SUMMARY
The p53 tumor suppressor is a key mediator of cellular responses to various stresses. Here we show that under conditions of basal physiologic and cell-culture stress, p53 inhibits expression of the CD44 cell-surface molecule via binding to a non-canonical p53-binding sequence in the CD44 promoter. This interaction enables an untransformed cell to respond to stress-induced, p53-dependent cytostatic and apoptotic signals that would otherwise be blocked by the actions of CD44. In the absence of p53 function, the resulting de-repressed CD44 expression is essential for the growth and tumor-initiating ability of highly tumorigenic mammary epithelial cells. In both tumorigenic and non-tumorigenic cells, CD44’s expression is positively regulated by p63, a paralogue of p53. Our data indicate that CD44 is a key tumor-promoting agent in transformed tumor cells lacking p53 function. They also suggest that the de-repression of CD44 resulting from inactivation of p53 can potentially aid the survival of immortalized, premalignant, cells.
INTRODUCTION
CD44 is a transmembrane cell-surface protein (Aruffo et al., 1990) that is synthesized in multiple isoforms due to alternative splicing of its pre-mRNA. While lacking its own signaling domain, it has recently been shown to be essential for the homing and stem-cell properties of leukemic stem cells (Jin et al., 2006; Krause et al., 2006). CD44 has also been found to support anchorage-independent growth in vitro as well as tumor growth and metastasis in experimental models of solid cancers (Barbour et al., 2003; Weber et al., 2002; Yu et al., 1997), whereas it inhibited tumor growth in yet other models (Gao et al., 1997; Schmits et al., 1997). The precise role played by CD44 in tumorigenesis has thus remained unclear.
The tumor-promoting functions of CD44 have been attributed to its association with and co-stimulation of signaling by a number of growth factor receptors, such as epidermal growth factor receptor-2 (Her2), epidermal growth factor receptor-1 (Her1), and Met (Ponta et al., 2003). In the case of breast cancer pathogenesis, for example, the most prominent of these receptors is Her2, which is over-expressed in 20–30% of these tumors and is responsible for releasing mitogenic and trophic signals into breast cancer cells (Yarden, 2001). These and other observations have suggested that CD44 confers a decided growth advantage on certain types of cancer cells. Moreover, the CD44 cell-surface antigen serves as a useful marker for detecting and enriching for several types of tumor-initiating cells (Al-Hajj et al., 2003; Dou et al., 2007; Hurt et al., 2008; Wright et al., 2008; Yang et al., 2008), which is consistent with its tumor-promoting capabilities cited above.
We hypothesized that signals regulating CD44 expression are essential for understanding the role of this protein in tumorigenesis. In studying this issue, we noted that immunohistochemical analyses of clinical samples of hepatocellular and renal carcinomas had demonstrated that CD44 protein is expressed at high levels together with elevated levels of p53 (Endo and Terada, 2000; Zolota et al., 2002). Since the expression of p53 protein in tumor samples indicates the presence of a mutant, functionally inactive p53 protein (Iggo et al., 1990; Sjogren et al., 1996), this suggested that CD44 might be repressed by wild-type p53.
In response to strong cellular stresses, such as DNA damage or oncogenic signals, the wild-type form of p53 regulates expression of a large cohort of genes that effect cell-cycle arrest, senescence and apoptosis (Levine, 1997; Levine et al., 2006). Recent findings have uncovered additional roles of the p53 protein that is expressed under basal physiologic and cell-culture stress conditions, notably regulation of mitochondrial respiration (Bensaad et al., 2006; Matoba et al., 2006), autophagy (Crighton et al., 2006), protection of the genome from reactive oxygen species (Sablina et al., 2005), and inhibition of the self-renewal capacity of neuronal stem cells (Meletis et al., 2006; Piltti et al., 2006). Under normal in vivo conditions, specifically in primary mouse mammary epithelial cells, p53 regulates the expression of ~40 transcripts, pointing to its potentially important physiologic role in the absence of any unusual cell-physiologic stresses (Aldaz et al., 2002).
The p53 paralogue, p63, has been recently shown to positively regulate CD44 mRNA expression in microarray-based gene expression analyses of the MCF10A immortalized non-tumorigenic breast epithelial cell line. This study demonstrated that ectopic expression of p63 leads to the upregulation of CD44 expression, whereas shRNA directed against p63 mRNA leads to loss of CD44 expression (Carroll et al., 2006). However, this work did not indicate whether p63, which is essential for the normal development of epithelial tissues (Mills et al., 1999; Yang et al., 1999), is also able to positively regulate CD44 expression at the protein level and in tumorigenic cells. These various observations provoked us to examine the mechanisms regulating the abundance of CD44 in cells and the functional consequences of its expression at various levels in such cells.
RESULTS
Repression of CD44 Expression by p53
To address the regulation and apparently important role of CD44 in mammary epithelial cell physiology and tumorigenesis, we chose to analyze its function in a novel type of early-passage, human mammary epithelial cells (BPECs) that were recently isolated and propagated in this laboratory (Ince et al., 2007). The experimentally transformed derivatives of these cells, termed BPLER cells, were created by the introduction of genes encoding the hTERT telomerase subunit, the SV40 large and small T antigens, and the H-Ras V12 oncoprotein (Hahn et al., 1999; Ince et al., 2007). These BPLER cells yield tumor xenografts that closely resemble, at the histopathological level, invasive ductal adenocarcinomas, the most common type of human breast cancer. In addition, injection of as few as 10–100 of these transformed cells suffices to induce tumors in immunocompromised mouse hosts, even in the absence of prior enrichment of tumor-initiating cells (Ince et al., 2007). This tumor-initiating efficiency is comparable to that of CD44high/CD24low cells that have been isolated directly from populations of human breast cancer cells (Al-Hajj et al., 2003). This experimental model of human breast pathogenesis allowed us to study the function and expression of p53 and CD44 in both primary BPEC cells and in the derived, experimentally transformed BPLER cells.
Prompted by the clinical evidence cited above (Endo and Terada, 2000; Zolota et al., 2002), we undertook to confirm that p53 mutant status is in fact associated with elevated CD44 expression in a series of breast cancer specimens whose p53mutant status and gene expression profiles were known (Miller et al., 2005). Since not all p53mutations are equally detrimental to p53 function, we chose to further sort these tumor specimens according to the expression of a p53-induced gene, p21Waf1. While CD44 is positively and negatively regulated by number of distinct signals (Ponta et al., 2003), we were surprised that p53-mutant/p21-low-expression patterns in human tumors were, on their own, able to predict high CD44 expression, doing so in a statistically significant manner (Figure S1). This correlation suggested that p53 could act as a negative regulator of CD44 expression in spontaneously arising human tumors.
We therefore undertook to investigate whether p53 indeed functions to repress CD44 expression in untransformed human mammary epithelial cells. Because the BPEC cells mentioned above have a limited life span in culture, they were initially immortalized by introduction of a vector expressing the catalytic subunit of human telomerase (hTERT), yielding BPEC-T cells. To visualize the CD44 protein expression at the single-cell level, we immunostained subconfluent monolayer cultures of these BPEC-T cells. Curiously, the CD44 expression in control BPEC-T cells was found to be elevated in the outer perimeters of BPEC-T cell clusters, with a gradual decrease toward the center of each cluster.
