Abstract
The marine bacterium Vibrio fischeri uses a biofilm to promote colonization of its eukaryotic host Euprymna scolopes. This biofilm depends on the symbiosis polysaccharide (syp) locus, which is transcriptionally regulated by the RscS-SypG two-component regulatory system. An additional response regulator (RR), SypE, exerts both positive and negative control over biofilm formation. SypE is a novel RR protein, with its three putative domains arranged in a unique configuration: a central phosphorylation receiver (REC) domain flanked by two effector domains with putative enzymatic activities (serine kinase and serine phosphatase). To determine how SypE regulates biofilm formation and host colonization, we generated a library of SypE domain mutants. Our results indicate that the N-terminus inhibits biofilm formation, while the C-terminus plays a positive role. The phosphorylation state of SypE appears to regulate these opposing activities, as disruption of the putative site of phosphorylation results in a protein that constitutively inhibits biofilm formation. Furthermore, SypE restricts host colonization: (1) sypE mutants with constitutive inhibitory activity fail to efficiently initiate host colonization, and (2) loss of sypE partially alleviates the colonization defect of an rscS mutant. We conclude that SypE must be inactivated to promote symbiotic colonization by V. fischeri.
Introduction
Bacteria possess the ability to adapt and thrive in a wide range of dynamic environments. To establish an ecological niche and prosper within that particular environment, bacteria must be able to sense diverse environmental conditions and then produce the appropriate cellular response. A common mechanism by which bacteria coordinate cellular responses with environmental stimuli is via two-component signal transduction (2CST) systems. The typical 2CST system consists of a histidine sensor kinase (SK) and a response regulator (RR). The sensing of an environmental stimulus by the SK initiates a signal transduction cascade by inducing the SK to autophosphorylate at a conserved histidine residue within the transmitter domain (Stock et al., 2000; West and Stock, 2001). The phosphoryl group is then transferred from the SK to a downstream RR. RRs catalyze phosphoryl transfer to a conserved aspartate residue located within the N-terminal regulatory receiver (REC) domain (Bourret et al., 1990). The phosphorylation state of the REC domain regulates the activity of an attached C-terminal effector domain, often a DNA binding domain (Stock et al., 2000; Gao and Stock, 2010). As a result, RR phosphorylation commonly results in a change in DNA binding and gene expression [reviewed in (Galperin, 2010)]. Alternatively, the RR REC domain may be attached to a large variety of other output domains, including those involved in protein-protein interaction, ligand-binding, or enzymatic activity (Galperin, 2006; Galperin, 2010).
2CST systems are particularly well-suited to allow bacteria to sense host signals and then elicit the appropriate response(s) necessary to promote colonization. The marine bacterium Vibrio fischeri utilizes multiple 2CST systems to promote symbiotic colonization of its host, the Hawaiian squid Euprymna scolopes (Whistler and Ruby, 2003; Hussa et al., 2007; Bose et al., 2007). In particular, 2CST regulators control one of the earliest stages of host colonization, the formation of a biofilm aggregate on the surface of the squid’s symbiotic light organ (Nyholm and McFall-Ngai, 1998; Nyholm et al., 2000). The formation of this biofilm depends upon the SK RscS and the RR SypG, which control transcription of the symbiotic polysaccharide locus (syp) (Yip et al., 2005; Yip et al., 2006; Hussa et al., 2008). The current model (Fig. 1) predicts that RscS, upon sensing an as-yet unidentified signal, initiates a phospho-relay and donates a phosphoryl group to SypG (Hussa et al., 2008). SypG is a syp-encoded, σ54-dependent RR with a DNA-binding output domain (Yip et al., 2005). Phosphorylated SypG is thought to directly activate transcription of the syp genes, whose products promote biofilm formation (Hussa et al., 2008) (Fig. 1).
Figure 1. SypE and syp-dependent biofilm regulation.

Model for the regulation of syp-dependent biofilms. (i) Transcription of the syp locus is controlled by the RscS-SypG two-component system, which results in expression of the 18 gene syp polysaccharide locus and, subsequently, biofilm formation and colonization. (ii) Under non-RscS activating conditions (i.e. upon induction with SypG), SypE functions downstream of syp transcription to inhibit biofilm formation. (iii) Upon expression of RscS, SypE inhibitory activity is inactivated, permitting biofilm formation and colonization. Phosphorylation of SypE is predicted to inactivate the inhibitory N-terminal effector domain, while promoting activation of the positive C-terminal effector domain. The dashed arrow indicates potential direct phosphorylation of SypE by RscS, resulting in inactivation of SypE inhibitory activity. (+) denotes positive activity. (−) denotes inhibitory activity.
Mutant analyses have shown that RscS and SypG are essential both for the formation of syp-dependent biofilms and for initiating host colonization (Visick and Skoufos, 2001; Hussa et al., 2008). Although the natural signals that activate the V. fischeri RscS-SypG system are not yet identified, overexpression of RscS or SypG promotes biofilm formation in laboratory culture and permits a dissection of the pathways leading to this phenotype. Overexpression constructs have been used to investigate the roles of regulatory components in a variety of bacterial systems, including Vibrio cholerae and Salmonella enterica serovar Typhimurium (Simm et al., 2004; Beyhan et al., 2006; Shikuma et al., 2009). In V. fischeri, the multi-copy expression of RscS or SypG results in activation of syp transcription and the production of robust biofilms. These syp-dependent biofilms can be observed by the formation of wrinkled colonies on solid media and pellicle formation at the air-liquid interface in static culture (Yip et al., 2005). Importantly, these in vitro phenotypes directly correlate with phenotypes observed in vivo. In the presence of juvenile squid, induction of the syp cluster via multi-copy expression of RscS results in enhanced aggregation on the surface of the host squid’s symbiotic light organ, which promotes subsequent colonization events (Yip et al., 2006).
In addition to SypG, the syp locus encodes a second putative RR, SypE [for a review see (Visick, 2009)]. The SypE protein is unusual both in its structure and in its apparent function in biofilm regulation and symbiosis. Bioinformatic analysis predicts a novel, multi-domain RR with the regulatory REC domain in a central position flanked by two terminal effector domains of putative opposing function: an N-terminal RsbW-like serine kinase and a C-terminal PP2C-like serine phosphatase domain (Fig. 2). The presence of a central REC domain flanked by two effector domains of apparent opposing function distinguishes SypE as a novel RR protein.
Figure 2. Domain structure of SypE and mutant derivatives.

SypE contains a central RR receiver (REC) domain flanked by two putative effector domains, including an N-terminal RsbW-like serine kinase domain and a C-terminal PP2C-like serine phosphatase domain. Conserved residues within the individual domains are indicated (see text for description) (Morris and Visick, 2010). Representations of the various SypE domain mutants generated in this study are presented below. See text for a full description. (Not shown are SypED495A and SypECTD, D495A).
Previous studies identified SypE as a potential regulator of V. fischeri biofilms. Hussa et al. (2008) demonstrated that overexpression of the SK RscS in a strain lacking sypE resulted in diminished biofilm phenotypes, indicated by a decrease in wrinkled colony formation relative to wild-type cells overexpressing RscS. These results suggested a positive regulatory role for SypE. In contrast, SypE appeared to strongly inhibit biofilms induced by SypG: overexpression of SypG in wild-type cells resulted in smooth colony formation and poor pellicle production, while its overexpression in a ΔsypE strain induced robust biofilms similar to that of an RscS-overexpressing wild-type strain. We hypothesize that SypE plays a novel role as a dual regulator of biofilm formation and that these activities must be controlled to permit biofilm formation and colonization (Fig. 1).
Here, we investigated the role of this unusual regulator in the control of biofilm formation and host colonization by V. fischeri. We found that the N- and C-terminal domains of SypE play negative and positive roles, respectively, in the control of biofilm formation. We further determined that the ability of SypE to switch to a biofilm-promoting activity (and away from an inhibitory activity) depends upon residue D192, the predicted site of phosphorylation within the central receiver domain. Finally, we found that the ability of SypE to inhibit biofilm formation in vitro correlated with its ability to inhibit host colonization and symbiotic aggregation. Together, our data suggest that inactivation of SypE is a critical step in symbiotic biofilm formation and colonization by V. fischeri.
Results
Deletion of sypE impairs RscS-induced biofilm phenotypes
sypE encodes an unusual, multi-domain RR protein consisting of a central receiver (REC) domain flanked by N- and C-terminal effector domains with sequence similarity to serine kinases and serine phosphatases, respectively (Fig. 2) (Morris and Visick, 2010). Consistent with the complex nature of this protein, SypE appears to both positively and negatively impact biofilm formation by V. fischeri (Hussa et al., 2008). In this work, our goal was to understand how the individual domains contribute to these opposing regulatory activities, and how these activities are regulated.
