Abstract
Objective
To determine whether genetic variants associated with diabetes and obesity predict gestational weight gain.
Study Design
960 participants in the Pregnancy, Infection and Nutrition cohorts were genotyped for 27 single-nucleotide polymorphisms (SNPs) associated with diabetes and obesity.
Results
Among white and black women (n=960), KCNQ1 risk allele carriage was directly associated with weight gain (p < 0.01). In Bayesian hierarchical models among white women (N=628), we found posterior odds ratios > 3 for inclusion of TCF2 and THADA SNPs in our models. Among black women (n=332), we found associations between risk allele carriage and weight gain for the THADA and INSIG2 SNPs. In Bayesian variable selection models, we found an interaction between the TSPAN8 risk allele and pre-gravid obesity, with lower weight gain among obese risk allele carriers.
Conclusion
We found evidence that diabetes and obesity risk alleles interact with maternal pre-gravid BMI to predict gestational weight gain.
Keywords: diabetes, gestational weight gain, genetics, obesity, single nucleotide polymorphisms
Introduction
Maternal weight gain during pregnancy is an important predictor of health outcomes for both mother and child1. Inadequate gestational weight gain is associated with preterm birth2, intrauterine growth restriction, low birth weight, and offspring obesity risk3, whereas mothers who gain excessively are more likely to deliver by cesarean section 4–7, have an unsuccessful trial of labor after c-section8, develop pre-eclampsia6, retain excessive weight after delivery9, 10, and become overweight or obese in later life11, 12. Infants born to women who gain excessively during pregnancy are more likely to be born preterm13, be macrosomic at birth (> 9 lbs)5, 14, 15, and become overweight or obese as toddlers16 and adults3. Based on these well-described epidemiologic associations, gestational weight gain has been targeted as a modifiable risk factor for metabolic disease in both mother and child.
While intervention strategies have targeted health behaviors that affect gestational weight gain, a mother’s genotype is also likely to influence her pattern of weight gain. Recent studies in non-pregnant populations have identified common genetic variants associated with diabetes and obesity. Family and twin studies suggest that 50% of obesity is attributable to genetic causes17. In a recent multicenter study, subjects homozygous for the FTO rs9939609 A allele had a 1.6-fold increased risk of obesity18. Variants in MC4R19 and INSIG220, as well as multiple gene regions recently identified by the GIANT consortium21, are also associated with body mass index (per-allele effect 0.06–0.33 kg/m2). In addition, association studies have identified common genes associated with type 2 diabetes22–26, such as PPARG and TCF7L2.
Elucidating the role of genetic variants in gestational weight gain has important implications for public health. If genetic variants associated with diabetes and obesity are also linked with inappropriate weight gain, then excessive or inadequate gain may a marker for genetic predisposition to metabolic disease. No studies to our knowledge have measured the association between genetic variants associated with diabetes and obesity and gestational weight gain. We hypothesized that such genetic variants predict a mother’s total weight gain during pregnancy. We further hypothesized that a woman’s complement of diabetes and obesity risk alleles predicts whether she will gain in excess of IOM guidelines. Finally, we hypothesized that genotype modifies the association between pregravid BMI and gestational weight gain. To test these hypotheses, we measured such associations in a subset of women enrolled in the Pregnancy, Infection and Nutrition Study, 1998–2005, a longitudinal pregnancy cohort study.
Materials and Methods
The Pregnancy, Infection and Nutrition Cohort study comprises three prospective cohorts of more than 5000 women enrolled in early to mid-pregnancy. Participants enrolled in PIN1 and PIN2 were 24–29 weeks gestation at study entry, and were recruited from University of North Carolina Resident and Private Physician Obstetrics Clinic and the Wake County Department of Human Services and Wake Area Health Education Center prenatal care clinics from August 1995 through June 2000. Subjects enrolled in PIN3 were less than 20 weeks gestation at study entry and were recruited from the prenatal clinics at UNC hospitals from January 2001 to June 2005.
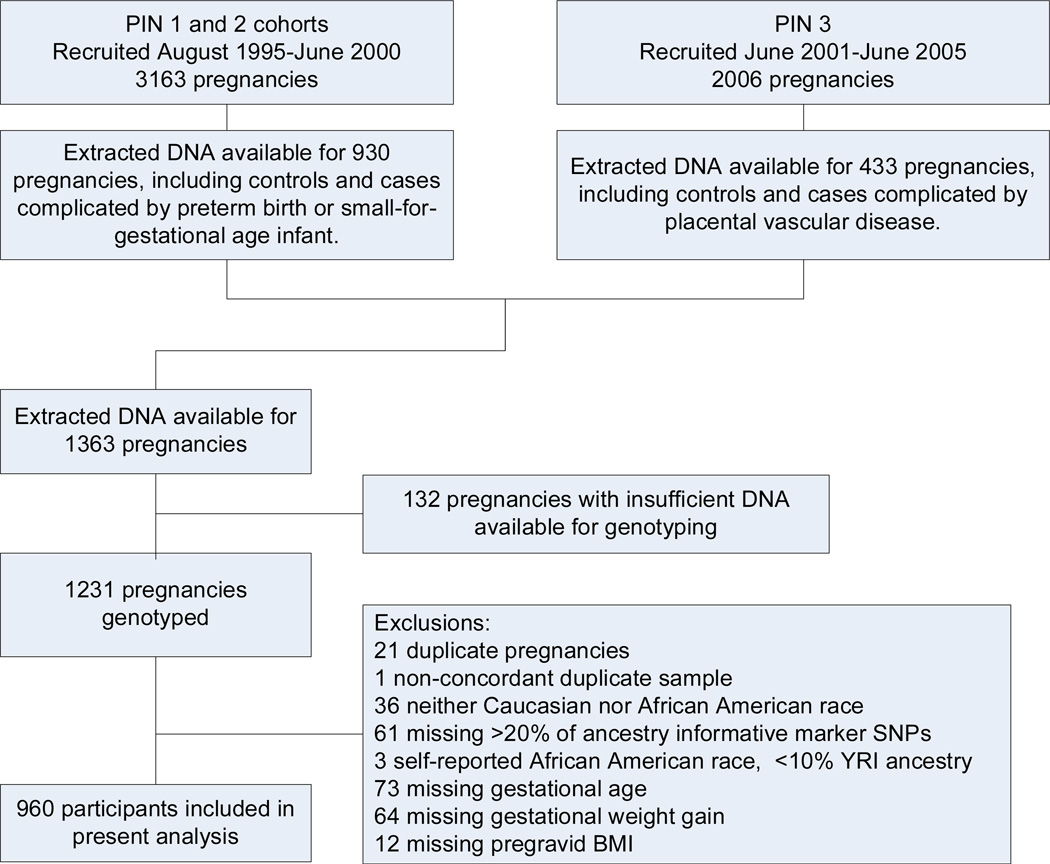
Extracted DNA was available for 1363 pregnancies that had undergone prior genotyping for case:control studies of preterm birth, small-for-gestational age birth weight, and placental vascular disease (Figure 1). For 132 pregnancies, there was insufficient DNA available for genotyping, leaving 1231 pregnancies eligible for our study. We allowed for only one pregnancy during the study period. If data were available for multiple pregnancies (N=21), we confirmed that the genotypes were concordant and included the pregnancy with the most complete SNP data. One pair of specimens was not concordant, so this subject was excluded. We further limited our analysis to self- identified black (n = 418) or white (n = 756) women, in order to avoid confounding by population stratification. As we considered ancestry, we removed women missing more than 20% of ancestral data (n = 61). We further removed black women for whom ancestry informative markers indicated a less than 10% probability of Yoruban ancestry (n = 3). Finally, we excluded women who were missing data on gestational age (n = 63), gestational weight gain (n = 64) or pre-gravid BMI (n =12, total removed = 149), leaving 960 women available for analysis (Figure 1).
Figure 1. Flow diagram.
Determination of pre-gravid BMI
Pre-gravid BMI was calculated based on self-reported pre-gravid weight and height at the first prenatal visit. Self-reported pre-gravid weights were examined for biological plausibility and imputed if deemed appropriate (<5% of weights were imputed). This imputed weight was calculated using the measured weight at the first prenatal visit (if taken prior to 15 weeks) minus the recommended amount of weight to be gained in the first and second trimesters as defined by the Institute of Medicine{Institute of Medicine, 1990 #1097}27.
Study covariates
The PIN datasets include information from telephone interviews, self-administered questionnaires, medical chart abstraction, and biological specimen collection. Information on race/ethnicity (non-Hispanic white, non-Hispanic black, and other) and maternal age was self-reported by the mother. Gestational age was estimated based on an algorithm that combined ultrasound dating with last menstrual period. If an ultrasound was done before 22 weeks gestation, it was used to date the pregnancy. If no ultrasound was done or it was done later in pregnancy, last menstrual period was used to date the pregnancy. In the PIN cohorts, 90.7% had an ultrasound that was used to date the pregnancy with the remaining 9.3% based on LMP.
Outcome assessment
Clinically obtained weights were recorded at each prenatal visit. We calculated gestational weight gain as the difference between pre-gravid self-reported weight and the last weight prior to delivery. We defined excessive or inadequate weight gain based on the 1990 Institute of Medicine (IOM) recommendations28; these were as follows: 28 to 40 lbs for low BMI (<19.8), 25–35 lbs for normal BMI (19.8 to 26.0), 15 to 25 lbs for overweight BMI (>26 to 29) and at least 15 pounds of gain for obese BMI (> 29). The IOM did not specify an upper limit for this group. For purposes of this analysis, excessive gain in the obese group was defined as greater than 18 lbs of gain, consistent with other analyses in the PIN cohorts. To calculate adequacy of gain for any given time point in pregnancy, the upper and lower limits of the weight gain intervals were extrapolated based on IOM-recommended rates of weight gain for the second and third trimesters29, consistent with earlier studies in this cohort30–32.
Genotyping
The Sequenom iPLEX platform33 was used to genotype 27 SNPs associated with obesity and diabetes19, 21–26, 34. For the purpose of quality control, 6 SNPs that had been previously assessed in the PIN cohorts were also genotyped. All SNPs were tested for Hardy-Weinberg equilibrium among self-identified white participants.
