Abstract
To define the molecular regulators required for differential pattern of H3K79 methylation by Dot1, we performed a GPS screen and discovered that the components of the cell cycle-regulated SBF complex were required for normal levels of H3K79 di- but not trimethylation. Genome-wide mapping revealed that H3K79 di- and trimethylation to present a mutually exclusive pattern on chromatin with M/G1 cell-cycle-regulated genes significantly enriched for H3K79 dimethylation. Since H3K79 trimethylation requires prior monoubiquitination of H2B, we performed genome-wide profiling of H2BK123 monoubiquitination and showed that H2BK123 monoubiquitination is excluded from cell cycle regulated genes and sites containing H3K79me2 but not from H3K79me3 containing regions. A genome-wide screen for factors responsible for the establishment/removal of H3K79 dimethylation resulted in the identification of several genes including NRM1 and WHI3, which both impact the transcription by the SBF, and MBF complexes, further linking the regulation of H3K79’s methylation status to the cell cycle.
INTRODUCTION
In eukaryotic cells, genomic DNA is packaged by the histone proteins, forming the fundamental repeating unit of chromatin, the nucleosome (Luger et al., 1997). Several residues within the histone tails and some within the histone core can be altered by post-translational modifications including acetylation, phosphorylation, ubiquitination and methylation (Shilatifard, 2006). Two classes of histone methylases have been identified to date. The first class includes the SET domain containing histone methylases capable of methylating many histones on different residues with varying biological outcomes (Shilatifard, 2006). The second class, the non-SET domain methylases, consists of only one member, Dot1, which is capable of methylating histone H3 on lysine 79 (H3K79). The H3K79 is one of the conserved core residues located in a loop within the histone-fold domain that can be methylated (Lu et al., 2008). Originally identified as a gene affecting the silencing of gene expression near telomeres in S. cerevisiae, Dot1 (disrupter of telomeric silencing 1) is an evolutionally conserved enzyme that catalyzes mono-, di-, and trimethylation of H3K79 (me1, me2 and me3, respectively) (Lacoste et al., 2002; Ng et al., 2002; van Leeuwen et al., 2002). We and others have demonstrated that similar to H3K4 methylation, efficient trimethylation of H3K79 by Dot1 requires monoubiquitination of K123 of histone H2B, catalyzed by the Rad6/Bre1 protein complex (Dover et al., 2002; Krogan et al., 2003; Shilatifard, 2006; Sun and Allis, 2002; Wood et al., 2003).
Dot1 methylates H3K79 only in the context of nucleosomes and can be regulated by several sites within other histones (Altaf et al., 2007; Onishi et al., 2007). The mammalian Dot1 has been directly implicated in leukemogenesis through the misregulation of HOX genes (Okada et al., 2005). Furthermore, it was shown that a MLL-AF4 fusion causes ectopic recruitment of Dot1, and the aberrant methylation of H3K79 at MLL target genes (Krivtsov et al., 2008). These studies point the way to the clinical significance of H3K79 methylation and to the pathogenesis of human cancer. We know very little about the functions of the different methylation states of H3K79 and the results of different studies are contradictory. H3K79me2 is suggested to be a mark of transcriptionally active genes in Drospohila (Schubeler et al., 2004) and in mammalian cells (Im et al., 2003; Martin and Zhang, 2005; Miao and Natarajan, 2005). However, a different study found that H3K79me1 and H3K79me2 did not have a significant preference toward either active or silent genes (Barski et al., 2007). Furthermore, H3K79me3 did not have a correlation with either active or silent genes in yeast (Pokholok et al., 2005), but is associated with transcriptionally repressed genes in human cells (Barski et al., 2007).
One of the major mechanisms driving cell cycle regulation is the orchestrated transcriptional program of gene expression. Genome-wide studies of cell cycle-regulated transcripts in budding yeast indicate that more than 800 genes are periodically expressed (Spellman et al., 1998). More than 300 of those, including the G1cyclins, CLN1 and CLN2, the S phase cyclins, CLB5 and CLB6, and many genes involved in DNA synthesis, show peak expression in G1. Two heterodimeric transcription factor complexes are required primarily for activation of gene expression at the G1-/S-phase transition of the cell cycle: SBF (SCB-binding factor) and MBF (MCB binding factor) (Breeden, 1996) (Harbison et al., 2004; Iyer et al., 2001; Simon et al., 2001). Both bind to repeated upstream regulatory sequences, SCB (for Swi4,6-dependent cell cycle box) or MCB (for Mlu1-dependent cell cycle box), respectively, in a broad range of targets. SBF and MBF each contain the regulator subunit Swi6 as well as a specific DNA binding factor, Swi4 in the case of SBF and Mbp1 in the case of MBF (Andrews and Herskowitz, 1989; Breeden and Nasmyth, 1987; Koch et al., 1993). Gene activation by SBF and MBF is subject to a variety of regulatory mechanisms, including phosphorylation of Swi6, cell cycle-regulated SWI4 expression and control of DNA binding. Despite the detailed knowledge of transcription factors involved in cell cycle control in budding yeast, fundamental questions about the role of chromatin structure in this process have not been answered so far. One exception involves the tightly timed transcription of canonical histone genes, which is restricted to S-phase in S. cerevisiae (Hereford et al., 1981).
To define the role of di- and trimethylation of H3K79, we screened the entire yeast gene deletion collection to identify proteins required for the establishment of different forms of H3K79 methylation by Dot1. We found that Swi4 and Swi6 were required for dimethylation, but not for trimethylation of H3K79, suggesting a link between cell cycle progression and chromatin modification at this site. Consistent with this possibility, we found that levels of H3K79me2 were decreased in the G1-phase and peak in G2/M. Furthermore, we showed that the loss of H3K79me2 in G1-phase requires Nrm1and Whi3. Nrm1 (negative regulator of MBF targets 1) is a co-repressor of MBF-regulated gene expression (de Bruin et al., 2006), and Whi3 is an RNA-binding protein involved in cell cycle control, and its loss results in the acceleration of the expression of genes controlled by the SBF and MBF complexes. Interestingly, high-resolution genome-wide mapping of H3K79 methylation by chromatin immunoprecipitation followed by microarray analysis (ChIP-on-Chip) showed that H3K79 di- and trimethylation were mutually exclusive and reside in different regions of the genome. Further supporting the link of H3K79 dimethylation to the cell cycle, we found M/G1 cell-cycle-regulated genes as well as Swi4/6 bound promoters to be significantly enriched for H3K79 di- but not trimethylation.
Since monoubiquitination of histone H2B is required specifically for H3K79 trimethylation, we have generated polyclonal antibodies to monoubiquitinated H2B on K123. Employing ChIP-on-Chip analysis, we demonstrated that H2B monoubiquitination was excluded from cell cycle regulated genes and sites containing H3K79me2 but not H3K79me3. Together, our studies presented in this manuscript provide a link between Rad6/Bre1 dependent H2B monoubiquitiation and H3K79me3 and its exclusion from H3K79me2 modified sites and provides a link between H3K79me2, the SBF- and MBF transcription complexes and the cell cycle.
RESULTS
Swi4 and Swi6 were required for normal levels of H3K79 di- but not trimethylation
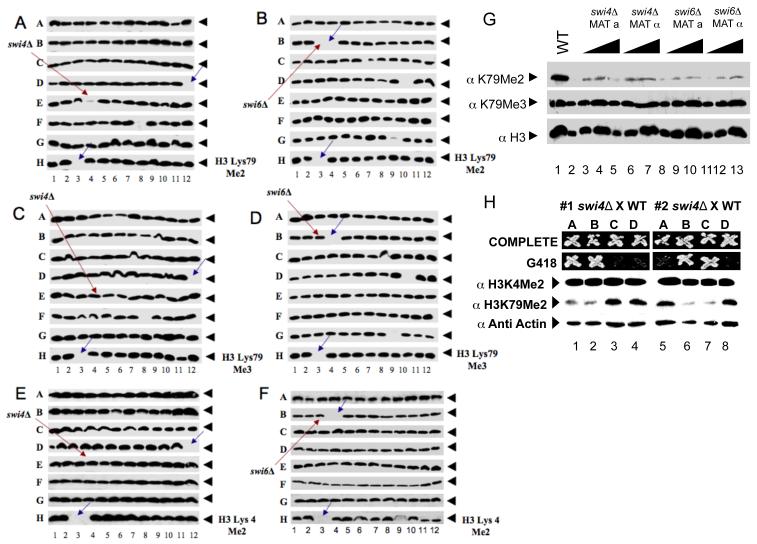
To identify proteins required for establishment of H3K79me2, we surveyed extracts of the collection of non-essential gene deletion mutants of S. cerevisiae for this modification by protein blotting, a method we call GPS (Global Proteomics in S. cerevisiae) (Schneider et al., 2004). This biochemical screen revealed that the loss of either Swi4 or Swi6 resulted in a decrease in the level of H3K79me2 (Figure 1A, B), without affecting either H3K79me3 or the H3K4me3 levels (Figure 1C-F). This defect was specifically due to the swi4Δ and swi6Δ deletions as both the MATa and MATα versions of the swi4Δ and swi6Δ mutants in the deletion collection had a reduced level of H3K79me2 (Figure 1G). Furthermore, the histone H3K79me2 defect co-segregated with the swi4Δ::kanMX mutation in spores from a cross of this mutant to a SWI4 (wild-type) strain (Figure 1H). The defect was complemented by a plasmid carrying either SWI4 or SWI6 (Figure 2A, B). Lastly, independent deletion of SWI4 in a different strain background gave the same result (data not shown).