Because the natural signal(s) to which RscS responds are not yet known, we overexpressed rscS to induce biofilm formation throughout these experiments. As previously discussed, biofilm formation appeared diminished by the deletion of sypE (Hussa et al., 2008). To better understand this putative role of SypE as a positive regulator, we examined biofilm development over time by wild-type (WT) and ΔsypE mutant cells containing the RscS-expressing plasmid, pARM7 (pRscS). In addition, to verify that any phenotypes of the ΔsypE mutant were due to the loss of sypE, and were not the result of a polar effect on downstream genes, we complemented the sypE null strain with a wild-type sypE allele (sypE+) inserted in the chromosome at the Tn7 integration site. As a control, we inserted an empty Tn7 cassette (EC) at the same site.
First, we evaluated the formation of wrinkled colonies using a spotted culture technique (Experimental Procedures). We found that RscS-expressing wild-type cultures consistently began to exhibit wrinkled colony morphology within 14 h of growth, while a sypE deletion strain consistently exhibited a 4–6 h delay, initiating wrinkled colony formation around 18–20 h post-spotting (Fig. 3A and data not shown). Complementation with sypE+ at the Tn7 site largely restored the timing and pattern of wrinkled colony development. We noted that at a late (48 h) time point, all the strains exhibited a similar degree of wrinkled colony morphology, indicating that deletion of sypE only results in a temporal delay in RscS-induced wrinkled colony development. As expected, cells carrying the empty vector (pKV282) never formed wrinkled colonies (Fig. 3A).
Figure 3. Impact of SypE on RscS-induced biofilm phenotypes.
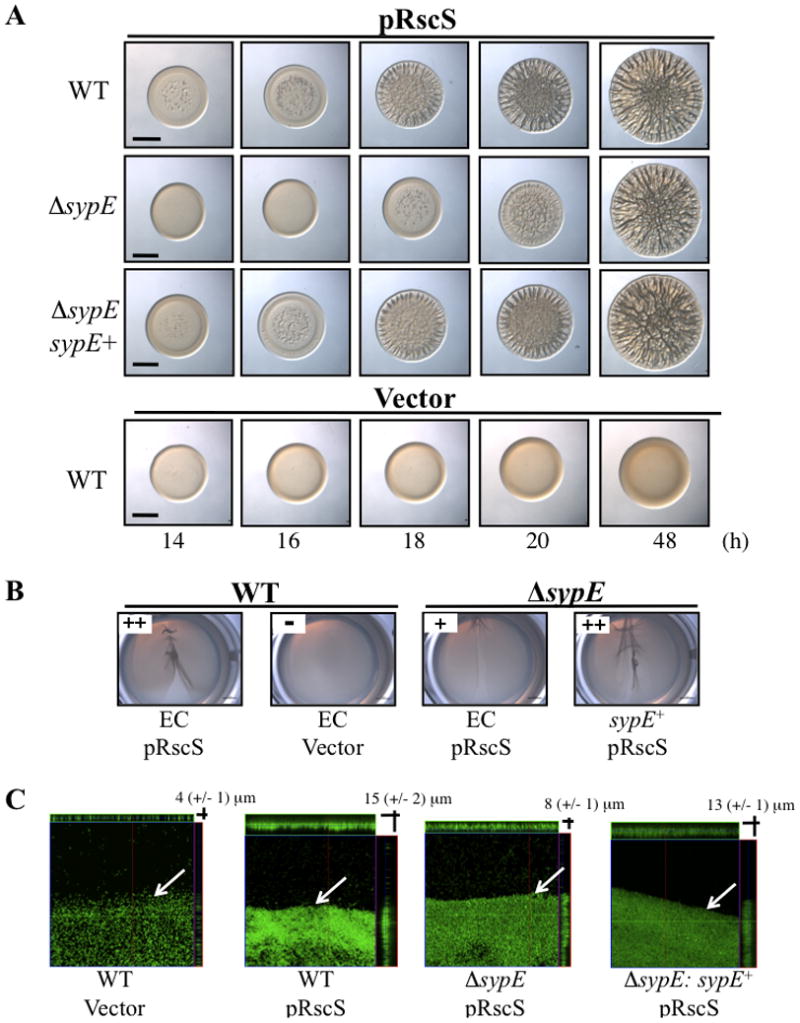
(A) Time course of RscS-induced wrinkled colony formation. Cultures of the following strains were spotted onto LBS medium at RT and wrinkled colony formation was assessed at various times up to 48 h post-spotting: wild-type (WT) cells containing the empty Tn7 cassette (EC) (KV4389) and carrying pRscS plasmid pARM7 or vector control (pKV282); ΔsypE cells carrying pARM7 and containing either EC (KV4390) or complemented with wild-type sypE+ (KV4819). Black bar represents 2 mm. (B) Pellicle formation in static culture. Cultures of the following strains were grown in HMM in 24-well plates at RT for 48 h: WT cells containing EC (KV4389) and carrying the vector control pKV282 or pARM7; pARM7-carryingΔsypE containing EC (KV4390) or complemented with sypE+ (KV4819). A pipette tip was dragged over the surface of the air-liquid interface to visualize the pellicle, and relative pellicle strength was determined as described in the Experimental Procedures. (C) Confocal microscopy of pellicles. The development of RscS-induced pellicle attachment to glass cover slips was visualized via confocal microscopy at 24h post inoculation. Representative views of the xy plane and z sections are shown for GFP-labeled derivatives of the strains described in panel B. White arrows indicate the air-liquid interface. Photographs are representative of at least 3 independent experiments.
Second, we evaluated the production of pellicles over time. When grown statically in HEPES minimal medium (HMM), RscS-expressing wild-type cells produced a structured pellicle at the air-liquid interface within 24 h of incubation, while vector control cells failed to form any observable pellicle (Fig. 3B) (Yip et al., 2005). The ΔsypE strain formed a diminished RscS-induced pellicle at 24 h that was easily disrupted relative to wild-type control cells (Fig. 3B). Complementation with sypE+ restored robust pellicle formation. Attempts to quantify the RscS-induced pellicles using crystal violet staining were not successful, as the crystal violet stained only the glass-attached cell mass and not the overall pellicle (Morris and Visick, unpublished data). Therefore, we utilized confocal microscopy to assess pellicle biofilms produced by wild-type and ΔsypE strains overexpressing both rscS and GFP at 24 h post inoculation. RscS-expressing wild-type cells exhibited increased pellicle attachment relative to vector-containing cells, with an average biofilm thickness of 15 (+/− 2) and 4 (+/− 1) μm, respectively (Fig. 3C). Compared to wild-type cells, the ΔsypE mutant strain consistently exhibited a 50% reduction in RscS-induced pellicle attachment with an average biofilm thickness of 8 (+/-1) μm (Fig. 3C). Complementation with sypE+ increased the average biofilm thickness to 13 (+/− 1) μm (Fig. 3C). These results confirm that loss of sypE impairs the development of biofilm formation, and support the hypothesis that under RscS-overexpressing conditions SypE exerts positive regulatory activity.
The SypE C-terminal domain is required for positive regulation of biofilm phenotypes
To determine the role(s) of the individual SypE domains in the positive regulation of RscS-induced V. fischeri biofilms, we constructed a library of SypE domain mutants (Fig. 2). We then assessed the ability of these sypE alleles to complement the sypE mutant by inserting these constructs into the Tn7 site of the ΔsypE strain and assaying for restoration of RscS-induced biofilm phenotypes (i.e., wrinkled colony morphology and pellicle formation). We performed time-course experiments using these complemented strains, but show here only the appropriate, representative time points.
As previously observed (Fig. 3), RscS-induced wild-type cells exhibited signs of wrinkled colony development within 16 h (Fig. 4A), whereas wild-type cells containing vector control fail to form a biofilm (Fig. 4B). At the early, 16 h time point, the ΔsypE strain (Fig. 4C) exhibited a delay in RscS-induced wrinkling relative to wild-type cells (Fig. 4A), but this delay could be complemented with a wild-type copy of sypE+ (Fig. 4D). Similarly, we found that expression of a SypE N-terminal deletion mutant (SypEΔNTD) (Fig. 2) fully complemented the sypE deletion for both wrinkled colony morphology and pellicle formation (Fig. 4E and Fig. S1). This mutant lacks the N-terminal 135 amino acids encoding the putative serine kinase domain but retains an intact C-terminal serine phosphatase domain and REC domain. We conclude from these data that the positive regulatory activity of SypE is retained in this deletion mutant and thus does not require the N-terminal domain.
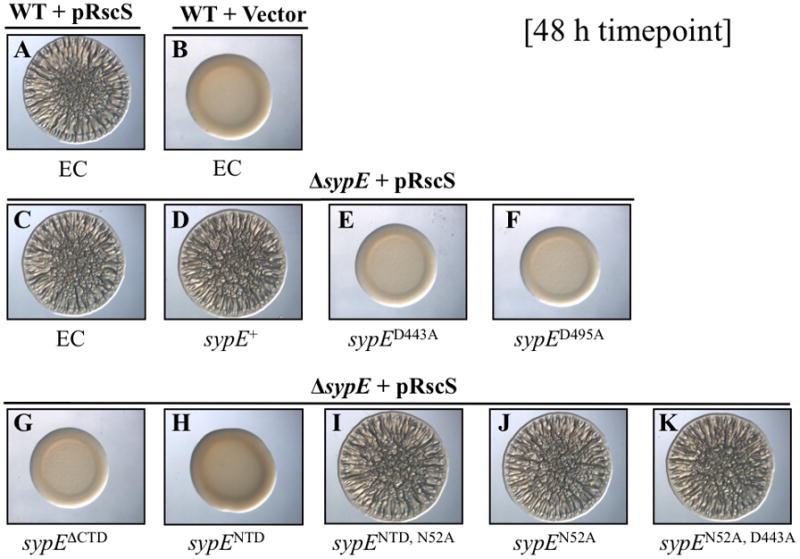
Figure 4. SypE C-terminal domain positively regulates RscS-induced biofilms.