Population stratification
In genetic association studies, differences in allele frequency among ethnic groups can confound relationships between genotype and disease outcome. To address population stratification in this cohort, genotyping was performed for 37 ancestry-informative markers that have been successfully in other genetic association studies35. STRUCTURE was used to infer population substructure and assign individuals to populations using probabilistic clustering methods36. We analyzed self-identified white and black participants separately, and we included probability of Yoruban ancestry as a covariate among self-identified black women.
Statistical analysis
The DNA used for this study had been extracted for previous case:control studies of SGA, preterm birth and placental vascular disease, so prevalence of these outcomes was high. In order to produce estimates of the association between genotype and outcome that would approximate what we would have observed for the full study cohort, we calculated the probability of each participant’s inclusion in our study population. We used inverse probability weights to adjust our findings in all regression analyses. We used the SAS 9.2 surveylogistic and surveyreg for these analyses.
We used linear regression to model associations between maternal genotype and total gestational weight gain, adjusting for maternal age, linear and quadratic gestational age at birth, as well as probability of Yoruban ancestry among self-identified black women. We similarly used logistic regression to model associations between maternal genotype and probability of excessive gestational weight gain. We did not include in our models reproductive and obstetric factors that may be affected by genotype and may also affect weight gain, such as gestational diabetes and preeclampsia. These factors are potential intermediates on the causal pathway between genotype and weight gain, and including them in our models would attenuate the true association between maternal genotype and the outcome of interest. Moreover, because genotype may impact parity, we did not include parity as a covariate in our models.
We next considered models incorporating pre-gravid BMI in addition to gestational age and maternal age. Because the association between pre-gravid BMI and gestational weight gain is non-linear, both linear and quadratic terms were included. We then used hierarchical selection to model 1) Quadratic models including the joint effects of SNP allele carriage and interactions between SNP carriage and both log BMI and log BMI squared; 2) Linear models including the effects of SNP carriage and interactions between SNP carriage and log BMI; and 3) main effect models including only SNP allele carriage. If the Wald chi square p value for model 1 was < 0.05, the wald chi square p value for the quadratic interaction term was determined. If this was < 0.1, then the quadratic interaction term was retained. We similarly evaluated the linear model, retaining the interaction term if the SNP and SNP * log BMI p was < 0.05 and the SNP * log BMI term was < 0.1. Finally, we retained the main effects model if the p for the SNP term was < 0.05. To avoid false-positive findings due to small cell sizes, we excluded SNPs with fewer than 5 homozygous low or high-risk allele participants from these interaction models.
We did not to adjust alpha levels for multiple comparisons in this analysis. We recognize that this approach is may produce false positive associations. However, the purpose of our pilot study was to investigate the strength and direction of associations between these diabetes and obesity SNPs and gestational weight gain. All results should be viewed as exploratory findings pending confirmation in larger cohorts.
We next considered the simultaneous effects of multiple SNPs, using Bayesian models. For these analyses, we analyzed the data for subjects with complete information on genotypes using additive parameterization for SNPs. We used linear regression with gestational weight gain as the response variable, and SNP carriage, obesity and their interactions as covariates, adjusting for maternal age, linear and quadratic terms for gestational age at birth, and probability of Yoruban ancestry among self-identified black women. We avoided using a model with interactions between SNPs and log(BMI), log(BMI) squared as it would lead to a much larger model space and induce high correlations in the design matrix. The Bayesian variable selection framework allows each covariate to be either included or excluded from the model with a pre-assigned prior probability, which we chose as 0.5. After observing the data, the idea is to search over the list of all models, which includes models with no covariates, 1 covariate, …., all covariates, to identify the models which explain the observed data the best. With our choice of prior distributions, the prior odds ratio of including a covariate versus excluding is 1. We report the covariates with posterior (after observing the data) odds ratios greater than 3.
Results
Of the 79 SNPs genotyped, 71 genotyped for more than 90% of samples and were in Hardy-Weinberg equilibrium for the self-described White/Caucasian population. We anticipated that some SNPs would not be in HWE for African American participants due to admixture within African American populations in the US, and we found that one diabetes SNP and 4 of 37 ancestry-informative marker SNPs were not in HWE in this group. Our results were 99.5% concordant for SNPs that had previously been genotyped in the PIN cohort. Compared with white participants in our cohort, African American women were younger, had slightly higher pre-gravid BMIs, lower gestational weight gain and lower birth weight infants than Caucasian women (Table 1).
Table 1.
Characteristics of participants in the PIN3 study, n=960, mean (SD).
| Caucasian | African American | |
|---|---|---|
| N | 628 | 332 |
| Maternal age | 27.7 (6.2) | 23.9 (5.3) |
| Pregravid BMI, kg/m2 | 25.0 (6.6) | 27.4 (8.1) |
| GA at delivery, wks | 40.1 (0.9) | 39.9 (1.1) |
| Infant birth weight, g | 3258 (672) | 2939 (753) |
| Gestational weight gain, kg | 15.3 (6.2) | 13.4 (7.8) |
| Glucose loading test, mg/dL | 108.4 (25.9) | 105.6 (31.0) |
| Gestational diabetes, % (n) | 7.2 (45) | 5.7 (19) |
| Small-for-gestational age, % (n) | 12.3 (77) | 18.7 (62) |
| Birth<37 wks, % (n) | 21.2 (133) | 28.0 (93) |
| Excessive weight gain, % (n) | 65.5 (411) | 58.4 (194) |
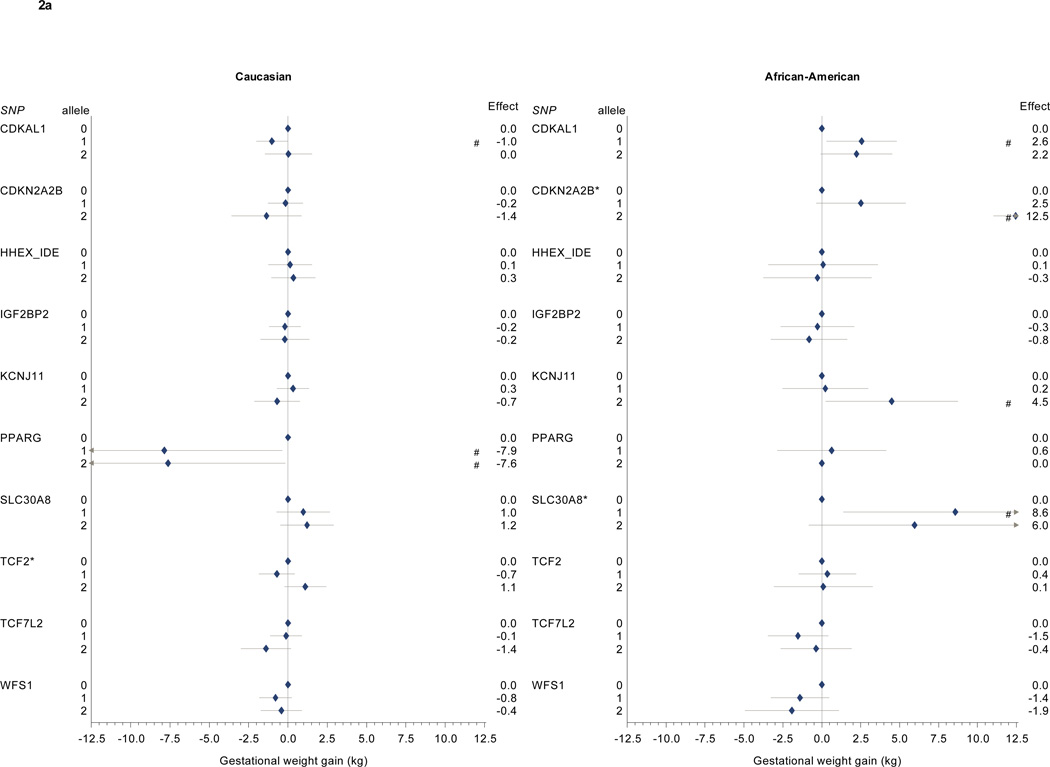
In linear regression analyses adjusted for maternal age and gestational age at birth and weighted to reflect the full PIN study population, we found associations between risk-allele carriage and gestational weight gain for several diabetes-associated variants (Table 2, Figure 1a and b). The KCNQ1 risk allele was associated with higher gestational weight gain (Caucasian 1 risk allele: 2.8 kg, 95% CI 0.4–5.1; 2 risk alleles: 2.9 kg, 95% CI 1.3 to 4.6; African American 1 risk allele: 3.4 kg, 95% CI 0.6 to 6.3; 2 risk alleles: 2.7 kg, 95% CI 0.5 to 4.8). Among Caucasian participants, PPARG risk allele carriage was associated with lower gestational weight gain (1 risk allele: −7.9 kg, 95% CI −15.4 to −0.4; 2 risk alleles: −7.6 kg, 95% CI −15.1 to −0.2). No African American participants were homozygous for the low-risk PPARG variant.
Table 2.
SNP frequencies and total gestational weight gain, kg, for Caucasian and African American participants in the PIN studies, n=960.