Figure 1. GPS reveals that SWI4 and SWI6 are required for H3K79 di- but not trimethylation.
(A - F) Cell extracts from each of the nonessential yeast gene deletion mutants were subjected to SDS-PAGE, Western blotted, and probed with antibodies specific for (A and B) H3K79 dimethylation or (C and D) trimethylation. (E and F) H3 K4 dimethylation was used as a control. Red arrows represent (A, C, E) swi4Δ and (B, D, F) swi6Δ. Blue arrows indicate empty wells that serve as plate markers. (G) Protein extracts from two different mating strains of swi4Δ and swi6Δ were analyzed as in A-F. Anti-acetyl H3 was probed for a loading control. (H) The swi4::kanMX mutant from the deletion collection was mated with a wild type strain and spores of the resulting tetrads were dissected and scored for the KanR and H3K79 methylation pattern.
Figure 2. Role of SWI4 and SWI6 in H3K79 methylation.
(A ,B) Cell extracts were subjected to SDS-PAGE blotted to a membrane, and probed with H3 Lys79 di- or trimethyl specific antibodies. Anti-acetyl H3 was probed as a loading control. H3 Lys79 dimethylation could be rescued by a plasmid carrying either (A) SWI4 or (B) SWI6 under control of the GAL1 promoter. (C) RT-PCR was performed using cDNA made from RNA extracted from SWI4, as well as, SWI6 deletion strains via reverse transcriptase (RT). Primers specific for DOT1 were used to detect its mRNA. (D) The effect of the loss of SWI4 on histone H2B monoubiquitination levels. Highly purified acid extracted histones from wild type strains (strains containing FLAG-tagged H2B as the only source of histone H2B) or strains deleted for SWI4 in the same background, were analyzed by Western blotting using (upper panel) antibodies against the FLAG epitope or (mid and lower panels) polyclonal antibodies raised against trimethylated H3K79. As indicated by the red arrows, monoubiquitinated H2B is the slower migrating, and the ubiquitinated species of histone H2B is the faster migrating form. rad6Δ cells lack the E2 ubiquitin-conjugating enzyme required for H2B ubiquitination.
Swi4 and Swi6 comprise the SBF complex that binds to the SCB cell cycle box (SCB) sequence and regulates expression of several genes involved in cell cycle progression (Harrington and Andrews, 1996). The possibility that the H3K79 dimethylation defect of the swi4Δ and swi6Δ mutants was due to reduced expression of DOT1 was ruled out by our observation that DOT1 expression is not affected in those mutants (Figure 2C). Furthermore, DOT1 does not appear to have SCB sequences in its promoter, but the DOT1 gene is under transcriptional control with peak expression at by the G1/S transition (Spellman et al., 1998). We also observed that loss of either Swi4 or Swi6 did not alter recruitment of Dot1 to chromatin (data not shown), suggesting that they do not affect H3K79me2 through Dot1 but rather alter some other aspect of dimethylation control.
Swi4 and Swi6 were not required for H2B monoubiquitination
Histone H2B monoubiquitination, catalyzed by the Rad6/Bre1 protein complex, is required for di and trimethylation of H3K4 and trimethylation of H3K79 catalyzed by COMPASS and Dot1, respectively (Dover et al., 2002; Hwang et al., 2003; Sun and Allis, 2002; Wood et al., 2003). We entertained the possibility that the methylation defect of swi4Δ and swi6Δ mutants was due to their lack of H2B monoubiquitination. However, we found normal levels of monubiquitinated histone H2B (tagged with the FLAG epitope) in swi4Δ (Figure 2D) and swi6Δ mutants (not shown). Therefore, Swi4 and Swi6 did not affect H3K79 methylation indirectly by regulating the level of H2B monoubiquitination.
Establishment of H3K79 dimethylation was cell cycle dependent
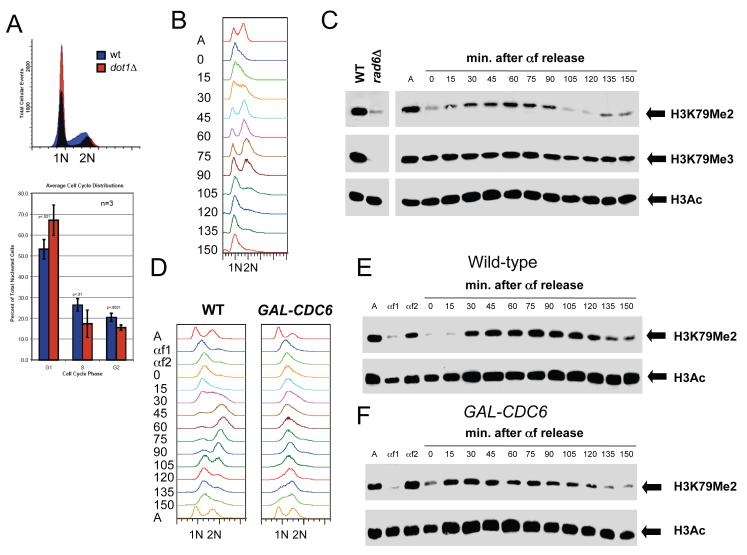
Due to the important role of Swi4 and Swi6 cell cycle progression, we next tested whether H3K79 methylation fluctuated during the cell cycle. Flow cytometric analysis of DNA content in dot1Δ mutants suggested that H3K79 methylation is important for normal entry into S-phase. Compared to wild-type cells, dot1Δ mutants displayed a statistically significant increase in the percentage of G1 cells (Figure 3A). To examine whether H3K79me2 is cell cycle-regulated, cells were synchronized in G1 using the mating pheromone α-factor, then cell cycle progression and H3K79me2 levels were monitored in a synchronous cell cycle (Figure 3B-C). Levels of H3K79 dimethylation were low in G1-arrested cells but increased as cells entered the S phase 15-30 min following release from α-factor, as measured by flow cytometric analysis of DNA (Figure 3B-C). H3K79 dimethylation peaked at G2/M around 60 min, and decreased at 105 min at the completion of mitosis (Figure 3B-C). In contrast, the H3K79me3 mark, as well as the acetylation of H3, was present throughout the cell cycle with no obvious variation in signal intensity (Figure 3C).
Figure 3. Cell cycle regulation of H3K79 dimethylation.
