Summary
Intracellular RIG-I-like receptors (RLRs, including RIG-I, MDA-5, and LGP-2) recognize viral RNAs as pathogen-associated molecular patterns (PAMPs) and initiate an antiviral immune response. To understand the molecular basis of this process, we determined the crystal structure of RIG-I in complex with double-stranded RNA. The dsRNA is sheathed within a network of protein domains that include a conserved “helicase” domain (regions HEL1 and HEL2), a specialized insertion domain (HEL2i), and a C-terminal regulatory domain (CTD). A V-shaped pincer connects HEL2 and the CTD by gripping an α-helical shaft that extends from HEL1. In this way, the pincer coordinates functions of all the domains and couples RNA binding with ATP hydrolysis. RIG-I falls within the Dicer-RIG-I clade of super family 2 of helicases and this structure reveals complex interplay between motor domains, accessory mechanical domains and RNA that has implications for understanding the nanomechanical function this protein family and other ATPases more broadly.
Keywords: RIG-I, RNA helicase, innate immunity, X-ray crystallography
Introduction
The innate immune system utilizes sensory molecules that detect and respond to pathogens. RIG-I-like receptors (RLRs: retinoic acid-inducible gene I, RIG-I; melanoma differentiation-associated gene 5, MDA-5; and laboratory of genetics and physiology 2, LGP-2) sense viral RNAs in the cellular environment and activate extensive host immunological responses against viral infection (Yoneyama et al., 2005; Yoneyama et al., 2004). The conserved “helicase” core of RIG-I and MDA-5 is connected to two caspase activation and recruitment domains (CARDs) at the N-terminus and a Zn2+-containing regulatory domain at the C-terminus. LGP-2 has similar domain architecture, although it contains no CARDs. RIG-I and MDA-5 share similar signaling pathways and adaptor molecules, although RIG-I and MDA-5 complement each other by responding to diverse types of non-self RNAs from different viruses (Yoneyama et al., 2005). The function of LGP-2 is less clearly defined, as it can act positively or negatively upon activation by different viruses (Saito et al., 2007; Satoh et al., 2010). Upon binding and activation by viral dsRNA or triphosphorylated RNA, RIG-I recruits the adaptor IPS-1 (also known as MAVS, CARDIF or VISA) on the outer membrane of the mitochondria through the CARDs (Fujita et al., 2007; Meylan et al., 2005). This activates several transcription factors, including IRF3, IRF7 and NF-κB, and leads to the production of type I interferon (IFN) and inflammatory cytokines (Fujita et al., 2007). Post-translational modifications of RIG-I, including ubiquitination, phosphorylation and SUMOylation, are reported to be important for its function (Gack et al., 2010; Gack et al., 2007; Mi et al., 2010). Moreover, RIG-I displays an apoptosis-inducing property in tumor cells (Kubler et al., 2010; Poeck et al., 2008). Effective therapeutic RIG-I agonists could serve as a new approach for the treatment of both viral infections and cancer (Zitvogel and Kroemer, 2009).
Proteins containing the conserved sequences typical of “helicase superfamily 2” (SF2, (Gorbalenya et al., 1989)) are involved in virtually all aspects of nucleic acid metabolism, displaying diverse functions that include translocation along nucleic acids, duplex unwinding, strand annealing, RNA chaperone activity, and protein displacement or recruitment (Fairman-Williams et al., 2010; Pyle, 2008). RIG-I belongs to an eponymous SF2 subfamily known as the “RIG-I-like Receptors”, or RLR proteins, which are also related to Dicer and orthologous motor proteins involved in gene silencing (the Dicer-RIG-I clade). RLRs have a conserved core architecture that is similar to SF1 and SF2 helicases, consisting of two RecA-like domains (a.k.a. Rossmann Fold with repeating α/β topology (Rao and Rossmann, 1973)) that form an active site for ATP binding and hydrolysis at their interface. Adjacent to the active site are conserved motifs that form a platform for recognition and remodeling of nucleic acids. The uniqueness of RLR helicases comes from their accessory domains, which include N-terminal CARDs, an insertion domain within the helicase core, and a C-terminal regulatory domain. In the case of RIG-I, the CARDs are responsible for downstream signal transduction (Fujita et al., 2007; Yoneyama et al., 2004), but the role of the insertion domain has remained unclear. The CTD helps recognize non-self RNAs within the cellular environment (Fujita et al., 2007; Wang et al., 2010b). It is generally accepted that RIG-I can be activated by 5’triphosphate (5’ppp) ssRNA or dsRNA (Hornung et al., 2006; Kato et al., 2006; Lu et al., 2010; Pichlmair et al., 2006; Wang et al., 2010b). Although blunt dsRNA is an effective activator of RIG-I, the presence of a 5’ppp increases the potency of dsRNA activation, highlighting the importance of both chemical and structural features of the substrate. Here, we report the first crystallographic structure of RIG-I (ΔCARDs) in complex with dsRNA. Analysis of the sequence and structure provides important insights into RNA induced RIG-I activation. In addition, this system expands our understanding of RNA recognition by SF2 proteins and reveals complex mechanical strategies for RNA signaling and remodeling.
Results
Overview of the RIG-I (ΔCARDs) dsRNA10 complex structure
To focus on the molecular basis for RNA recognition by RIG-I, we designed a deletion construct (RIG-I (ΔCARDs)) lacking the N-terminal CARDs (residues 1-229). We co-crystallized the protein with a 10-mer palindromic duplex RNA that has 5’OH blunt ends (5’ GCGCGCGCGC 3’) (Fig.1A) and determined the structure of the complex to a resolution of 2.5 Å (Table S1, Fig. S1).
Figure 1. The RIG-I (ΔCARDs) dsRNA complex.
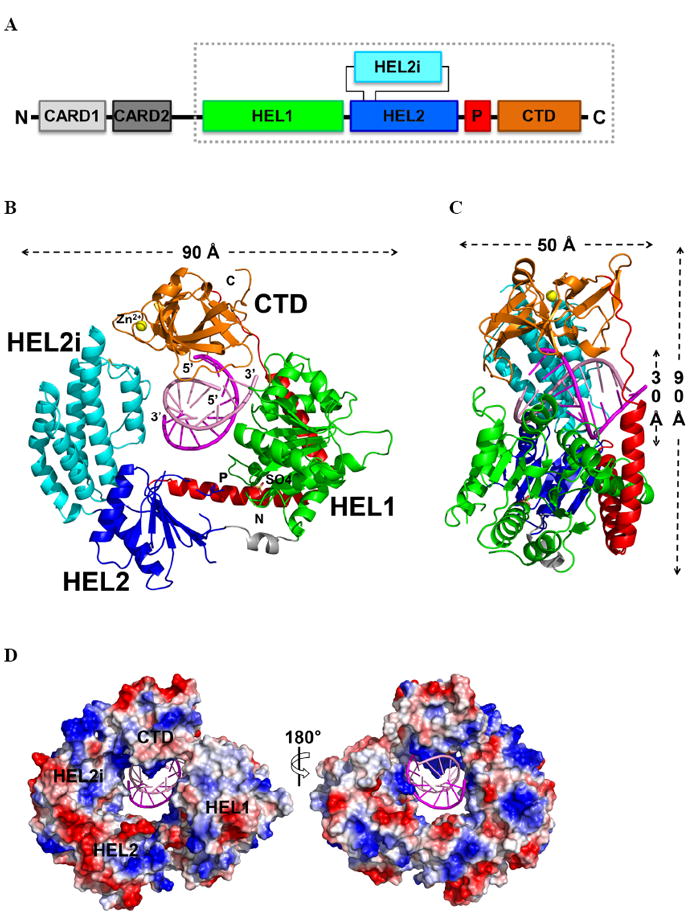
(A) Schematic representation of the RIG-I protein. The RIG-I (ΔCARDs) construct used for this study is boxed. (B) Overall structure of the RIG-I (ΔCARDs) dsRNA complex. Starting from the N-terminus: the helicase domain 1 (HEL1) is in green, the bridge between the two helicase domains grey, the helicase domain 2 (HEL2) blue, the insertion domain (HEL2i) cyan, the pincer region (P) red, and the C-terminal domain (CTD) orange. The top strand of the RNA duplex (dsRNA10) is in magenta and the bottom strand pink. Data collection and refinement statistics are provided in Table S1. See also the 2Fo-Fc map, shown in Fig. S1. (C) Side view showing the RNA interface with the central groove of RIG-I (ΔCARDs). A 90° rotation along the y-axis relative to the orientation shown in (B). (D) Solvent accessible electrostatic surface views of RIG-I (ΔCARDs), shown with ±10 kbT/ec. See also SAXS data provided in Fig. S3.
