C. The aqueous layer was separated and extracted with Et2O (3 × 25 mL). The combined organic layers were washed with saturated aqueous NaCl (25 mL), dried (MgSO4) and concentrated. Purification by flash chromatograph afforded the title compound (1.00 g, 87%) as a colorless oil: Rf 0.25 (15% EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 0.05 (s, 6H), 0.88 (s, 9H), 1.56 (p, J = 6.0 Hz, 2H), 2.07 (q, J = 6.6 Hz, 2H), 3.56 (t, J = 6.6 Hz, 2H), 4.05 (d, J = 3.5 Hz, 2H), 5.50–5.75 (m, 2H); 13C NMR (150 MHz, CDCl3) δ –5.1, 18.3, 25.9, 28.5, 32.2, 62.5, 63.5, 129.5, 133.5. All data are consistent with reported values.9
(E)-6-(tert-Butyldimethylsilyloxy)hex-2-en-1-yl 2,2,2-trichloroacetimidate (10)
To a solution of S2 (920 mg, 4 mmol, 1.0 equiv) in CH2Cl2 (20 mL) at 0 °C, DBU (0.72 mL, 4.8 mmol, 1.2 equiv) and CCl3CN (0.6 mL, 6 mmol, 1.5 equiv) were sequentially added dropwise. The reaction was stirred 10 min at 0 °C then warmed to 23 °C and stirred for 2 h. The reaction was concentrated in vacuo and directly purified by flash chromatograph to afford the title compound (1.42 g, 95%) as colorless oil: Rf 0.70 (10% EtOAc/hexanes); 1H NMR (600 MHz, CDCl3) δ 0.04 (s, 6H), 0.88 (s, 9H), 1.63 (p, J = 6.9 Hz, 2H), 2.14 (td, J = 6.9, 6.6 Hz, 2H), 3.62 (t, J = 6.9 Hz, 2H), 4.73 (d, J = 6.6 Hz, 2H), 5.68 (td, J = 15.0, 6.6 Hz, 1H), 5.88 (td, J = 15.0, 6.6 Hz, 1H), 8.26 (br s, 1H); 13C NMR (150 MHz, CDCl3) δ –5.05, 18.5, 26.2, 28.8, 32.1, 62.6, 70.1, 91.8, 123.5, 136.7, 162.8; HRMS (ESI+): calcd for C14H27Cl3NO2Si [M + H]+ 374.0871, found 374.0859 (error 3.2 ppm).
(R)-N-[6-(tert-Butyldimethylsilyloxy)hex-1-en-3-yl]-2,2,2-trichloroacetamide (11)
To a solution of 10 (1.12 g, 3 mmol, 1.0 equiv) in CH2Cl2 (15 mL) at 23 °C was added [(R)-COP-Cl]2 (219 mg, 0.15 mmol, 0.05 equiv). The reaction was stirred at 38 °C for 36 h, then cooled to 23 °C and concentrated in vacuo. Purification by flash chromatograph afforded the title compound (1.04 g, 93%) as light yellow oil: Rf 0.66 (10% EtOAc/hexane); (c 0.5, CH2Cl2); 1H NMR (600 MHz, CDCl3) δ 0.04 (s, 6H), 0.88 (s, 9H), 1.55–1.59 (m, 2H), 1.61–1.69 (m, 1H), 1.73–1.79 (m, 1H), 3.61–3.67 (m, 2H), 4.43 (dt, J = 13.8, 6.9 Hz, 1H), 5.18 (d, J = 10.8 Hz, 1H), 5.23 (d, J = 17.4 Hz, 1H), 5.80 (ddd, J = 17.4, 10.8, 6.9 Hz, 1H), 6.66 (br d, J = 6.0 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ –5.3, 18.3, 25.9, 28.7, 30.8, 53.3, 62.3, 92.9, 116.0, 136.6, 161.2; HRMS (ESI–): calcd for C14H25Cl3NO2Si [M – H]– 372.0726, found 372.0734 (error 2.2 ppm).