In order to explore the possible connection of this behavior with p53 function, we infected populations of BPEC-T cells with a lentiviral construct encoding an shRNA that causes degradation of p53 mRNA or, alternatively, with a retroviral construct encoding the human papillomavirus E6 oncoprotein, which is known to trigger degradation of the p53 protein. The expression gradient described above was absent in cells expressing the p53-shRNA construct or in cells in which the E6 oncoprotein was deployed to degrade p53 (Figure 1A). In both cases, these cells exhibited elevated levels of CD44 protein nearly uniformly, regardless of the cells’ positions within a cluster (Figure 1A). This suggested that the uneven expression of CD44 by control BPEC-T cells was a reflection of p53 function. Control experiments in BPEC-T cells treated with a genotoxic agent – doxorubicin – indicated that the p53 gene and protein expressed in all cellular locations within these clusters were equally capable of responding to genotoxic stress (Figure S2).
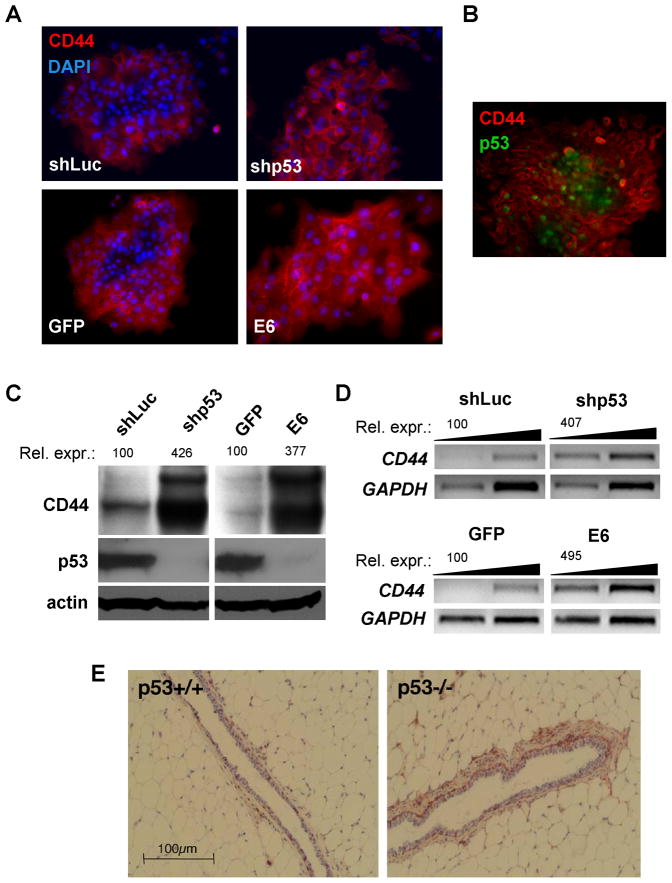
Figure 1. CD44 Expression in BPEC-T Cells and Mammary Fat Pad is Dependent on p53.
(A) Immunofluorescent detection of CD44 and nuclei (DAPI) in BPEC-T cells stably infected with the control shRNA against firefly luciferase (shLuc), p53 shRNA (shP53), E6 oncoprotein- or GFP-expressing constructs in subconfluent cultures.
(B) Immunofluorescent analysis of CD44 and p53 protein co-expression in BPEC-T cells.
(C–D) Western blotting analysis of total CD44 protein (C) and semi-quantitative RT-PCR analysis of total CD44 mRNA expression (D) in confluent BPEC-T cells with normal and inhibited p53 expression. The cDNAs for RT-PCR were PCR amplified from 3.3 and 1.1ng of respective cDNA.
(E) Immunohistochemical analysis of CD44 expression in mammary fat pads. The epithelial and non-epithelial compartments of mammary glands from p53−/− or p53+/+ Balb/c mice were immunostained for the epitope present in all CD44 known isoforms by the IM7 antibody and counterstained with hematoxylin.
These findings suggested heterogeneous activation of p53 protein expression within the cell clusters of BPEC-T cells growing in monolayer culture. To test this possibility, we performed double immunofluorescent staining for both p53 and CD44. As anticipated, the control BPEC-T cells revealed largely mutually exclusive localizations of the two proteins (Figure 1B), consistent with the notion of p53-dependent repression of CD44. Further analysis of protein expression in these cells using immunoblotting revealed that CD44 expression was significantly increased in the confluent cells in which p53 expression had been suppressed by either the shRNA construct or E6 oncoprotein (Figure 1C). At present, the nature of the contextual signals causing differential activation of p53 expression within the BPEC-T cell clusters is obscure. Nonetheless, these results provided an initial indication that high CD44 expression is a signature of relatively low p53 activity.
To determine whether p53 regulates CD44 expressionat the transcriptional or posttranscriptional level, total CD44 mRNA expression was measured by semi-quantitative RT-PCR using primers to the CD44 sequences common to all CD44 mRNA isoforms. As shown in Figure 1D, the mRNA levels for CD44 paralleled the results from the protein expression analyses, i.e., CD44 mRNAs were upregulated 4-to-5-fold and CD44 protein 4-fold in BPEC-T cells in which p53 function had been largely abolished. Hence, CD44 expression was suppressed by p53 largely through effects on CD44 mRNA levels. We also examined how the levels of p53 expression were correlated with those of CD44 in a living tissue. To do so, we undertook immunohistochemical analyses of the mammary glands of 10 week-old wild-type and p53−/− isogenic mice. These analyses demonstrated significantly higher CD44 protein expression in the mammary glands of p53−/− mice compared with those of wild-type animals. The difference in CD44 signal was most prominent in the basal epithelial cells, but was also apparent in occasional luminal epithelial cells (Figure 1E).
To exclude the possibility that the observed repression was a consequence of genomic instability in these p53−/− mice, we took advantage of a recently published conditional mouse model, in which p53 expression is restored in otherwise p53-null tissues of mice by administration of tamoxifen to activate the Cre recombinase (Ventura et al., 2007). When p53 expression was restored in the tissues of adult mice (with 50–80% efficiency in mammary epithelial cells), we observed downregulation of CD44 protein levels specifically in the mammary epithelium of these mice, but not in the mammary epithelium of mice treated only with oil vehicle (Figure S3). As before, these observations indicated that the expression of p53 varied inversely with that of CD44. Moreover, they indicated that p53 antagonism of CD44 expression can operate in normal tissues, where p53 is expressed at low levels and in the apparent absence of any unusual cell-physiologic stresses.
Mechanism of CD44 Repression by p53
In order to explore the possibly direct influence of p53 on the promoter of the CD44 gene, a 2kb DNA segment located upstream of its transcription initiation site was introduced into a luciferase reporter vector. Since p53 can regulate transcription either directly by binding to a promoter sequence or indirectly, via protein-protein interactions (Ho and Benchimol, 2003), we distinguished between these alternative mechanisms by using two p53mutants that are defective in DNA binding. As shown in Figure 2A, in contrast to their wild-type counterpart, which strongly repressed the activity of the CD44 promoter reporter construct, both p53 mutants failed to do so. Hence, the actions of p53 on the CD44 gene promoter appeared to require the intact DNA-binding activity of p53.