A striking feature of the complex is that RIG-I completely surrounds the RNA and encloses it within an elaborate network of interactions (Fig. 1B). The central cavity of the protein has dimensions of 30 × 30 × 50 Å and buries the entire dsRNA10 (Fig. 1B,C). HEL1, the first Rec-A like domain, contains seven α-helices and seven β-strands. HEL1 faces the minor groove of the RNA and interacts with the RNA backbone of both strands (Fig. 2A). A sulfate ion is present at the ATP binding site and stabilizes the otherwise flexible Walker A motif (a.k.a. phosphate binding loop, P-loop). There is a relatively open conformation between HEL1 and HEL2. Almost half of HEL2, including the conserved motifs (IVa, Va, Vb, and VI), is disordered in the present structure. This may be attributable to the lack of direct interactions between HEL2 and RNA, or to the absence of ATP. An intriguing insertion domain (HEL2i) is present between the β21 strand and the α22 helix of HEL2 (Fig. 1). Similar to the insertion domain that is present in Pfu Hef (Nishino et al., 2005), HEL2i consists of five α-helices and adopts an α-helical bundle structure. HEL2i is an important component of the ring that grips dsRNA by directly interacting with the minor groove of the RNA backbone. Strikingly, HEL2 and the CTD are connected through a V-shaped structure that is composed of two long α helices (αP1 - αP2) (Fig. 1 and Fig. 3). This V-shaped pincer grips an α-helix that projects from HEL1 like a shaft (α17), thereby establishing extensive interactions and a mechanical connection between HEL1, HEL2 and the CTD. The CTD interacts with RNA in a manner similar to previously-reported CTD domain structures. However, the orientation of the dsRNA relative to the CTD varies in all of these cases (Fig. 4A,B). The inner face of the CTD cavity is positively charged in order to accommodate the dsRNA (Fig. 1D). The dsRNA10 maintains an A-form conformation and there is no evidence of destabilization or partial unwinding.
Figure 2. Double-stranded RNA recognition.
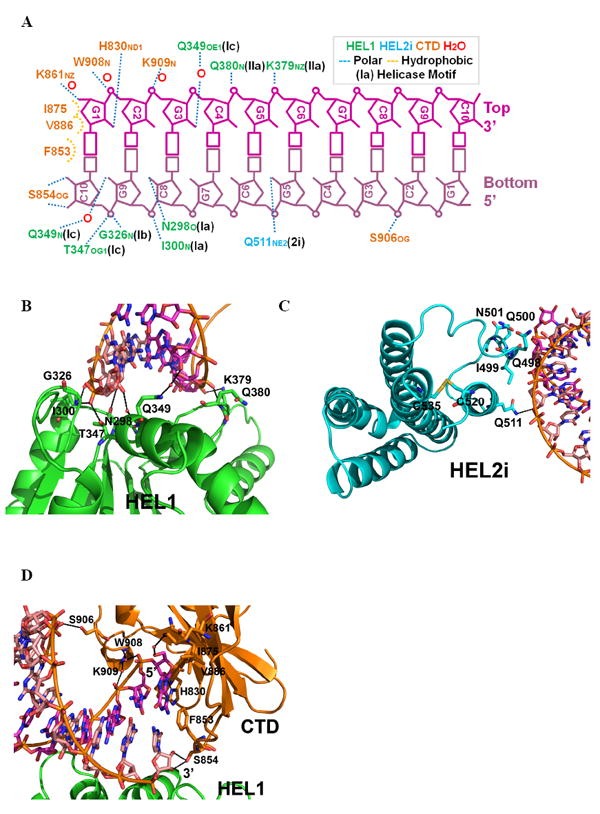
(A) The interactions between RIG-I (ΔCARDs) and dsRNA10, determined with a 3.3 Å cutoff for hydrogen bonding. Close-up views of the (B) HEL1-RNA, (C) HEL2i-RNA, and (D) CTD-RNA interfaces. See also Fig. S4.
Figure 3. Sequence comparison of RIG-I orthologs and related RLR proteins.
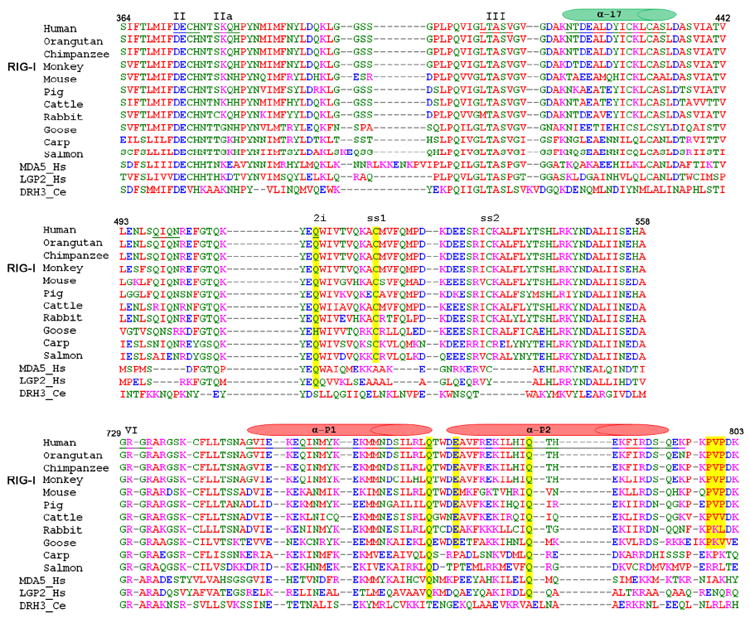
Selected sequence alignments of all available RIG-I orthologs (top 11 rows) and comparison with closely related RLR proteins MDA-5 and LGP-2 from Homo sapiens, Dicer-related-Helicase 3 (DRH-3) from Caenorhabditis elegans. Top panel shows conservation of Motif IIa and the a17 helical shaft (green cylinder). Middle panel shows conservation of insertion domain HEL2i. Bottom panel shows conservation of residues in Pincer domain, and helices of the pincer are indicated (red cylinders). Yellow lines indicate positions of mutated residues. A complete alignment of RLR sequences is provided as Fig. S2.