(A) Cells lacking DOT1 accumulate in G1. Wild-type and dot1Δ cells were grown to mid-log phase at 30°C in YPD and DNA content was analyzed by flow cytometry. The percentage of cells in G1 with 1N DNA content, S phase with intermediate DNA content and G2/M with 2N DNA content was quantitated using 3 independent samples. Error bars represent standard deviation of the mean. (B-C) H3K79 dimethylation increases during S phase. Wild-type cells (SLJ001) were arrested in G1 with α-factor at 30°C for 3 hr. They were released into fresh YPD at 30°C, and α-factor was added back when small buds appeared, to prevent cells from entering the next cell cycle. Samples were taken at the indicated times following release from α-factor; asynchronous cells (A) were also collected. (B) DNA content was analyzed by flow cytometry to estimate cell cycle position. Entry into S phase begins around 15-30 min. based on the shift of the 1N DNA peak, while cells have entered G2/M approximately 45-60 min. after α-factor release. By 90 min., cells have exited M phase and re-arrested in G1. (C) Cell cycle-dependent modification of H3 was analyzed by immunoblotting with anti-acetyl-H3 and anti-H3K79me2 and anti-H3K79me3 antibodies. Equal protein concentrations were loaded in all lanes as judged by total H3 levels (data not shown). (D-F) DNA replication is not required for cell cycle oscillations in H3K79 dimethylation. Wild-type and GAL-CDC6 cells were grown in YEP containing 2% raffinose and 4% galactose at 30°C then arrested in G1 with α-factor (αf1). Cells were released from G1 into YEP containing 2% raffinose and 4% galactose to allow CDC6 expression to initiate DNA replication. After 20 min., cells were transferred into YPD to repress GAL-CDC6 and α-factor was added at 45 min. to arrest cells in G1 (αf2). Cells were then released from G1 in glucose-containing media to analyze cell cycle progression in cells lacking Cdc6 protein. α-factor was added back when small buds appeared, to prevent cells from entering the next cell cycle. Samples were taken at the indicated times following release from the second α-factor release; asynchronous cells (A) were also collected. (D) DNA content was analyzed by flow cytometry. While wild-type cells replicate DNA (30 min.), progress through mitosis and rearrest in G1, similar to our results in (B), cells lacking Cdc6 protein do not undergo DNA replication. Peak drift in this sample is likely due to mitochondrial DNA since cells continue to increase in size. Cell cycle-dependent modification of H3 was analyzed by immunoblotting with anti-H3K79me2 and anti-acetyl-H3 antibodies in wild-type (E) and GAL-CDC6 (F) cells. Equal protein concentrations were loaded in all lanes as judged by total H3 levels (data not shown).
To exclude the possibility that cell cycle-dependent changes in H3K79 dimethylation are the result of a global turnover of histones during DNA replication, we used a strain containing the replication initiation factor CDC6 under the regulatable GAL1 promoter (Figure 3D-F). In glucose, CDC6 expression is repressed and DNA replication does not occur, yet other cell cycle events such as Cdk activation/inactivation, securin degradation and spindle formation still occur (Biggins and Murray, 2001; Stern and Murray, 2001). Lack of tension at kinetochores activates the spindle checkpoint in budding yeast (Biggins and Murray, 2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of the S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae (Piatti et al., 1995). H3K79 dimethylation also oscillates during the cell cycle in GAL-CDC6 cells that fail to replicate DNA just as it does in wild-type cells that undergo S phase (Figure 3D-E). Thus, cell cycle changes in H3K79me2 are likely regulated at the level of its establishment and/or removal and may be important for progression through the cell cycle.
Genome-wide localization of H3K79me2 and H3K79me3 were mutually exclusive
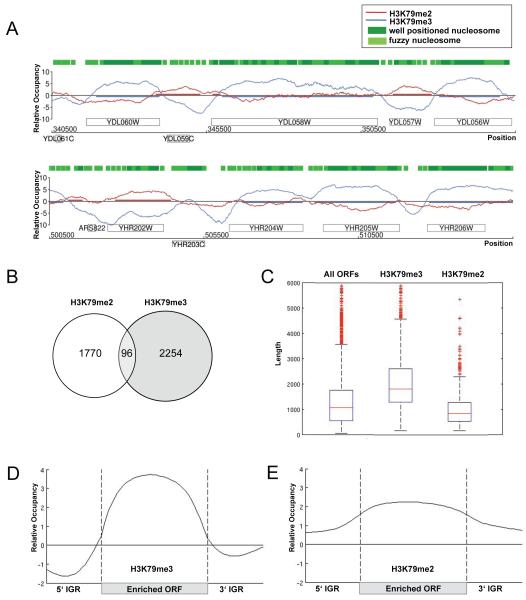
Little is known about the distinct functions of H3K79me2 and me3 marks in the cell, and a redundancy in their roles was proposed in budding yeast (Frederiks et al., 2008; Shahbazian et al., 2005). To better understand the relationship of H3K79 di- and trimethylation as well as elucidate a possible role for H3K79me2 in cell cycle progression, a comprehensive genome-wide map of these modifications was established via ChIP-on-chip experiments using high-resolution tiling microarrays. We first tested the specificity of our antibodies using ChIP-Chip (Supplementary Figure 1). Protein-DNA complexes containing either di- or trimethylated forms of histone H3K79 were specifically immunoprecipitated with antibodies against H3K79me2 and H3K79me3, respectively. In order to reliably detect enriched regions, we applied an adapted version of the Model-based analysis of tiling arrays algorithm (MAT) (Schulze, Gottardo, Kobor, manuscript in preparation) (Johnson et al., 2006), comparing signal intensities between ChIP and genomic DNA to calculate the protein binding profile. Spearman rank correlation coefficients of r=0.9 in average indicated high reproducibility and robustness of the performed replicates. Intriguingly, we found that H3K79me2 and me3 are localized to different regions of the genome and have distinct and mutually exclusive patterns on chromatin (Figure 4A). In total, H3K79me2 covered ~ 22% and H3K79me3 ~ 35% of the genome with only 2% overlap, suggesting that these H3K79 methyl marks were associated with distinct genomic regions. To compare our localization data with known genome-wide nucleosome occupancy data, we overlayed our H3K79 methylation profiles with nucleosome position data predicted by an Hidden Markov Model (Lee et al., 2007) (Figure 4A). Despite the slight differences in resolution, we found enriched regions co-localizing with regions of known nucleosome occupancy. To further ensure that our profiles truly reflected specific H3K79 methylation marks recognized by the two antibodies, control experiments were performed in a strain lacking the H3K79 methyltransferase Dot1, in which H3K79 methylation is completely eliminated. The genome-wide control profiles showed randomly scattered background peaks and a trend towards occupancy of repetitive regions (Figure S1), demonstrating that the antibodies were specific for their respective H3K79 methylation state.
Figure 4. High-resolution profile of H3K79me2 and H3K79me3 across the yeast genome with global occupancy analysis.
(A) H3K79me2 and H3K79me3 profiles. Sample genomic positions for chromosome 4 and 8 were plotted along the x-axis against the relative occupancy of H3K79me2 and H3K79me3 on the y-axis. ORFs are indicated as rectangles, above the axis for Watson genes and below the axis for Crick genes. Green boxes represent HMM-predicted well-positioned and fuzzy nucleosome positions derived from Lee et al. (Lee et al., 2007) (B) Venn diagram comparing the number of H3K79me2 and -me3-enriched ORFs. (C) Average lengths of H3K79me3 and -me2-enriched genes. Boxplots showing the lengths of 6576 ORFs in yeast, as well as, the lengths of H3K79me3 and H3K79me2-enriched ORFs. (D-E) Average profile of H3K79me3 (D) and H3K79me2 (E)-enriched ORFs. A gene was considered to be enriched if at least 50% of its ORF was covered by the modification. ORFs were aligned according to their translational start and stop sites similar to an approach by the Young lab (Pokholok et al., 2005). Each ORF was divided into 40 bins of equal length, probes were assigned accordingly, and average enrichment values were calculated for each bin. Probes in promoter regions (500 base pairs upstream of transcriptional start site) and 3′UTR (500 base pairs downstream of stop site) were assigned to 20 bins, respectively. The average enrichment value for each bin was plotted.
Differential association of H3K79 di- and trimethylation with promoters and ORFs
Having established the detailed maps of H3K79 di- and trimethylation, we wanted to understand the general features of the occupied regions. Genome-wide analysis revealed that H3K79me2 and H3K79me3 covered 1866 and 2350 of 6576 total open reading frames (ORFs), respectively. As expected, the two sets of ORFs enriched with either H3K79 di- or trimethylation overlapped in very few genes (Figure 4B). Interestingly, H3K79me3-enriched ORFs were longer (median 1815 base pairs) while H3K79me2-enriched ORFs were shorter (median 848 base pairs) relative to the average ORF (median 1067 base pairs) (Figure 4C). To visualize the average profile of all ORFs occupied by H3K79me3 and H3K79me2, enriched ORFs were aligned according to the location of translation initiation and termination sites similar to an earlier published analysis (Pokholok et al., 2005) (Figure 4D-E). Consistent with previous studies (Pokholok et al., 2005), H3K79me3 was uniformly enriched within the protein-coding region of the genes (Figure 4D). In contrast, H3K79me2-enriched ORFs showed that H3K79 dimethylation was not only found in the protein-coding regions of genes, but also covered their promoter region (Figure 4E). In general, genes enriched for H3K79me2 in their promoter are also enriched for H3K79me2 in their ORFs (Figure S2A), but in addition, there were genes whose promoter region was occupied without extending into the down-stream ORF (Figure S2B). Overall, H3K79me3 was found in only a few promoters, whereas H3K79me2 covered promoter regions more frequently. Promoters comprise a fraction of the intergenic region that we defined as regions that do not encode protein. Consistent with the higher occupancy of H3K79me2 in promoter regions, we found that H3K79me2 covered ~ 20% of the intergenic regions, whereas H3K79me3 covered less than 4%.