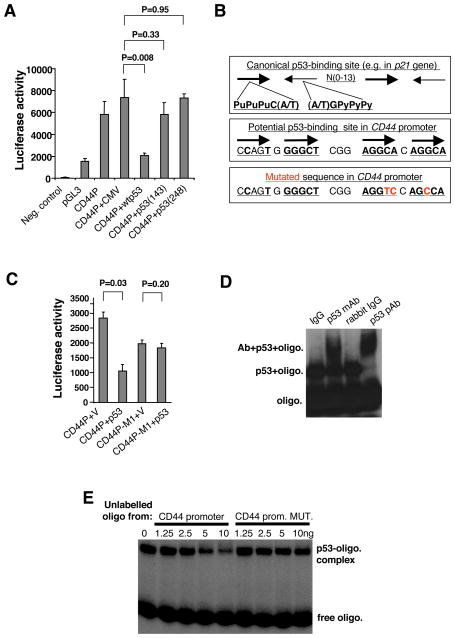
Figure 2. Mechanism of p53-mediated Repression of CD44.
(A) Repression of CD44 promoter activity with p53 in the human osteosarcoma cell line SAOS-2 lacking endogenous p53 expression. Human CD44 promoter (CD44P, 0- 2021 bp upstream of translation initiation codon) was fused to a firefly luciferase gene in pGL3 vector. The CD44P construct was co-transfected with CMV-vector-based constructs bearing either normal or specific-DNA-binding-defective p53 genes into the SAOS-2 cells. The cells were lysed 30h after transfection and analyzed for luciferase activity (mean±SD, n=4).
(B) The consensus sequence of p53-mediated transcriptional regulation (upper sequence), a potential non-canonical p53-binding site in the CD44 promoter, 239–263bp upstream from the first transcription initiation site (lower sequence), and the introduced point mutations (in red, lower sequence) used to inactivate the potential p53-binding site.
(C) Promoter reporter assay with normal CD44 promoter or mutated CD44P-M1 constructs in SAOS-2 cells co-transfected with or without the p53 expression vector (mean±SD, n=4). The basal activity of promotorless construct was subtracted from the presented values of CD44 promoter activity.
(D) Gel-shift analysis of p53 protein interaction with CD44P-derived, 32P-labeled oligonucleotides with normal or mutated p53-binding site.
(E) Competition analysis of p53 protein interaction between labeled CD44P-derived oligonucleotide and non-labeled oligonucleotides bearing either normal or mutated p53-binding site sequence.
It remained unclear, however, whether the observed repression was mediated by direct binding of p53 to the CD44 promoter. To address this issue, we attempted to identify a p53-response element in this promoter. Indeed, sequence analyses revealed a region within the CD44 promoter that exhibits strong sequence similarity to the non-canonical p53-binding site found in the MDR1 gene (Figure 2B) (Johnson et al., 2001), which is known to contain four p53-binding elements. The canonical p53-binding site implicated in activation of transcription by p53 is comprised of two copies of the sequence PuPuPuC(A/T) arranged head-to-head and separated by 0–13 nucleotides, as is seen, for example, in the promoter of the p21Waf1 gene. The non-canonical p53-binding site in the MDR1 gene consists of four p53-binding sites that are oriented in an alternating head-to-tail arrangement (Johnson et al., 2001). Accordingly, we created a mutant CD44 reporter construct bearing three point mutations in two of the four putative p53-response elements (Figure 2B). As shown in Figure 2C, the resulting mutant CD44 promoter was no longer repressed by ectopic expression of p53 in otherwise p53-null SAOS-2 osteosarcoma cells. In addition, in the absence of ectopically expressed p53, the basal activity of the mutant promoter was about 20% lower than of the wild-type promoter, suggesting the presence of some type of transcription-promoting site within the p53-response elements (Figure 2C).
To prove that the CD44 reporter construct was also susceptible to p53-dependent repression when chromosomally integrated, we introduced constructs encoding the wild-type CD44 promoter, the mutant CD44 promoter, or a promoterless reporter, each driving a luciferase gene, into lentiviral vectors and used these to infect the BPEC-T cells with normal or experimentally suppressed p53 levels. The promoter activities of the resulting chromosomally integrated constructs closely paralleled those obtained from transient transfections of SAOS-2 cells (Figures 2A and 2C) and suggested that the above-described non-canonical p53-binding site is involved in repression of CD44 promoter activity in the presence of the basal p53 expression in BPEC-T cells (Figure S4). These results agreed with deletion analyses of the CD44 promoter performed in parallel (Figure S5).
In order to confirm the direct physical interaction of p53 with sequences in the CD44 promoter, we extended these studies by performing gel-shift analyses and chromatin immunoprecipitation. Using DNA oligonucleotides mimicking the potential p53-binding site, we demonstrated direct binding of p53 protein to the CD44 promoter sequence, as indicated by the retardation of electrophoretic migration in the presence of added purified p53 (Figure 2D). To analyze the specificity of this binding, we performed a competition assay with either an unlabelled wild-type oligonucleotide or an oligonucleotide containing the base substitutions previously introduced into the promoter in the promoter-reporter assays described above. As shown in Figure 2E, the wild-type oligonucleotide was more efficient in displacing the labeled oligonucleotide from the complex with p53 protein than the mutated one.
In order to determine whether the interaction between CD44 promoter and p53 protein also occurs in vivo, we undertook chromatin immunoprecipitation of chromatin complexes containing p53 and analyzed them for the presence of a p53-response site in CD44 promoter. As shown in Figure S6, we were able to detect physical association of p53 with the CD44 promoter sequence in BPEC-T cells. These results provide evidence that p53 represses CD44 transcription via direct interaction with a specific site within the CD44 promoter located at position -239 to -263 bp relative to the transcription start site of the human CD44 gene (Shtivelman and Bishop, 1991).
CD44 Interferes with p53 Function in Immortalized Non-tumorigenic Cells
Having found that p53 functions as a repressor of CD44 expression, we attempted to discover the cell-physiologic rationale of this repression. CD44 is known to act predominantly in a growth-promoting and anti-apoptotic fashion (Ponta et al., 2003). Because the tumor-suppressing function of p53 relies on inhibition of these processes, we hypothesized that p53 downregulates CD44 expression in order to prevent CD44 from compromising its growth-inhibitory and pro-apoptotic functions.
At the biochemical level, CD44 is known to stimulate EGF-induced signaling (Bourguignon et al., 1997; Ponta et al., 2003). We therefore asked whether CD44 could facilitate EGF signaling in the BPEC-T cells used in our experiments and, if so, whether such signaling influenced these cells’ responses to p53-directed inhibition of cell proliferation and induction of apoptosis. To address these questions, we analyzed p53 function in the BPEC-T cells expressing normal levels of p53 (shLuc/GFP), in BPEC-T cells overexpressing the “standard” form of CD44 [which does not contain variable exons (shLuc/CD44s)] and in BPEC-T cells expressing an shRNA directed against p53 (shp53/GFP; Figure 3A). The first of these cells served as controls, since they were forced to express GFP and an shRNA directed against luciferase.