(R)-6-(tert-Butyldimethylsilyloxy)hex-1-en-3-amine (12)
To a solution of 11 (935 mg, 2.5 mmol, 1.0 equiv) in toluene (10 mL) at –78 °C, was added a solution of DIBAL-H (12.5 mL, 1 M in toluene, 12.5 mmol, 5 equiv) over 15 min. The solution was stirred for 1 h at –78 °C, then quenched with MeOH (0.5 mL). Next, EtOAc (25 mL) and Rochelle's salt solution (25 mL) were added and the reaction mixture was stirred at 23 °C for 30 min. The aqueous layer was separated and extracted with EtOAc (3 × 10 mL). The combined organic extracts were washed with saturated aqueous NaCl (30 mL), dried (MgSO4) and concentrated. Purification by flash chromatograph afforded the title compound (384 mg, 67%) as colorless oil: Rf 0.15 (10% MeOH/CH2Cl2); (c 0.4, CH2Cl2); 1H NMR (600 MHz, CDCl3) δ 0.04 (s, 6H), 0.88 (s, 9H), 1.56–1.61 (m, 2H), 1.71–1.76 (m, 2H), 3.56–3.64 (m, 3H), 5.26 (d, J = 10.2 Hz, 1H), 5.36 (d, J = 17.4 Hz, 1H), 5.84 (ddd, J = 17.4, 12.5, 10.2 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ –5.31, 18.3, 26.0, 28.7, 31.0, 54.4, 62.5, 118.6, 136.2; HRMS (ESI+): calcd for C12H28NOSi [M + H]+ 230.1935, found 230.1941 (error 2.6 ppm).
tert-Butyl ((S)-1-{[(R)-6-(tert-butyldimethylsilyloxy)hex-1-en-3-yl]amino}-1-oxobut-3-en-2-yl)carbamate (14)
To a solution of 12 (687 mg, 3.0 mmol, 1.0 equiv) and (S)-N-Boc-vinylglycine 133 (600 mg, 3.0 mmol, 1.0 equiv) in THF (20 mL) at 0 °C, was added NaHCO3 (756 mg, 9.0 mmol, 3.0 equiv) and DEPBT (2.69 g, 9.0 mmol, 3.0 equiv) sequentially. The mixture was stirred at 0 °C for 1 h then warmed to 23 °C and stirred an additional 48 h. The reaction was cooled down to 0 °C and quenched by the addition of saturated NH4Cl aqueous solution (20 mL). The aqueous layer was separated and extracted with EtOAc (3 × 10 mL). The combined organic layers were washed with saturated aqueous NaCl (30 mL), dried (MgSO4) and concentrated. Purification by flash chromatograph afforded the title compound (877 mg, 71%) as a colorless oil: Rf 0.60 (40% EtOAc/hexanes); (c 3.2, CH2Cl2); 1H NMR (600 MHz, CDCl3) δ 0.03 (s, 6H), 0.88 (s, 9H), 1.44 (s, 9H), 1.45–1.58 (br m, 3H), 1.61–1.67 (br m, 1H), 3.60 (br d, J = 4.8 Hz, 2H), 4.44 (br s, 1H), 4.61 (br s, 1H), 5.11 (d, J = 9.6 Hz, 1H), 5.14 (d, J = 16.8 Hz, 1H), 5.28 (d, J = 9.6 Hz, 1H), 5.36 (br s, 1H), 5.39 (br s, 1H), 5.71–5.77 (m, 1H), 5.83–5.88 (m, 1H), 5.89–5.97 (m, 1H); 13C NMR (150 MHz, CDCl3) δ –5.3, 18.3, 25.9, 28.3, 28.9, 31.2, 51.4, 57.3, 62.5, 80.1, 115.3, 118.2, 134.3, 137.9, 155.4, 169.2; HRMS (APCI+): calcd for C21H41N2O4Si [M + H]+ 413.2830, found 413.2847 (error 4.1 ppm).