| Gestational Weight Gain, kg Caucasian |
African-American | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene / SNP | Effect allele / other allele |
Risk alleles |
N* | Mean (SD) |
Adjusted† effect estimate (95% CI) |
N* | Mean (SD) |
Adjusted† effect estimate (95% CI) |
| N | 628 | 332 | ||||||
| PPARG | C/G | 0 | 7 | 21.7 (9.5) | 0.00 (ref) | 0 | - | - |
| rs1801282 | 1 | 118 | 14.8 (5.9) | −7.9 (−15.4, −0.4) | 20 | 13.4 (8.5) | 0.6 (−2.9, 4.1) | |
| 2 | 500 | 15.4 (6.2) | −7.6 (−15.1, −0.2) | 309 | 13.4 (7.8) | 0.00 (ref) | ||
| KCNJ11 | C/T | 0 | 243 | 15.5 (6.3) | 0.00 (ref) | 295 | 13.4 (7.8) | 0.00 (ref) |
| rs5215 | 1 | 291 | 15.5 (6.1) | 0.3 (−0.7, 1.3) | 34 | 13.7 (7.8) | 0.2 (−2.5, 3.0) | |
| 2 | 94 | 14.3 (6.6) | −0.7 (−2.1, 0.8) | 3 | 17.6 (4.1) | 4.5 (0.2, 8.7) | ||
| TCF7L2 | T/C | 0 | 285 | 15.5 (6.1) | 0.00 (ref) | 109 | 14.0 (7.4) | 0.00 (ref) |
| rs7901695 | 1 | 277 | 15.4 (6.4) | −0.1 (−1.1, 0.9) | 152 | 12.8 (8.1) | −1.5 (−3.5, 0.4) | |
| 2 | 65 | 14.2 (6.0) | −1.4 (−3.0, 0.2) | 71 | 13.9 (7.7) | −0.4 (−2.6, 1.9) | ||
| TCF2 | A/G | 0 | 153 | 15.0 (5.9) | 0.00 (ref) | 145 | 13.1 (7.1) | 0.00 (ref) |
| rs4430796 | 1 | 294 | 14.9 (6.2) | −0.7 (−1.8, 0.4) | 150 | 13.5 (8.3) | 0.4 (−1.5, 2.2) | |
| 2 | 179 | 16.3 (6.5) | 1.1 (−0.2, 2.4) | 37 | 14.7 (8.0) | 0.1 (−3.1, 3.3) | ||
| WFS1 | A/G | 0 | 236 | 15.7 (6.0) | 0.00 (ref) | 136 | 14.2 (7.7) | 0.00 (ref) |
| rs10010131 | 1 | 286 | 15.1 (6.5) | −0.8 (−1.8, 0.2) | 158 | 12.9 (7.3) | −1.4 (−3.3, 0.5) | |
| 2 | 105 | 14.9 (6.1) | −0.4 (−1.7, 0.9) | 38 | 12.8 (9.7) | −1.9 (−4.9, 1.1) | ||
| HHEX_IDE | C/T | 0 | 108 | 15.3 (6.6) | 0.00 (ref) | 17 | 13.3 (6.1) | 0.00 (ref) |
| rsllll875 | 1 | 297 | 15.2 (6.6) | 0.1 (−1.3, 1.5) | 127 | 14.2 (7.9) | 0.1 (−3.4, 3.6) | |
| 2 | 221 | 15.5 (5.5) | 0.3 (−1.0, 1.7) | 188 | 12.9 (7.8) | −0.3 (−3.7, 3.2) | ||
| SLC30A8 | C/T | 0 | 55 | 14.3 (7.2) | 0.00 (ref) | 3 | 12.3 (8.0) | 0.00 (ref) |
| rs13266634 | 1 | 273 | 15.4 (6.2) | 1.0 (−0.7, 2.7) | 61 | 15.0 (8.5) | 8.6 (1.4, 15.8) | |
| 2 | 300 | 15.4 (6.1) | 1.2 (−0.5, 2.9) | 268 | 13.1 (7.6) | 6.0 (−0.8, 12.8) | ||
| CDKAL1 | C/A | 0 | 285 | 15.7 (6.7) | 0.00 (ref) | 45 | 12.0 (6.6) | 0.00 (ref) |
| rs10946398 | 1 | 289 | 14.9 (5.9) | −1.0 (−2.0, 0.0) | 162 | 13.9 (8.5) | 2.6 (0.3, 4.8) | |
| 2 | 52 | 15.6 (5.3) | 0.0 (−1.5, 1.5) | 121 | 13.4 (7.2) | 2.2 (−0.1, 4.5) | ||
| CDKN2A2B | C/T | 0 | 425 | 15.5 (6.2) | 0.00 (ref) | 287 | 13.1 (7.7) | 0.00 (ref) |
| rs10811661 | C/T | 1 | 172 | 15.0 (6.3) | −0.2 (−1.3, 1.0) | 42 | 15.1 (7.8) | 2.5 (−0.4, 5.4) |
| 2 | 21 | 13.6 (5.9) | −1.4 (−3.6, 0.8) | 1 | 26.8 (.) | 12.5 (11.0, 13.9) | ||
| IGF2BP2 | T/G | 0 | 276 | 15.6 (6.2) | 0.00 (ref) | 68 | 14.3 (7.0) | 0.00 (ref) |
| rs4402960 | 1 | 275 | 15.2 (6.3) | −0.2 (−1.2, 0.8) | 176 | 13.2 (8.3) | −0.3 (−2.6, 2.1) | |
| 2 | 74 | 14.8 (6.3) | −0.2 (−1.8, 1.4) | 81 | 13.3 (7.4) | −0.8 (−3.3, 1.6) | ||
| JAZF1 | T/C | 0 | 152 | 14.8 (6.3) | 0.00 (ref) | 18 | 12.9 (8.0) | 0.00 (ref) |
| rs864745 | 1 | 303 | 15.4 (6.0) | 0.2 (−1.0, 1.4) | 126 | 13.7 (7.7) | 1.6 (−1.9, 5.1) | |
| 2 | 169 | 15.7 (6.5) | 0.5 (−0.9, 1.9) | 188 | 13.3 (7.8) | 1.2 (−2.2, 4.6) | ||
| CDC123 | G/A | 0 | 424 | 15.3 (6.3) | 0.00 (ref) | 242 | 12.9 (7.8) | 0.00 (ref) |
| rs12779790 | 1 | 176 | 15.5 (5.9) | 0.4 (−0.7, 1.4) | 82 | 14.8 (7.9) | 2.0 (−0.1, 4.1) | |
| 2 | 18 | 15.2 (6.8) | −0.4 (−3.2, 2.5) | 4 | 15.0 (7.1) | 3.0 (0.4, 5.6) | ||
| TSPAN8 | C/T | 0 | 323 | 15.3 (6.0) | 0.00 (ref) | 220 | 13.5 (7.6) | 0.00 (ref) |
| rs7961581 | 1 | 239 | 15.6 (6.6) | 0.1 (−0.9, 1.2) | 97 | 13.5 (8.0) | −1.3 (−3.2, 0.7) | |
| 2 | 56 | 13.5 (6.2) | −2.0 (−3.6, −0.4) | 12 | 10.3 (9.1) | −1.6 (−6.2, 3.1) | ||
| THADA | T/C | 0 | 6 | 15.5 (2.5) | 0.00 (ref) | 24 | 15.5 (6.3) | 0.00 (ref) |
| rs7578597 | 1 | 118 | 15.7 (6.8) | 1.7 (−1.2, 4.5) | 109 | 13.1 (7.9) | −3.3 (−5.9, −0.7) | |
| 2 | 496 | 15.2 (6.1) | 0.7 (−2.0, 3.4) | 197 | 13.4 (7.8) | −2.5 (−5.0, −0.1) | ||
| ADAMTS9 | C/T | 0 | 42 | 13.0 (7.1) | 0.00 (ref) | 28 | 12.6 (5.2) | 0.00 (ref) |
| rs4607103 | 1 | 247 | 15.8 (5.9) | 1.3 (−0.8, 3.4) | 139 | 14.0 (9.2) | 1.2 (−1.3, 3.8) | |
| 2 | 339 | 15.3 (6.3) | 1.2 (−0.9, 3.3) | 165 | 13.1 (6.8) | −0.6 (−2.9, 1.8) | ||
| NOTCH2 | T/G | 0 | 506 | 15.3 (6.2) | 0.00 (ref) | 153 | 13.8 (8.1) | 0.00 (ref) |
| rs10923931 | 1 | 115 | 15.4 (6.5) | 0.3 (−0.8, 1.5) | 140 | 13.2 (7.5) | −0.9 (−2.8, 1.0) | |
| 2 | 4 | 16.0 (3.5) | −0.6 (−2.9, 1.7) | 39 | 12.8 (7.8) | −1.1 (−4.3, 2.0) | ||
| KCNQ1 | C/T | 0 | 4 | 13.4 (1.6) | 0.00 (ref) | 2 | 5.9 (1.3) | 0.00 (ref) |
| rs2237892 | 1 | 61 | 14.6 (6.6) | 2.8 (0.4, 5.1) | 51 | 14.1 (7.6) | 3.4 (0.6, 6.3) | |
| 2 | 542 | 15.4 (6.2) | 2.9 (1.3, 4.6) | 273 | 13.3 (7.7) | 2.7 (0.5, 4.8) | ||
| G6PC2 | A/G | 0 | 309 | 15.3 (6.1) | 0.00 (ref) | 286 | 13.2 (8.0) | 0.00 (ref) |
| rs560887 | 1 | 262 | 15.1 (6.5) | 0.4 (−0.6, 1.4) | 42 | 14.7 (6.5) | 2.1 (−0.1, 4.2) | |
| 2 | 52 | 15.9 (5.9) | 1.3 (−0.2, 2.9) | 0 | - | - | ||
| INSIG2 | C/G | 0 | 301 | 15.2 (6.6) | 0.00 (ref) | 183 | 12.8 (7.5) | 0.00 (ref) |
| rs7566605 | 1 | 255 | 15.5 (6.1) | 0.2 (−0.8, 1.2) | 126 | 14.9 (8.0) | 2.0 (0.2, 3.9) | |
| 2 | 72 | 15.4 (5.2) | 0.2 (−1.1, 1.6) | 22 | 10.5 (7.7) | −2.5 (−6.0, 1.1) | ||
| FTO | A/T | 0 | 246 | 15.5 (6.8) | 0.00 (ref) | 83 | 12.9 (8.5) | 0.00 (ref) |
| rs9939609 | 1 | 270 | 14.9 (5.7) | −0.4 (−1.5, 0.6) | 158 | 13.4 (8.0) | 0.5 (−1.7, 2.7) | |
| 2 | 94 | 15.9 (6.4) | 0.4 (−1.1, 1.9) | 81 | 13.8 (7.0) | 0.9 (−1.4, 3.3) | ||
| MC4R | C/T | 0 | 369 | 15.7 (6.1) | 0.00 (ref) | 162 | 12.5 (7.0) | 0.00 (ref) |
| rs17782313 | 1 | 219 | 14.9 (6.5) | −0.7 (−1.7, 0.3) | 135 | 14.2 (8.4) | 1.1 (−0.8, 2.9) | |
| 2 | 34 | 13.9 (6.6) | −1.6 (−3.6, 0.4) | 33 | 14.8 (8.7) | 3.8 (0.0, 7.7) | ||
| TMEM18 | C/T | 0 | 19 | 14.4 (3.4) | 0.00 (ref) | 7 | 13.9 (6.1) | 0.00 (ref) |
| rs6548238 | 1 | 167 | 15.2 (7.1) | 1.1 (−0.8, 2.9) | 58 | 15.1 (8.0) | 2.9 (−2.4, 8.2) | |
| 2 | 441 | 15.4 (6.0) | 1.4 (−0.3, 3.0) | 266 | 13.0 (7.7) | 0.7 (−4.4, 5.8) | ||
| GNPDA2 | G/A | 0 | 195 | 15.5 (6.6) | 0.00 (ref) | 185 | 13.0 (8.2) | 0.00 (ref) |
| rs10938397 | 1 | 308 | 15.5 (5.7) | 0.2 (−0.9, 1.3) | 131 | 14.1 (7.4) | 0.9 (−1.0, 2.7) | |
| 2 | 114 | 14.8 (6.4) | 0.0 (−1.4, 1.5) | 15 | 12.8 (5.9) | −1.0 (−4.1, 2.1) | ||
| SH2B1 | G/A | 0 | 249 | 15.5 (5.9) | 0.00 (ref) | 178 | 14.1 (7.8) | 0.00 (ref) |
| rs7498665 | 1 | 280 | 15.2 (6.3) | −0.3 (−1.3, 0.8) | 121 | 12.9 (7.3) | −0.5 (−2.4, 1.4) | |
| 2 | 98 | 15.4 (6.7) | −0.1 (−1.6, 1.3) | 33 | 11.9 (9.2) | −2.3 (−5.5, 0.9) | ||
| MTCH2 | G/A | 0 | 257 | 15.8 (6.3) | 0.00 (ref) | 267 | 13.1 (7.8) | 0.00 (ref) |
| rs10838738 | 1 | 289 | 14.9 (6.1) | −0.9 (−1.9, 0.2) | 60 | 14.6 (7.4) | 1.9 (0.0, 3.9) | |
| 2 | 82 | 15.1 (6.4) | −0.3 (−1.9, 1.3) | 5 | 15.1 (9.1) | 0.4 (−6.6, 7.3) | ||
| KCTD15 | G/A | 0 | 73 | 15.5 (6.1) | 0.00 (ref) | 50 | 11.3 (9.1) | 0.00 (ref) |
| rs11084753 | 1 | 288 | 15.2 (6.6) | −0.1 (−1.6, 1.5) | 155 | 13.7 (7.6) | 1.1 (−1.8, 3.9) | |
| 2 | 262 | 15.3 (5.8) | 0.0 (−1.5, 1.6) | 126 | 13.9 (7.4) | 1.3 (−1.6, 4.2) | ||
| NEGR1 | T/C | 0 | 97 | 16.1 (6.9) | 0.00 (ref) | 87 | 13.3 (6.6) | 0.00 (ref) |
| rs2815752 | 1 | 285 | 15.1 (6.0) | −0.7 (−2.2, 0.7) | 169 | 13.7 (8.0) | 0.9 (−1.0, 2.8) | |
| 2 | 244 | 15.3 (6.2) | −0.4 (−1.9, 1.1) | 76 | 13.0 (8.6) | 0.9 (−1.7, 3.4) | ||
Frequencies sum to less than 628 or 332 because participants were excluded from an analysis if their samples did not genotype successfully.