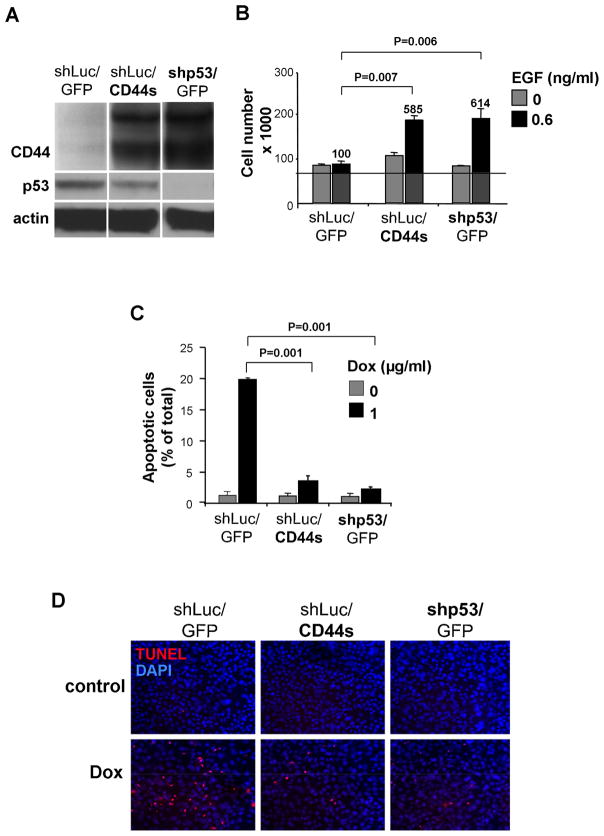
Figure 3. Effect of CD44 Expression on p53-dependent Proliferation and Apoptosis in BPEC-T Cells.
(A) BPEC-T cells stably infected with retroviral vectors expressing either the standard form of CD44 (CD44s), control GFP, or lentiviral vectors expressing an shRNA directed against p53 (shp53) or control shRNA (shLuc) were analyzed for expression levels of p53 and CD44 by western blotting. The CD44 with higher Mw than CD44s is either a product of differential post-translational modification of CD44s or an alternatively spliced variant form of endogenous CD44, whose expression is up-regulated by ectopic expression of CD44s.
(B) Cell growth (mean±SD, n=3) of BPEC-T cells variants in the presence or absence of EGF analyzed after 72h.
(C and D) Apoptosis (mean±SD, n=3) of cells exposed to doxorubicin in the presence EGF (0.6ng/ml), analyzed by TUNEL assay after 24h.
We exposed these various BPEC-T populations to chemically defined serum- and growth-factor-free media with or without added EGF. In the absence of added EGF, these three cell populations (shLuc/GFP, shLuc/CD44s and shp53/GFP) were largely quiescent. The presence of added EGF stimulated proliferation of shp53 cells (5.9-fold relative to the control) and shLuc/CD44s cells (6.1-fold relative to the control), but not the control shLuc/GFP cells expressing basal p53 levels. Hence, the basal levels of p53 present in the control cells sufficed to inhibit EGF-stimulated proliferation. Moreover, the ectopic constitutive expression of the standard form of CD44 in cells with basal levels of p53 increased their EGF-stimulated proliferation rate to a level comparable to that of cells in which p53 expression had been knocked down: this similarity in proliferation rate correlated with comparable expression levels of CD44 protein between these two cell lines (Figure 3A). We concluded that any growth-inhibitory effects that p53 imposed on EGF-stimulated cell proliferation could be reversed by ectopic CD44s expression (Figure 3B).
To analyze effects of CD44 on p53-dependent apoptosis, we challenged the various BPEC-T derivatives with the genotoxic drug doxorubicin. Many of the cells expressing wild-type p53 responded to genotoxic stress by entering into apoptosis (19.5% apoptotic, Figures 3C–D), while cells expressing the p53 shRNA construct were largely protected from this fate (3.9% apoptotic). Moreover, cell populations with wild-type p53 plus ectopically expressed CD44s were significantly more resistant to doxorubicin-induced apoptosis (2.3% apoptotic) than were cells expressing only endogenous levels of CD44 (19.5% apoptotic, Figure 4C). Lastly, in doxorubicin-treated BPEC-T cell clusters growing in monolayer culture (see above), the apoptotic cells were recruited from those that expressed low levels of CD44 and high levels of p53 and were localized to the interior of each cluster; this provided further support for the notion that cells with high p53 expression are more susceptible to apoptosis (Figure S7). We note that the inhibition of apoptosis by CD44 was paralleled by increased signaling by the anti-apoptotic PI3-kinase pathway in both shLuc/CD44s and shp53/GFP cells, as evidenced by higher levels of phospho-Akt (Figure S8). In sum, these proliferation and apoptotic assays provided a clear functional rationale of the p53-CD44 interaction: p53 must repress CD44 expression in order to reduce the anti-apoptotic and mitogenic effects of CD44.
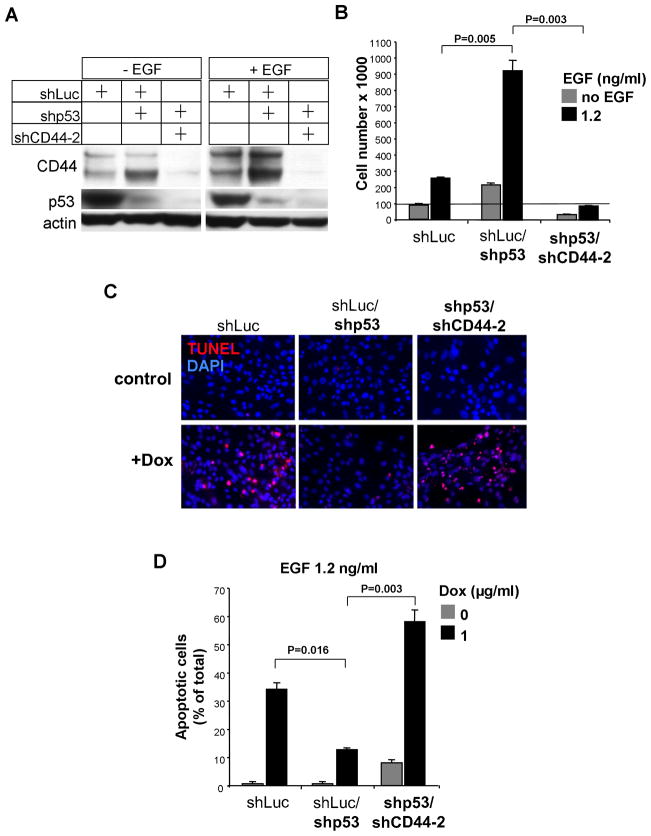
Figure 4. Effect of inhibition of CD44 Expression on Proliferation, Apoptosis and EGF-dependent signaling in BPEC-T Cells with Inhibited p53 Levels.
(A) Western blotting analysis of CD44 and p53 in cytokine-starved cells cultured in the presence or absence or EGF for 4h.
(B) Cell growth (mean±SD. n=3) of BPEC-T cells stably infected with lentiviral constructs expressing shRNA against CD44 or p53 were analyzed after 72h incubation in the presence or absence of EGF.