H3K79 di- and trimethylation associated with genes that tend to be transcriptionally less active
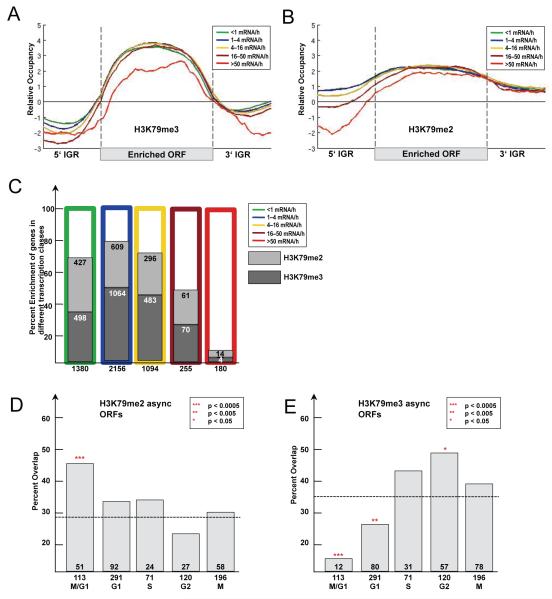
In order to examine the correlation between H3K79 di- and trimethylation and gene expression, enriched genes were assigned into five different classes according to their transcription rate (Holstege et al., 1998), and a composite occupancy profile for each class was determined (Figure 5A-B). Genes enriched for H3K79 dimethylation had a tendency to be present at higher levels in transcriptionally less active genes mainly towards their 5′end and promoter region (Figure 5B). Consistent with previous findings (Pokholok et al., 2005), no clear correlation to transcriptional activity of genes and enrichment for H3K79 trimethylation was found (Figure 5A). Genes with lower expression level were enriched for H3K79 di- and trimethylation with similar ratio (Figure 5C). An exception, however, was the small group of the most highly expressed genes. These genes had very low levels of H3K79 di- and trimethylation in their promoter as well as in their ORFs. Gene expression has been reported to correlate inversely with nucleosome occupancy in promoters (Lee et al., 2007), so it is not surprising to find low H3K79 di- and trimethylation in these promoters. However, coding regions of highly expressed genes have been shown to be more occupied by nucleosomes than lower expressed genes (Lee et al., 2007). Therefore, ORFs of highly expressed genes are devoid of H3K79 di- and trimethylation despite the dense occupancy with nucleosomes.
Figure 5. Functional characterization of H3K79 dimethylated genes.
(A-B) Average profiles of H3K79me3 (A) and H3K79me2 (B)-enriched ORFs according to transcriptional activity. All genes for which information was available (Holstege et al., 1998) (Transcriptome 2005) were divided into five classes according to their transcriptional rate. Average gene profiles were computed and plotted as described in Figure 4D. (C) Percent enrichment of H3K79 di- and trimethylated ORFs in different transcriptional classes. As before genes were divided into five classes according to their transcriptional activity (Holstege et al., 1998) and the percent overlap with H3K79 di- and trimethylated ORFs was plotted. (D-E) Overlap of H3K79me2 and H3K79me3-enriched ORFs with transcriptionally regulated genes for each cell cycle stage (Spellman et al., 1998). Numbers below the x-axis represent total number of genes with periodic transcription. Numbers above x-axis represent the overlap of these genes with those enriched for H3K79me2 and H3K79me3. The percentage of the overlap was plotted on the y-axis. (D) H3K79me2-enriched ORFs in asynchronous cells. Expected by chance are 28% (1866 H3K79me2-enriched ORFs out of 6576 total). (E) H3K79me3-enriched ORFs in asynchronous cells. Expected by chance are 36% (2350 H3K79me3-enriched ORFs out of 6576 total). The p-values were calculated using the hypergeometric test.
M/G1-regulated genes were significantly enriched for H3K79me2
Because of the cell cycle dependence of H3K79me2, we tested if H3K79 dimethyl-marked genes were regulated during the cell cycle. In budding yeast, approximately 800 genes change their transcriptional profile and peak in certain stages of the cell cycle (Spellman et al., 1998). We compared ORFs enriched for H3K79 di- and trimethylation with the different classes of cell cycle-regulated genes and asked if the overlap was significant using a hypergeometric test (Tavazoie et al., 1999). Indeed, we found that M/G1-regulated genes were significantly enriched for H3K79me2, but were not enriched for H3K79me3 (Fig 5D-E). In contrast, genes regulated in the G2-phase showed a significant occupancy with H3K79me3, but are not marked by H3K79me2 (Figure 5D-E). These results suggest that H3K79 methylation is not random, but rather that it is regulated in conjunction with progression through the cell cycle and might be involved in periodic transcription of genes during distinct cell cycle phases.
Genome-wide association of H3K79 di- but not trimethylation was altered during the cell cycle
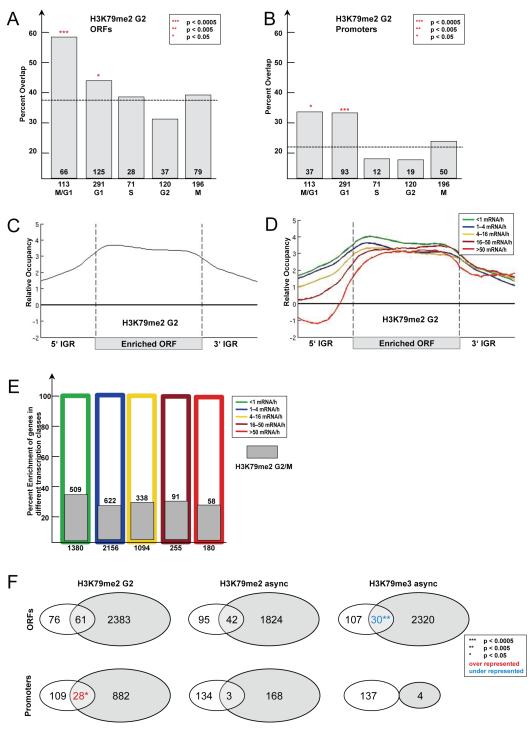
Our findings suggested that bulk levels of H3K79me2, but not H3K79me3, changed during the progression of the cell cycle, with reduced levels in G1 and elevated levels in G2/M. To test if levels of H3K79me2 fluctuated at the level of single genes during the cell cycle, we performed ChIP-on-Chip assays using chromatin of nocodazole arrested yeast cells and compared it to asynchronous cells. In nocodazole, cells are arrested in the G2/M-phase of the cell cycle and should have the highest level of H3K79me2, while asynchronous cells have a mixed distribution of G1-, S- and G2/M-phase cells. While H3K79me2 profiles in asynchronous and G2/M-arrested cells were overall similar with a Spearman rank correlation coefficient of r=0.69 (Figure S3A), a detailed analysis revealed important differences between them. In asynchronous cells, ORFs enriched for H3K79me2 significantly overlapped with genes whose expression is regulated in M/G1. Interestingly, ORFs enriched for H3K79me2 in G2/M-arrested cells significantly overlapped not only with M/G1 but also with G1 regulated genes (Fig 6A). Moreover, this effect expanded into the promoter region of genes since promoter regions of M/G1 and G1 regulated genes were also significantly enriched for H3K79me2 in G2/M arrested cells (Figure 6B). This result suggests that M/G1- and G1-regulated genes are marked in their ORF and promoters by H3K79me2 during cell cycle stages (G2/M) when these genes are inactive.
Figure 6. Genome-wide characteristics of H3K79me2-enriched genes in G2/M.