tert-Butyl {(3S, 6R)-6-[3-(tert-butyldimethylsilyloxy)propyl]-2-oxo-3,6-dihydropyridin-3-nonyl}carbamate (15)
To a solution of diene 14 (82.4 mg, 0.20 mmol, 1.0 equiv) in CH2Cl2 (20 mL) at 23 °C was added the Hoveyda-Grubbs-II catalyst (6.2 mg, 0.01 mmol, 0.05 equiv). The solution was then stirred at reflux for 12 h. The reaction was cooled to room temperature and concentrated in vacuo. Purification by flash chromatography afforded the title compound (63.7 mg, 83%) as a brown solid: Rf 0.30 (50% EtOAc/hexane); mp 108–109.5 °C; (c 0.4, CH2Cl2); 1H NMR (600 MHz, CDCl3) δ 0.04 (s, 3H), 0.05 (s, 3H), 0.88 (s, 9H), 1.45 (s, 9H), 1.55–1.63 (m, 3H), 1.66–1.68 (m, 1H), 3.62–3.65 (m, 2H), 4.08 (br s, 1H), 4.59 (br s, 1H), 5.29 (br s, 1H), 5.72 (d, J = 12 Hz, 1H), 5.93 (d, J = 12 Hz, 1H), 6.65 (br s, 1H); 13C NMR (150 MHz, CDCl3) δ –5.39, –5.37, 18.3, 25.9, 28.0 28.3, 32.5, 49.1, 52.8, 62.5, 79.8, 126.8, 126.9, 155.9, 168.7; HRMS (APCI+): calcd for C19H37N2O4Si [M + H]+ 385.2517, found 385.2518 (error 0.3 ppm).
tert-Butyl [(3S, 6R)-6-(3-hydroxypropyl)-2-oxo-3,6-dihydropyridin-3-nonyl] carbamate (S3)
To a solution of 15 (57.6 mg, 0.15 mmol) in THF (5 mL) at 0 °C was added HF·Py solution (0.5 mL) dropwise over 10 min and the reaction mixture stirred at 0 °C for an additional 1.5 h. The solution was diluted with EtOAc (20 mL) and saturated aqueous NaHCO3 (10 mL) was added slowly to neutralize the mixture. The aqueous layer was separated and extracted with EtOAc (3 × 10 mL). The combined organic extracts were washed with saturated aqueous NaCl (30 mL), dried (MgSO4) and concentrated. Purification by flash chromatograph afforded the title compound (30.3 mg, 75%) as colorless oil: Rf 0.30 (10% MeOH/CH2Cl2); (c 0.8, CH2Cl2); 1H NMR (600 MHz, CDCl3) δ 1.46 (s, 9H), 1.63–1.67 (m, 3H), 1.77–1.83 (m, 1H), 2.17 (br s, 1H), 3.68 (br s, 2H), 4.12 (br s, 1H), 4.60 (br s, 1H), 5.35 (br s, 1H), 5.73 (d, J = 12 Hz, 1H), 5.93 (d, J =12 Hz, 1H), 6.98 (br s, 1H); 13C NMR (150 MHz, CDCl3) δ 27.5, 28.3, 32.1, 48.8, 52.9, 61.9, 79.9, 126.3, 127.1, 156.0, 169.4; HRMS (APCI+): calcd for C13H23N2O4 [M + H]+ 271.1652, found 271.1649 (error 1.1 ppm).
(3S, 6R)-3-Amino-6-(3-hydroxypropyl)-3,6-dihydropyridin-2(3H)-one hydrochloride salt (1)
A solution of HCl in dioxane (1.5 mL, 3 M) was added to S3 (54 mg, 0.2 mmol) at 0 °C and the solution stirred for 1 h at 0 °C, then the reaction was concentrated in vacuo. Recrystallization with 1:20 EtOH/Et2O (6 mL) afforded the title compound (37 mg, 88%) as an off-white solid: mp 182–183.5 °C; (c 0.5, MeOH); 1H NMR (600 MHz, CD3OD) δ 1.55–1.63 (m, 2H), 1.77–1.81 (m, 2H), 3.58–3.62 (m, 2H), 4.24 (br s, 1H), 4.43 (br s, 1H), 5.93 (dt, J = 10.8, 2.4 Hz, 1H), 6.14 (dt, J = 10.8, 2.4 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 28.4, 32.6, 54.7, 62.6, 70.1, 120.7, 133.1, 166.8; HRMS (ESI+): calcd for C8H15N2O2 [M + H]+ 171.1128, found 171.1124 (error 2.3 ppm).