(C–D) Apoptosis (mean±SD, n=3) of BPEC-T cells exposed to doxorubicin at in the absence of 1.2 ng/ml EGF, analyzed by TUNEL assay after 24h.
CD44 is an Effector of Growth-supporting and Anti-apoptotic Effects of Low p53 Expression
The experiments described above provided evidence that CD44 can counteract p53 functions. However, it was unclear whether the various responses to downregulated p53 expression could be attributed solely to the actions of the derepressed CD44 observed in, for example, the shp53/GFP BPEC-T cells (Figure 3).
To address this issue, we inhibited CD44 expression in shp53 BPEC-T cells by an shRNA construct that was designed to target the expression of all known CD44 isoforms (Ponta et al., 2003) (Figure 4A). As a consequence, these shp53/shCD44-2 cells grew more slowly (93% decrease) and had a >4-fold higher apoptotic index than their shp53 counterparts (Figure 4B–D). In this respect they resembled control cells with normal p53 levels and p53-repressed CD44 expression. Hence, the anti-apoptotic effects deriving from suppression of p53 synthesis largely disappeared if CD44 expression was also blocked. This indicated that CD44 is a key effector of anti-apoptotic and mitogenic signals of shp53 cells. We also note that the shp53/shCD44-2 cells were more apoptotic than the cells with normal p53 expression, suggesting that the anti-apoptotic functions of CD44 extend beyond its ability to antagonize the pro-apoptotic function of p53.
On the other hand, suppression of CD44 expression in cells with normal p53 expression did not sensitize them to doxorubicin-induced apoptosis to the same extent as it sensitized the shp53 cells (Figure S9).
The Role of Standard Part of CD44 in CD44 Protein Functions
CD44 has multiple isoforms arising from alternative splicing of its mRNA (Ponta et al., 2003). However, in the experiments described here, the CD44 standard form (CD44s), whose sequences are present in all known CD44 isoforms, was found to suffice to inhibit p53-dependent apoptosis and proliferation. To address whether or not a CD44 isoform encoded by mRNAs containing additional variable exons is also able to inhibit p53-dependent functions more or less effectively than CD44s, we transiently transfected the SAOS-2 cells with vectors expressing either the CD44s or CD44VE isoforms together with apoptosis-inducing amounts of p53 (Figure S10A–B). The mRNA encoding CD44VE isoform contains 8 out of the 10 variable CD44 exons (v3–v10). We found that both CD44s and CD44VE isoforms inhibited p53-dependent apoptosis to approximately the same extent (Figure S10C), indicating that inclusion of sequences encoded by the variable exons v3–v10 does not substantially influence the anti-apoptotic activity of CD44.
CD44 Function and Expression in Tumorigenic BPLER Cells with Inactivated p53
The interactions described above shed light on the role of CD44 expression in untransformed BPEC-T cells possessing wild-type p53 function. We wondered whether CD44 could also exert similar functions in the transformed, tumorigenic derivatives of BPEC-T cells, termed BPLER cells, in which p53 is inactivated through sequestration by the SV40 large T antigen. To analyze CD44 function in this tumor xenograft model, BPLER cells were first infected with lentiviral constructs expressing shRNAs directed against the mRNA encoding the standard form of CD44, or as control, against the firefly luciferase gene. As indicated in Figure 5A, introduction of CD44-specific shRNAs decreased CD44 protein expression by more than 95%.
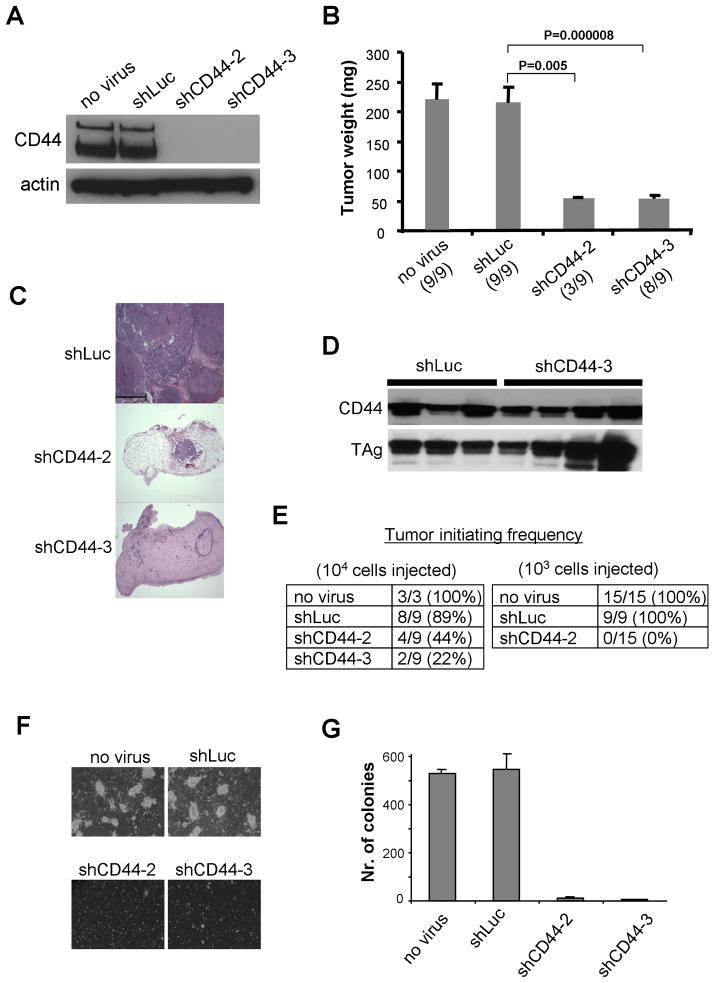
Figure 5. Tumor Growth of Human Mammary Epithelial BPLER Cells with Normal and Suppressed CD44 Expression.
BPLER cells were infected with a lentiviral vector expressing an shRNA to firefly luciferase (shLuc) or to CD44 (shCD44-2 and shCD44-3). The infection efficiency was more than 95% for all constructs (data not shown).
(A) The level of CD44 expression in individual infected cell populations in vitro was analyzed by western blotting 72h after the infection.
(B) Individual lentivirus-infected cells (106 cells per injection) were injected in nude mice and tumor weights (mean±SEM) were analyzed 4 weeks after the injection. Tumor incidence per injection is indicated in parentheses.
(C) Hematoxylin and eosin staining of the resulting tumors. Scale = 500μm.
(D) The level of CD44 expression in tumors arising from individual infected cell populations was analyzed by western blotting 72h after the infection.
(E) Tumor-initiating frequency of individual infected cell populations.
(F, G) Soft agar colony assay of individual lentivirus-infected cells. The cells (105/6cm dish) were plated in culture medium in soft agar and cultured for 4 weeks. The assay was terminated when colonies of control cells reached 1mm in diameter, at which point they were counted (mean±SEM, n+3).
BPLER cells expressing these CD44 shRNAs formed fewer tumors (Figures 5A and 5E), and those that did appear were quite small (50mg or less); this contrasted with the size of large tumors (average weight 220mg) formed by the BPLER cells in which CD44 expression had not been suppressed by the shRNA vector (Figure 5B–C). Significantly, immunoblotting analysis of those few tumors that did emerge from cells infected with CD44-specific shRNA constructs demonstrated that these tumors arose from the minority of BPLER cells whose CD44 expression failed to be suppressed by the shRNA (Figure 5D).