Figure 4. Superposition of RLR CTD-dsRNA complexes.
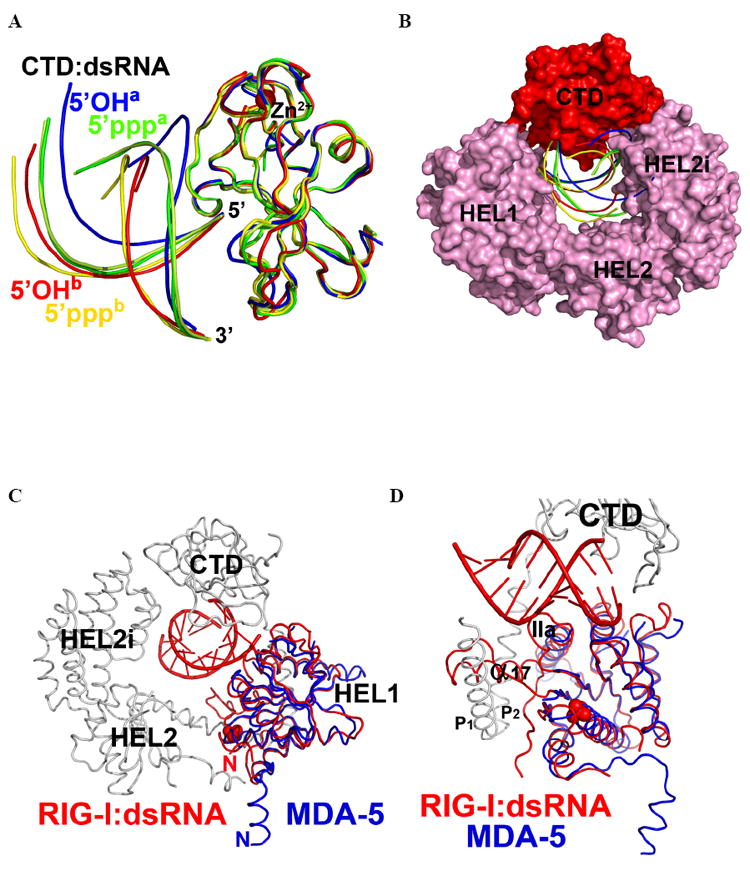
Comparison of the RNA duplex orientation in RIG-I (ΔCARDs):dsRNA with the available CTD:dsRNA structures. (A). CTDs of all complexes are superimposed and color-coded as follows. Red: CTD of RIG-I (ΔCARDs) and its complex with 5’OH-dsRNA10 complex, designated “5’OHb”. Green: RIG-I CTD and 5’ppp-dsRNA12 (PDB: 3LRR), designated 5’pppa. Pale Green: RIG-I CTD and 5’ppp-dsRNA12 (PDB: 3NCU), labeled as “5’pppa”. Blue: RIG-I CTD and 5’OH-dsRNA14 (PDB: 3OG8), labeled as “5’OHa”. Yellow: RIG-I CTD and 5’ppp-dsRNA14 (PDB: 3LRN), labeled as “5’pppb”. (B). Space-filling representation of RIG-I (ΔCARDs):dsRNA showing variation in RNA position among superimposed structures (C) Superposition of the RIG-I (ΔCARDs) and 5’OH-dsRNA10 complex and the MDA-5 HEL1 (blue ribbon) (PDB: 3B6E). (E) Close-up view of (D). See also Fig. S4.
Duplex RNA recognition by RIG-I
Three different RIG-I domains (HEL1, HEL2i and the CTD) clasp the duplex RNA, enfolding it within a network of interactions that are dominated by polar contacts (Fig. 2A). Numerous contacts are formed between 2’-hydroxyl groups on the RNA and protein amide groups, located either on the main chain or side chains of conserved Q and N residues at the protein interface. Less numerous are highly electrostatic contacts between nonbridging phosphoryl oxygens of the RNA backbone and charged residues, such as lysine. The pattern of molecular contacts observed here thus explains the specific role of RIG-I in recognizing duplex RNA and it is consistent with previous studies showing that stimulatory RNAs bearing 2’-modifications are inhibitory to RIG-I function (Uzri and Gehrke, 2009; Wang et al., 2010b).
HEL1 is of particular interest because it interacts with both strands at the same time, in a manner that has never been observed previously for RNA recognition (Fig. 2B). Motifs Ia, Ib, and Ic interact with the bottom RNA strand, displaying the 3’ to 5’ directionality and a binding mode that are common to all SF2 helicases (Appleby et al., 2010; Bono et al., 2006; Gu and Rice, 2010; Luo et al., 2008; Pyle, 2008). A specialized motif (motif IIa, located three amino acids after motif II, Fig. 3, Fig. S2) interacts with the top strand of the dsRNA. The Q380 residue in motif IIa interacts with RNA through main chain atoms of the peptide backbone, which may explain the low sequence conservation of this motif (Fig. 2A and Fig. 3, top panel). The close proximity between motif II (essential for divalent metal ion binding and ATP hydrolysis) and motif IIa (important for second-strand RNA binding) also suggests direct mechanical coupling between these two activities of RNA helicase enzymes (Schutz et al., 2010). It is worth noting that, in the absence of dsRNA, motif IIa is completely disordered in MDA-5 HEL1 (Fig. 4D) (Karlberg et al., 2007).
HEL2i plays an important role in the specific recognition of RNA (Fig. 2C). One hydrogen bond is established between Q511 and the 2’OH of residue G5 on the RNA bottom strand. Q511 is conserved among RLRs (Fig. 3), suggesting that it plays a critical role in RNA recognition by RLR proteins in general (Fig. S2). Indeed, when Q511 is mutated to an alanine, the RNA-stimulated interferon response is greatly reduced (Fig. 5A). Together with Q511 on the α2i2 helix, a stretch of amino acids (Q498-I499-Q500-N501) unique to RIG-I creates a concave binding surface for the bottom strand RNA (Fig. 2C). Interestingly, a disulfide bond is formed between C520 on α2i2 and C535 on α2i3 helix, potentially providing additional structural support for the RNA binding site (Fig. 2C). The cysteines comprising this disulfide bond are invariant among all RIG-I orthologs (Fig. 3), but not related RLR proteins (Fig. S2). A stretch of sequence unique to MDA-5 HEL2i (with characteristic two-thirds acidic N-terminus and one-third basic C-terminus) is inserted between α2i4 and α2i5 (Fig. S2). The homologous domain from Hef has been suggested to be responsible for recognition of forked DNA substrates (Nishino et al., 2005).
Figure 5. Mutational effects on RIG-I interaction interfaces.