Cultures of the following strains were spotted onto LBS medium at RT and wrinkled colony formation was assessed at an early time point (16 h) post-spotting: WT cells containing EC (KV4389) and carrying either pRscS plasmid pARM7 (A) or vector control pKV282 (B), and ΔsypE mutants carrying pARM7 and containing EC (KV4390) (C) or complemented with WT sypE+ [KV4819] (D), sypEΔNTD [KV5124] (E), sypECTD [KV5204] (F), sypECTD, D443A [KV5314] (G), sypECTD, D495A [KV5345] (H), sypED443A [KV4886] (I), sypED495A [KV4887] (J), sypEΔCTD [KV5315] (K), or sypEN52A, D192A [KV5205] (L). Photographs are representative of at least 3 independent experiments.
We hypothesized that the positive regulatory activity retained in the SypEΔNTD mutant protein may reside in the C-terminal effector domain. Indeed, a deletion derivative that expresses only the C-terminal 245 amino acids (SypECTD) (Fig. 2) fully complemented the ΔsypE deletion, restoring wrinkled colony morphology (Fig. 4F) and pellicle production (Fig. S1) to wild-type levels. We also observed that expression of SypECTD in the ΔsypE mutant was sufficient to restore pellicle attachment to near wild-type levels (16 μm +/− 1μm) as determined by confocal microscopy (data not shown). These data thus demonstrate that the C-terminal effector domain of SypE alone is active and can positively regulate RscS-induced biofilms.
The positive activity of the C-terminal domain of SypE requires conserved aspartates
The C-terminal effector domain of SypE exhibits weak sequence similarity to serine phosphatases of the PP2C family, including RsbU and SpoIIE of Bacillus subtilis (Morris and Visick, 2010). Proteins of the PP2C family are Mg2+/Mn2+-dependent phosphatases, and possess several invariant aspartate residues required for coordination of divalent cation binding and catalytic activity (Adler et al., 1997; Jackson et al., 2003). SypE contains several of these residues, including the conserved aspartates (Asp-443 and Asp-495) (Morris and Visick, 2010). We therefore reasoned that these conserved residues were likely required for the observed positive regulatory activity. We generated alanine substitutions in the context of both the C-terminal domain alone (SypECTD, D443A and SypECTD, D495A) and of the full-length protein (SypED443A and SypED495A) (Fig. 2 and not shown). Mutation of either conserved aspartate in the context of the C-terminal domain alone (SypECTD, D443A and SypECTD, D495A) resulted in failure to complement the ΔsypE defect: these mutants exhibited delayed wrinkled colony morphology and pellicle formation similar to the uncomplementedΔsypE control (Fig. 4G, 4H and Fig. S1). To ensure that both the SypECTD, D443A and SypECTD, D495A proteins were indeed expressed, we FLAG-tagged these proteins and confirmed their expression by Western blot analysis (Fig. S2A). We conclude that the positive regulatory activity of the SypE C-terminal domain requires the conserved aspartate residues (D443 and D495).
In the context of the full-length SypE, the SypED443A and SypED495A mutants again failed to complement the sypE deletion as observed at the early (16 h) time point (see Fig. 4I and 4J, respectively), but unexpectedly also completely inhibited biofilm formation, as indicated by smooth colony morphology (Fig. 5E and 5F, respectively) and lack of pellicle even at the 48 h time point (Fig. S1). This result was not due to aberrant protein expression, as these proteins were present at levels similar to the wild-type SypE control (Fig. S2B).
Figure 5. SypE N-terminal domain inhibits RscS-induced biofilms.
Cultures of the following strains were spotted onto LBS agar medium at RT and wrinkled colony formation was assessed at a late time point (48 h) post-spotting: WT cells containing EC (KV4389) and either pRscS plasmid pARM7 (A) or vector control pKV282 (B); ΔsypE cells carrying pARM7 and containing EC (KV4390) (C) or complemented with WT sypE+ [KV4819] (D), sypED443A [KV4886] (E), sypED495A [KV4887] (F), sypEΔCTD [KV5315] (G), sypENTD [KV5129] (H), sypENTD, N52A [KV5143] (I), sypEN52A [KV5142] (J), sypEN52A, D443A [KV5379] (K). Photographs are representative of at least 3 independent experiments.
We hypothesized that loss of function in SypE’s C-terminus “locked” SypE into an inhibitory state. Consistent with that interpretation, we found that a SypEΔCTD mutant, which contains the N-terminus and the central REC domain, but lacks the C-terminal domain, exhibited the same phenotype: this strain failed to form wrinkled colonies and pellicles even at late time points (48 h post-incubation) (Fig. 4K, 5G and Fig. S1). Taken together, these results demonstrate that an intact C-terminal domain is required for the positive regulatory activity of SypE and that loss of the C-terminal function results in a protein with constitutive inhibitory activity.
The N-terminal domain of SypE mediates negative regulation of biofilm phenotypes
We hypothesized that the inhibitory activity observed upon disruption of the C-terminal domain likely resides in the protein’s N-terminal effector domain. To test this hypothesis, we evaluated biofilm formation at a late time point (48 h) in a ΔsypE strain that expressed only the 140 amino acids comprising the N-terminal domain (SypENTD). This strain failed to form biofilms, as indicated by the smooth colony morphology and lack of pellicle production at 48 h (Fig. 5H and Fig. S3). These results demonstrate that the isolated SypE N-terminal effector domain is active and sufficient to inhibit the development of RscS-induced biofilm phenotypes.
The N-terminal domain of SypE exhibits sequence similarity to HPK-like serine kinases of the Gyrase, Hsp90, histidine kinase, MutL (GHKL) superfamily, including RsbW of B. subtilis (Morris and Visick, 2010). Serine kinases of the GHKL superfamily are demonstrated to form an ATP-binding Bergerat fold consisting of 4 conserved motifs termed the N, G1, G2, and G3 boxes (Bergerat et al., 1997, Dutta and Inouye, 2000). In particular, a conserved asparagine residue located within the N box motif is required for coordination of a Mg2+ ion involved in ATP binding (Dutta and Inouye, 2000). The N-terminal domain of SypE possesses these conserved ATP-binding residues, including an invariant asparagine residue (N52) (Morris and Visick, 2010) (Fig. 2).
We asked whether the conserved asparagine N52 is required for the inhibitory activity of SypE by substituting alanine at that position. In the context of the inhibitory NTD alone (SypENTD, N52A), this substitution resulted in loss of inhibitory activity: expression of SypENTD, N52A in the ΔsypE strain resulted in delayed wrinkled colony morphology and reduced pellicles similar to empty cassette (EC) control (Fig. 5I and Figs. S3 and S4E). Western blot analysis confirmed that the SypENTD, N52A protein was expressed at levels similar to the SypENTD control, indicating that the loss of inhibitory activity results from the N52A mutation (Fig. S2A). As expected, the N52A mutation in the context of the full-length protein (SypEN52A) permitted complementation of the sypE deletion; the SypEN52A strain exhibited wrinkled colony morphology (Fig. 5J and Fig. S4F) and pellicle formation (Fig. S3) similar to the wild-type and sypE+complemented strains. These results suggest that the N52A mutation disrupts the inhibitory activity of the SypE protein, but does not impact its positive regulatory activity when a functional C-terminal domain is present.
The central REC domain modulates SypE regulatory activity
Our results demonstrate that SypE exerts both negative and positive control over biofilm formation through the opposing activities of its N- and C-terminal effector domains, respectively. In general, the activity of a RR effector domain is regulated by the phosphorylation of a conserved aspartate residue located in the attached REC domain (Stock et al., 2000). We therefore hypothesized that the phosphorylation state of the central REC domain likely controls the opposing activities of SypE’s terminal effector domains. To test this hypothesis, we evaluated biofilm formation by expressing a SypE mutant containing an alanine substitution at the conserved asparate residue (D192) predicted to be the site of phosphorylation (Hussa et al., 2008). This substitution in other RRs results in a protein that cannot become phosphorylated, and whose activity therefore mimics that of the non-phosphorylated state (e.g., (Freeman and Bassler, 1999)). We found that a SypED192A mutant not only failed to complement the sypE deletion but also completely inhibited wrinkled colony and pellicle formation as observed at the late (48 h) time point (Fig. 6E and Fig. S5). The inhibitory activity of the SypED192A mutant is not likely due to aberrant protein expression as Western blot analysis indicated that the SypED192A protein was expressed at levels similar to wild-type SypE (Fig. S2B). These data indicate that the conserved Asp residue (D192) in the REC domain is required for the positive regulatory activity of SypE. Thus, the phosphorylation state of this residue is likely important in controlling SypE regulatory activity.