Model covariates include log pregravid BMI, log pregravid BMI squared, gestational age at birth, gestational age at birth squared, and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
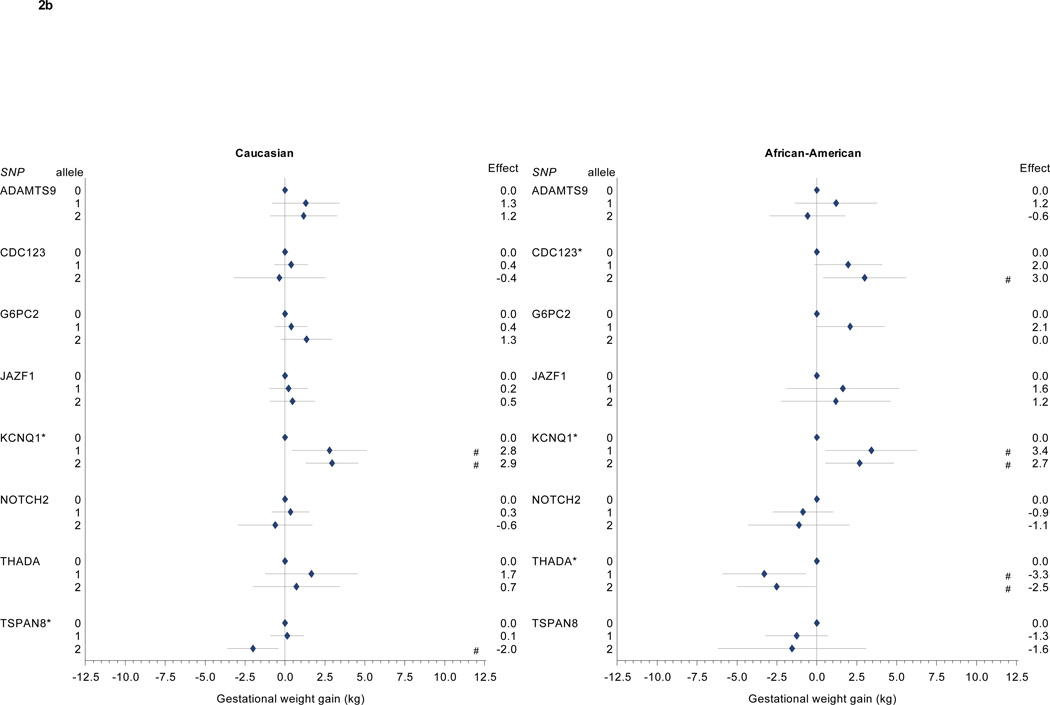
We found different patterns of association for Caucasian and African American participants for several other diabetes-associated SNPs (Table 2, Figure 2a and 2b). Among Caucasian participants, we further identified associations between gestational weight gain and CKDAL1 as well as TSPAN8. Among African American participants, we found associations between gestational weight gain and CDKAL1, CDKN2A2B, KCNJ11, SLC30A8, CDC123, and THADA.
Figure 2. a and 2b: Diabetes-associated SNPs and gestational weight gain.
Multivariate-adjusted§ effect estimate (95% CI) for change in gestational weight gain associated with risk allele carriage for single nucleotide polymorphisms associated with diabetes in non-pregnant populations. Caucasian participants (n=628) on left, African-American participants (n=332) on right.
* p < 0.05 for partial F test for this SNP. # p < 0.05 for comparison with 0 risk alleles (referent).
§ Model covariates include log pregravid BMI, log pregravid BMI squared, gestational age at birth, gestational age at birth squared, and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
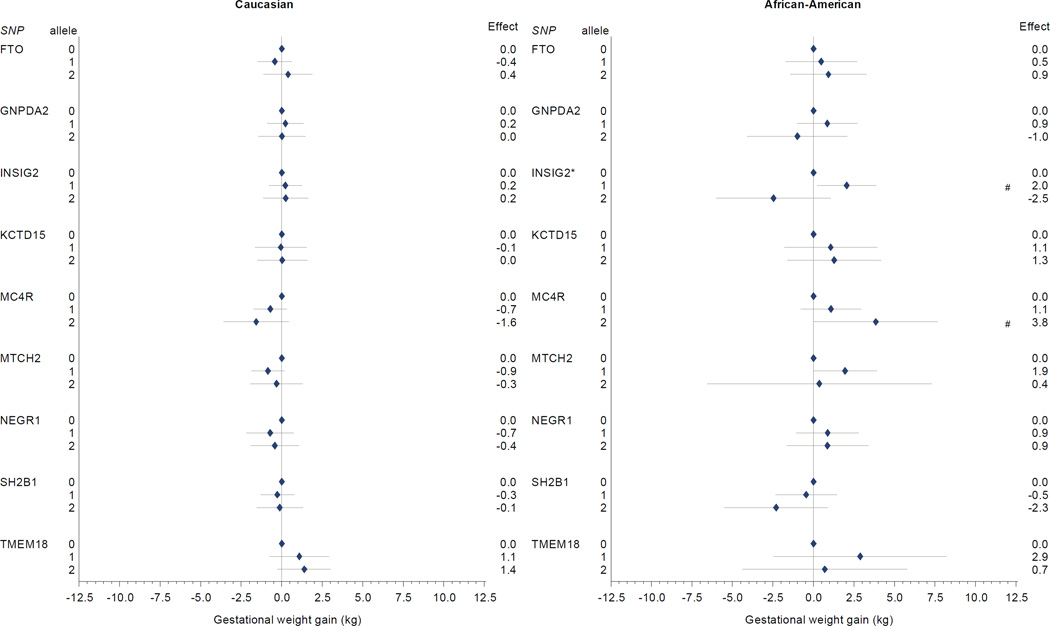
In our adjusted models for obesity-related risk variants (Table 2, Figure 3), we found higher gestational weight gain for African American participants with 2 MC4R risk alleles (3.8 kg, 95% CI 0.01 to 2.9), compared with women with no MC4R risk alleles. We found no other consistent patterns of association between obesity risk variants and gestational weight gain among African-American women, although individuals with one risk allele for INSIG2 had higher weight gain than women with 0 or 2 risk alleles. In contrast to our findings for MC4R among African American women, among Caucasian women, MC4R carriage was inversely associated with weight gain (1 risk allele: −0.7 kg, 95% CI −1.7 to 0.3; 2 risk alleles: −1.6 kg, 95% CI −3.6 to 0.4), although confidence intervals were wide.
Figure 3. Obesity-associated SNPs and gestational weight gain.
Multivariate-adjusted§ effect estimate (95% CI) for change in gestational weight gain associated with risk allele carriage for single nucleotide polymorphisms associated with obesity in non-pregnant populations. Caucasian participants (n=628) on left, African-American participants (n=332) on right.
* p < 0.05 for comparison with 0 risk alleles (referent).