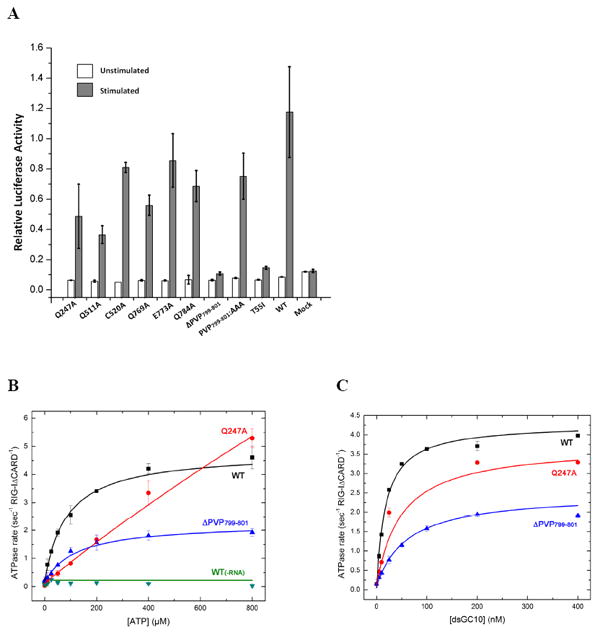
(A) IFN-β reporter assays for analysis of RIG-I function in vivo. (B) ATP hydrolysis by RIG-I (ΔCARDs), mutants Q247A and ΔPVP799-801 at different ATP concentrations and 200 nM RNA. The Km values for WT, Q247A, and ΔPVP799-801 are 85.1 ± 9.4, 1760 ± 546, and 122.0 ± 19.0 μM. (C) RNA stimulated ATP hydrolysis by RIG-I (ΔCARDs), mutants Q247A and ΔPVP799-801 at varying dsGC10 concentrations and 500 μM ATP. The Km values are 18.0 ± 2.30, 49.3 ± 10.4, and 59.9 ± 17.5 nM. Q247 is the Q motif that is responsible for recognizing the adenine base of ATP. Error bars represent the standard deviation. Data showing calibration of the IFN-β luciferase reporter assay for RIG-I signaling is provided as Fig. S5
The CTD of RIG-I (ΔCARDs) interacts with dsRNA in a manner similar to that observed previously for the isolated CTD (Fig. 2, Fig. 4AB) (Lu et al., 2011; Lu et al., 2010; Wang et al., 2010b). For example, hydrophobic stacking between residue F853 and the terminal base pair are observed, together with polar contacts involving H830, S854, K861, S906-K907-W908-K909 and the dsRNA backbone (Lu et al., 2011; Lu et al., 2010; Wang et al., 2010b).
Given that the RNA binding interface involves at least three separate protein domains (Fig. 2), free RIG-I is expected to undergo large conformational changes upon binding to dsRNA, as reflected by limited proteolysis experiments (Takahasi et al., 2008). To experimentally evaluate the degree of RNA-induced conformational change in the protein, and to determine whether the solution conformation of RIG-I (ΔCARDs):dsRNA is consistent with that observed in the crystal structure, we conducted small angle X-ray scattering experiments on RIG-I (ΔCARDs) and on the RIG-I (ΔCARDs):dsRNA complex (Fig. S3). We observe that the radius of gyration (Rg) of the RIG-I (ΔCARDs):dsRNA10 complex in solution is 33.2 ± 0.4 Å, as determined by the Guinier approximation (blue curve, Fig. S3). The distance distribution function (P(r), the distribution of all interatomic distances in the sample) has the typical shape for a compact, globular protein with a single major center of mass (blue curve, Fig. S3). These data are in good agreement with the crystal structure, as the simulated curve for the RIG-I (ΔCARDs):dsRNA10 crystal structure has a calculated Rg value of 30.5 (Fig. S3A, black curve). By contrast, the radius of gyration for the isolated RIG-I (ΔCARDs) protein is much larger than the complex (Rg = 39.6 ± 0.1 Å, red curve, Fig. S3A). Furthermore, the P(r) function shows that isolated RIG-I (ΔCARDs) has a bimodal shape (red curve, Fig. S3B), indicating that the protein has multiple lobes and at least two large centers of mass that define its shape. Taken together, these data indicate that the RIG-I (ΔCARDs):dsRNA complex has approximately the same size and shape in solution as in the crystal structure. Most importantly, they demonstrate that free RIG-I (ΔCARDs) has an extended, multi-part shape that collapses into a compact form upon binding of dsRNA. Double-stranded RNA, therefore, provides the template for RIG-I domain assembly.
HEL2: a module for coupling ATP hydrolysis and signaling
In the structure of RIG-I (ΔCARDs), HEL2 is partially disordered and it does not form contacts with HEL1 or dsRNA10 (Fig. 1B). This contrasts with the structures of other SF2 proteins, in which HEL2 is involved in extensive protein-nucleic acid interactions (Fig. S4) and plays key roles in function of the protein (Bono et al., 2006; Del Campo and Lambowitz, 2009; Appleby et al., 2010; Gu and Rice, 2010; Luo et al., 2008; Myong et al., 2007). The HEL2 of RLRs is known to be important for RNA binding and ATP hydrolysis, as mutations of helicase motifs IV-VI on HEL2 are detrimental for enzymatic activity and antiviral signaling (Bamming and Horvath, 2009). Therefore, additional conformation(s) of RIG-I probably exists, in which HEL2 participates directly in RNA binding and/or ATP hydrolysis. In this structure, while the N-terminal tail of RIG-I (ΔCARDs) points towards the ATPase active site and HEL2, the N-terminal tail of MDA-5 HEL1 is pointing away (Fig. 4CD). In this construct, the lack of CARDs may therefore contribute to the partial disordering of HEL2. It is therefore tempting to speculate that the CARDs play a direct role in modulating ATPase activity of the HEL1/2 domains in RIG-I (Cui et al., 2008; Gee et al., 2008; Myong et al., 2009).
The pincer motif coordinates RIG-I domains during ligand binding
The pincer motif (also described as a “bridge” in the accompanying manuscript by Kowalinski et al.) connecting HEL2 and the CTD is composed of two α-helices (αP1 − αP2), that emerge from the last β-strand of HEL2 (β26) (Fig. 6). The first helix spans across HEL1 and then makes a 65° turn that connects it with a second long helix, which extends to the CTD through a stretch of proline-rich sequence (residues 796-804, Fig. 3). That these sequences contribute to communication between the CTD and HEL2 is supported by mutagenesis studies showing that their conversion to alanine reduces the RNA-stimulated interferon response, and their deletion eliminates the response altogether (Fig. 5 and Fig. S5). The role of this region in transmitting information between domains is further supported by direct enzymological studies of ligand coupling, showing that P799-P801 are essential for linking ATPase activity to the binding of RNA (Fig. 5B/C). Thus, while the pincer is not part of the ATPase core domains, nor is it a direct RNA binding site, it transmits information between these sites, resulting in behavior that is essential for RNA sensing by RIG-I (Fig. 5).
Figure 6. The Pincer domain and linkage between HEL and CTD upon RNA binding.