Anti-TB activities in BioA-suppressed Mtb mutant (bioA-TetON-1)
Mtb bioA-TetON-1 was grown at 37 °C in Sauton's medium containing 1 μM biotin to an OD580 of 0.2 to 0.4, harvested by centrifugation, washed twice with biotin-free Sauton's medium, and diluted in fresh biotin-free Sauton's medium to an OD580 of ~0.07. 50 μL of the bacterial culture were used to inoculate 96-well plates and 150 μL of compound 1 were added to give final concentrations of 500 to 1.95 mM using 2-fold serial dilutions. When added, anhydrotetracycline (ATC) was used at a concentration of 10 ng/ml. Wells containing no compound 1 were used as controls. 96-well plates were incubated in a humidified incubator at 37 °C. Optical densities were measured after 6 to 28 days.
Coupled continuous assay to evaluate the time-dependent inhibition of BioA
A. Assay procedure
BioA (2 μM) was combined with compound 1 (0–1 mM) in 2× reaction buffer (200 mM bicine pH 8.6, 100 mM NaHCO3, 2 mM MgCl2, 0.005 % Igepal CA-630, 10 mM ATP, 200 μM PLP) at 0, 10, 15, 20, 25, and 27.5 min, which correspond to pre-incubation times of 30, 20, 15, 10, 5, and 2.5 min. At 30 min, 2.5 μL of these BioA–1 incubation mixtures were added to 47.5 μL of a reaction solution, resulting in a final concentration of 100 nM BioA, 320 nM BioD, 12.5 μM KAPA, 20 nM Fl-DTB, 185 nM streptavidin, 1 mM TCEP, 1 mM SAM in 1× reaction buffer in duplicate. Addition of the substrates KAPA and SAM at these concentrations was shown to prevent measurable binding of 1 or 2 to BioA over the time period used to analyze the data. Reactions were read in a 384 well black plate (Corning #3575) using an excitation of 485 nm, emission at 530 nm and cutoff also at 530 nm. The initial velocities were determined as the time to produce 70 nM dethiobiotin.
B. Data processing
Inhibitor 1 showed time dependent inhibition of BioA while 2 was not active under the assay condition. The residual activity curve for 1 shown in Figure 3A, which describes the initial velocity, vi as a function of pre-incubation time, t, was fit to equation 1 by nonlinear regression analysis using Graphpad Prism 4.0 to obtain values for kobs at each inhibitor concentration:
| (1) |
where P is the concentration of product formed at time t. The secondary plot of kobs as a function of inhibitor concentration, [I], was fit to equation (2) to provide values for kinact and KI by nonlinear regression analysis using Graphpad Prism 4.0.
| (2) |
MS/MS Analysis of PLP-inhibitor adduct
A. Sample preparation
A solution of inhibitor 400 μM 1, 26 μM BioA, 50 mM Tris–HCl (pH 8.0), 10 mM 2-mercaptoethanol and 100 μM PLP was incubated at 37 °C for 2 h in a total volume of 100 μL. Excess inhibitor and PLP were removed by gelfiltration chromatography on a Zeba desalting column (Thermo Fisher). The columns were washed 3 times with 300 μL 1 mM Tris–HCl buffer pH 7.5, after which 100 μL of the reaction mixture was applied to the column and eluted by centrifugation. The purified protein solution was diluted 10-fold with deionized H2O.
B. MS procedure and data analysis
Enzyme samples were introduced into the ESI source by infusion with a syringe pump (120 μL/h). Optimal ESI conditions (source temperature 250 °C; drying gas (N2) flow rate 7 mL/min; neblulizing gas 7 psi; capillary voltage 400 V; capillary offset 70 V; skimmer 27 V; octopole RF 120 Vpp) were based on similar studies by Mann et al.10b For MS/MS experiments, the observed quasimolecular molecular ion at m/z 400.4 was selected and a collision energy of 40 was applied. The resulting MS/MS spectrum is shown in Figure S2.