(A-B) Overlap of H3K79me2-enriched ORFs in G2/M-arrested cells with transcriptionally regulated genes for each cell cycle stage (Spellman et al., 1998). (A) H3K79me2-enriched ORFs in G2/M-arrested cells. Expected by chance are 38% (2444 H3K79me2-enriched ORFs out of 6576 total). (B) H3K79me2-enriched promoters in G2/M. Expected by chance are 23% (1483 H3K79me2-enriched promoters out of 6576 total). (C) Average profile of H3K79me2-enriched ORFs in G2/M-arrested cells. The profile for the average enriched ORF was determined as explained in Figure 4D. (D) Average profile of H3K79me2-enriched ORFs in G2/M-arrested cells according to their transcriptional activity. The profile was determined as described in Figure 5A. (E) Percent enrichment of H3K79 dimethylated ORFs in different transcriptional classes. As before genes were divided into five classes according to their transcriptional activity (Holstege et al., 1998) and the percent overlap with H3K79 dimethylated ORFs in G2/M- arrested cells was plotted. (F) Venn diagram showing overlap of the 137 Swi4-bound genes (Iyer et al., 2001) with H3K79me2 and H3K79me3-enriched ORFs and promoters, respectively. H3K79me2-enriched promoters in G2/M-arrested cells showed significant overlap with Swi4-bound genes. H3K79me3-enriched ORFs and promoters show significant under-representation of Swi4-bound genes. Promoters were called enriched when 450bp upstream of the ORF were covered by the methyl mark.
In contrast to H3K79me2, global levels of H3K79me3 were not altered during the cell cycle (Figure 3C). In order to confirm this observation on a single gene level, the H3K79me3 profile in G2/M-arrested cells was determined. As expected, the profiles of the asynchronous and G2/M-arrested cells were similar, occupied the same regions, and correlated with a Spearman rank correlation coefficient of r=0.91 (Figure S3B). Consistent with the observations in asynchronous cells, H3K79 di- and trimethylation had distinct and mutually exclusive patterns on chromatin in the G2/M-phase and overlapped in only 1% of the genome (Figure S3C).
H3K79me2 was found in intergenic regions and covered ARS in G2/M-arrested cells
To further characterize similarities and differences between H3K79me2 in asynchronous and G2/M-arrested cells, we concentrated on typical genomic features. It is known that nucleosome occupancy does not exhibit large, global variation between cell cycle phases (Hogan et al., 2006), and observed changes in H3K79 dimethylation pattern during the cell cycle were most likely not due to global changes in nucleosome occupancy. We visualized the average profile of all genes occupied by H3K79me2 in G2/M-arrested cells. Similar to asynchronous cells, we found that H3K79 dimethylation was not only found in the protein-coding regions of genes, but it also covered their promoters (Figure 6C and S2). The trend towards occupancy of less transcribed genes was weaker in G2/M-arrested cells compared to the asynchronous dataset (Figure 6E), but the relative height of the H3K79 dimethylation profile followed precisely the decreasing order of transcriptional activity (Figure 6D). Furthermore, H3K79me2 was enriched more frequently in the promoter regions in the G2/M-arrested cells compared to the asynchronous cells (Figure S2). Not only promoters were enriched more frequently with H3K79me2 in G2/M-arrested cells, but also intergenic regions in general. Indeed, we found them to be covered to ~50% in G2/M-arrested cells compared to ~20% in the asynchronous dataset.
To further characterize additional chromosomal feature for their enrichment with H3K79 di- and trimethylation, we focused on autonomously replicating sequences (ARS) and centromeres. Intriguingly, we found that ARSs were significantly enriched (131 out of 274 ARS, p-value = 3e-14) for H3K79me2 in the G2/M-phase. In contrast, low ARS occupancy of H3K79 di- and trimethylation (17 and 3 ARS out of 274) was detected in asynchronous cells. The significant overlap of ARS with H3K79me2 in G2/M arrested cells indicates a potential role of H3K79me2 in regulating ARS in their inactive state. In yeast, the single centromeric nucleosome contains a specialized H3-variant, Cse4, in place of canonical histone H3. Consistent with replacement of H3 at the centromere, neither H3K79me2 nor H3K79me3 antibodies enriched for CEN sequences in either the asynchronous or G2/M dataset. The differences in association of H3K79 di- and trimethylation with telomeres and the rDNA locus will be reported elsewhere.
Swi4-regulated genes were significantly enriched for H3K79me2 in their promoter region during G2/M-phase
Genome-wide analysis of SBF binding sites by ChIP-on-Chip using the DNA-binding subunit Swi4 revealed that the SBF binds to promoters of genes expressed in G1/S (Harbison et al., 2004; Iyer et al., 2001; Simon et al., 2001). A comparison of H3K79me2-enriched promoters to the 137 promoters bound by Swi4 (Iyer et al., 2001) gave a significant overlap in G2/M-arrested cells (Figure 6E). Curiously, this overlap was not significant for H3K79me2-enriched promoters and ORFs in asynchronous cells, perhaps indicating that H3K79me2 is a consequence of SBF binding/transcription earlier in the cell cycle. In contrast, ORFs and promoters enriched with H3K79me3 showed significantly lower overlap than expected by chance and no overlap with Swi4-bound genes, respectively (Figure 6F). This analysis showed that a significant number of SBF-regulated genes were H3K79 dimethylated in their promoter during G2/M.
Moreover, most known cell cycle key regulators of G1/S transition, some of them bound by the SBF-complex, were clearly enriched for H3K79me2 (Figure S4). These included the G1-cyclins, CLN1 and CLN2, and the S-phase cyclin, CLB5 and CLB6. Furthermore, several histone genes (HHT2, HTZ1, and HHO1) were marked with H3K79me2 (Figure S4). Intriguingly, H3K79me2 mainly marks these genes during the cell cycle stage (G2/M) in which these genes are inactive.
Genome-wide colocalization of H2BK123 monoubiquitination with H3K79 tri- but not dimethylation
Given our results demonstrating that the pattern of H3K79 di-and trimethylation on chromatin are mutually exclusive (Figure 4B), we next sought to understand the mechanism by which a single enzyme Dot1 can distinguish between sites of di- vs trimethylation. The appearance of H3K79me2 after replication in S-phase could be explained by either the de-novo establishment of the methyl mark or by the demethylation of an existing H3K79me3 mark resulting in H3K79 dimethylation. The latter would require regions that are dimethylated in G2/M to be trimethylated during G1/S transition. We ruled out this possibility, because ChIP-on-Chip of H3K79me3 in G1-arrested cells showed that regions that are H3K79 dimethylated in G2/M were not trimethylated in G1 (Figure S5A) and the profiles of H3K79 trimethylation were similar in G1- and G2/M-arrested cells with a Spearman rank correlation coefficient of r=0.82 (Figure S5B). Based on these observations, it seems to us that demethylation of H3K79me3 may not be the main method by which yeast cells regulate the pattern of H3K79me2 and Me3, however, it remains possible that the trimethyl state could be achieved transiently and quickly removed in G1. Given our data, we hypothesize that H3K79 di- and trimethylation are established independently and additional factors control the distribution of di- vs trimethylation. One major candidate is histone H2B K123 monoubiqitination mediated by Rad6/Bre1 that is known to be required for proper H3K79 trimethylation. Since ChIP-Chip studies indicated that the patterns of H3K79 di- and trimethylation are mutually exclusive and have ~ 2% overlap throughout the yeast genome, we hypothesize that the pattern of H2B monoubiquitination could control distribution of H3K79 di- vs trimethylation on chromatin. Based on this hypothesis, we predict that factors required for H3K79me3 should function through the regulation of H2B monoubiquitination. To address this possibility, we set our biochemical screen (GPS) to identify factors that are solely required for H3K79me3 establishment with no effect on H3K79me2. Our survey of extracts of the collection of gene deletion mutants for the presence of H3K79me3 resulted in the identification of several known and novel factors including which are required for proper H2B monoubiquitination and H3K79 trimethylation (data not shown).