§ Model covariates include log pregravid BMI, log pregravid BMI squared, gestational age at birth, gestational age at birth squared, and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
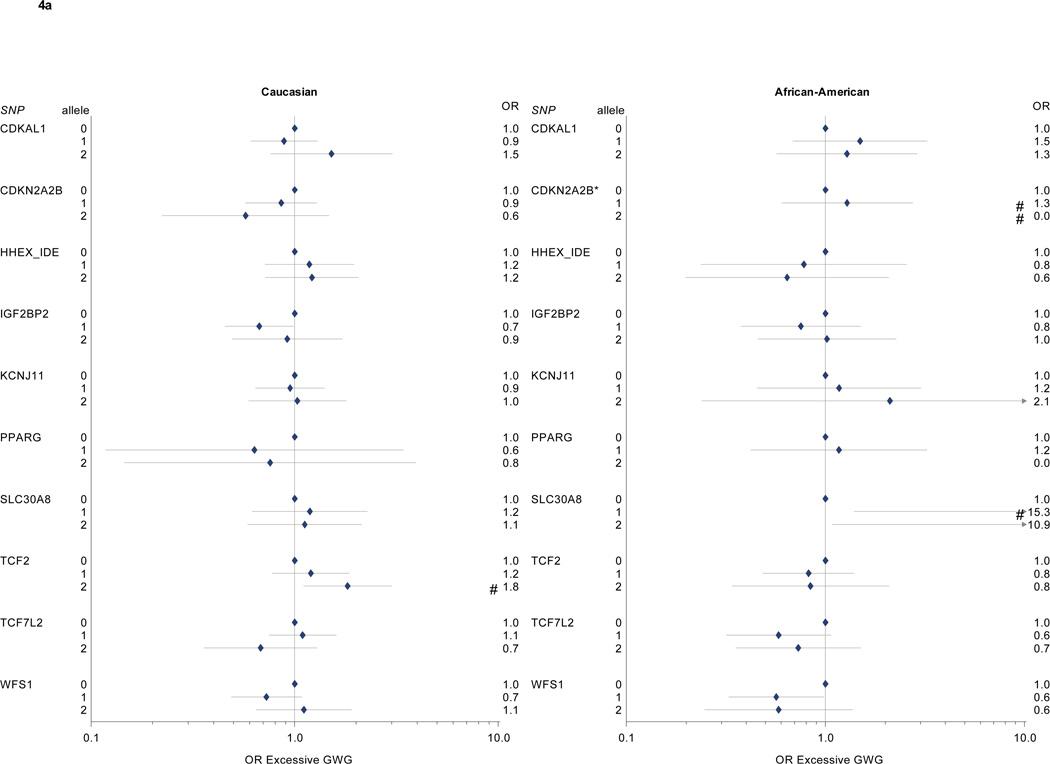
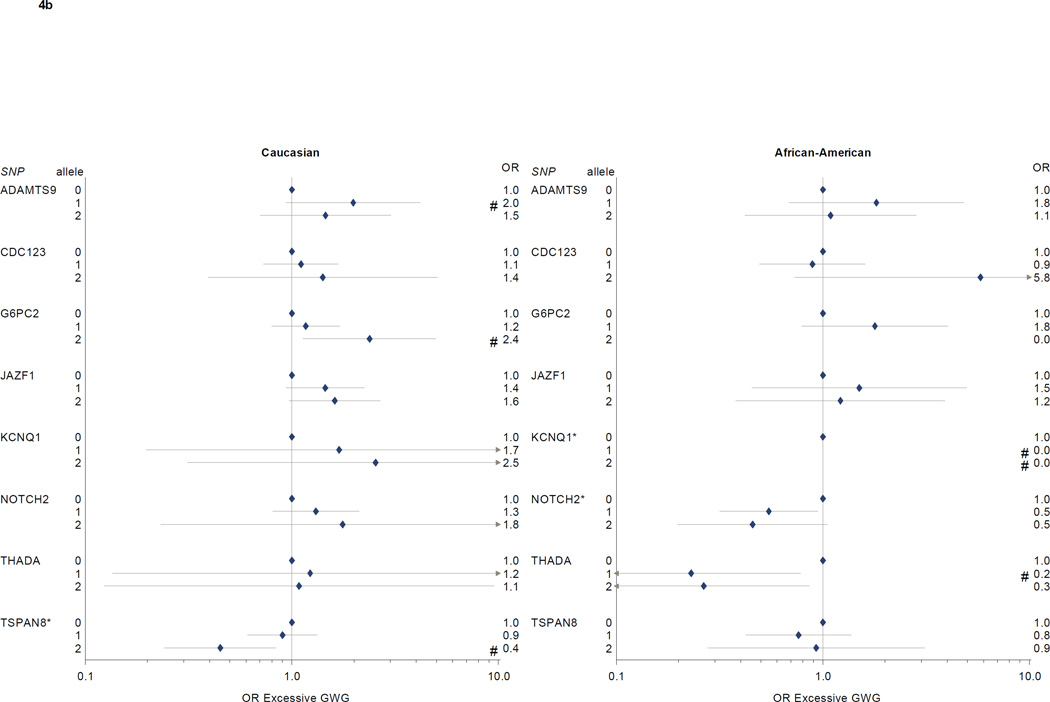
In logistic regression models of associations between diabetes SNPs and excessive gestational weight gain (Table 3, Figure 4a and 4b), we found higher risks for excessive gain among Caucasian participants with 2 copies of the TCF2 risk allele (OR 1.8, 95% CI 1.1–3.0, vs. 0 copies of the risk allele) or 2 copies of the G6PC2 risk allele (OR 2.4, 95% CI 1.1–5.0), and lower risks among participants with 2 copies of the TSPAN8 risk allele (OR 0.4, 95% CI 0.2–0.8, vs. 0 copies). Among African American participants, NOTCH2 risk allele carriage was associated with reduced risk for excess gain (OR for 1 risk allele 0.5, 95% CI 0.3–0.9; OR for 2 risk alleles, 0.5, 95% CI 0.2–1.1, vs 0 risk alleles). THADA risk allele carriage was also associated with less excessive gain (OR for 1 risk allele 0.2, 95% CI 0.1–0.8, and for 2 risk alleles, 0.3, 95% CI 0.1–0.9, vs. 0 risk alleles), but confidence intervals were wide. SLC30A8 risk allele carriage was associated with markedly increased excessive weight gain, but there were only 3 participants with 0 risk alleles in our study population, leading to imprecise effect estimates.
Table 3.
Risk allele carriage and odds of excessive gestational weight gain* for Caucasian and African American participants in the PIN studies, n=960.
| Caucasian | African-American | |||||||
|---|---|---|---|---|---|---|---|---|
| Effect allele / other allele |
Risk alleles |
Excessive / Total† |
Excessive weight gain, % |
Adjusted‡ OR excessive gain (95% CI) |
Excessive / Total† |
Excessive weight gain, % |
Adjusted‡ OR excessive gain (95% CI) |
|
| 411/628 | 65.5 | 194/332 | 58.4 | |||||
| PPARG | C/G | 0 | 5/7 | 71.4 | 1.0 (ref) | ./. | . | |
| rs1801282 | 1 | 72/118 | 61.0 | 0.6 (0.1, 3.4) | 12/20 | 60.0 | 1.0 (ref) | |
| 2 | 332/500 | 66.4 | 0.8 (0.1, 3.9) | 180/309 | 58.3 | 1.2 (0.4, 3.2) | ||
| KCNJ11 | C/T | 0 | 160/243 | 65.8 | 1.0 (ref) | 171/295 | 58.0 | 1.0 (ref) |
| rs5215 | 1 | 188/291 | 64.6 | 0.9 (0.6, 1.4) | 21/34 | 61.8 | 1.2 (0.5, 3.0) | |
| 2 | 63/94 | 67.0 | 1.0 (0.6, 1.8) | 2/3 | 66.7 | 2.1 (0.2, 18.6) | ||
| TCF7L2 | T/C | 0 | 188/285 | 66.0 | 1.0 (ref) | 68/109 | 62.4 | 1.0 (ref) |
| rs7901695 | 1 | 184/277 | 66.4 | 1.1 (0.7, 1.6) | 83/152 | 54.6 | 0.6 (0.3, 1.1) | |
| 2 | 38/65 | 58.5 | 0.7 (0.4, 1.3) | 43/71 | 60.6 | 0.7 (0.4, 1.5) | ||
| TCF2 | A/G | 0 | 92/153 | 60.1 | 1.0 (ref) | 85/145 | 58.6 | 1.0 (ref) |
| rs4430796 | 1 | 190/294 | 64.6 | 1.2 (0.8, 1.9) | 87/150 | 58.0 | 0.8 (0.5, 1.4) | |
| 2 | 129/179 | 72.1 | 1.8 (1.1, 3.0) | 22/37 | 59.5 | 0.8 (0.3, 2.1) | ||
| WFS1 | A/G | 0 | 159/236 | 67.4 | 1.0 (ref) | 87/136 | 64.0 | 1.0 (ref) |
| rs10010131 | 1 | 179/286 | 62.6 | 0.7 (0.5, 1.1) | 87/158 | 55.1 | 0.6 (0.3, 1.0) | |
| 2 | 73/105 | 69.5 | 1.1 (0.6, 1.9) | 20/38 | 52.6 | 0.6 (0.2, 1.4) | ||
| HHEX_IDE | C/T | 0 | 67/108 | 62.0 | 1.0 (ref) | 12/17 | 70.6 | 1.0 (ref) |
| rsllll875 | 1 | 195/297 | 65.7 | 1.2 (0.7, 2.0) | 79/127 | 62.2 | 0.8 (0.2, 2.6) | |
| 2 | 149/221 | 67.4 | 1.2 (0.7, 2.1) | 103/188 | 54.8 | 0.6 (0.2, 2.1) | ||
| SLC30A8 | C/T | 0 | 37/55 | 67.3 | 1.0 (ref) | 1/3 | 33.3 | 1.0 (ref) |
| rs13266634 | 1 | 182/273 | 66.7 | 1.2 (0.6, 2.3) | 41/61 | 67.2 | 15.3 (1.4–167.5) | |
| 2 | 192/300 | 64.0 | 1.1 (0.6, 2.1) | 152/268 | 56.7 | 10.9 (1.1–110.8) | ||
| CDKAL1 | C/A | 0 | 185/285 | 64.9 | 1.0 (ref) | 25/45 | 55.6 | 1.0 (ref) |
| rs10946398 | 1 | 188/289 | 65.1 | 0.9 (0.6, 1.3) | 100/162 | 61.7 | 1.5 (0.7, 3.3) | |
| 2 | 38/52 | 73.1 | 1.5 (0.8, 3.0) | 67/121 | 55.4 | 1.3 (0.6, 2.9) | ||
| CDKN2A2B | C/T | 0 | 284/425 | 66.8 | 1.0 (ref) | 165/287 | 57.5 | 1.0 (ref) |
| rs10811661 | 1 | 109/172 | 63.4 | 0.9 (0.6, 1.3) | 27/42 | 64.3 | 1.3 (0.6, 2.7) | |
| 2 | 11/21 | 52.4 | 0.6 (0.2, 1.5) | 1/1 | 100.0 | - | ||
| IGF2BP2 | T/G | 0 | 190/276 | 68.8 | 1.0 (ref) | 43/68 | 63.2 | 1.0 (ref) |
| rs4402960 | 1 | 169/275 | 61.5 | 0.7 (0.5, 1.0) | 96/176 | 54.5 | 0.8 (0.4, 1.5) | |
| 2 | 51/74 | 68.9 | 0.9 (0.5, 1.7) | 52/81 | 64.2 | 1.0 (0.5, 2.3) | ||
| JAZF1 | T/C | 0 | 91/152 | 59.9 | 1.0 (ref) | 10/18 | 55.6 | 1.0 (ref) |
| rs864745 | 1 | 202/303 | 66.7 | 1.4 (0.9, 2.2) | 76/126 | 60.3 | 1.5 (0.5, 5.0) | |
| 2 | 115/169 | 68.0 | 1.6 (1.0, 2.7) | 108/188 | 57.4 | 1.2 (0.4, 3.9) | ||