When analyzing tumor-initiating ability, another feature of tumorigenic cells, we found that it was substantially reduced in cells with suppressed CD44 expression (56–100% decrease, depending on the numbers of implanted cells and on the particular CD44 shRNA, Figure 5E) than their control counterparts. This indicated that, in addition to serving as a useful marker for breast tumor-initiating cells, CD44 positively regulates the functions of these cells (Figure 5E). These results reinforced yet other observations using an in vitro surrogate assay for tumorigenicity – the soft agar colony-forming assay – which also demonstrated that BPLER cells deprived of CD44 expression, unlike their control counterparts, failed to form anchorage-independent colonies (Figure 5F–G). Our results, taken together with those of others (Weber et al., 2002; Yu et al., 1997), demonstrate that by fostering tumorigenic growth in the absence of functional p53, CD44’s functions extend beyond antagonizing the proapoptotic and antiproliferative actions of p53.
CD44 is an Effector of Enhanced Tumor Growth in Tumor Cells with Low p53 Expression
The experiments described above provided evidence that CD44 contributes importantly to the tumor-initiating ability and tumor growth of BPLER cells with suppressed p53 function. However, it was unclear whether the suppression of p53 expression in cancer cells exhibiting wild-type, normal p53 expression would permit increased CD44 expression, and if so, whether the resulting elevating CD44 levels were important to any increases in tumor growth observed following p53 suppression.
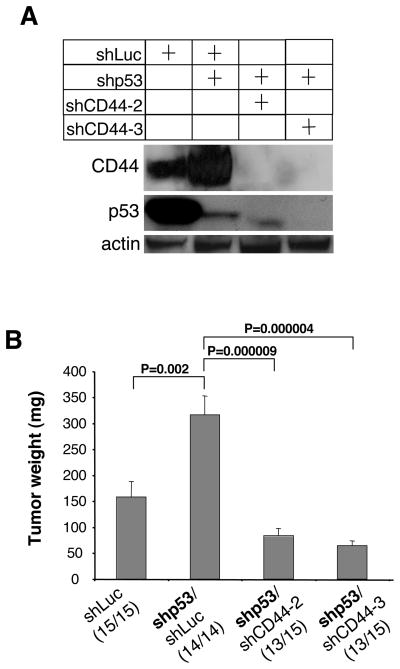
To address these questions, we examined cells of the A549 human lung adenocarcinoma line, which express wild-type p53. In particular, we constructed A549 cell populations with either suppressed p53 expression (shp53/shLuc) or suppressed expression of both p53 and CD44 (shp53/shCD44-2 and shp53/shCD44-3); as controls, we used cells expressing basal levels of both p53 and CD44 proteins (shLuc). These cells were first analyzed for their expression of p53 and CD44. As anticipated, cells with suppressed p53 expressed higher levels of CD44 than did the control cells with normal p53 levels. In addition, both shRNA constructs directed against CD44 were able to efficiently suppress CD44 protein expression (Figure 6A).
Figure 6. Effect of Suppression of CD44 Expression on Tumor Growth in A549 Cells with Suppressed p53 Levels.
A549 cells were infected with a lentiviral vector expressing an shRNA to firefly luciferase (shLuc), to p53 (shp53) or to CD44 (shCD44-2 and shCD44-3). The infection efficiency was more than 95% for all constructs (data not shown).
(A) The level of p53 and CD44 expressions in individual infected cell populations in vitro was analyzed by western blotting 72h after the infection.
(B) Individual lentivirus-infected cells (106 cells per injection) were injected in nude mice and tumor weights (mean±SEM) were analyzed 4 weeks after the injection. Tumor incidence per injection is indicated in parentheses.
Upon injection into immunocompromised mice, A549 cells with suppressed p53 gave rise to tumors that were two times larger than tumors induced by control cells. However, the A549 cells with suppressed p53 and CD44 expression yielded tumors three- to four-fold smaller than those induced by cells that had only suppressed p53 expression (Figure 6B). Taken together with earlier results, these observations indicate that CD44 is an essential effector of tumor growth caused by suppressed p53 expression and that CD44 supports tumor growth through both p53-dependent and p53-independent mechanisms.
Regulation of CD44 Expression by Proteins Related to P53-Induced Signaling
In order to obtain a more detailed view of the possible roles of other proteins in the p53 pathway that might modulate CD44 expression, we analyzed the potential involvement of the p21Waf1 protein and the p53 paralogue, p63. For example, p21 has been reported to mediate certain types of p53-induced transcription regulation (Gottifredi et al., 2001; Taylor et al., 2001). In fact, knockdown ofp21Waf1 expression, achieved by expression of two independent siRNAs to p21, did not influence CD44 protein levels in BPEC-T cells Figure S11A, indicating that p21 is not an essential component of the p53-dependent repression of CD44. Conversely, suppression of CD44 expression in BPEC-T cells or its ectopic expression in MCF7Ras cells did not influence the p21 levels in these cells (Figure S11B–C).
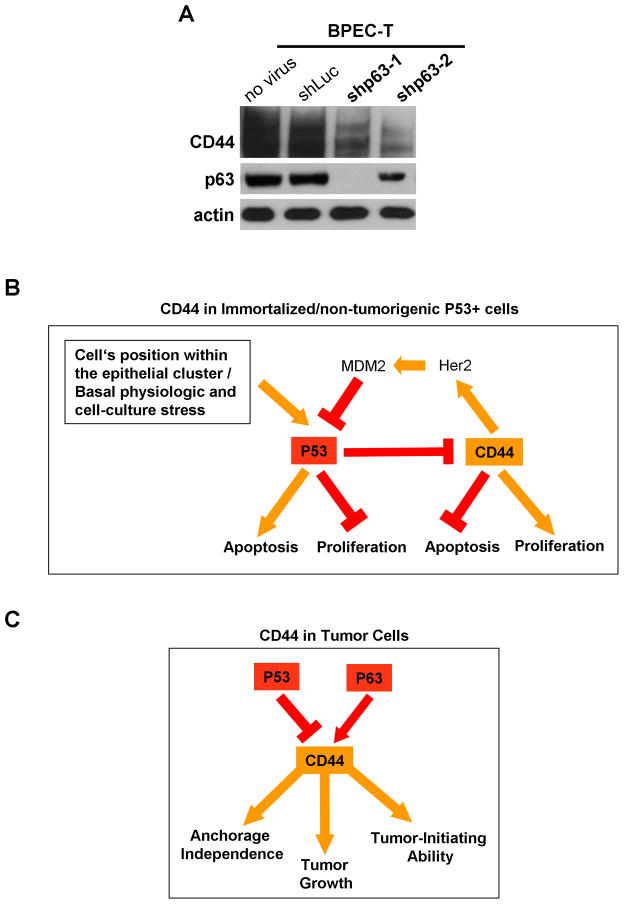
We also tested whether the p63, a p53 paralogue known to have partially overlapping promoter-binding specificities with p53 (Gottifredi et al., 2001), is essential for repression of CD44 expression. As we discovered, shRNA-mediated knockout of p63 expression did not result in activation but instead in inhibition of CD44 protein expression in BPEC-T as well as BPLER and lung adenocarcinoma A549 cells (Figure 7A and Figure S12A–B). Hence, p63 acts on the CD44 protein in a fashion opposite to that of p53 by stimulating CD44 expression, a finding that was also recently reported by others (Boldrup et al., 2007; Carroll et al., 2006).