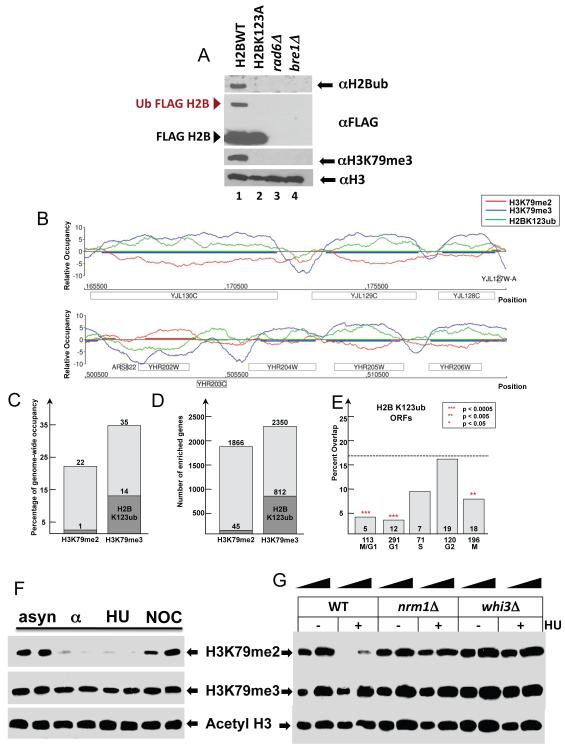
To further test the above hypothesis that H2BK123 monoubiquitination determines the pattern of H3K79 trimethylation and co-localizes with H3K79 tri- but not dimethylation , we developed polyclonal antibodies specifically recognizing K123 monoubiquitinated H2B (Figure 7A). Employing this antibody, we determined a comprehensive genome-wide map of H2BK123 ubiquitination via ChIP-on-Chip (Figure 7B). To ensure the specific enrichment of H2BK123ub over un-modified H2B, non-specific binding was blocked with H2B peptide during the immunoprecipitation step. In addition, H2BK123ub enrichment data were normalized using an H2BubK123A mutant profile obtained from an identical ChIP-on-Chip reaction. Intriguingly, we found that H2BK123ub co-localized with H3K79 trimethylation at many genomic loci, but showed distinct and mutually exclusive patterns with H3K79 dimethylation (Figure 7B). Overall, high confidence regions co-localized with H3K79 trimethylation, and only a very minor fraction overlapped with H3K79 dimethylation (Figure 7C). As expected from this analysis, very similar distributions existed at the open reading frame (ORF) level where 812 out of 2350 H3K79 trimethylated ORFs were marked by H2BK123 ubiqitination, but only 45 out of 1866 H3K79 dimethylated ORFs were enriched for H2BK123 ubiqitination (Figure 7D). Since H3K79 dimethylation and H2B monoubiquitination were mutually exclusive on a genome-wide level, we predicted the link of H3K79me2 to genes expressed specifically during the cell cycle to be independent of H2B monoubiquitination pattern. Indeed, cell cycle regulated genes, especially those regulated in M/G1, were not marked by H2B ubiquitination (Figure 7E). Taken together, these findings suggest that the regulation of H2BK123 monoubiquitination is linked to H3K79 tri- but not dimethylation and could play role in distinguishing the genome-wide establishment of H3K79 di- vs trimethylation.
Figure 7. Genome-wide profile of H2B monoubiqitination demonstrates an association with H3K79 trim but not dimethylation.
(A) Development of polyclonal antibodies specific to monoubiquitinated H2B. H2B specific antibodies generated in rabbit were affinity purified and used for testing extracts from strains either carrying either Flag::H2B (lane 1) Flag::H2BK123R (lane 2), or wild type H2B in strains deleted for RAD6 (lane 3) or BRE1 (lane4). (B) Overlay of H2BK123ub, H3K79me2, and H3K79me3 profiles. Sample genomic positions for chromosome 8, and 10 were plotted along the x-axis against the relative occupancy of the indicated histone modifications on the y-axis. ORFs are indicated as rectangles, above the axis for Watson and below the axis for Crick strand. (C) Diagram summarizing the percentage of genome-wide occupancy of H3K79me2 and H3K79me3 and their overlap with H2BK123ub. (D) Diagram illustrating the overlap of H3K79me2 and H3K79me3 enriched with H2BK123ub enriched genes. (E) Overlap of H2BK123 ubiquitinated ORFs with transcriptionally regulated genes for each cell cycle stage (Spellman et al., 1998). Numbers below and above x-axis represent total number of genes in each cell cycle class and their overlap with H2B ubiquitinated genes, respectively. The percentage of overlap is plotted on the y-axis with a dashed line indicating the percentage expected by chance. The p-values were calculated using the hypergeometric test. (F) Yeast cell arrest arrested in the G1 and S phase of the cell cycle. To better understand the role of factors responsible for the implementation/removal of H3K79 dimethylation during G1/S stages of cell cycle, we performed a biochemical screen with the entire collection of viable yeast gene deletion mutants arrested with HU (Supplementary Figure 6). (G) This screen resulted in the identification of Nrm1 and Whi3, which is required for exit from the G1 phase of the cell cycle, as a factor required for the removal of dimethylated H3K79, further linking H3K79 methylation status to cell cycle.
Nrm1 and Whi3 were required for the loss of H3K79 dimethylation in the S-phase of cell cycle
Our data indicate that the establishment of the H3K79 dimethylation pattern requires the Swi4/Swi6 complex, and this mark is removed during the G1- and S-phase of the cell cycle. Cells arrested in G1 with alpha factor or in early S phase with hydroxyurea contain low levels of H3K79me2 compared to asynchronous cells or cells arrested in the G2-/M-phase with nocodazole (Figure 7F). To better understand molecular processes required for the establishment of the H3K79me2 mark and how this posttranslational modification is regulated during certain cell cycle stages, we performed a biochemical screen using the viable yeast gene deletion mutants arrested in the early S phase with hydroxyurea. Extracts from the arrested collection were prepared and levels of H3K79me2 were analyzed by Western blotting (Supplementary Figure 6). By arresting cells in hydroxyurea, we eliminated mutants from our screen that simply had low or high levels of H3K79me2 due to the fact that they were enriched in a particular cell cycle stage.
We identified several factors including Nrm1 (negative regulator of MBF targets 1) and Whi3 as required for the cell cycle pattern of H2K79me2 during early S phase (Figure 7G). Nrm1 functions as a co-repressor with the MBF transcription factor that is required for exit from the G1 phase of the cell cycle (de Bruin et al., 2006). Whi3 is an RNA-binding protein involved in cell cycle control and its loss results in the acceleration of the expression of genes controlled by the SBF and MBF complexes. Identification of both Nrm1 and Whi3 in our biochemical screen as specific regulators of H3K79me2 further links the H3K79 methylation pattern to cell cycle control.
DISCUSSION
One major aspect of cell cycle control is the regulatory network of interconnected transcriptional activators (Simon et al., 2001). Often, transcription factors function specifically during one cell cycle stage and control the expression of transcriptional activators for the subsequent one, thereby forming a feed-forward regulatory circuit to ensure an ordered progression through cell division. In addition to transcriptional regulation, cell cycle progression also employs proteolysis, phosphorylation, localization and other regulatory mechanisms to ensure completion of one event before entry into the next. The role of chromatin modifications in transcription control has been intensively studied in recent years. However, little is known about cell cycle dependent changes in chromatin structure and the role that chromatin alterations play in normal cell cycle progression and transcriptional regulation.
Here, we report five fundamental findings, which point toward a connection between the chromatin modification H3K79 methylation and the cell cycle. First and foremost, employing a Global Proteomic Screen (GPS) approach (Schneider et al., 2004), we found that swi4Δ and swi6Δ mutants are defective in establishing H3K79me2. Swi4 and Swi6 compose the SBF transcription factor that regulates the expression of genes involved in the G1/S-transition. Secondly, we showed that global levels of H3K79me2 fluctuate during the cell cycle, in contrast to H3K79me3, which was not altered. Thirdly, ChIP-on-chip analysis revealed an overlap between the genes bound by H3K79me2 and the SBF, suggesting coordination between cell cycle-dependent transcription and dimethylation. Consistent with this idea, H3K79me2-bound genes were those that showed increased expression during the M/G1-phase of the cell cycle. Fourth, analysis of the entire yeast deletion collection arrested in the G1/S-phase pointed to a role for Nrm1 and Whi3, which both impact the transcription by the SBF, and a related transcription complex, MBF in the process of H3K79me2 establishment/removal. Finally, we have demonstrated via genome-wide studies that H2BK123 monoubiquitination pattern colocalizes with H3K79 tri- but not dimethylation indicating that the regulation of H2BK123 monoubiquitination pattern could distinguish the genome-wide establishment of H3K79 di- vs trimethylation.
Based on these observations, we propose the following model of how H3K79me2 and cell cycle control relate to one another. Dephosphorylation of Swi6 and synthesis of SWI4 during mitosis allows binding of SBF to its target genes in the late M/early G1-phase of the cell cycle. However, inhibitory factors such as Whi5 prevent transcriptional activation at these targets until early G1, after cells have reached a critical size threshold. At this point in the cell cycle, known as START in budding yeast, activation of the G1 cyclin-dependent kinase Cln3-Cdk1 inside the nucleus renders the SBF functionally active and a positive feedback loop results in amplification of SBF, as well as, MBF-dependent transcription and synthesis of genes required for progression through G1 and entry into the S-phase. The SBF is inactivated upon entry into the S-phase through phosphorylation of Swi6, which disrupts its binding to Swi4 and results in its export to the cytoplasm. Without Swi6, Swi4 can no longer bind to DNA. Our observations that levels of H3K79me2 increase during the S-phase and remain high during the G2/M-phase, combined with our genome-wide location analysis showing that H3K79me2-occupied genes overlap extensively with those expressed specifically in the G1-phase (or bound by Swi4) suggests that H3K79me2 marks cell cycle-specific genes during G2/M.