Structural characterization of BioA·1 complex
Crystallization of BioA
Yellow crystals of BioA were grown within 24–48 hrs at 20 °C by microseeding in hanging drops using the vapor diffusion method. Crystals were obtained by mixing 2.0 μL of 10 mg mL-1 protein solution (25 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, 0.1 mM TCEP), 2.0 μL of microseed-containing reservoir solution (8-12% PEG 8000, 100 mM MgCl2, 100 mM HEPES pH 7.5). For the pre-reaction complex, red crystals were obtained by soaking the yellow, PLP-loaded crystals in reservoir solution containing 15% PEG 400 and 10 mM 1 for 5 seconds before freezing in liquid nitrogen. For the post-reaction complex, colorless crystals were obtained by soaking the yellow, PLP-loaded crystals in 2 mM 1 for 30 minutes at 23 °C followed by a 3-second soak in reservoir solution containing 2 mM 1 and 15% PEG 400 and immediately freezing in liquid nitrogen.
Structure determination
Diffraction data were collected at 100 K on GM/CA-CAT beamline 23ID-D at the Advanced Photon Source in the Argonne National Laboratory (Argonne, IL). Data were integrated and scaled with d*TREK.19 The structure was solved by molecular replacement using the program Phaser within CCP4 and PDB code 3BV0 as a search model.20, 21 Initial refinement and model building was done using REFMAC5 and Coot, respectively.22, 23 Topology and parameter files for 7 were generated using the PRODRG server.24 Final refinement was carried out using Phenix.25 The structures were evaluated using the MOLPROBITY server.26 Images and figures were prepared using PyMOL.19, 27 Coordinates and structure factors have been deposited at the Protein Data Bank with accession codes of 3TFT (pre-reaction) and 3TFU (post-reaction).
Supplementary Material
ACKNOWLEDGMENT
This research was supported by a grant from the Bill and Melinda Gates foundation and the Wellcome Trust through the Grand Challenges in Global Health Initiative (to Douglas Young, Imperial College), the National Institutes of Health (AI091790 to D.S. and C.C.A.) and the Intramural Research Program of the NIAID, NIH (to C.E.B.). The authors would like to thank Dr. Lorraine Anderson for assistance in MS/MS experiments, Kathryn Gustafson for assistance in purification and crystallization experiments, and Dr. Anja Meißner for performing cytotoxicity assays. We are grateful for resources from the University of Minnesota Supercomputing Institute.
Footnotes
ASSOCIATED CONTENT
Supporting information Available. Completed citation for reference 25, characterization data for 2 and all synthetic intermediates, general description of the coupled enzyme assays for the other transaminases evaluated, cell cytotoxicity assays, MS/MS data of PLP-inhibitor adduct, data collection and refinement statistics for BioA structural characterization, figures of pre- and post-reaction complex, and 1H NMR and 13C NMR spectra for all compounds. This material is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.a World Health Organization. Global tuberculosis control. [04/13/11];2010 http://www.who.int/tb/-publications/global_report/2010/en/index.html. [Google Scholar]; b Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. Nature. 2011;469:483. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]; c Barry CE, 3rd, Blanchard JS. Curr. Opin. Chem. Biol. 2010;14:456. doi: 10.1016/j.cbpa.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrich CC, Boshoff HI, Remmel RP. Antitubercular Agents. In: Rotella D, Abraham DJ, editors. Burgers Medicinal Chemistry. 7th Edition Wiley; 2010. p. 713. [Google Scholar]