| CDC123 | G/A | 0 | 275/424 | 64.9 | 1.0 (ref) | 142/242 | 58.7 | 1.0 (ref) |
| rs12779790 | 1 | 119/176 | 67.6 | 1.1 (0.7, 1.7) | 47/82 | 57.3 | 0.9 (0.5, 1.6) | |
| 2 | 13/18 | 72.2 | 1.4 (0.4, 5.1) | 3/4 | 75.0 | 5.8 (0.7, 45.9) | ||
| TSPAN8 | C/T | 0 | 217/323 | 67.2 | 1.0 (ref) | 128/220 | 58.2 | 1.0 (ref) |
| rs7961581 | 1 | 154/239 | 64.4 | 0.9 (0.6, 1.3) | 57/97 | 58.8 | 0.8 (0.4, 1.4) | |
| 2 | 32/56 | 57.1 | 0.4 (0.2, 0.8) | 6/12 | 50.0 | 0.9 (0.3, 3.1) | ||
| THADA | T/C | 0 | 4/6 | 66.7 | 1.0 (ref) | 18/24 | 75.0 | 1.0 (ref) |
| rs7578597 | 1 | 78/118 | 66.1 | 1.2 (0.1, 11.1) | 63/109 | 57.8 | 0.2 (0.1, 0.8) | |
| 2 | 322/496 | 64.9 | 1.1 (0.1, 9.5) | 112/197 | 56.9 | 0.3 (0.1, 0.9) | ||
| ADAMTS9 | C/T | 0 | 23/42 | 54.8 | 1.0 (ref) | 14/28 | 50.0 | 1.0 (ref) |
| rs4607103 | 1 | 174/247 | 70.4 | 2.0 (0.9, 4.2) | 89/139 | 64.0 | 1.8 (0.7, 4.8) | |
| 2 | 214/339 | 63.1 | 1.5 (0.7, 3.0) | 91/165 | 55.2 | 1.1 (0.4, 2.8) | ||
| NOTCH2 | T/G | 0 | 326/506 | 64.4 | 1.0 (ref) | 101/153 | 66.0 | 1.0 (ref) |
| rs10923931 | 1 | 79/115 | 68.7 | 1.3 (0.8, 2.1) | 74/140 | 52.9 | 0.5 (0.3, 0.9) | |
| 2 | 3/4 | 75.0 | 1.8 (0.2, 13.4) | 19/39 | 48.7 | 0.5 (0.2, 1.1) | ||
| KCNQ1 | C/T | 0 | 2/4 | 50.0 | 1.0 (ref) | 2/2 | 100.0 | 1.0 (ref) |
| rs2237892 | 1 | 37/61 | 60.7 | 1.7 (0.2, 14.5) | 34/51 | 66.7 | - | |
| 2 | 357/542 | 65.9 | 2.5 (0.3, 20.6) | 154/273 | 56.4 | - | ||
| G6PC2 | A/G | 0 | 197/309 | 63.8 | 1.0 (ref) | 164/286 | 57.3 | 1.0 (ref) |
| rs560887 | 1 | 174/262 | 66.4 | 1.2 (0.8, 1.7) | 28/42 | 66.7 | 1.8 (0.8, 4.0) | |
| 2 | 38/52 | 73.1 | 2.4 (1.1, 5.0) | ./. | . | |||
| INSIG2 | C/G | 0 | 200/301 | 66.4 | 1.0 (ref) | 101/183 | 55.2 | 1.0 (ref) |
| rs7566605 | 1 | 160/255 | 62.7 | 0.9 (0.6, 1.3) | 82/126 | 65.1 | 1.6 (0.9, 2.7) | |
| 2 | 51/72 | 70.8 | 1.3 (0.7, 2.3) | 11/22 | 50.0 | 0.6 (0.2, 1.7) | ||
| FTO | A/T | 0 | 162/246 | 65.9 | 1.0 (ref) | 43/83 | 51.8 | 1.0 (ref) |
| rs9939609 | 1 | 172/270 | 63.7 | 1.1 (0.7, 1.6) | 93/158 | 58.9 | 1.2 (0.6, 2.1) | |
| 2 | 65/94 | 69.1 | 1.2 (0.7, 2.2) | 50/81 | 61.7 | 1.4 (0.7, 2.8) | ||
| MC4R | C/T | 0 | 251/369 | 68.0 | 1.0 (ref) | 88/162 | 54.3 | 1.0 (ref) |
| rs17782313 | 1 | 137/219 | 62.6 | 0.7 (0.5, 1.1) | 84/135 | 62.2 | 1.2 (0.7, 2.1) | |
| 2 | 21/34 | 61.8 | 0.9 (0.4, 2.0) | 21/33 | 63.6 | 2.2 (0.9, 5.5) | ||
| TMEM18 | C/T | 0 | 13/19 | 68.4 | 1.0 (ref) | 5/7 | 71.4 | 1.0 (ref) |
| rs6548238 | 1 | 107/167 | 64.1 | 0.8 (0.3, 2.4) | 36/58 | 62.1 | 1.1 (0.2, 8.3) | |
| 2 | 290/441 | 65.8 | 0.8 (0.3, 2.4) | 152/266 | 57.1 | 0.7 (0.1, 4.9) | ||
| GNPDA2 | G/A | 0 | 127/195 | 65.1 | 1.0 (ref) | 103/185 | 55.7 | 1.0 (ref) |
| rs10938397 | 1 | 200/308 | 64.9 | 1.0 (0.7, 1.5) | 82/131 | 62.6 | 1.4 (0.8, 2.3) | |
| 2 | 77/114 | 67.5 | 1.0 (0.6, 1.8) | 8/15 | 53.3 | 0.9 (0.3, 2.8) | ||
| SH2B1 | G/A | 0 | 160/249 | 64.3 | 1.0 (ref) | 100/178 | 56.2 | 1.0 (ref) |
| rs7498665 | 1 | 180/280 | 64.3 | 1.0 (0.7, 1.5) | 74/121 | 61.2 | 1.2 (0.7, 2.2) | |
| 2 | 70/98 | 71.4 | 1.4 (0.8, 2.4) | 20/33 | 60.6 | 1.1 (0.5, 2.7) | ||
| MTCH2 | G/A | 0 | 174/257 | 67.7 | 1.0 (ref) | 148/267 | 55.4 | 1.0 (ref) |
| rs10838738 | 1 | 182/289 | 63.0 | 0.9 (0.6, 1.3) | 43/60 | 71.7 | 3.1 (1.5, 6.5) | |
| 2 | 55/82 | 67.1 | 1.1 (0.6, 2.1) | 3/5 | 60.0 | 0.7 (0.1, 5.7) | ||
| KCTD15 | G/A | 0 | 53/73 | 72.6 | 1.0 (ref) | 24/50 | 48.0 | 1.0 (ref) |
| rs11084753 | 1 | 184/288 | 63.9 | 0.8 (0.4, 1.4) | 95/155 | 61.3 | 1.2 (0.5, 2.5) | |
| 2 | 170/262 | 64.9 | 0.8 (0.4, 1.5) | 74/126 | 58.7 | 1.3 (0.6, 2.9) | ||
| NEGR1 | T/C | 0 | 65/97 | 67.0 | 1.0 (ref) | 49/87 | 56.3 | 1.0 (ref) |
| rs2815752 | 1 | 184/285 | 64.6 | 0.8 (0.5, 1.3) | 103/169 | 60.9 | 1.1 (0.6, 2.1) | |
| 2 | 161/244 | 66.0 | 0.8 (0.5, 1.5) | 42/76 | 55.3 | 1.1 (0.5, 2.3) | ||
1990 Institute of Medicine (IOM) recommendations
Frequencies sum to less than 628 or 332 because participants were excluded from an analysis if their samples did not genotype successfully.
Model covariates include log pregravid BMI, log pregravid BMI squared and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
Figure 4. a and 4b: Diabetes-associated SNPs and excessive gestational weight gain.
Multivariate-adjusted§ odds ratio (95% CI) for excessive weight gain by risk allele carriage for single nucleotide polymorphisms associated with diabetes in non-pregnant populations. Caucasian participants (n=628) on left, African-American participants (n=332) on right.
* p < 0.05 for Wald Chi Square test; # p <0.05 for comparison with 0 risk alleles (referent).