Figure 6. Role of the central REC domain in biofilm regulation.
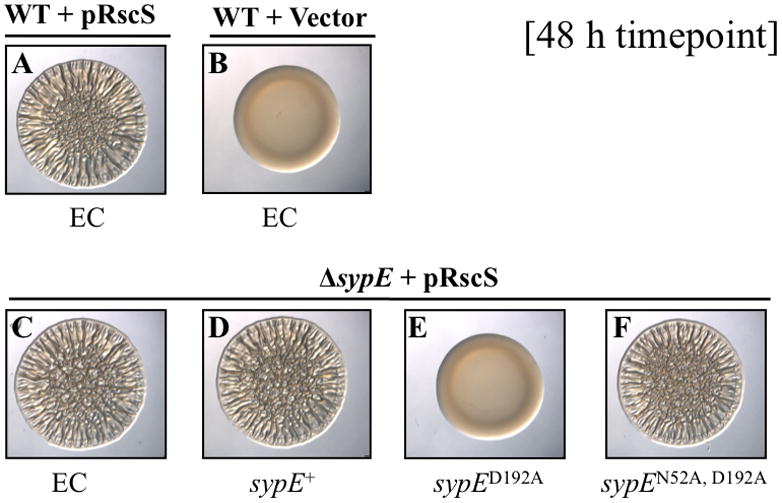
Cultures of the following strains were spotted onto LBS agar medium at RT and wrinkled colony formation was assessed at a late time point (48 h) post-spotting: WT cells containing EC (KV4389) and either pRscS plasmid pARM7 (A) or vector control pKV282 (B); and ΔsypE mutants carrying pARM7 and containing EC (KV4390) (C) or complemented with WT sypE+ [KV4819] (D), sypED192A [KV4885] (E), sypEN52A, D192A [KV5205] (F). Photographs are representative of at least 3 independent experiments.
We hypothesized that the D192A substitution “locks” SypE into a conformation that favors the inhibitory state. Earlier in this report, we demonstrated that the inhibitory activity of SypE requires an intact N-terminal domain, specifically the conserved residue N52. We predicted that if the D192A mutation “locks” SypE in a constitutive inhibitory state, then this activity should still require residue N52. To test this, we combined the D192A and N52A mutations (SypEN52A, D192A) (Fig. 2). In contrast to the biofilm-inhibitory SypED192A mutant, cells carrying SypEN52A, D192A exhibited biofilm formation indistinguishable from the uncomplemented (EC) ΔsypE control (Fig. 6F and S5). This result could not be attributed to a defect in protein production of the double mutant (Fig. S2C). These data demonstrate that the N52A mutation is epistatic to the D192A mutation, and that the constitutive inhibitory activity of the D192A mutant requires an intact N-terminal domain. We further noted that while the SypEN52A, D192A mutant lost inhibitory activity, the protein remained incapable of complementing the sypE deletion despite the fact that the positive-acting CTD remained intact (Fig 4L). We conclude from these data that residue D192 is therefore also required for positive regulatory activity. These data suggest that SypE phosphorylation may function both to activate the positive CTD and to inactivate the inhibitory NTD.
We noted that the constitutive inhibitory activity of the SypED192A mutant mimicked that observed previously in a C-terminal domain mutant, SypED443A. We predicted that the inhibitory activity of the SypED443A mutant would similarly depend upon residue N52, and tested this by generating a SypEN52A, D443A double mutant (Fig. 2). As expected, a SypEN52A, D443A mutant failed to inhibit biofilm formation, and exhibited delayed wrinkled colony morphology and pellicle formation similar to uncomplemented ΔsypE control (Fig. 5K and Figs. S3 and S4G). Western blot analysis demonstrated that the SypEN52A, D443A protein was expressed at levels similar to wild-type SypE+ (Fig. S2C). These data demonstrate that the N52A mutation is also epistatic to the D443A mutation, and suggest that in all cases SypE inhibitory activity requires an intact N-terminal domain.
The negative activity of SypE inhibits host colonization
Earlier studies demonstrated that the ability to produce syp-dependent biofilms is essential for V. fischeri cells to efficiently colonize host squid (Visick and Skoufos, 2001; Yip et al., 2005; Hussa et al., 2007). Because SypE exerts a subtle positive effect and a strong negative effect over biofilm formation, we therefore asked if either or both of these activities were important to host colonization. We evaluated the role of SypE in colonization of juvenile E. scolopes using both single strain inoculations and mixed strain competitions. We found that deletion of sypE did not substantially impact the ability of V. fischeri to colonize squid, when in competition or presented alone (Fig. 7A and 7B, respectively), although there was a small advantage for wild-type cells to out-compete the ΔsypE mutant (mean Log RCI, −0.21 +/− 0.63). Similar results were also observed when juvenile squid were exposed to a mixture of wild-type and ΔsypE cells both overexpressing rscS: the squid contained roughly equal numbers of both strains (data not shown). We conclude from these results that loss of sypE does not greatly impact colonization fitness. Furthermore, these data suggest that during colonization the minor, positive activity SypE exerts over biofilm formation is not likely critical.
Figure 7. Impact of SypE positive activity on host colonization.

(A) Competitive colonization with WT and ΔsypE mutant strains. Newly hatched squid were exposed to a mixed inoculum of WT (ES114) and ΔsypE cells, in which one strain carried an erythromycin resistance (EmR) marker. Reciprocal experiments in which the other strain carried the EmR marker were also performed and yielded similar results. The Log relative competitive index (Log RCI) is plotted on the x-axis. The log RCI was calculated as the Log ratio of EmR bacteria in the homogenate divided by the ratio of EmR bacteria in the inoculum. Each circle represents a single animal (n=30). The position of the circles on the y-axis is merely for spacing. The black diamond and error bars indicate the average Log RCI and standard deviation for 30 animals. (B) Single-strain colonization with WT and ΔsypE strains. Newly hatched squid were exposed for 18 h to either WT or ΔsypE mutant cells. Each circle represents the number of V. fischeri cells recovered from an individual animal (n=15). The black bar indicates the average CFU for 15 animals. The data shown in panel B are from one experiment and are representative of at least 3 independent experiments.
Our in vitro studies suggested that, of the two roles of SypE, the inhibitory activity appears to play a more important role with respect to biofilm formation. We therefore asked whether the inhibitory activity of SypE impacts symbiotic colonization. To address this, we focused our attention on the SypED192A mutant, which exhibits constitutive inhibitory activity. Because rscS expressed from the chromosome is active to promote biofilm formation and colonization during symbiotic initiation, we performed these experiments in the absence of rscS overexpression. We exposed squid to a mixture of wild-type and ΔsypE mutant cells complemented with either wild-type sypE+ or the inhibitory sypED192A allele. We found that, while the SypE+ strain competed well for colonization with the wild-type strain (mean Log RCI, −0.07 +/− 0.44), the SypED192A mutant exhibited a strong colonization defect (mean Log RCI, −1.59 +/− 0.52), with all squid colonized predominantly by the wild-type cells (Fig. 8A and 8B, respectively). These data suggested that the inhibitory activity of SypE is detrimental to colonization.
Figure 8. SypE-mediated inhibition of host colonization.
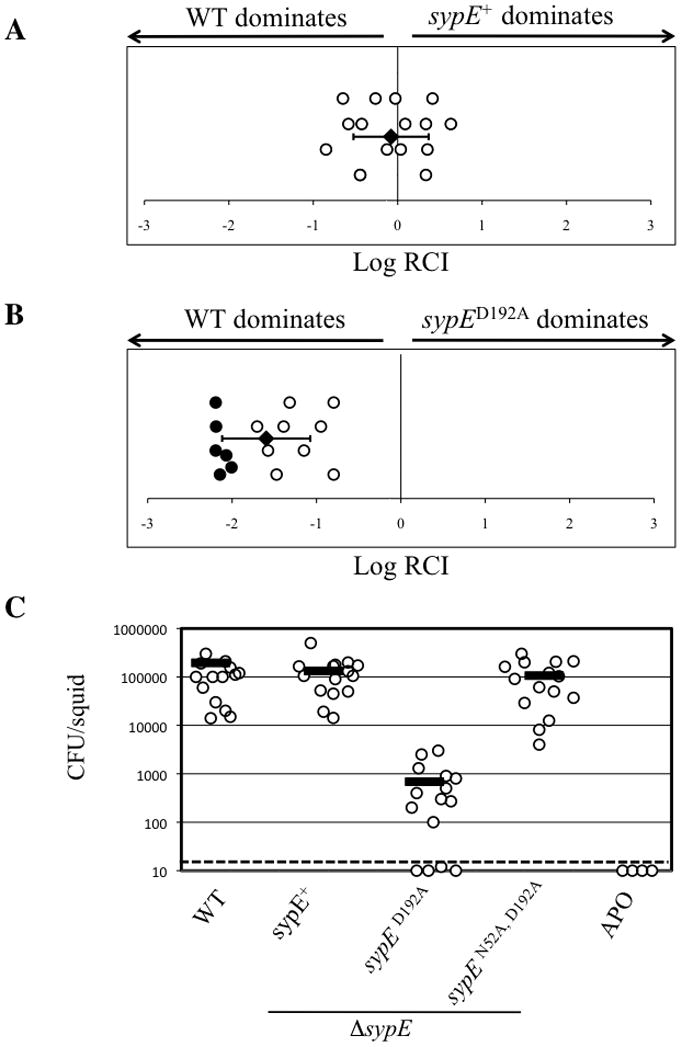
Competitive colonization with WT and sypE mutant strains. Newly hatched squid were exposed to a mixed inoculum of WT (ES114) and ΔsypE cells complemented with either WT sypE+ (A) or sypED192A (B). The Log RCI is plotted on the x-axis. The log RCI was calculated as in Figure 7. The position of the circles on the y-axis is merely for spacing. Closed circles indicate squid containing no sypE mutant cells. The black diamond and error bars indicate the average Log RCI and standard deviation for 15 animals. Data shown are representative of at least 3 independent experiments. (C) Single-strain colonization by WT ES114 and sypE strains. Newly hatched squid were exposed for 18 h to WT (ES114) or ΔsypE strains complemented with either WT sypE+, sypED192A, or the sypEN52A, D192A double mutant. As a negative control, aposymbiotic (APO) juvenile squid were maintained in bacteria free water. Each circle represents the number of V. fischeri cells recovered from an individual animal. The dashed line indicates the limit of detection. The black bar indicates the average CFU and standard deviation for 15 animals. The data shown above are from one experiment and are representative of at least 3 independent experiments.