- 3.Marquet A, Bui BTS, Florentin D. Vit. Horm. 2001;61:51. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- 4.a Dey S, Lane JM, Lee RE, Rubin EJ, Sacchettini JC. Biochemistry. 2010;49:6746. doi: 10.1021/bi902097j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Sassetti CM, Rubin EJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12989. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SW, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE, 3rd, Aldrich CC, Ehrt S, Schnappinger D. PloS Pathog. 2011;7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Bhor VM, Dev S, Vasanthakumar GR, Kumar P, Sinha S, Surolia A. J. Biol. Chem. 2006;281:25076. doi: 10.1074/jbc.M604477200. [DOI] [PubMed] [Google Scholar]; b Mann S, Ploux O. Biochim. Biophys. Acta. 2011 in press, DOI: 10.1016/j.bbapap.2010.12.004. [Google Scholar]; c Mann S, Ploux O. FEBS J. 2006;273:4778. doi: 10.1111/j.1742-4658.2006.05479.x. [DOI] [PubMed] [Google Scholar]; d Mann S, Colliandre L, Labesse G, Ploux O. Biochimie. 2009;91:826. doi: 10.1016/j.biochi.2009.03.019. [DOI] [PubMed] [Google Scholar]; e Escalettes F, Florentin D, Bui BTS, Lesage D, Marquet A. J. Am. Chem. Soc. 1999;121:3571. [Google Scholar]; f Farrar CE, Siu KKW, Howell PL, Jarrett JT. Biochemistry. 2010;49:9985. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S, Hanson RE, Cronan JE. Nat. Chem. Biol. 2010;6:682. doi: 10.1038/nchembio.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a Okami Y, Kitahara T, Hamada M, Naganawa H, Kondo S. J. Antibiot. 1974;27:656. doi: 10.7164/antibiotics.27.656. [DOI] [PubMed] [Google Scholar]; b Kitahara T, Hotta K, Yoshida M, Okami Y. J. Antibiot. 1975;28:215. doi: 10.7164/antibiotics.28.215. [DOI] [PubMed] [Google Scholar]; c Hotta K, Kitahara T, Okami Y. J. Antibiot. 1975;28:222. doi: 10.7164/antibiotics.28.222. [DOI] [PubMed] [Google Scholar]; d Poetsch M, Zahner H, Werner RG, Kern A, Jung G. J. Antibiot. 1985;38:312. doi: 10.7164/antibiotics.38.312. [DOI] [PubMed] [Google Scholar]
- 9.Mann S, Carillon S, Breyne O, Marquet A. Chem. Eur. J. 2002;8:439. doi: 10.1002/1521-3765(20020118)8:2<439::AID-CHEM439>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.a Sandmark J, Mann S, Marquet A, Schneider G. J. Biol. Chem. 2002;277:43352. doi: 10.1074/jbc.M207239200. [DOI] [PubMed] [Google Scholar]; b Mann S, Florentin D, Lesage D, Drujon T, Ploux O, Marquet A. Helv. Chim. Acta. 2003;86:3836. [Google Scholar]; c Orisadipe A, Boshoff HI, Barry CE., 3rd unpublished results.
- 11.a Olson GT, Fu M, Lau S, Rinehart KL, Silverman RB. J. Am. Chem. Soc. 1998;120:2256. [Google Scholar]; b Fu M, Nikolic D, Van Breemen RB, Silverman RB. J. Am. Chem. Soc. 1999;121:7751. [Google Scholar]; c Fu M, Silverman RB. Bioorg. Med. Chem. 1999;7:1581. doi: 10.1016/s0968-0896(99)00081-4. [DOI] [PubMed] [Google Scholar]; d Fu M, Silverman RB. Bioorg. Med. Chem. Lett. 2004;14:203. doi: 10.1016/j.bmcl.2003.09.065. [DOI] [PubMed] [Google Scholar]; e Wang Z, Yuan H, Nikolic D, Van Breemen RB, Silverman RB. Biochemistry. 2006;45:14513. doi: 10.1021/bi061592m. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Liu D, Pozharski E, Lepore BW, Fu M, Silverman RB, Petsko GA, Ringe D. Biochemistry. 2007;46:10517. doi: 10.1021/bi700663n. [DOI] [PubMed] [Google Scholar]; g Lepore BW, Liu D, Peng Y, Fu M, Yasuda C, Manning JM, Silverman RB, Ringe D. Biochemistry. 2010;49:3138. doi: 10.1021/bi902052x. [DOI] [PubMed] [Google Scholar]; h Liu D, Pozharski E, Fu M, Silverman RB, Ringe D. Biochemistry. 2010;49:10507. doi: 10.1021/bi101325z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Anderson CE, Overman LE. J. Am. Chem. Soc. 2003;125:12412. doi: 10.1021/ja037086r. [DOI] [PubMed] [Google Scholar]; b Perez-Fuertes Y, Kelly AM, Johnson AL, Arimori S, Bull SD, James TD. Org. Lett. 2006;8:609. doi: 10.1021/ol052776g. er determination protocol. [DOI] [PubMed] [Google Scholar]
- 13.Vinylglycine syntheses, (S)-enantiomer: Daniel RN, Melancon BJ, Wang EA, Crews BC, Marnett LJ, Sulikowski GA, Lindsley CW. J. Org. Chem. 2009;74:8852. doi: 10.1021/jo902115s.Berkowitz DB, Maiti G. Org. Lett. 2004;6:2661. doi: 10.1021/ol049159x. (R)-enantiomer: Hallian KO, Crout DHG, Errington W. J. Chem. Soc. Perkin Trans, 1. 1994;24:3537.