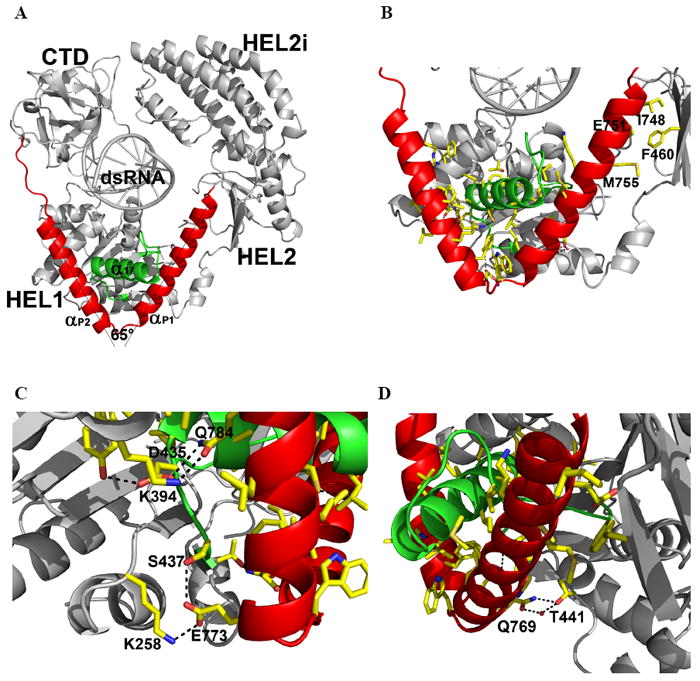
(A) Pincer motif (red) connecting the HEL and CTD. The two arms of the Pincer are separated by 65°. Helix α17 from HEL1 is in green. (B) Interaction network between the Pincer domain and the helicase domain. (C) and (D), Polar contacts. See also Fig. S5.
Inserted within the resultant V-shaped structure is an α-helix that protrudes like a shaft from the RecA core of HEL1 (α17), suggesting a mechanical assembly that looks much like the camshaft in an engine (Fig. 6A, B). The central axle emerges from motif III in HEL1, where it is part of a turn-α17-turn structure (residues 409-438). The core interface between the V-shaped pincer and the α-helical shaft is dominated by hydrophobic interactions (Fig. 6B), which may allow smooth rotational motion of the V-shaped substructure around the shaft. Specific hydrogen bonds control the spatial orientation of the HEL1 RecA core, the shaft, and the pincer (K258-E773-S437, K394-Q784-D435, and Q769-T441; residues underlined are conserved among human RLRs) (Fig. 6C,D). This extensive interaction network could serve to lock the pincer in a fixed orientation relative to HEL1. That this network is functionally important is underscored by the phylogenetic conservation of the constituent amino acids (Fig. 3, bottom panel), and the fact that alanine substitution of even single residues in this region results in diminished RNA-stimulated interferon response (Fig. 5). There is a small hydrophobic interface between a beta strand in HEL2 (β26) and αP1 of the pincer (I748-F460-M755), which may anchor that portion of αP1 to the helicase domains (Fig. 6B). Although the pincer motif by itself does not interact with dsRNA, it connects functional domains of the protein (HEL1, HEL2 and CTD) and facilitates communication between them (Fig. 6), potentially acting as a mechanical transmitter for coordinating quaternary structural rearrangements within RIG-I. The pincer may function by opening and closing its two arms in response to movements of the CTD and/or HEL2; or it may rotate around the α-helical shaft, thereby adjusting the relative orientations of HEL2 to HEL1, CTD to HEL1, or both, in a dynamic fashion. Similar mechanical coordination by rigid helical units was recently suggested by the crystal structure of mitochondrial respiratory complex I (Efremov et al., 2011; Hunte et al., 2011).
Double-stranded RNA: a potent activating ligand for RIG-I
Double-stranded RNA lacking a 5’ppp readily stimulates the IFN response and is therefore a biologically-relevant PAMP (Lu et al., 2010; Schlee et al., 2009). However, short dsRNAs with a 5’ppp blunt end are the most potent activators of IFN response and they are the highest affinity ligands for RIG-I (Lu et al., 2010; Marq et al., 2010; Marques et al., 2006; Wang et al., 2010b). There are several structures of the RIG-I CTD in complex with various dsRNA substrates (Cui et al., 2008; Lu et al., 2011; Lu et al., 2010; Pippig et al., 2009; Wang et al., 2010b). Overlaying all structures of RIG-I CTD dsRNA complexes reveals multiple orientations of the bound dsRNA (Fig. 4). In all of these structures, the 5’ end of the top strand is highly superimposable, as F853 of the CTD stacks on the terminal base pair, and H830 forms a hydrogen bond with the 2’OH of the top strand. However, the structures then begin to diverge, revealing four distinct orientations of the CTD, including two for complexes with 5’OH dsRNA and two for 5’ppp dsRNA (Fig. 4A, 5’OHa/b and 5’pppa/b; See Fig. 4 legend for the nomenclature). Docking these orientations into the structure of RIG-I (ΔCARDs) shows that only one of the RNA orientations observed in the isolated CTD structures (which also contains a 5’ppp, Fig. 4AB, 5’pppb) is compatible with the RIG-I (ΔCARDs) structure (Fig. 4B). That the structure of RIG-I (ΔCARDs):dsRNA superimposes best with a CTD:dsRNA complex containing a 5’-ppp indicates that we have captured an activated form of the complex. Steric clashes are observed between the other two RNAs and HEL2i (Fig. 4AB, 5’OHa and 5’pppa), suggesting that the insertion domain plays a role in specifying the orientation of the duplex RNA.
Oligomerization of RIG-I is triggered by RNA and is dependent on RNA length
Dimerization or oligomerization of RIG-I has been suggested to be essential for its activation (Saito et al., 2008), and a model for the RIG-I dimer has been proposed (Cui et al., 2008). To elucidate the nature of RIG-I oligomerization, and to understand the implications of our structure for the molecular architecture of RIG-I oligomerization states, we examined the RNA site size and multimerization of the protein using RNase footprinting and analytical size exclusion chromatography. We first performed limited RNase digestion on the RIG-I (ΔCARDs):polyIC complex and showed that the minimum length of RNA protected by RIG-I (ΔCARDs) is 10-15 bases (Fig. S6). We also performed size exclusion chromatography (SEC) to study the multimerization of RIG-I (ΔCARDs) as a function of RNA length (Fig. S7). Examining the RIG-I (ΔCARDs):dsRNAs complexes (10, 12, 14, 18 and 22 base pairs), we observe a monomeric site size of ~10 base-pairs, consistent with the footprinting experiments (Fig. S6) and with the functional titration and microscopy experiments reported in Binder et al (Binder et al., 2011). These experiments also indicate that dimerization of RIG-I (ΔCARDs) is induced by increasing dsRNA length (Fig. S7). Significant dimerization is only observed when dsRNA is 18bp or longer (Fig. S7), indicating that each incoming RIG-I molecule requires a full RNA site in order to bind and that the RNA surface, rather than an extensive protein-protein interface, drives multimerization of the protein. The observed dimerization was not accompanied by apparent enhancements in ATPase activity (data not shown), but this may be attributable to the presence of a second 5’-terminus on the 18 and 22-mer duplexes, which may differ from internal sites on long RNAs. Collectively, these data indicate that dimerization or oligomerization of RIG-I is triggered by RNA and is dependent on RNA length. Thus, the RIG-I “dimerization” observed upon binding of long RNAs may reflect RNA-templated multimerization rather than a functional protomer-protomer interface of the protein.
The central groove of RIG-I (ΔCARDS) is completely engaged in contacts to dsRNA10 (Fig. 1B,C), which corresponds well with the biochemical data of Binder, et al., suggesting that RIG-I units bind duplex RNA like beads on a string (Binder et al., 2011). Furthermore, correspondence between the two studies indicates that RIG-I (ΔCARDs) binds RNA in the same way as full-length RIG-I protein, indicating that RIG-I extends the CARDS away from RNA and into solution for signaling. The RNA length-dependent oligomerization of RIG-I may appear to stimulate activation by increasing the local concentration of activated RIG-I in the cell and activate IPS-1 by promoting IPS-1 oligomerization (Baril et al., 2009; Hou et al., 2011; Tang and Wang, 2009).