Figure 7. Scheme of p53-CD44-p53 Axis in Untransformed and Transformed Cells.
(A) The p63 protein expression was inhibited by two independent shRNAs in pLKO1-Puro lentiviral vector in BPEC-T cells. The cells were lysed and analyzed for p63 and CD44 protein expression by western blotting one week after infection with viral shRNA constructs.
(B) The scheme of the function of p53-CD44-p53 axis in immortalized BPEC-T cells. Unknown signals, dependent on a cell’s position within an epithelial cell cluster, trigger induction of p53 expression, which leads, in turn, to CD44 repression, slower proliferation and increased apoptosis in response to a strong genotoxic stress. Conversely, increased CD44 expression can inhibit p53 stability by stimulating Her2-dependent activation of MDM2 protein expression (Mayo and Donner, 2001; Zhou et al., 2001).
(C) The summary of CD44 function in tumor cells. In highly tumorigenic BPLER cells with SV40 LTg-inactivated p53, CD44 is essential for anchorage-independent growth, tumor growth kinetics as well as tumor-initiating ability. In A549 cells, suppression of p53 expression accelerates tumor growth, which is dependent on elevated CD44 expression resulting from its derepression occuring in the absence of p53. The expression of CD44 is positively regulated by p63 in BPEC-T cells, in BPLER cells and in A549 cells, but the detailed molecular mechanism of this regulation in not known.
DISCUSSION
Inhibition of cell proliferation, and induction of apoptosis and senescence, are thought to be the major biological outputs of the p53 pathway in response to various types of cell-physiologic stresses (Levine, 1997; Levine et al., 2006). We present evidence revealing an additional mechanism of p53 action: under conditions of minimal stress in vitro and in vivo, the basal levels of p53 that are present suffice to repress expression of CD44 mRNA. Several types of observations persuade us that the cultured BPEC cells studied here are experiencing very low levels of cell-physiologic stress in the chemically defined WIT medium. Under these conditions of tissue culture, the BPEC cells express minimal levels of p53 over extended periods of time in culture, significantly lower than the levels expressed when human mammary epithelial cells from reduction mammoplasties are propagated in the MEGM medium that is commonly used for human mammary epithelial cells (Ince et al., 2007). These low levels of p53 expression correlate with the ability of BPEC cells to proliferate for at least 40 doublings in vitro (Ince et al., 2007). This notion that p53 functions at basal levels in the absence of unusual stress is further supported by our in vivo observation that CD44 expression is upregulated in the mammary fat pad of virgin p53−/− mice (relative to wild-type mice) that have not been exposed to any stress beyond the stresses that attend normal development.
Taken together, these observations indicate that p53 exerts regulatory functions that are dissociated from its normal role in programming responses to various types of cell-physiologic stress, and that p53 can do so when expressed at the low, basal levels that are usually depicted as its inactive, non-signaling state. Indeed, the recent report of p53’s ability to regulate the neuronal stem cell pool (Meletis et al., 2006) is in consonance with this thinking.
The observations presented here suggest that high CD44 expression, by opposing p53 function, can serve as an important growth-promoting and survival factor in early stages of tumor progression, when its expression may counteract p53’s tumor-suppressing functions. Acting in the opposite direction, p53 represses CD44 expression, doing so even when present at basal levels and in the absence of any apparent cell-physiologic stress. When placed in the context of previous reports, the present observations suggest that p53 and CD44 may establish a self-amplifying positive feedback loop, in which p53 represses CD44 expression, which results in suppression of growth receptor signaling and a resulting decrease in MDM2 activity, which permits, in turn, further increases in p53 levels and function (Figure 7A) (Bourguignon et al., 1997; Mayo and Donner, 2001; Zhou et al., 2001).
A variety of observations indicate that the CD44 molecule is an important factor for the progression of acute myeloid leukemia (Jin et al., 2006; Krause et al., 2006) as well as for the growth of both primary and metastatic tumors (Ahrens et al., 2001; Yu et al., 1997). We note that the CD44 in the presently described breast tumor model is indispensable not only for tumor growth but also for tumor-initiating ability, which correlates with its critical role in fostering anchorage-independent growth (Figure 7C). While CD44 has been considered only as a marker of breast cancer stem cells, the present observations indicate that it also contributes in functionally important ways to the maintenance of the tumor-initiating ability of transformed cells (Al-Hajj et al., 2003).
In a fashion similar to its actions in untransformed breast epithelial cells, p53 operating in lung carcinoma cells suppresses CD44 protein expression, thereby precluding it from antagonizing p53 function. Most studies of p53 have suggested that the loss of p53 function enables cancer cells to escape p53-induced cytostasis and/or apoptosis that would otherwise be triggered by the multiple cell-physiologic stresses encountered at various stages of tumor formation. The present results indicate another important benefit conferred on cancer cells by p53 loss – an increased resistance to apoptosis and responsiveness to mitogenic signals resulting from elevated CD44 levels.
In contrast to the ability of p53 to repress CD44 expression, its paralogue, p63, acts in an opposing fashion to stimulate CD44 expression, as recently reported by others in the context of a human head-and-neck carcinoma cell line (Boldrup et al., 2007). We note that suppression of p63 levels resulted in significant loss of CD44 expression in BPEC-T cells expressing primarily the deltaN (DN) isoform of p63, in their tumorigenic BPLER derivatives, as well as in human lung adenocarcinoma cells (Figure 7A and Figure S12A–C). Hence, p63 stimulation of CD44 expression operates in multiple cell types and in both normal and transformed cells. The mechanism of this activation by p63 and its physiologic relevance require further investigation. Nonetheless, it is already apparent that CD44 protein expression is positively regulated, by a transcription factor that is known to be essential for normal epithelial development (Mills et al., 1999; Yang et al., 1999) and for the proliferative potential of epithelial stem cells (Senoo et al., 2007).
EXPERIMENTAL PROCEDURES
Plasmids and constructs
The human CD44 promoter (2021bp fragment upstream of transcription initiation site) was PCR amplified from chromosomal DNA of HMLE cells (Elenbaas et al., 2001) using primers 5′-AGCTCCTGAATCCATGCTGT-3′ (forward) and 5′-CTTCGCAGACAGCTCACTTG-3′ (reverse), re-amplified with primers introducing NheI (5′-ACTATGCTAGCCTGAATCCATGCTG-3′) or XhoI (5′-ATCAACTCGAGGGTGTCCGGAGCGAA-3′) restriction sites. The resulting fragment was cloned into a pGL3 luciferase reporter vector (Promega) and sequence was verified. The potential p53-binding site was mutated by QuickChange site-directed mutagenesis kit (Stratagene), introducing the primer 5′-AGTGGGGCTCGGAGGTCCAGCCACCCCGCGACA-3′ and the resulting construct was verified by sequencing. The standard form of CD44 (CD44s) was subcloned from CDM8 construct into retroviral vector pWZL-blasticidin.