To explore this possibility further, we performed single-strain colonization experiments. Squid inoculated with wild-type cells contained levels of bacteria between 104 and 105 colony-forming units per animal, and those inoculated with the ΔsypE mutant complemented with wild-type sypE+ were similarly proficient at colonization (Fig. 8C). However, squid inoculated with the ΔsypE mutant complemented with sypED192A exhibited a dramatic defect in host colonization relative to wild-type cells: multiple squid remained completely uncolonized, while the others had a severe decrease in their level of colonization (2–3 log decrease) (Fig. 8C). These data indicate that residue D192 within the central REC domain, is necessary to promote efficient host colonization.
Finally, to confirm that the colonization disadvantage of the SypED192A strain is due to the activity of the inhibitory domain, we assessed colonization by the SypEN52A, D192A expressing strain. Whereas the SypED192A mutant inhibited host colonization, the SypEN52A, D192A double mutant achieved colonization to near wild-type levels (Fig. 8C). These data support our earlier observations that the inhibitory activity of a SypED192A mutant resides in the N-terminal effector domain. These results are consistent with the biofilm phenotypes observed in culture, and suggest that the constitutive inhibition of colonization resulting from disruption of the predicted site of phosphorylation (D192) is mediated by the N-terminal effector domain. As such, this inhibitory activity can in turn be disrupted by mutation of the critical N52 site. Together, these data indicate that the inhibitory activity of SypE is physiologically relevant and the ability of the cell to turn off this activity is critical to permit symbiotic colonization.
SypE impacts aggregation outside the symbiotic light organ
syp-dependent biofilm formation is required for the initial stages of host colonization, when V. fischeri cells aggregate in the squid-secreted mucus on the light organ surface. Previously, Yip et al. (2006) reported that rscS and syp mutants exhibit impaired aggregation and subsequently fail to initiate host colonization, while cells overexpressing rscS exhibit enhanced aggregation. To determine if SypE regulates this early stage in symbiotic colonization, we assessed aggregate formation by wild-type or sypE mutant strains expressing rscS and GFP. In agreement with previous data (Yip et al., 2006), wild-type cells exhibited large aggregates (green color) on the light organ surface (blue color) (Fig. 9A), as did the ΔsypE strain complemented with wild-type sypE+ (Fig. 9B). In contrast, the SypED192A mutant strain formed no observable aggregates, with only single cells occasionally observed on the light organ surface (dashed arrow) (Fig. 9C). As expected, the aggregation defect of this mutant was dependent upon the inhibitory N-terminal domain, as a SypEN52A, D192A double mutant formed large aggregates similar to control (SypE+) cells (Fig. 9D). These data suggest that the colonization defect of the SypED192A mutant likely results from a loss of bacterial aggregation at the initiation stage. Thus, SypE appears to impact bacterial aggregation, a critical stage in the initiation of host colonization. Finally, these results support our earlier conclusion that the inhibitory activity of SypE must be deactivated to promote efficient aggregation and host colonization.
Figure 9. SypE impacts RscS-induced aggregation on the light organ surface.

Newly hatched juvenile squid were inoculated with GFP-labeled derivatives of the following strains: (A) pARM7 (pRscS) in WT cells [KV4389] or ΔsypE cells complemented with WT sypE+ [KV4819] (B), sypED192A [KV4885] (C), or sypEN52A, D192A [KV5205] (D). After 3 h, animals were stained with CellMask stain (blue color) and the light organs were examined by confocal microscopy. Representative images of V. fischeri cells aggregating on the light organ surface are shown. White arrows indicate the surface of the light organ appendages. The white, dashed arrow indicates single V. fischeri cells on the light organ surface.
Deletion of sypE improves colonization by an rscS mutant
Our current data demonstrate that the conserved site of phosphorylation in the SypE REC domain is necessary to inactivate the inhibitory activity of the N-terminus and promote its positive activity. Furthermore, although SypE expressed from its native site in the chromosome inhibits biofilm formation induced by the overexpression of the RR SypG (Hussa et al. 2007), it does not inhibit RscS-induced biofilm formation. These data indicate that upon RscS induction, this inhibitory activity of SypE must be turned off. We therefore hypothesized that in a strain lacking rscS, SypE is likely to exhibit constitutive inhibitory activity and negatively impact host colonization (Fig. 1). This phenomenon could account, at least in part, for the severe colonization defect of an rscS mutant (Visick and Skoufos, 2001). To determine whether this is the case, we generated a ΔrscS ΔsypE double mutant and assessed the ability of this strain to compete with the ΔrscS parent to colonize juvenile squid. Indeed, the ΔrscS ΔsypE mutant cells dramatically outcompeted the ΔrscS single mutant (mean Log RCI, 1.76 +/− 0.66) with many animals exclusively colonized by the double mutant (open circles) (Fig. 10). These results indicate that SypE contributes to the colonization defect observed in an rscS mutant, supporting our hypothesis and linking SypE-mediated regulation of colonization to the activity of RscS. Together, these data demonstrate that SypE can function as a key regulator of host colonization, restricting colonization to conditions in which RscS is expressed or activated.
Figure 10. Impact of SypE on host colonization in rscS mutant cells.
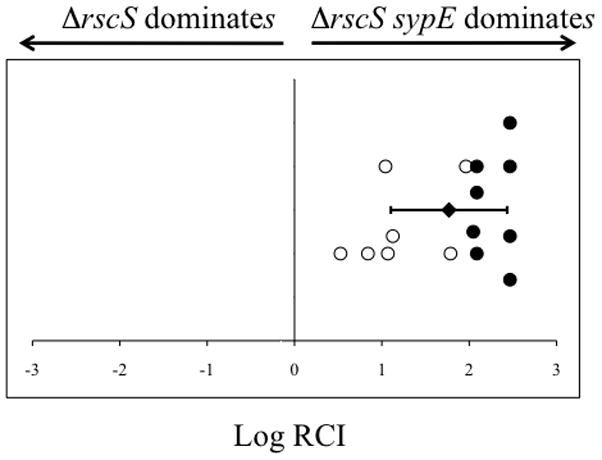
Competitive colonization assay with ΔrscS and ΔrscS ΔsypE mutant cells. Colonization assays were performed and Log RCI of the competing bacterial strains was calculated as described in Figure 7. Each circle represents a single animal. Closed circles represent animals in which no ΔrscS cells were present in the homogenate. The position of the circles on the y-axis is merely for spacing. The black diamond and error bars indicate the average Log RCI and standard deviation for 15 animals. The data shown above are from one experiment and are representative of at least 3 independent experiments.
Discussion
In this study, we uncovered a novel and critical role for the RR SypE in the inhibition of biofilm formation and host colonization by Vibrio fischeri. Our interest in characterizing SypE was prompted both by its unusual domain architecture and by earlier studies that suggested that SypE regulates V. fischeri biofilm regulation (Hussa et al., 2008; Darnell et al., 2008). Bioinformatics indicate that SypE is a novel RR protein, with its three putative domains arranged in a unique configuration: a central REC domain flanked by two effector domains with apparently opposing activities. We hypothesized that SypE could exert dual regulatory control over syp biofilm formation, depending on which of the terminal effector domains was active.