§ Model covariates include log pregravid BMI, log pregravid BMI squared, and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
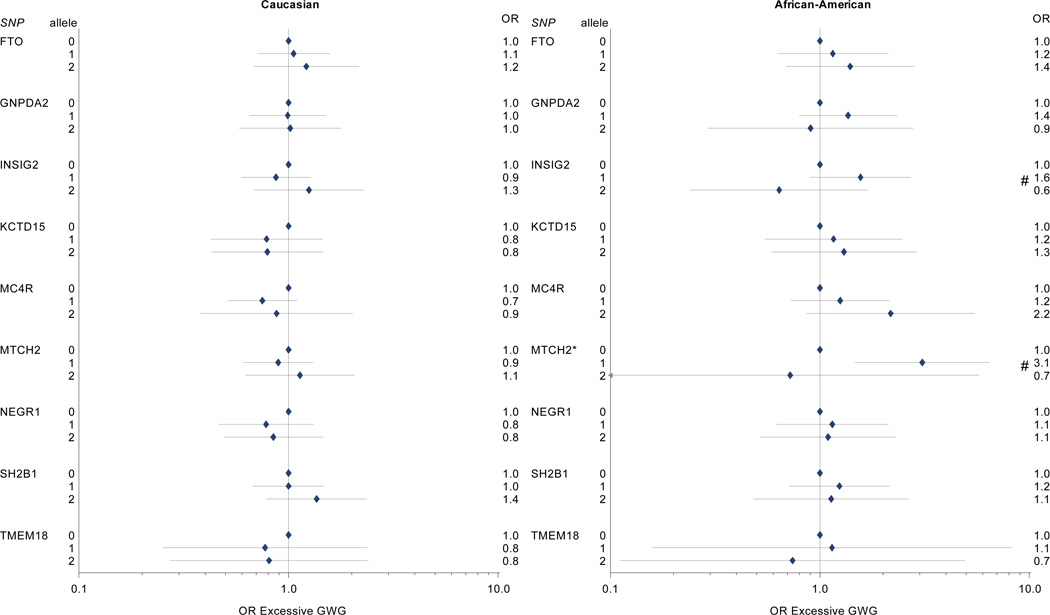
In our analysis of obesity SNPs and excessive weight gain (Table 3, Figure 5), carriage of one copy of the MTCH2 risk allele was associated with excessive gain among YRI participants (OR 3.1, 95% CI 1.5–6.5, vs 0 risk alleles). There were no statistically significant associations between obesity risk allele carriage and excessive weight gain risk among CEU participants (Figure 5).
Figure 5. Obesity-associated SNPs and excessive gestational weight gain.
Multivariate-adjusted§ odds ratio (95% CI) for excessive weight gain by risk allele carriage for single nucleotide polymorphisms associated with obesity in non-pregnant populations. Caucasian participants (n=628) on left, African-American participants (n=332) on right.
* p < 0.05 for Wald Chi Square test; # p <0.05 for comparison with 0 risk alleles (referent).
§ Model covariates include log pregravid BMI, log pregravid BMI squared, and maternal age. Effect estimates among self-identified African American participants further adjusted for probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
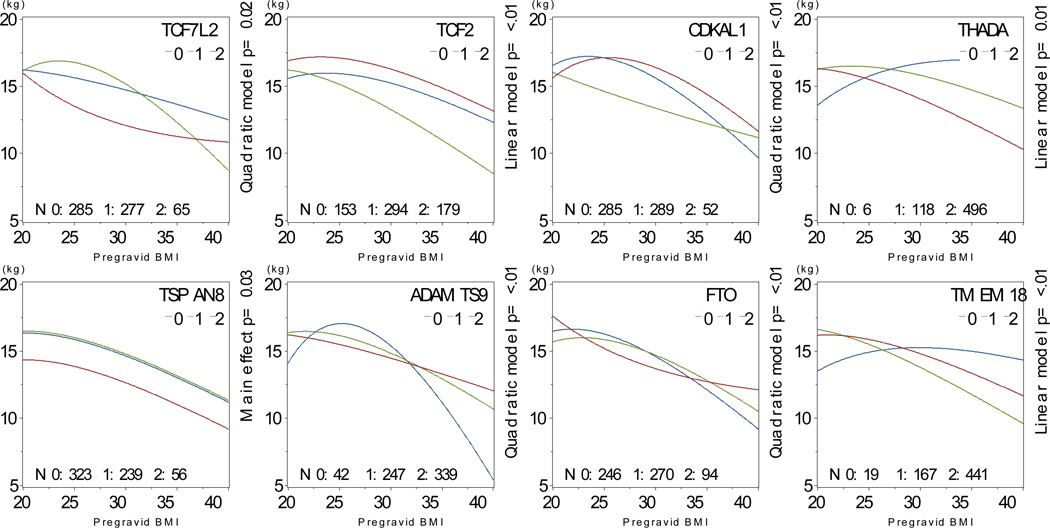
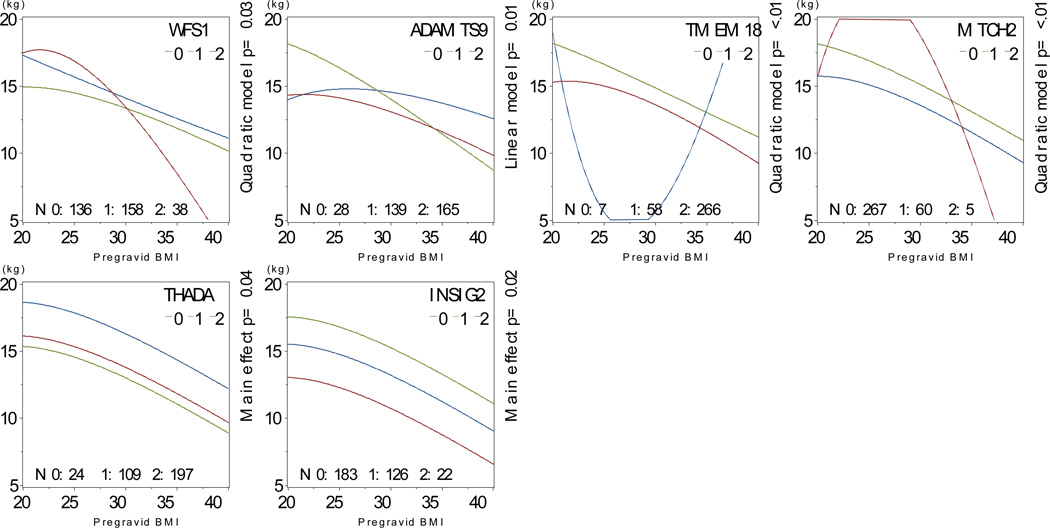
We found interactions between risk allele carriage and pre-gravid BMI for TCF7L2, TCF2, CDKAL1, THADA, ADAMTS9, NOTCH2, FTO and TMEM18, as well as main effects for TSPAN8 (Figure 6). Among African-American participants, we found interactions between risk allele carriage and pre-gravid BMI for WFS1, ADAMTS9, TMEM18 and MTCH2 and main effects for THADA and INSIG2 (Figure 7).
Figure 6. Interactions between pre-gravid BMI and SNP carriage, Caucasian participants.
Multivariate-adjusted§ mean predicted gestational weight gain as a function of body mass index and risk allele carriage among self-identified Caucasian participants, N=628.
§ Model covariates include log pregravid BMI, log pregravid BMI squared, and maternal age. All models weighted to reflect the composition of the full PIN studies population.
Figure 7. Interactions between pre-gravid BMI and SNP carriage, African-American participants.
Multivariate-adjusted§ mean predicted gestational weight gain as a function of body mass index and risk allele carriage among self-identified African-American participants, N=332.
§ Model covariates include log pregravid BMI, log pregravid BMI squared, and maternal age, and probability of Yoruban ancestry. All models weighted to reflect the composition of the full PIN studies population.
Finally, we used Bayesian variable selection models to test the additive effect of multiple SNPs on gestational weight gain. Among Caucasian women, we found a posterior odds ratio > 3 for greater weight gain with carriage of the TCF2 risk allele (1.1 kg per allele, 95% CI 0 to 2.7, posterior OR 4.0) and for lower weight gain among obese women carrying the THADA risk allele (−1.7 kg per allele, 95% CI −4.2 to 0.07, posterior OR 4.26). Among African-American women, we found a posterior odds ratio >3 for an interaction between the TSPAN8 risk allele and obesity, with lower weight gain among obese risk allele carriers (−2.4 kg per allele, 95% CI −6.7 to 0.3, posterior OR 3.2).
Comment
In this prospective longitudinal study of pregnant women, we found several associations between diabetes and obesity SNPs and gestational weight gain. The effect of risk allele carriage varied with pre-gravid body mass index, suggesting that genotype may modify the effect of a woman’s body composition prior to pregnancy on weight gain trajectory.
Our findings confirm and extend earlier work on associations between diabetes and obesity SNPs and weight trajectory. We found greater weight gain among women with the Ala12Ala PPARG genotype than those with Pro12Pro genotype, although the number of Ala12Ala participants was small (N=7), and our findings may be sensitive to two of the seven Ala12Ala participants with weight gains of 28 and 40 kg. In a small study of women with gestational diabetes (N=62), Tok et al found greater weight gain among women with the Pro12Ala genotype than women with the Pro12Pro genotype. PPARG is expressed primarily in adipocytes, and this gene appears to regulate triglyceride storage in adipose tissue37–39. It is plausible that Ala12Ala carriers have increased capacity for triglyceride storage, leading to greater gestational weight gain.
We also found greater gestational weight gain among women with one or two copies of the KCNQ1 risk allele, compared to homozygous low-risk women, although the number of low-risk homozygotes was low (4 Caucasian women and 2 African-American women). The low-risk KCNQ1 rs2237892 variant has been associated with increased c-peptide levels at 30 minutes after oral glucose load 40. It is plausible that more efficient insulin release lowers postprandial glucose concentrations and thus reduces gestational weight gain.
Among African-American participants, we found greater weight gain among MC4R high-risk homozygotes. The MC4R rs17782313 variant is associated with increased energy intake, increased total fat and protein intake, and greater weight gain over time in the predominantly-Caucasian Nurses’ Health Study cohort41. Interestingly, MC4R risk allele carriage was not associated with obesity among African-American children in a large case:control study42.
We also found interactions between pre-gravid BMI and genotype for several diabetes and obesity risk alleles in our population. These results suggest that a woman’s pre-gravid BMI as well as her genotype influence gestational weight gain. For example, in analyses of FTO rs9939609 risk allele carriage among Caucasian women, thin or obese women homozygous for the high risk allele gained more weight than low-risk allele carriers, but among women of average pre-gravid BMI, weight gain was similar regardless of allele carriage. These results suggest that it may be important to consider baseline BMI in longitudinal studies of genetic determinants of weight trajectory.