Discussion
Insights into the molecular mechanism of RIG-I activation
For RIG-I (ΔCARDs), the process of binding RNA is expected to be highly complex as multiple RNA binding sites are involved. Furthermore, ATP binding and hydrolysis may provide additional changes to the mode of RNA recognition. The structure suggests that substantial quaternary movements in the RIG-I domains are required for RIG-I to optimize its fit with various substrates. This process is probably necessary for biological function, as the activation of RIG-I must be tightly controlled. We surmise that the current structure may be an intermediate state of dsRNA recognition and RIG-I activation (Fig. 7). Efficient and effective activation of RIG-I could well be the consequence of many factors, including the chemical nature and local concentration of the substrate, post-translational modification state, sub-cellular localization, and the availability of the downstream signaling proteins.
Figure 7. Model for RIG-I activation.
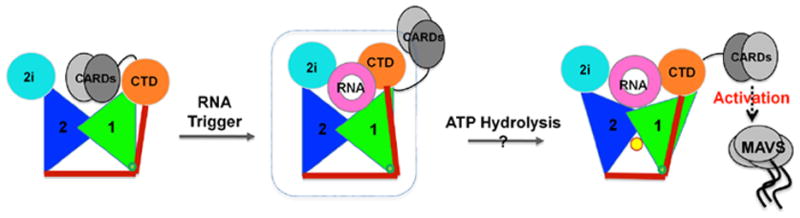
(1) In the absence of RNA, RIG-I is in autoinhibited state. (2) Upon binding of duplex RNA (pink ring), the helicase-CTD form a compact structure that surrounds the RNA (dotted square circles the crystal structure). (3) Upon binding and/or hydrolysis of ATP (yellow dot), the protein is fully activated for catalysis and signaling. Note that the pincer motif (red lines) is likely to transmit conformational information among multiple protein domains. Color of the domains follows Fig. 1. See also Fig. S6 and S7.
Based on the structure of RIG-I (ΔCARDs) in complex with dsRNA10, along with currently available experimental data, we propose a simple molecular model for RIG-I activation (Fig. 7). In this model (Left, Fig. 7), RIG-I, in the absence of an RNA trigger, is in the auto-inhibited state (Fujita et al., 2007), in which the CARDs fold back to the C-terminal portion of RIG-I and interact with either HEL, the CTD or both (Saito et al., 2007). Upon binding of non-self duplex RNA (Center, Fig. 7), RIG-I undergoes extensive structural rearrangements, resulting in the core structure that has been visualized crystallographically. Fully-activated RIG-I (Right, Fig. 7) releases its N-terminal CARDs for display in solution and initiates the antiviral signaling cascade (Fujita et al., 2007; Yoneyama et al., 2004). ATP binding and hydrolysis, along with multimerization along the RNA lattice, are likely to participate extensively in the process and provide an additional means of regulation (Right, Fig. 7). The precise role of ATP hydrolysis in RIG-I signaling remains a major question, and perspective on this problem may come from both the fields of G-proteins and SF2 proteins. Like a G-protein (Oldham et al., 2008), RIG-I may hydrolyze ATP in order to adopt a conformational state that is optimal for presentation of the CARDS or for maintaining active signaling. Alternatively (but not exclusively), RIG-I may function like other SF2 motor proteins (Pyle, 2008), utilizing ATP hydrolysis to move along the RNA lattice and thereby optimize its spatial position for downstream signaling. The latter mechanism is consistent with recent studies demonstrating that RIG-I can translocate on RNA duplexes in an ATP-dependent manner (Myong et al., 2009).
Comparative strategies for RNA recognition
The RNA recognition strategy of RIG-I differs from that of other proteins that are important for sensing or remodeling RNA. The toll-like receptors (TLRs) are a structurally distinct family of proteins that sense the invasion of pathogenic molecules and TLR3 is a variant that binds and recognizes dsRNA, providing a system complementary to RIG-I and MDA-5 in the cell. The remarkable molecular structure of TLR3 bound to dsRNA reveals a strategy for duplex recognition. TLR3 is crescent-shaped molecule consisting of rigid leucine-rich repeats that bind to RNA and form obligate dimers (Liu et al., 2008). Unlike RIG-I, which surrounds the RNA duplex, TLR3 monomers lie along one side of the RNA duplex, with each terminus of the crescent forming a patch of two colinear RNA-protein contacts. The molecular contacts of TLR3 are distinct from the neutral, polar contacts in RIG-I in that they are dominated by electrostatic interactions between histidine side chains and phosphate oxygens, resulting in pH-sensitive RNA signaling as in the endosomal environment (Leonard et al., 2008; Liu et al., 2008; Wang et al., 2010a).
The DEAD-box proteins are SF2 cousins of the RLR proteins and, thus far, they have only been captured in a form that is bound to ssRNA (Fig. S4). In these structures, represented by Vasa, eIF4a3 and Mss116, the ssRNA is crimped and bent in a manner incompatible with an A-form configuration (Andersen et al., 2006; Bono et al., 2006; Del Campo and Lambowitz, 2009; Sengoku et al., 2006). While it is possible that DEAD-box proteins sample other conformational states when bound to RNA (vida infra), the available structures underscore the importance of RNA distortion in their molecular mechanism. This behavior contrasts sharply with that of the related viral DExH-box helicases such as the NS3 proteins of the Flaviviridae (Appleby et al., 2010; Gu and Rice, 2010; Luo et al., 2008). In these cases, ssRNA is not distorted and it is bound in an elongated form that is compatible with A-form geometry. However, the common feature uniting all of these proteins is that they form primary contacts with the sugar-phosphate backbone.
Functional significance of the RIG-I interaction network
The structure of RIG-I (ΔCARDs):dsRNA reveals an elaborate network of interactions that appear to control the RNA specificity and mechanical function of the protein. Although many of the amino acids involved in these networks are invariant among RIG-I orthologs (Fig. 3, Fig. S2), suggesting key roles in function, it was nonetheless important to mutate them and directly test their effects on function of the protein. To this end, we created mutations along the RNA binding interface of the insertion domain (for example, Q511A, Fig. 2A,C, Fig. 6), the conserved interface between HEL1 and helices of the pincer domain (for example, Q769A and Q784A, Fig. 6C,D), and the conserved connector between the pincer and the CTD (for example, the ΔPVP799-801 mutant, Fig. 5). The overall influence of these mutations on RNA stimulation of the interferon response was studied in-vivo, by using a well-established transfection approach (Fig. S5, (Lu et al., 2011; Lu et al., 2010; Wang et al., 2010b)). For the mutant that produced the most severe effect on interferon response (ΔPVP799-801), we expressed it in recombinant form and examined its ability to allosterically couple RNA binding (sensing) with ATPase function in-vitro. All these classes of mutation influence the function of RIG-I (Fig. 5), providing important mechanistic insights into behavior and role of RIG-I. For example, the effect of Q511A on the RNA-stimulated interferon response indicates that the HEL2i insertion domain plays a key role in specifically recognizing duplex RNA, thereby mediating one of the earliest molecular events during innate immune response. In addition, residues Q769 and Q784 form bifurcated hydrogen bonds that appear to anchor the pincer domain to HEL1. When these are mutated to alanine, significant defects in interferon response are observed, suggesting that spatial organization of the mechanical domains is tightly controlled. Finally, residues P799-P801 connect the Pincer domain with the CTD and when these are mutated, large effects on RNA-stimulated interferon response in-vivo and RNA-stimulated ATPase activity in-vitro are observed. This establishes that the pincer domain is essential for relaying information between the CTD sensor and mechanical/catalytic components of the protein (HEL1 and HEL2).