In this report, we performed a genetic analysis of SypE to determine how the individual domains contributed to the protein’s apparent dual regulatory activities. We found that SypE indeed possesses both negative and positive regulatory activities that are mediated by the N- and C- terminal effector domains, respectively. Although each domain was necessary and sufficient for the respective activities, our data suggested that one domain could influence the activity of another. In particular, we found that mutations within the C-terminal domain not only resulted in loss of SypE positive activity, but also shifted the protein’s activity to an inhibitory state. For example, SypED443A and SypED495A, which were predicted to lack the positive activity and thus behave similar to the uncomplemented (empty cassette) control, instead completely inhibited biofilm formation (Figs. 5E and 5F). This inhibitory activity depended on an intact N-terminal domain, as a SypEN52A, D443A double mutant lost its inhibitory activity (Fig. 5K). We interpret these data to suggest that the SypE protein structure is sensitive to molecular perturbations. In support of this, we found that the fusion of certain epitope tags to the C-terminus of SypE protein also resulted in aberrant protein activity in which the protein appeared “locked” in an inhibitory state (Morris and Visick, unpublished data). We speculate that disruption of the C-terminal effector domain likely alters the protein conformation to favor activation of the inhibitory N-terminal domain, perhaps indirectly by impacting phosphorylation of the protein. This would not be unprecedented: mutations within the C-terminal DNA-binding effector domain of the RR OmpR not only affect DNA-binding, but also impact the phosphorylation state of the N-terminal REC domain (Tran et al., 2000).
Our genetic analysis supports the model that the central REC domain functions to regulate SypE activity. In general, RR activity is controlled through the phosphorylation of a conserved aspartate residue located in the REC domain (Stock et al., 2000). REC domains are proposed to exist in an equilibrium of conformational states, which either activate or inactivate an effector domain (Stock and Guhaniyogi, 2006). Phosphorylation of the REC domain is proposed to shift this equilibrium and promote a particular conformational state, resulting in RR activation or inactivation (Stock and Guhaniyogi, 2006). Our results indicate that an intact REC domain is required to promote the positive regulatory activity of SypE. We found that a SypE mutant that mimics the unphosphorylated state (SypED192A) exhibits constitutive inhibitory activity, thus preventing biofilm formation (Fig. 6E). We interpret these data to indicate that phosphorylation of SypE at residue D192 is required to overcome SypE default inhibitory activity (Fig. 1). In support of this hypothesis, the inhibitory activity of the D192A mutant depended upon an intact N-terminal domain. Furthermore, the loss of inhibitory activity in the SypEN52A, D192A mutant failed to permit the protein to exert its positive activity (see Fig. 4L), suggesting that phosphorylation at residue D192 is still required for activation of the C-terminal domain and, thus, positive activity. Similarly, we found that the conserved aspartate was also required for positive regulatory activity displayed by a SypE mutant lacking the N-terminus (SypEΔNTD) (Morris and Visick unpublished data). We propose that REC phosphorylation shifts the equilibrium of the SypE conformational states such that the inhibitory N-terminus is inactivated, while the positively acting C-terminus is activated (Fig. 1).
In our assessment of the SypE REC domain, we attempted to identify mutations that would effectively mimic the phosphorylated state of SypE, resulting in “constitutively active” SypE and dominant positive activity. However, we found that several mutations reported to mimic the phosphorylated state in select RRs, such as a SypED192E (Klose et al., 1993) or a SypED192E, D150K (Bourret et al., 1993) mutant, failed to result in the constitutive activation of the positive C-terminal domain (Morris and Visick, unpublished data). The inability of these mutations to result in a SypE mutant that mimics the phosphorylated state is not surprising, as few mutations have been found to universally activate all RRs (Smith et al., 2004). Indeed, these common mutations have failed to activate several well-characterized RRs (Bourret et al., 1990; Pazour et al., 1992; Stewart et al., 1990). We hypothesize that, due to the complex domain structure of SypE, these mutations likely fail to mimic the conformational changes induced and/or stabilized by phosphorylation.
An earlier study reported that wild-type SypE inhibited biofilms induced by the RR SypG: overexpression of sypG in wild-type cells induced weak biofilm formation, while its overexpression in a ΔsypE strain resulted in robust biofilms similar to RscS-induced cells (Hussa et al., 2008). In contrast, we have shown here that upon rscS overexpression wild-type SypE exhibits positive regulatory activity. These data indicate that while SypG overexpression is capable of inducing syp biofilms, it is not sufficient to mimic the natural regulation of SypE and syp-dependent biofilms. These data can be explained by a model in which RscS modulates SypE activity (Fig. 1). As such, SypE would promote biofilms under conditions in which RscS is activated, and would prevent aberrant biofilm formation induced under other conditions (such as SypG overexpression). We hypothesize that RscS directs phosphorylation of SypE, as a SypED192A mutant that cannot become phosphorylated fails to promote biofilm formation (Fig. 6E). Whether RscS can directly phosphorylate SypE, or whether RscS influences the activity of another regulator that acts upon SypE, remains unknown. The possibility that RscS serves as a phosphodonor to both SypG and SypE would not be unprecedented, as other studies have reported a single SK interacting with multiple RRs [e.g., (Biondi et al., 2006)].
In this study, we provide compelling evidence that, in addition to a role in biofilm regulation, SypE also impacts host colonization in vivo. Our current data indicates that the minor positive role of SypE in promoting biofilm formation is relatively unimportant, as deletion of sypE did not substantially affect host colonization (Fig. 7 and data not shown). It remains possible that the positive activity of SypE does indeed contribute to colonization fitness, but that this impact may not be readily evident given our current experimental conditions. In contrast to the positive activity, we show that inhibition by SypE is a significant activity that must be overcome during the colonization process. We found that cells expressing a SypE mutant disrupted at the predicted site of phosphorylation (SypED192A) exhibited a substantial defect in host colonization (Fig. 8), a defect that could be attributed to the inability of these cells to efficiently aggregate in the squid-derived mucus on the light organ surface, an essential stage in the initiation of host colonization (Fig. 9C). Thus, residue D192 within the central REC domain is absolutely required to inactivate the inhibitory activity of SypE. We propose that phosphorylation at residue D192 is necessary to shift the activity of SypE away from the inhibitory N-terminal domain, and permit activation of the positive C-terminal domain. Consistent with our biofilm results, this inhibition of host colonization and bacterial aggregation by the SypED192A mutant depended upon an intact N-terminal domain, as cells expressing the SypEN52A, D192A double mutant exhibited near wild-type levels of colonization and aggregation (Fig. 8C and 9D). These data underscore the utility of intra-molecular epistasis as a powerful approach to understanding the function of a multi-domain protein.
Together, these results—that (1) SypE inhibitory activity must be inactivated for colonization to proceed, (2) inactivation of this inhibitory activity depends upon the REC domain and likely phosphorylation at D192, and (3) the SK RscS promotes this inactivation—permitted us to generate a model for the severe colonization defect observed in an rscS mutant (Visick and Skoufos, 2001) (Fig. 1). In this model, RscS would play roles in both inducing syp transcription and inactivating the inhibitory activity of SypE. We hypothesized that if this were true, then a ΔrscS ΔsypE double mutant should colonize better than an rscS mutant. Indeed, we found that the ΔrscS ΔsypE mutant dramatically outcompeted the ΔrscS mutant in competitive colonization assays (Fig. 10). Colonization by the ΔrscS ΔsypE mutant did not fully mimic that of wild-type cells (Morris and Visick, unpublished data); this is likely due to the additional role of RscS in activating SypG-dependent transcription of the syp locus and biofilm formation (Hussa et al., 2008). These experiments support the model that the inactivation of SypE inhibitory activity is critical for successful symbiotic colonization. Intriguingly, sypE is conserved in the genome of several syp-containing Vibrios that lack the SK rscS, including V. fischeri strain MJ11. Previously, Mandel et al. (2009) demonstrated that the inability of strain MJ11 to proficiently colonize E. scolopes could be attributed to the absence of rscS. It is possible that the inhibitory activity of SypE contributes to the inability of MJ11 to colonize E. scolopes. A role for SypE in restricting host colonization in V. fischeri strain MJ11 or other Vibrios awaits future studies.
The mechanism by which SypE controls biofilm formation remains an area of active study. Current evidence indicates that SypE exerts its regulatory control at some unknown level downstream of SypG activation and syp transcription (Hussa et al., 2008; Morris and Visick, unpublished data). Our genetic and bioinformatic analyses suggest that the N- and C-terminal effector domains likely control biofilm formation through the phosphorylation and dephosphorylation of an as-yet unknown downstream target protein. The N- and C-terminal effector domains of SypE share weak sequence similarity to the serine kinase, RsbW, and the serine phosphatase, RsbU, of Bacillus subtilis, respectively. In B. subtilis, these proteins function as regulators of σB activity in the general stress response pathway (Dufour and Haldenwang, 1994; Voelker et al., 1996). In this pathway, both RsbW and RsbU interact with and mediate the phosphorylation and dephosphorylation, respectively, of RsbV, an anti-sigma factor antagonist (Dufour and Haldenwang, 1994; Voelker et al., 1996). Intriguingly, the syp locus encodes a putative anti-sigma factor antagonist, SypA, which shares sequence similarity to RsbV (Yip et al., 2005; Morris and Visick, 2010). Current studies are underway to determine if SypE’s effector domains indeed possess serine kinas and serine phosphatase activity, and whether SypA serves as the target of SypE regulatory activity.