Whether H3K79me2 is a consequence of gene inactivation or if it actively causes transcriptional inactivation remains to be determined. Here we provide evidence that the modification is created de novo, that is through the addition of two methyl groups to an unmodified H3K79, and is not generated by demethylation of an existing H3K79me3 residue.
How does Swi4/Swi6 control H3K79me2 oscillations? Based on our GPS screen showing that swi4Δ and swi6Δ mutants are unable to establish the H3K79me2 mark (Figure 1-2), it is formally possible that the SBF could directly establish dimethylation. However, sequence and motif analysis of Swi4 and Swi6 do not indicate either protein contains enzymatic domains that might catalyze dimethylation. Instead, the SBF might activate genes that are required for H3K79me2 formation. One candidate could be DOT1. SBF could regulate H3K79me2 establishment through the regulation of DOT1, but we were able to rule out this possibility by showing that Dot1 expression (Figure 2C) is not altered in swi4 and/or swi6 deletion mutants.
The SBF could also control H3K79me2 by recruiting factors required for establishment of this modification to target genes. This scenario is supported by our observation that H3K79me2 is higher at cell cycle-regulated ORFs and promoters that are also bound by Swi4 and is consistent with our genome-wide ChIP-on-chip studies. It also raises the interesting situation where transcription in the preceding G1 could establish a chromatin mark that could be stably inherited and recognized in subsequent cell cycles via an epigenetic mechanism. The cell cycle oscillation of H3K79me2 could also solve an outstanding question in the field of how SCB-less genes are regulated via the SBF complex. Cross and colleagues have reported a considerable overlap in SBF and MBF target genes, as well as, cell cycle-dependent genes whose expression requires one or more of these factors (Bean et al., 2005). However, many of these identified genes lack the canonical SCB and MCB binding sites in their promoter elements. The existence of H3K79me2 in the promoter regions of SBF-bound genes and their possible propagation through epigenetic mechanisms could in part explain how these genes are regulated during the cell cycle.
Why is the pattern of H3K79 di- and trimethylation are mutually exclusive? Our ChIP-Chip analysis of H3K79 methylation has indicated that there is only ~ 2% overlap between H3K79me2 and H3K79me3 throughout the yeast genome (Figure 4B). We found this observation to be quite unexpected. Histone H2B monoubiquitination pattern may in part explain this observation, since H3K79me3 is dependent on H2B monoubiquitination. Therefore, the pattern of H2B monoubiquitination could control distribution of H3K79 di- Vs trimethylation on chromatin. Indeed, via our GPS, we have identified several factors required for the proper establishment of H3K79 tri- but not dimethlyation and have demonstrated that all of these factors exert their H3K79me3 regulatory activity through the regulation of H2B monoubiquitination levels (data not shown). Furthermore, our genome wide studies have demonstrated that the pattern of H2B monoubiquitination appears to be excluded from cell cycle regulated gene and genes containing H3K79me2 but to be associated with H3K79me3 containing regions.
Previous analysis of H3K79 methylation during the S. cerevisiae cell cycle using an antibody that recognized both H3K79 di- and trimethylation suggested that this modification increased weakly during the late S-phase and remained constant during the G2/M-phase (Zhou et al., 2006). Our results extend this finding since we were able to distinguish between the di- and trimethylated forms of H3K79. In contrast to previous findings that implicated di- and trimethylation of H3K79 in redundant functions (Frederiks et al., 2008; Shahbazian et al., 2005), our results clearly show they have distinct, non-overlapping roles. ChIP-on-chip assays revealed mutually exclusive patterns of H3K79 di- and trimethylation across the yeast genome. In addition, factors influencing H3K79me2 levels had no effect on H3K79me3 levels and only H3K79me2 levels were cell cycle regulated. Mitosis specific dimethylation of H3K79 in human cells and of the synonymous residue H3K76 in Trypanosoma brucei have also been observed. (Feng et al., 2002; Janzen et al., 2006). Given the similarity of cell cycle regulation across species and the evolutionary conservation of H3K79 methylation, it seems probable that dimethylation, transcriptional control and cell cycle progression are also coupled in higher eukaryotes as they appear to be in yeast. With this study, we reveal a novel link between histone modifications and cell cycle control. The discovery of a functional relationship between H3K79me2 and the transcriptional cell cycle activator SBF gives insight into the complex network of cell cycle control and chromatin regulation.
MATERIALS AND METHODS
Yeast Strains
Strains used in this study were derived from BYXXX (genotype of the YKO library) or from W303 (a bar ura3-1 trp1-1 ade2-1 his3-11,15 leu2-3,112 can1-100).
Global Proteomic Analysis of Histone Modifications
GPS analyses were carried out as described previously (Schneider et al., 2004) with the use of antibodies specific for dimethylated Lys 79 (Millipore, 04-835) or trimethylated Lys 79 (Abcam, ab2621) and for dimethylated Lys 4 of histone H3 or acetylated histone H3 .
RT-PCR
Cells were grown in YPD to an OD600 of 0.8 – 1.0. RNA was extracted by resuspending the cells in 500ml of TES buffer (10mM Tris, 10mM EDTA, 0.5% SDS) and 500ml of 65°C acidic phenol. Cells were then placed at 65°C for 1 hour and vortexed every 10 minutes. After phenol-chloroform extraction of the cell extract, RNA was precipitated with 100% ethanol, and treated with RNase-free DNase I. 1μg of RNA was used in a reverse transcriptase (RT) reaction (Clontech). 5μl of cDNA was used in a 50μl PCR with 26 cycles of amplification.
Cell Synchronization and FACS Analysis
α-factor was used at a final concentration of 1 μg/mL. The GAL-CDC6 experiment was carried out as described (Biggins and Murray, 2001). Analysis of DNA content by flow cytometry was carried out as previously described
Chromatin Immunoprecipitation and Genome-wide ChIP-on-Chip
Chromatin immunoprecipitation and genome-wide location analyses were performed as described previously, using the adapted linear amplification method that involves two rounds of T7 RNA polymerase amplification (van Bakel et al., 2008). In brief, 500 ml yeast cells were grown in a rich medium to an OD600 0.8–0.9 and were cross-linked with 1% formaldehyde for 20 min before chromatin was extracted. The chromatin was sonicated (Bioruptor, Diagenode: 10 cycles, 30s on/off, high setting) to yield an average DNA fragment of 500 bp. 4 μl of H3K79me2 or H3K79me3 antibodies were coupled to 60 μl of protein A magnetic beads (Invitrogen). After reversal of the crosslinking and DNA purification, the immunoprecipitated and input DNA were amplified to about 6 μg aRNA using T7 RNA polymerase in two rounds. Samples were labeled with biotin, and the immunoprecipitated and input sample were hybridized to two Affymetrix 1.0R S. cerevisiae microarrays, which are comprised of over 3.2 million probes covering the complete genome. 25-mer probes are tiled at an average of 5 base pair resolution, creating an overlap of approximately 20 base pairs between adjacent probes.
Data Analysis
We used an adapted version of the Model-based Analysis of Tiling-arrays (MAT) algorithm to reliably detect enriched regions (Schulze, Gottardo, Kobor, manuscript in preparation) (Johnson et al., 2006). MAT was applied to corresponding immunoprecipitated and input sample array, and the probe behavior model was estimated by examining the signal intensity, sequence, and copy number of all probes on an array. After probe behavior model fitting, the residuals between the model and observation were normally distributed and centered at 0. MAT uses a score function to identify regions of ChIP enrichment, which allows robust p-value and false discovery rate calculations. MATscores are calculated from all probes within a 300-bp sliding window and returns a MATscore for each probe. The MAT scores, as a measure for relative enrichment, were visualized along the whole genome using custom-written scripts in the language and statistics environment R.
Annotations for ORFs, ARS and centromeres were derived from the SGD database. An open reading frame (ORF) was termed enriched if at least 50% of all probes had a MATscore above a threshold of 1.5. Promoters were defined as enriched if all probes 300 base-pairs upstream of the transcriptional start site were above the MATscore cutoff. Promoters which overlap with ORFs of other genes were not considered.