Implications for the function of related SF2 proteins
Dicer and the Dicer-related helicases that play key roles in RNA interference are the most closely related RNA “helicases” to the RLR family (Matranga and Pyle, 2010; Tabara et al., 2002; Zou et al., 2009). The helicase domain of Dicer appears to be responsible for differential recognition and processing of precursor duplex RNA during siRNA / miRNA biogenesis (Ma et al., 2008; Soifer et al., 2008; Welker et al., 2011; Welker et al., 2010). Like RIG-I, Dicer has an insertion domain which, together with the helicase domains, is likely to play a key role in duplex recognition. Thus, the structure of RIG-I (ΔCARDs) is likely to have important implications for the function of Dicer and its orthologs.
Although DEAD-box helicases are phylogenetically distinct from RLR proteins and they appear to bind ssRNA, DEAD-box proteins have a distinct motif IIa element that is similar to that observed in RLRs, and which contributes to dsRNA binding in the RIG-I structure (Fairman-Williams et al., 2010; Pyle, 2008). This motif may mark a point of functional divergence between single- and double-strand recognition by proteins during the evolution of SF2 helicases. Alternatively, the presence of motif IIa may indicate that certain DEAD-box proteins retain dsRNA binding properties and that this contributes to their mechanism. The functional diversity of DEAD-box proteins may be explained by this versatility of substrate selection and recognition (Linder, 2006; Pyle, 2008; Yang and Jankowsky, 2006).
Concluding remarks
The structure of RIG-I (ΔCARDs) in complex with RNA reveals the molecular basis for RIG-I sensing and recognition of duplex RNA. RNA binding is mediated by multiple protein domains, including a novel insertion domain. In addition, the structure reveals the molecular basis for mechanical communication among the various components of the protein, and it contains a new type of nanomechanical device that may function like a camshaft, containing V-shaped domain that appears to revolve or pinch around a central, α-helical shaft. The structure suggests coordinated motion of the RIG-I domains, which self-organize through the binding of duplex RNA. Going forward, studies of full length RIG-I in complex with ligands or in the presence of post-translational modifications will provide more insights into the multi-stage process of RIG-I activation. Finally, RIG-I represents an important host target for both antiviral and anticancer drugs and the crystal structure reveals multiple strategies for therapeutic design.
Experimental Procedures
Cloning, expression and purification
The N-terminal CARDs (residues 1-229) deletion construct of human RIG-I, hereafter named RIG-I (ΔCARDs), was cloned and expressed using standard procedures (see Supplemental Extended Experimental Procedures) Mutations were generated by PCR using corresponding primers (Table S2). Coding region of the gene was sequenced to confirm the presence of the designed mutations. Recombinant mutant proteins were expressed and purified as described above.
Crystallization and data collection
The RIG-I (ΔCARDs) complex with dsRNA10 was preassembled by incubating at a protein:RNA molar ratio 1:1.5 on ice for 1 hour and then purified by using a HiPrep 16/60 Superdex 200 column (Amersham Bioscience). Crystals of the RIG-I (ΔCARDs) dsRNA10 complex were grown at 13 °C by mixing equal volumes of precipitating solution (0.1 M Bicine, pH 9.0, 22.5 % polyethylene glycol 6,000) and RIG-I (ΔCARDs) dsRNA10 complex (2-3 mg ml-1). Crystals grew into needle clusters within a week and were harvested within two weeks. Crystals were soaked in a cryoprotecting solution containing 0.1 M Bicine, pH 9.0, 30 % polyethylene glycol 6,000, 50 mM MgSO4 for 12 hours before being flash frozen with liquid nitrogen. Diffraction intensities were recorded at NE-CAT beamline ID-24 at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois). Integration, scaling and merging of the intensities were carried out by using the programs XDS (Kabsch, 2010) and SCALA (Evans, 2006).
Structure determination and refinement
The structure was determined through molecular replacement with the program Phaser (McCoy, 2007) by using the CTD of RIG-I (Wang et al., 2010b), duplex RNA (generated by using Coot (Emsley and Cowtan, 2004)), HEL1 of MDA-5 (Karlberg et al., 2007), and HEL2i of Hef (Nishino et al., 2005) as search probes. Refinement cycles were carried out by using Phenix Refine (Adams et al., 2010) and REFMAC5 (Murshudov et al., 1997) with the TLS (translation, liberation, screw-rotation displacement) refinement option with four TLS groups (HEL1: aa 236-455, HEL2-HEL2i: aa 456-795, CTD: aa 796-922, and dsRNA10). Refinement cycles were interspersed with model rebuilding by using Coot (Emsley and Cowtan, 2004). The quality of the structures was analyzed by using PROCHECK (Laskowski et al., 1993). A summary of the data collection and structure refinement statistics is given in Table 1. Figures were prepared by using the program Pymol (DeLano, 2002).
SAXS data collection and analysis
Synchrotron X-ray scattering data were collected at the National Synchrotron Light Source (NSLS) beamline X9. SAXS data were collected by using a Mar CCD 165 located at 3.4 m distance from the sample. Wide angle X-ray scattering (WAXS) data were collected simultaneously with SAXS data by using a Photonic Science CCD located at 0.47 m from the sample. Scattering data were collected at concentrations of 4.7 mg ml-1, 3.1 mg ml-1 and 2.35 mg ml-1 for RIG-I (ΔCARDs) and 6.25 mg ml-1, 5 mg ml-1 and 2.5 mg ml-1 for RIG-I (ΔCARDs):dsRNA10 complex. All samples were in buffer consisting of 20 mm Hepes (pH 7.4), 150 mm NaCl, 5% Glycerol, 5 mM β-ME. Each sample was measured by continuously pushing 20 μl of sample through a 1 mm diameter capillary for 30 s of exposure time. Three independent measurements were averaged for each sample and subtracted with buffer scattering by using the pyXS package (NSLS beamline X9). No aggregation was observed for samples at the three concentrations, thus scattering data from the highest concentration sample were used for calculations. All data analysis and calculations were performed by using the ATSAS software suite (Konarev et al., 2006). Radius of gyration (Rg) was estimated by using AutoRg (Konarev et al., 2003). The distance distribution function, P(r), of the particle was computed by using GNOM (Svergun, 1992). Theoretical scattering curve for the RIG-I (ΔCARDs):dsRNA10 crystal structure was calculated by using CRYSOL (Svergun et al., 1995).