We conclude from this study that initiation of host colonization by V. fischeri is controlled through the inactivation of a negative regulator. Such a mechanism—inactivation of an inhibitory response regulator—is rarely observed in other characterized models of colonization. This is also an unusual mechanism for the control of biofilm formation. The data we present here also provide insights into the previously established requirement for the SK RscS in initiating colonization (Visick and Skoufos, 2001) by demonstrating that this regulator is necessary for the inactivation of SypE. That this inactivation is necessary for symbiotic aggregation and colonization, fully correlating with its requirement for biofilms outside the host, further demonstrates the utility of the V. fischeri-squid symbiosis as a model for the role of biofilm formation in host colonization. Interestingly, bioinformatic analyses suggest that SypE-like proteins may be present in diverse bacterial species, including Vibrios, such as Aliivibrio salmonicida, and non-Vibrios, such as Aeromnas veronii. What function(s), if any, these SypE-like proteins possess in these other organims remains unknown. Additional characterization of this non-DNA binding RR with its unusual structure of apparently opposing enzymatic activities will undoubtedly add to our understanding of RR function and diversity, as well as control of biofilm formation and host colonization.
Experimental Procedures
Bacterial strains, plasmids and media
The bacterial strains and plasmids used in this study are listed in Table 1. V. fischeri strain ES114, a bacterial isolate from E. scolopes, was used as the parental strain in this study (Boettcher and Ruby, 1990). All V. fischeri derivatives were generated by conjugation, as previously described (Visick and Skoufos, 2001). The RscS-expressing plasmid, pARM7, was derived from plasmid pKG11 (Yip et al., 2006), in which an EcoRI partial digest was performed to remove the chloramphenicol resistance cassette. E. coli strains Tam1 pir (Active Motif, Carlsbad, CA), DH5α (Invitrogen, Carlsbad, CA), and GT115 (Invivogen, San Diego, CA) were used for cloning and conjugative purposes. E. coli strains were grown in Luria Bertani media (LB) (Davis et al., 1980). V. fischeri strains were grown in complex media [Sea Water Tryptone (SWT) (Yip et al., 2005) or LBS (Graf et al., 1994)] or in HEPES minimal medium [HMM (Ruby and Nealson, 1977)] containing 0.3% Casamino acids and 0.2% glucose (Yip et al., 2005). Agar was added to a final concentration of 1.5% for solid media. The following antibiotics were added to V. fischeri media, where necessary, at the indicated concentrations: chloramphenicol (Cm) 5 μg mL−1 and tetracycline (Tc), 5μg mL−1 in LBS, and 30 μg ml 1 in SWT and HMM. The following antibiotics were added to E. coli media, where necessary, at the indicated concentrations: Cm at 25 mg/ml−1, kanamycin (Kan) at 50 μg ml−1, Tc at 15 μg/ml−1, or ampicillin (Ap) at 100 μg ml−1.
Table 1.
Strains and plasmids used in this study
| Relevant Genotype | Reference | |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 (rK-mK+) supE44 thi-1 recA1 relA Δ(lacIZYA-argF)U169 phoA [φ80dlacΔ(lacz)M15] | Invitrogen |
| TAM1λ pir | mcrA Δ(mrr-hsdRMS-mcrBC) Φ80Δ(lacz)M15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG attλ::pir+ | Active Motif |
| GT115 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80laczΔM15 ΔlacX74 recA1 endA1 Δdcm uidA(ΔMluI)::pir-116 ΔsbcC-sbcD | InvivoGen |
| V. fischeri | ||
| ES114 | Wild-type V. fischeri | (Boettcher and Ruby, 1990) |
| KV3299 | ΔsypE | (Hussa et al., 2008) |
| KV4389 | ES114 attTn7::erm | This Study |
| KV4390 | ΔsypE attTn7::erm | This Study |
| KV4819 | ΔsypE attTn7:: Placz-sypE+ (erm) | This Study |
| KV4885 | ΔsypE attTn7:: Placz-sypED192A (erm) | This Study |
| KV4886 | ΔsypE attTn7:: Placz-sypED443A (erm) | This Study |
| KV4887 | ΔsypE attTn7:: Placz-sypED495A (erm) | This Study |
| KV5124 | ΔsypE attTn7:: Placz-sypEΔNTD (erm) | This Study |
| KV5129 | ΔsypE attTn7:: Placz-sypENTD (erm) | This Study |
| KV5142 | ΔsypE attTn7:: Placz-sypEN52A (erm) | This Study |
| KV5143 | ΔsypE attTn7:: Placz-sypENTD, N52A (erm) | This Study |
| KV5204 | ΔsypE attTn7:: Placz-sypECTD (erm) | This Study |
| KV5205 | ΔsypE attTn7:: Placz-sypEN52A, D192A (erm) | This Study |
| KV5314 | ΔsypE attTn7:: Placz-sypECTD, D443A (erm) | This Study |
| KV5315 | ΔsypE attTn7:: Placz-sypEΔCTD (erm) | This Study |
| KV5345 | ΔsypE attTn7:: Placz-sypECTD, D495A (erm) | This Study |
| KV5379 | ΔsypE attTn7:: Placz-sypEN52A, D443A (erm) | This Study |
| Plasmids | ||
| pARM7 | EcoRI partial digest of pKG11 (rscS); tetR | This study |
| pARM47 | pEVS107 containing 1.5 kb sypE | This study |
| pARM54 | pEVS107 containing 1.5 Kb sypED192A | This study |
| pARM55 | pEVS107 containing 1.5 Kb sypED443A | This study |
| pARM56 | pEVS107 containing 1.5 Kb sypED495A | This study |
| pARM81 | pEVS107 containing 1.2 Kb sypEΔNTD | This study |
| pARM82 | pEVS107 containing 700 bp sypECTD | This study |
| pARM89 | pEVS107 containing 500 bp sypENTD | This study |
| pARM90 | pEVS107 containing 1.5 kb sypEN52A | This study |
| pARM91 | pEVS107 containing 500 bp sypENTD, N52A | This study |
| pARM101 | pEVS107 containing 1.5 kb sypEN52A, D192A | This study |
| pARM109 | pEVS107 containing 700 bp sypECTD, D443A | This study |
| pARM110 | pEVS107 containing 1.0 kb sypEΔCTD | This study |
| pARM112 | pEVS107 containing 700 bp sypECTD, D495A | This study |
| pARM125 | pEVS107 containing 1.5 kb sypEN52A, D443A | This study |
| pEVS104 | Conjugal helper plasmid (tra trb), KanR | (Stabb and Ruby, 2002) |
| pEVS107 | Mini-Tn7 delivery plasmid; oriR6K, mob; KanR, EmR | (McCann et al., 2003) |
| pJET1.2 | Commercial cloning vector; ApR | Fermentas |
| pKV282 | Mobilizable vector; TetR | This Study |
| pVSV105 | Mobilizable vector, R6Kori ori(pES213) RP4 oriT cat | (Dunn et al., 2006) |
Construction of sypE alleles
The sypE alleles used in this study were generated by PCR amplification using the primers listed in Table 2. The PCR products were cloned into the pJET1.2 cloning vector (Fermentas, Glen Burnie, MD), and then subcloned into mobilizable plasmid pVSV105 (Dunn et al., 2006) using standard molecular techniques. For chromosomal insertion at the Tn7 site, the sypE genes were subcloned with the upstream PlacZ promoter into the mini-Tn7 delivery vector pEVS107 (McCann et al., 2003). The sypE alleles were then inserted into the chromosomal Tn7 site of V. fischeri using a tetraparental mating as previously described (McCann et al., 2003). To generate site-directed mutations in sypE, we utilized the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Plasmid pCLD48 (Table 1) served as the template for the primer Phos-lacZ-up-rev and select sypE mutagenic primers (Table 2). Generation of the desired mutations was confirmed by sequence analysis using the Genomics Core Facility at the Center for Genetic Medicine at Northwestern University (Chicago, IL) and ACGT, inc (Wheeling, IL). FLAG epitope fusions to C-terminus of full-length SypE proteins were generated by standard PCR using the forward primer, SypE-F, and reverse primer, SypE-Full-R FLAG (Table 2). FLAG epitope fusions were generated to the C-terminus of the SypENTD protein using the primers forward primer SypE-F and the reverse primer SypE-RsbW-R FLAG (Table 2). Forward primer, SypECTD-F, and reverse primer, SypE-Full-R FLAG, were used to generate FLAG fusions to the C-terminus of the SypECTD proteins (Table 2).
Table 2.