ARS were defined to be enriched when they were completely covered by the methyl mark. Nucleosome position data were derived from Lee et al (Lee et al., 2007).
Analysis of published genome-wide expression and binding data sets
To examine the overlap between H3K79 methylated genes and cell cycle-regulated genes, the Spellman et al. (1998) analysis was chosen because it incorporates several types of yeast cell cycle expression data. To compare our data sets with SBF binding data from previously published genome-wide localization analyses, we used the supplementary data available from Iyer et al. (Iyer et al., 2001). The figure3_data.xls file was used to identify SBF-bound, and SBF +MBF-bound ORFs.. For all data sets, any duplicate ORFs and ORFs with highly repetitive sequence were removed and the systematic yeast gene names were used. MATLAB scripts were used to compare the different data sets.
Statistics
In order to explore the statistical significance of the overlap between different sets of genes, we used the hypergeometric test as described (Tavazoie et al., 1999).
Supplementary Material
Figure S1 H3K79me2 and H3K79me3 antibody is specific. (A) Comparison of H3K79me2 profiles in wildtype and dot1 deletion strain. In the dot1 deletion strain, H3K79me2 was eliminated and the ChIP-Chip profile showed only background peaks. (B) Comparison of H3K79me3 profiles in wildtype and dot1 deletion strain. In dot1 deletion strain H3K79me3 was eliminated, and the ChIP-Chip profile showed only background peaks. (C) Comparison of H3K79me2 and H3K79me3 background profiles from dot1 deletion strain. The background was independent of the antibody.
Figure S2 H3K79me2 enriched promoters and their global occupancy across the average gene. Average profiles of H3K79me2 enriched promoters in asynchronous and G2/M-arrested cells. Promoters were considered to be enriched if the 250bp region upstream of their transcriptional start site was dimethylated and their promoter did not overlap with any other gene. The average profile was calculated as described in Figure 4D. (A) Average profile of genes with H3K79me2 enriched promoters in asynchronous (578 out of 6576) and G2/M arrested cells (1782 out of 6576). (B) Average profile of genes with H3K79me2 enriched promoters and non-enriched associated ORFs in asynchronous (197 out of 6576) and G2/M arrested cells (641 out of 6576).
Figure S3 Genome-wide H3K79me2 and me3 profile in G2/M-arrested cells. (A) Comparison of genome-wide H3K79me2 profiles in asynchronous and G2/M-arrested cells. (B) H3K79me3 profiles in asynchronous and in G2/M-arrested cells. (C) H3K79me2 and H3K79me3 profiles of cells arrested in G2/M.
Figure S4 Cell cycle regulated genes bound by Swi4 are highly enriched for H3K79me2. Table with characteristic cell cycle regulators and histone genes, showing the enrichment of those for Swi4 binding (Iyer et al., 2001) and H3K79me2 occupancy.
Figure S5 Genome-wide H3K79me3 profile in G1-arrested cells was similar to H3K79me3 profile in G2/M- arrested cells.
(A) Partial genome plot comparing H3K79me3 profile in G1-arrested cells with H3K79me2 profile in G2/M-arrested cells.
(B) Partial genome plot comparing H3K79me3 profiles in G1-arrested with G2/M-arrested cells.
Figure S6 GPS analyses of the entire yeast deletion collection arrested in G1/S with HU. Cell extracts from each of the nonessential yeast gene deletion mutants arrested with HU were subjected to SDS-PAGE, Western blotted, and probed with antibodies specific for H3K79 dimethylation Red arrows (D5) represent signal from negative control and red arrow (H3) represent signal in the absence of HU.
ACKNOWLEDGMENTS
We thank Dr. Edwin Smith for critical reading of this manuscript and Laura Shilatifard for editorial assistance. Furthermore, we thank Dr. Raphael Gottardo for providing the MAT package to analyse the ChIP-Chip data, Dr. Hunter Fraser for help with the statistical analysis as well as Dr. Frank Holstege and Dr. Harm van Bakel for detailed ChIP-Chip protocols using double-round T7 amplification. JMS is supported by a fellowship from the German Academic Exchange Service and a graduate studentship from the Child and Family Research Institute. MSK is a Scholar of Michael Smith Foundation for Health Research and the Canadian Institute for Advanced Research. Research for this study in the laboratory of MSK was supported by an operating grant from the Canadian Institute of Health Research (MOP-79442). The work in Shilatifard’s laboratory was supported by a grant from the National Institute of Health (GM069905) to ASH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Molecular cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews BJ, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. Start-specific transcription in yeast. Current topics in microbiology and immunology. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and transacting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, Russell P, Wittenberg C. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Molecular cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. The Journal of biological chemistry. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nature structural & molecular biology. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LA, Andrews BJ. Binding to the yeast SwI4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic acids research. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford LM, Osley MA, Ludwig TR, 2nd, McLaughlin CS. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS genetics. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Molecular cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Im H, Park C, Feng Q, Johnson KD, Kiekhaefer CM, Choi K, Zhang Y, Bresnick EH. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. The Journal of biological chemistry. 2003;278:18346–18352. doi: 10.1074/jbc.M300890200. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Molecular cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science (New York, NY. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. The Journal of biological chemistry. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nature genetics. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nature structural & molecular biology. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature reviews. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Molecular and cellular biology. 2005;25:4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes & development. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Molecular cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Schneider J, Dover J, Johnston M, Shilatifard A. Global proteomic analysis of S. cerevisiae (GPS) to identify proteins required for histone modifications. Methods in enzymology. 2004;377:227–234. doi: 10.1016/S0076-6879(03)77013-X. [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & development. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Molecular cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Molecular biology of the cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nature genetics. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, Holstege FC, Timmers HT. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic acids research. 2008;36:e21. doi: 10.1093/nar/gkm1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 H3K79me2 and H3K79me3 antibody is specific. (A) Comparison of H3K79me2 profiles in wildtype and dot1 deletion strain. In the dot1 deletion strain, H3K79me2 was eliminated and the ChIP-Chip profile showed only background peaks. (B) Comparison of H3K79me3 profiles in wildtype and dot1 deletion strain. In dot1 deletion strain H3K79me3 was eliminated, and the ChIP-Chip profile showed only background peaks. (C) Comparison of H3K79me2 and H3K79me3 background profiles from dot1 deletion strain. The background was independent of the antibody.
Figure S2 H3K79me2 enriched promoters and their global occupancy across the average gene. Average profiles of H3K79me2 enriched promoters in asynchronous and G2/M-arrested cells. Promoters were considered to be enriched if the 250bp region upstream of their transcriptional start site was dimethylated and their promoter did not overlap with any other gene. The average profile was calculated as described in Figure 4D. (A) Average profile of genes with H3K79me2 enriched promoters in asynchronous (578 out of 6576) and G2/M arrested cells (1782 out of 6576). (B) Average profile of genes with H3K79me2 enriched promoters and non-enriched associated ORFs in asynchronous (197 out of 6576) and G2/M arrested cells (641 out of 6576).
Figure S3 Genome-wide H3K79me2 and me3 profile in G2/M-arrested cells. (A) Comparison of genome-wide H3K79me2 profiles in asynchronous and G2/M-arrested cells. (B) H3K79me3 profiles in asynchronous and in G2/M-arrested cells. (C) H3K79me2 and H3K79me3 profiles of cells arrested in G2/M.
Figure S4 Cell cycle regulated genes bound by Swi4 are highly enriched for H3K79me2. Table with characteristic cell cycle regulators and histone genes, showing the enrichment of those for Swi4 binding (Iyer et al., 2001) and H3K79me2 occupancy.
Figure S5 Genome-wide H3K79me3 profile in G1-arrested cells was similar to H3K79me3 profile in G2/M- arrested cells.
(A) Partial genome plot comparing H3K79me3 profile in G1-arrested cells with H3K79me2 profile in G2/M-arrested cells.
(B) Partial genome plot comparing H3K79me3 profiles in G1-arrested with G2/M-arrested cells.
Figure S6 GPS analyses of the entire yeast deletion collection arrested in G1/S with HU. Cell extracts from each of the nonessential yeast gene deletion mutants arrested with HU were subjected to SDS-PAGE, Western blotted, and probed with antibodies specific for H3K79 dimethylation Red arrows (D5) represent signal from negative control and red arrow (H3) represent signal in the absence of HU.