IFN-β reporter assays for analysis of RIG-I function in vivo
Huh-7.5 cells were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) containing 10% heat-inactivated fetal calf serum (Hyclone) and non-essential amino acids (Invitrogen). On the day before transfection, cells were seeded in 24-well plates at 1.6 × 105 cells per well. Cells were co-transfected with 3 ng per well pUNO-RIG-I or an empty vector, 178 ng per well of an IFN-ß/Firefly luciferase reporter plasmid (a kind gift of Dr. Jürg Tschopp), and 30 ng per well pRL-TK (Promega) as an internal transfection control. Transfections were performed by using TransIT LT1 (Mirus) reagent according to the manufacturer’s instructions. Transfected cells were incubated for 24 hrs to allow RIG-I expression, then stimulated by transfecting them with 1 μg per well poly(I:C) (0.2-1 kb; Invivogen) and TransIT-mRNA transfection reagent (Mirus). After a further 16 hrs incubation, cells were lysed and analyzed by using the Dual Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions. Firefly and Renilla luciferase activities were assayed on a Centro LB 960 plate reader (Berthold). The ratios of firefly luciferase activity to Renilla luciferase activity were determined, and normalized to the ratio obtained with WT-RIG-I stimulated with poly(I:C). For studies of RIG-I mutants, the pUNO-RIG-I plasmid was mutated by PCR using corresponding primers (Table S2). Coding region of the gene was sequenced to confirm the presence of the designed mutations. Data are representative of experiments performed at least twice.
ATPase assays for analysis of RIG-I enzymatic function and coupling in-vitro
The ATPase activity of the RIG-I (ΔCARDs) and it mutant proteins was monitored by using an ATP/NADH linked assay as previously described (De La Cruz et al., 2000; Lindsley, 2001). Briefly, 50 μl reaction mixtures containing 100 nM of protein, 5x NADH enzyme buffer, 25 mM MOPS pH 7.4, 10 mM Mg(OAc)2, 30 mM K(OAc), and 2 mM DTT were mixed with varying amounts of ATP and dsGC10 at 25 °C. The 5x enzyme buffer contained 1 mM NADH, 100 U of lactic dehydrogenase/ml, 500 U of pyruvate kinase/ml, and 2.5 mM phosphoenolpyruvate. Fluorescence readings (excitation, 340 nm; emission, 450 nm) were collected in Corning 96 well black half area flat bottom plates in a SpectraMax 250 plate reader. Initial velocities were calculated from a linear regression of each time course and corrected for background ATP hydrolysis and NADH oxidation. The initial velocities (v0) at various RNA concentrations and ATP concentrations were plotted and fit to the following Briggs-Haldane equation: y = y0+(amp*(([Mt]+[St]+Km)-sqrt(([Mt]+[St]+Km)^2-4*[Mt]*[St]))/(2*[Mt])), where [Mt] is the total protein concentration, [St] is the total [RNA] or [ATP], y0 is the basal activity, and Km is the apparent Michaelis constant for substrate activation.
Supplementary Material
Highlights.
Structural analysis of RIG-I shows how it recognizes and encases double stranded RNA
A pincer motif formed by the helicases domain organizes RIG-I for RNA binding
Functional analyses confirm the use of the pincer motif in vitro and in vivo
Structure offers mechanistic insight into SF2 family and Dicer-related helicases
Acknowledgments
DL and AMP designed the experiments. DL performed protein expression, purification, crystallization, X-ray diffraction data collection and analysis, structure determination and refinement. DL carried out RNAse digestion and SEC. DL and SCD collected SAXS data. SCD performed mutagenesis and purified the mutant proteins. ASK and SCD performed the ATPase assays. AV and SCD carried out the cloning, the initial expression and purification studies. DL and BDL performed the cell-based assay. DL and AMP wrote the manuscript with input from all authors.
We thank Dr Marco Marcia, Dr Kevin Keating, Dr Vivien Nagy, Dr Olga Fedorova, and other members of Pyle Lab for their generous help and insightful discussions. We thank scientists (Drs. Kanagalaghatta Rajashankar, Frank Murphy, Kay Perry, Narayanasami Sukumar, and Malcolm Capel) from APS NECAT 24-ID and scientists (Drs Marc Allaire and Lin Yang) from NSLS X9 for the beam line access and technical support. We also like to thank Kevin Smith, Ganes C. Sen, Jürg Tschopp, Wang Penghua and Erol Fikrig for providing us with reagents. We also like to thanks Cao Wenxiang and Dr De La Cruz, E. M. for advice on ATPase assay. BDL was supported by NIH grant AI089826. This research was funded by Howard Hughes Medical Institute. DL is a postdoctoral associate and AMP is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Accession code The atomic coordinate and structure factor of RIG-I (ΔCARDs) dsRNA complex have been deposited with the RCSB Protein Data Bank under the accession codes 2ykg.
Competing financial interests The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, Brendza KM, Somoza JR. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol. 2010;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid inducible gene-I (RIG-I) J Biol Chem. 2011 doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys J. 2000;79:1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2011;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Onoguchi K, Onomoto K, Hirai R, Yoneyama M. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 2007;89:754–760. doi: 10.1016/j.biochi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gee P, Chua PK, Gevorkyan J, Klumpp K, Najera I, Swinney DC, Deval J. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J Biol Chem. 2008;283:9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Zhijian JC. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011 doi: 10.1016/j.cell.2011.1006.1041. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2011;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg T, Welin M, Arrowsmith CH, Berglund H, Busam RD, Collins R, Dahlgren G, Edwards AM, Flodin S, Flores A, et al. Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain. Protein Data Bank 2007 [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, S Dmitri I. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Cryst. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, S DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- Kubler K, Gehrke N, Riemann S, Bohnert V, Zillinger T, Hartmann E, Polcher M, Rudlowski C, Kuhn W, Hartmann G, Barchet W. Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Res. 2010;70:5293–5304. doi: 10.1158/0008-5472.CAN-10-0825. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley JE. Use of a real-time, coupled assay to measure the ATPase activity of DNA topoisomerase II. Methods Mol Biol. 2001;95:57–64. doi: 10.1385/1-59259-057-8:57. [DOI] [PubMed] [Google Scholar]
- Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5’ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5’ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq JB, Hausmann S, Veillard N, Kolakofsky D, Garcin D. Short double-stranded RNAs with an overhanging 5’ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J Biol Chem. 2010;286:6108–6116. doi: 10.1074/jbc.M110.186262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- Matranga C, Pyle AM. Double-stranded RNA-dependent ATPase DRH-3: insight into its role in RNAsilencing in Caenorhabditis elegans. J Biol Chem. 2010;285:25363–25371. doi: 10.1074/jbc.M110.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Mi Z, Fu J, Xiong Y, Tang H. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell. 2010;1:275–283. doi: 10.1007/s13238-010-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5’-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G-protein activation by G-protein coupled receptors. Nature Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, Kirschnek S, Gaffal E, Landsberg J, Hellmuth J, et al. 5’-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annual Review of Biophysics. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schutz P, Karlberg T, van den Berg S, Collins R, Lehtio L, Hogbom M, Holmberg-Schiavone L, Tempel W, Park HW, Hammarstrom M, et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, Collingwood MA, Behlke MA, Rossi JJ. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, MHJ K. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules fromAtomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Tang ED, Wang CY. MAVS self-association mediates antiviral innate immune signaling. J Virol. 2009;83:3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu L, Davies DR, Segal DM. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J Biol Chem. 2010a;285:36836–36841. doi: 10.1074/jbc.M110.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010b;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. Rna. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Kroemer G. Anticancer immunochemotherapy using adjuvants with direct cytotoxic effects. J Clin Invest. 2009;119:2127–2130. doi: 10.1172/JCI39991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol. 2009;9:85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