Primers used in this study
| Primer Name | Sequence (5′-3′) |
|---|---|
| SypE-F | GTCCAAAGAAACCGATTTTTATC |
| SypE-R | TTTTTCTGCACTTATTGATTCTCAATTAACAGC |
| SypEΔNTD-F | GTGGTGTAATCATGGAGCGTTCCCCTTCCCAT |
| SypENTD-R | CTTAATGGGAAGGGGAACGCTC |
| SypECTD-F | GTGGTGTAATCATGGCCCATACTCTATTACCACAA |
| SypEN52A | TCTGAATGGAGCACCGCTCTAGTTTTGCACCCT |
| SypED443A | TTAGCATTATTTACTGCTGGGTTATTTGAATCA |
| SypED495A | GGGACACCACCCAATGCCGATGCATTATTGCTG |
| SypED192A | GATTTAATTATCTCAGCTATTCAAATGCCTGAA |
| SypED192E | GATTTAATTATCTCAGAGATTCAAATGCCTGAA |
| SypE-Full-R FLAG | ACCCGGGTTATTTATCATCATCATCTTTATAATCTTGATTCTCAATTAACAG |
| SypE-RsbW-R FLAG Phospho-lacZ-R | ACCCGGGTTATTTATCATCATCATCTTTATAATCATGGGAAGGGGAACGCTCCCTGTGTGAAATTGTTATCCG |
Wrinkled colony assays
To observe wrinkled colony formation, strains were streaked onto LBS agar plates containing Tc. Single colonies were then cultured with shaking in LBS broth with Tc overnight at 28°C and then sub-cultured to an OD600 of 0.1 in 5 mL of fresh medium. Cells were spun down, washed twice in 70% artificial seawater (ASW) (280 mM MgSO4, 56 mM CaCl2, 1.68 M NaCl, 56 mM KCl), and re-suspended in 70% ASW and diluted to an OD of 0.1. 10 μL of re-suspended cultures were spotted onto LBS agar plates and grown for 48 h at 22°C. Images of the spotted cultures were acquired at the indicated time points using the Zeiss stemi 2000-C dissecting microscope. For RscS-induced wrinkling time course assays, spot development was followed over a course of 48 h and images taken at indicated time points.
Pellicle Assay
Strains were grown with shaking in HMM with Tc at 28°C overnight and then subcultured to an OD of 0.1 in 1.5mL of fresh medium in 24-well microtiter dishes. Cultures were then grown at room temperature for up to 48 h. The strength of each pellicle was evaluated by disrupting the air-liquid interface with a sterile pipette tip after 48 h of incubation. A pellicle is observed as a disruption at the culture surface. Cultures with no pellicle were scored as (−); cultures with an intermediate/weak pellicle were scored as (+); cultures with a strong pellicle that was able to be lifted from the culture intact were scored as (++).
Confocal microscopy
Cells expressing green fluorescent protein (GFP) were grown statically in HMM containing Tc in 12 well microtiter plates with glass cover slips partially submerged into the culture medium. Cover slips were incubated with bacteria at room temperature for 24 h and removed for biofilm examination. A Zeiss LSM 510 confocal microscope (40X objective) was used to collect xy plane and z sections (xz and yz plane) images of the biofilms. Representative images were prepared using the Zeiss LSM Image Browser software.
Squid colonization assays
To perform single-strain colonization assays, juvenile squid were placed into artificial seawater (ASW) (Instant Ocean; Aquarium Systems, Mentor, OH) containing roughly 1,000–1,500 cells per mL of seawater. Colonization assays were allowed to proceed for 16 h post inoculation, at which time the animals were washed twice in ASW and homogenized to release the light organ contents. Serial dilutions of the light organs were plated and colony-forming units (CFU) calculated. For competitive colonization assays, juvenile squid were placed into ASW containing approximately 1,000 V. fischeri cells per mL of seawater. Juvenile squid were inoculated with an approximate 1:1 ratio of mutant and wild-type cells, and colonization was allowed to proceed for 18 h. For these assays, sypE+ cells were marked with an erythromycin resistance (EmR) cassette within the chromosome at the Tn7-site. Reciprocal experiments were also performed in which ΔsypE cells contained the EmR marker. The ratio of bacterial strains within the light organs of the animals was assessed through luminescence and homogenization/plating assays as described previously (Yip et al., 2006). The competitive colonization data are reported as the log-transformed relative competitive index (Log RCI). This index is generated by dividing the ratio of mutant to wild-type in the homogenate by the ratio present in the inoculum and calculating the log10 value of that number.
Symbiont aggregation assays
Log-phase cells (OD600 0.3 to 0.6) were grown in 2 ml SWT medium at 22°C. Bacterial cells were then inoculated into filtered ASW at a concentration of 106 cells per ml. Juvenile squid were then placed into inoculated seawater and the two organisms were allowed to associate at room temperature for 3 h prior to dissection. The juvenile squid were removed to vials containing 2 ml filter-sterilized seawater and 5 μM of the counter-stain, CellMask (Invitrogen, Eugene, OR). The animals were then anesthetized in filtered artificial seawater containing 2% ethanol. Each squid was placed ventral side up on a depression well slide and dissected to remove the mantle and funnel and expose the light organ. Fluorescently labeled light organs (blue) and GFP-labeled (green) bacteria were viewed using a Zeiss LSM 510 confocal microscope.
SypE FLAG protein expression
FLAG epitope fusions were generated to the C-terminus of wild type sypE or mutant sypE derivatives as described earlier. FLAG-sypE alleles were cloned into the expression vector pVSV105 and subsequently conjugated into ΔsypE cells containing the rscS expressing plasmid (pARM7). Biofilm assays were performed to confirm that the FLAG epitope fusions did not disrupt SypE regulatory activity. Strains were cultured in LBS containing Tc and Cm for 18h at 28°C and subsequently sub-cultured to an OD600 of 0.2. 250 mL of culture was pelleted by centrifugation, lysed in 250 mL 4x SDS sample buffer, and resolved on 10–15% SDS-PAGE gels. Duplicate gels were stained with Coomassie brilliant blue in order to ensure roughly equal loading of protein samples. Protein transfer to PVDF membrane was performed using standard Tris-Glycine transfer buffer containing 20% methanol. Western blotting was performed using standard protocols with rabbit anti-FLAG primary antibody and donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase. Blots were imaged by chemiluminescent detection (SuperSignal West Pico Chemiluminescent Substrate, Pierce Biotechnology, Inc., Rockford, IL.)
Supplementary Material
Acknowledgments
We thank Ned Ruby and Margaret McFall-Ngai for the use of E. scolopes juveniles and a confocal microscope, and Elizabeth Heath-Heckman and Valerie Ray for assistance in the bacterial aggregation assays. We also thank Alan Wolfe, Jonathan Visick, and members of the Visick and Wolfe labs for critical reading of the manuscript. This work was supported by NIH grant GM59690 awarded to K.L.V and a grant from the Conservation Medicine Center of Chicago awarded to A.R.M.
References
- Adler E, Donella-Deana A, Arigoni F, Pinna LA, Stragler P. Structural relationship between a bacterial developmental protein and eukaryotic PP2C protein phosphatases. Mol Microbiol. 1997;23:57–62. doi: 10.1046/j.1365-2958.1997.1801552.x. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Drake SK, Chervitz SA, Simon MI, Falke JJ. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Hess JF, Simon MI. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell CL, Hussa EA, Visick KL. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. Advanced bacterial genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1980. [Google Scholar]
- Dufour A, Haldenwang WG. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp-and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol. 2010;13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J Bacteriol. 2007;189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MD, Fjeld CC, Denu JM. Probing the function of conserved residues in the serine/threonine phosphatase PP2Calpha. Biochemistry. 2003;42:8513–8521. doi: 10.1021/bi034074+. [DOI] [PubMed] [Google Scholar]
- Klose KE, Weiss DS, Kustu S. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J Mol Biol. 1993;232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Visick KL. Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ Microbiol. 2010;12:2051–2059. doi: 10.1111/j.1462-2920.2010.02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Ta CN, Das A. Constitutive mutations of Agrobacterium tumefaciens transcriptional activator virG. J Bacteriol. 1992;174:4169–4174. doi: 10.1128/jb.174.12.4169-4174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Nealson KH. Pyruvate production and excretion by the luminous marine bacteria. Appl Environ Microbiol. 1977;34:164–169. doi: 10.1128/aem.34.2.164-169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Fong JC, Odell LS, Perchuk BS, Laub MT, Yildiz FH. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Smith JG, Latiolais JA, Guanga GP, Pennington JD, Silversmith RE, Bourret RB. A search for amino acid substitutions that universally activate response regulators. Mol Microbiol. 2004;51:887–901. doi: 10.1046/j.1365-2958.2003.03882.x. [DOI] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stewart RC, Roth AF, Dahlquist FW. Mutations that affect control of the methylesterase activity of CheB, a component of the chemotaxis adaptation system in Escherichia coli. J Bacteriol. 1990;172:3388–3399. doi: 10.1128/jb.172.6.3388-3399.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Guhaniyogi J. A new perspective on response regulator activation. J Bacteriol. 2006;188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, V, Robinson L, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Tran VK, Oropezaand R, Kenney LJ. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J Mol Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- Visick KL. An intricate network of regulators controls biofilm formation and colonizationby Vibrio fischeri. Mol Microbiol. 2009;74:782–789. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Skoufos LM. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol. 2001;183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker U, Voelker A, Haldenwang WG. Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Whistler CA, Ruby EG. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J Bacteriol. 2003;185:7202–7212. doi: 10.1128/JB.185.24.7202-7212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