- 14.a Li H, Jiang X, Ye Y, Fan C, Romoff T, Goodman M. Org. Lett. 1999;1:91. doi: 10.1021/ol990573k. [DOI] [PubMed] [Google Scholar]; b Jiang W, Wanner J, Lee RJ, Bounaud P-Y, Boger DL. J. Am. Chem. Soc. 2002;124:5288. doi: 10.1021/ja020237q. [DOI] [PubMed] [Google Scholar]
- 15.Kingsbury JS, Harrity JPA, Bonitatebus PJ, Jr., Hoveyda AH. J. Am. Chem. Soc. 1999;121:791. [Google Scholar]
- 16.Wilson DJ, Shi C, Duckworth BP, Muretta JM, Manjunatha U, Sham YY, Thomas DD, Aldrich CC. Anal. Biochem. 2011;416:27. doi: 10.1016/j.ab.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. Wiley & Sons; Hoboken, N. J.: 2005. Irreversible Enzyme Inactivators. p. 214. [Google Scholar]
- 18.Garcia-Fandino R, Aldegunde MJ, Codesido EM, Castedo L, Granja JR. J. Org. Chem. 2005;70:8281. doi: 10.1021/jo050568w. [DOI] [PubMed] [Google Scholar]
- 19.Pflugrath JW. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J. Appl. Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4 Acta Crystallogr. D Biol. Crystallogr. 1994;50:760. [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Schuttelkopf AW, van Aalten DM. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, et al. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis IW, Murray LW, Richardson JS, Richardson DC. Nucleic Acids Res. 2004;32:W615. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano WL. The PyMOL molecular graphics system: DeLano Scientific. Palo Alto, CA, USA: 2002. [Google Scholar]
- 28.Cook MJ, Katritzky AR, Linda P, Tack RD. Chem. Commun. 1971:510. [Google Scholar]
- 29.a Singh J, Petter RC, Baillie TA, Whitty A. Nat. Rev. Drug Discov. 2011;10:307. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]; b Lu H, Tonge P. J. Curr. Opin. Chem. Biol. 2010;14:467. doi: 10.1016/j.cbpa.2010.06.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a Cerqueira NMFSA, Fernandes PA, Ramos MJ. J. Chem. Theory Comput. 2011;7:1356–1368. doi: 10.1021/ct1002219. [DOI] [PubMed] [Google Scholar]; b Oliveira EFO, Cerqueira NMFSA, Fernandes PA, Ramos MJ. J. Am. Chem. Soc. 2011 doi: 10.1021/ja204229m. in press, DOI: 10.1021/ja204229m. [DOI] [PubMed] [Google Scholar]
- 31.Nanavati SM, Silverman RB. J. Med. Chem. 1989;32:2413. doi: 10.1021/jm00131a001. [DOI] [PubMed] [Google Scholar]
- 32.Lowther J, Yard BA, Johnson KA, Carter LG, Bhat VT, Raman MC, Clarke DJ, Ramakers B, McMahon SA, Naismith JH, Campopiano DJ. Mol. Biosyst. 2010;6:1682. doi: 10.1039/c003743e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a Bird CW. Tetrahedron. 1985;41:1409. [Google Scholar]; b Bird CW. Tetrahedron. 1986;42:89. [Google Scholar]
- 34.Nishino T, Murao S, Wada H. J. Biochem. 1984;95:1283. doi: 10.1093/oxfordjournals.jbchem.a134733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.