Our results must be interpreted within the context of the study design. Strengths of this study include our prospective collection of gestational weight gain data and the use of ancestry informative markers to adjust for population stratification. In addition, we used innovative techniques to model the role of pre-gravid BMI in modifying the effect of genotype on pregnancy phenotype. Our study also has several limitations. This is a secondary analysis of data collected over a 10-year period, and secular changes in medical recommendations, diet and physical activity, as well as the evolving nature of the PIN studies, may have modified the association between genotype and weight gain. Bias is also a concern, because we used samples from a subset of a larger cohort. Our use of sampling weights allowed us to produce estimates that approximate what we may have observed from a complete data set, but we were unable to adjust for the possibility that a woman’s willingness to allow genetic analysis or provide a blood sample may be non-random. Moreover, this is a pilot study, and our sample size was small, reducing our ability to detect differences among women with differing genotypes. In GWAS studies of obesity risk alleles and body mass index, risk allele carriage has been associated with differences of 0.10 to 0.33 kg/m221, which are considerably smaller than what we could detect in our population. In addition, pregnancy weight gain was measured in a clinical setting, without assessment of water weight vs. adipose tissue. Associated measurement error may further reduce our power to detect associations between genotype and outcome. At the same time, multiple testing is a concern. To address this issue, we limited our analysis to candidate SNPs that have been validated in multiple large studies. Nevertheless, we recognize that some of our findings may be false positives. With 18 diabetes SNPs and 2 comparisons for each SNP, we would expect to find 1.8 significant associations by chance alone, and with 9 obesity SNPs, we would expect to find 0.9 significant association by chance alone, if we conservatively assume independence of all SNPs.
In conclusion, we found evidence that maternal diabetes- and obesity-risk allele genotype interact with pre-gravid BMI to affect gestational weight gain. These results suggest that excessive or inadequate gain may be marker for maternal genotype, and these differences in genetic risk may explain some observed associations between gestational weight gain and long term health outcomes for mothers and infants. Further studies in larger cohorts will be needed to delineate further the role of genotype in maternal weight gain during pregnancy.
Acknowledgements
We would like to thank all of the PIN investigators that are not coauthors (David Savitz, John Thorp, Kelly Evenson, and Nancy Dole) for their help in obtaining funding and designing the study, and we would like to thank Kathryn Carrier for technical assistance. Additionally, we would like to thank the field collection staff for their dedication and hard work, and we would like to thank all the families involved in the PIN studies for their participation.
Funding: This study received support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD28684, HD28684A, HD37584, HD39373), the National Institute of Diabetes and Digestive and Kidney Diseases (DK61981, DK56350), the National Cancer Institute (CA109804-01), National Institute of Health General Clinical Research Center (RR00046), the Carolina Population Center, the UNC University Research Council and the Nutrition and Obesity Research Council (DK056350). Dr. Stuebe receives support from the WRHR Career Development Center at UNC (5K12HD050113-04). Dr. Ghosh receives support from NIEHS 5T32ES007018 and NIEHS P30 ES10126.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Location where study was conducted: Central North Carolina.
Prior presentation: Preliminary findings were presented at the 30th Annual Meeting of the Society for Maternal-Fetal Medicine on February 5, 2010, Chicago, IL, as poster presentation #592.
References
- 1.Viswanathan M, Siega-Riz AM, Moos MK, et al. Outcomes of maternal weight gain. Evidence report/technology assessment. 2008:1–223. [PMC free article] [PubMed] [Google Scholar]
- 2.Siega-Riz AM, Adair LS, Hobel CJ. Maternal underweight status and inadequate rate of weight gain during the third trimester of pregnancy increases the risk of preterm delivery. J Nutr. 1996;126:146–153. doi: 10.1093/jn/126.1.146. [DOI] [PubMed] [Google Scholar]
- 3.Stuebe AM, Forman MR, Michels KB. Maternal-recalled gestational weight gain, pre-pregnancy body mass index, and obesity in the daughter. Int J Obes (Lond) 2009;33:743–752. doi: 10.1038/ijo.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stotland NE, Hopkins LM, Caughey AB. Gestational Weight Gain, Macrosomia, and Risk of Cesarean Birth in Nondiabetic Nulliparas. Obstetrics and gynecology. 2004;104:671–677. doi: 10.1097/01.AOG.0000139515.97799.f6. [DOI] [PubMed] [Google Scholar]
- 5.Bianco AT, Smilen SW, Davis Y, Lopez S, Lapinski R, Lockwood CJ. Pregnancy outcome and weight gain recommendations for the morbidly obese woman. Obstetrics and gynecology. 1998;91:97–102. doi: 10.1016/s0029-7844(97)00578-4. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg TJ, Garbers S, Lipkind H, Chiasson MA. Maternal Obesity as Diabetes as Risk Factors for Adverse Pregnancy Outcomes: Differences Among 4 Racial / Ethnic Groups. American journal of public health. 2005;95:1544–1661. doi: 10.2105/AJPH.2005.065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuebe AM, Zera C, Ecker JL, McElrath TM. A prospective assessment of metabolic function, compliance with gestational weight gain guidelines, and associated pregnancy outcomes. American journal of obstetrics and gynecology. 2006;195:S82. [Google Scholar]
- 8.Juhasz G, Gyamfi C, Gyamfi P, Tocce K, Stone JL. Effect of body mass index and excessive weight gain on success of vaginal birth after cesarean delivery. Obstetrics and gynecology. 2005;106:741–746. doi: 10.1097/01.AOG.0000177972.32941.65. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson E, Abrams B, Selvin S. The relative importance of gestational gain and maternal characteristics associated with the risk of becoming overweight after pregnancy. International Journal of Obesity & Related Metabolic Disorders: Journal of the International Association for the Study of Obesity. 2000;24:1660–1668. doi: 10.1038/sj.ijo.0801456. [DOI] [PubMed] [Google Scholar]
- 10.Siega-Riz AM, Herring AH, Carrier K, Evenson KR, Dole N, Deierlein A. Sociodemographic, Perinatal, Behavioral, and Psychosocial Predictors of Weight Retention at 3 and 12 Months Postpartum. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rooney B, Schauberger C. Excess pregnancy weight gain and long-term obesity: one decade later. Obstetrics and gynecology. 2002;100:245–252. doi: 10.1016/s0029-7844(02)02125-7. [DOI] [PubMed] [Google Scholar]
- 12.Rooney BL, Schauberger CW, Mathiason MA. Impact of Perinatal Weight Change on Long-Term Obesity and Obesity-Related Illnesses. Obstetrics and gynecology. 2005;106:1349–1356. doi: 10.1097/01.AOG.0000185480.09068.4a. [DOI] [PubMed] [Google Scholar]
- 13.Schieve LA, Cogswell ME, Scanlon KS. Maternal weight gain and preterm delivery: differential effects by body mass index. Epidemiology (Cambridge, Mass. 1999;10:141–147. doi: 10.1097/00001648-199903000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Edwards LE, Hellerstedt WL, Alton IR, Story M, Himes JH. Pregnancy complications and birth outcomes in obese and normal-weight women: effects of gestational weight change. Obstetrics and gynecology. 1996;87:389–394. doi: 10.1016/0029-7844(95)00446-7. [DOI] [PubMed] [Google Scholar]
- 15.Sewell MF, Huston-Presley L, Super DM, Catalano P. Increased neonatal fat mass, not lean body mass, is associated with maternal obesity. American journal of obstetrics and gynecology. 2006;195:1100–1103. doi: 10.1016/j.ajog.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Oken E, Taveras EM, Kleinman KP, Rich-Edwards JW, Gillman MW. Gestational weight gain and child adiposity at age 3 years. American journal of obstetrics and gynecology. 2007;196:322, e1–e8. doi: 10.1016/j.ajog.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyon HN, Hirschhorn JN. Genetics of common forms of obesity: a brief overview. The American journal of clinical nutrition. 2005;82:215S–217S. doi: 10.1093/ajcn/82.1.215S. [DOI] [PubMed] [Google Scholar]
- 18.Frayling TM, Timpson NJ, Weedon MN, et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loos RJF, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 21.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nature reviews. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 23.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008 doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008 doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 26.Bouatia-Naji N, Rocheleau G, Van Lommel L, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 27.Saldana TM, Siega-Riz AM, Adair LS. Effect of macronutrient intake on the development of glucose intolerance during pregnancy. The American journal of clinical nutrition. 2004;79:479–486. doi: 10.1093/ajcn/79.3.479. [DOI] [PubMed] [Google Scholar]
- 28.Institute of Medicine. Nutrition during pregnancy. Washington, DC: National Academies Press; 1990. [Google Scholar]
- 29.Siega-Riz AM, Adair LS, Hobel CJ. Institute of Medicine maternal weight gain recommendations and pregnancy outcome in a predominantly Hispanic population. Obstetrics and gynecology. 1994;84:565–573. [PubMed] [Google Scholar]
- 30.Saldana TM, Siega-Riz AM, Adair LS, Suchindran C. The relationship between pregnancy weight gain and glucose tolerance status among black and white women in central North Carolina. American journal of obstetrics and gynecology. 2006;195:1629–1635. doi: 10.1016/j.ajog.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Deierlein AL, Siega-Riz AM, Herring A. Dietary energy density but not glycemic load is associated with gestational weight gain. The American journal of clinical nutrition. 2008;88:693–699. doi: 10.1093/ajcn/88.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb JB, Siega-Riz AM, Dole N. Psychosocial Determinants of Adequacy of Gestational Weight Gain. Obesity (Silver Spring) 2008 doi: 10.1038/oby.2008.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell CD, Lyon HN, Nemesh J, et al. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes. 2007;56:1460–1467. doi: 10.2337/db06-1051. [DOI] [PubMed] [Google Scholar]
- 34.Lyon HN, Emilsson V, Hinney A, et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gajdos ZK, Butler JL, Henderson KD, et al. Association studies of common variants in 10 hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab. 2008;93:4290–4298. doi: 10.1210/jc.2008-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma AM, Staels B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue-understanding obesity-related changes in regulation of lipid and glucose metabolism. J Clin Endocrinol Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 39.Catalano PM, Nizielski SE, Shao J, Preston L, Qiao L, Friedman JE. Downregulated IRS-1 and PPARgamma in obese women with gestational diabetes: relationship to FFA during pregnancy. American journal of physiology. 2002;282:E522–E533. doi: 10.1152/ajpendo.00124.2001. [DOI] [PubMed] [Google Scholar]
- 40.MussiG K, Staiger H, Machicao F, et al. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes. 2009;58:1715–1720. doi: 10.2337/db08-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet. 2008;17:3502–3508. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant SF, Bradfield JP, Zhang H, et al. Investigation of the locus near MC4R with childhood obesity in Americans of European and African ancestry. Obesity (Silver Spring) 2009;17:1461–1465. doi: 10.1038/oby.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]