Antibodies
For immunohistochemistry we used CD44 antibody (Becton Dickinson, IM7). For immunofluorecent detection we used phycoerythrin (PE)-conjugated CD44 antibody (Becton Dickinson, Nr.550989). For western blotting we used CD44 antibody MEM-85 (from V. Horejsi) and anti-CD44H antibody (R&D Systems). For p53 and p21 immunofluorescence and/or western blotting analyses we used rabbit anti-p53 antibody (Santa Cruz, FL-393) and mouse monoclonal antibody to p21 (Santa Cruc, sc-817). For western blotting detection of anti-Akt and anti-P-Akt we used antibodies from Cell Signaling.
Cell culture
HMEC-T cells were cultured in MEGM media with bovine pituitary extract. BPEC-T and BPLER cells were cultured in chemically-defined WIT media (Ince et al., 2007). The human osteosarcoma cell line SAOS-2 (ATCC no.HTB85) and lung adenocarcinoma A549 cells were maintained in DME medium supplemented with 10% fetal calf serum, 100units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine at 37 °C in 5% CO2. For western blotting analysis of CD44 and p53 expressions, the A549 cells were cultured in WIT medium. For promoter reporter assays, the SAOS-2 cells were cultured in WIT or DME medium.
Immunofluorescence
Cultured cells were fixed with 100% methanol, blocked with 10% calf serum and stained sequentially with primary rabbit antibody against either p53 or p21, incubated with secondary antibodies conjugated to Alexa-488, and followed by incubation with pan CD44 antibody directly conjugated to phycoerythrin (BD Pharmingen).
Semi-quantitative RT-PCR
Complementary DNA from BPEC-T cells was synthesized from 1μg of total RNA and diluted to 100μl. The cDNA solution (0.25-0.06μl) was amplified using primers to the standard region of CD44 gene (5′-CCACGTGGAGAAAAATGGTC-3′ from exon 2 and 5′-CATTGGGCAGGTCTGTGAC-3′ from exon 3) and to the control GAPDH gene (5′-ACCCAGAAGACTGTGGATGG-3′ and 5′-TCTAGACGGCAGGTCAGGTC-3′). The amplified products were resolved by agarose gel electrophoresis.
Promoter reporter assay
SAOS-2 cells or HMEC-T cells were plated 12h prior to transfection in 48-well plates (3 × 104 cells) in culture medium. Cells were transfected with Fugene transfection reagent (Roche) with 200ng of reporter plasmid and/or 10ng of p53 plasmid. 30h after transfection, cell extracts were prepared and luciferase activity was determined according to the vendor instructions (Promega).
Gel shift assay
Gel shift assays were performed using 50 ng of recombinant human p53 (Calbiochem) in 20 μl of binding buffer (10 mM HEPES (pH 7.5), 5 mM NaCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 20% glycerol). When indicated, cold competitor or antibody was added in the concentrations noted. Reactions were incubated at room temperature for 20 min prior to the addition of 0.2 ng of double-stranded oligonucleotides (~6×104 cpm) (sequences shown below), then incubated for additional 20 min at room temperature. The antibodies used for supershift assays were pAb421 (epitope: aa 371–380), and sc-6243 (Santa Cruz). Reactions were electrophoresed on a 4% nondenaturing polyacrylamide gel, which was dried and exposed to film for 16 h at −80°C.
Soft agar assay
A layer of 0.6% agar noble in DME without serum was placed onto 6-cm dishes. BPLER cells were then seeded in 0.3% top agar containing WIT medium atop the first layer. Fresh top agar was added after 1.5 wk, and colonies were counted after 4 wks.
Apoptotic assay
BPEC-T cells, 80% confluent, were incubated 48h with the DNA-damaging agent doxorubicin in cytokine-free M199/F12 (1:1) culture medium supplemented with 100units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine and various concentrations of EGF. Cells were than analyzed by TdT-mediated dUTP nick end labeling (TUNEL) assay or by Hoechst-33342 dye.
Proliferation assay
BPLER or BPEC-T cells (5×104) were plated on 6-well plates in cytokine-free M199/F12 (1:1) culture medium supplemented with 100units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine and various concentrations of EGF. Cells were allowed to proliferate for three days and counted.
Tumorigenic assays
Subcutaneous tumorigenicity assays were performed as described (Elenbaas et al., 2001) with modification. Six- to eight-week-old immuno-compromised athymic nude mice (Ncr-Nu, Taconic) were irradiated with 400 rad 4 hr prior to injection. Cells (106) were resuspended in 100 μl of culture medium, mixed with Matrigel (Becton Dickinson) and injected with a 25-gauge needle into anaesthetized mice. Tumor size was measured every 3–4 days. The time of initial tumor formation was defined as the time when the tumor had reached a diameter of 3 mm. Mice were sacrificed when the tumors grew to 1 cm in diameter or after 5 wk of monitoring. Tumors were fixed in 10% formalin and paraffin-embedded for histological examination or minced and lysed in HEPES-beffered saline containing 1% NP-40 detergent and protease inhibitor mix (Roche).
Statistical analysis
Statistical significance was analyzed by Student’s t test and expressed as p value.
Supplementary Material
Acknowledgments
We thank C. Kuperwasser for tissues from p53-deficient mice, Vaclav Horejsi for anti-CD44 antibodies, B. Seed for CD44s cDNA, Ch. Chen for CD44VE cDNA, S.A. Stewart for lentiviral and retroviral shRNA expression plasmids, N. Albright, A. Godarova, R. Goldsby, W. Guo and P. Raifor reading the manuscript and valuable comments, K. W. Scotto and R. Johnson for p53 mutant constructs and Maki Saitoh for excellent technical assistance.
R.A.W. is an American Cancer Society research professor and a Daniel K. Ludwig Foundation cancer research professor. J.B.’s research group at CCK, KI, is supported by grants from the Swedish Cancer Society, Swedish Research Council and Linne grant, the research funds at Radiumhemmet. This work was funded by grant from Ludwig Center for Molecular Oncology at MIT (R.A.W.), Breast Cancer Research Foundation (R.A.W.), NIH P01 CA080111 (R.A.W.), Elsa U. Pardee Foundation (S.G.), Alexander and Margaret Stewart Trust (R.A.W./S.G.), Advanced Medical research Foundation (R.A.W.), Breast Cancer Research Foundation, NY (T.A.I.), NCI (CA092013) KO8 Mentored Clinical Scientist Development Award (T.A.I) and Dana-Farber/Harvard SPORE in Breast Cancer (CA 089393) Career Development Award (T.A.I).
Footnotes
Competing interest statement
The authors declare that they have no competing financial interest.
References
- Ahrens T, Sleeman JP, Schempp CM, Howells N, Hofmann M, Ponta H, Herrlich P, Simon JC, Morrison H, Sherman LS, et al. Soluble CD44 inhibits melanoma tumor growth by blocking cell surface CD44 binding to hyaluronic acid. Oncogene. 2001;20:3399–3408. doi: 10.1038/sj.onc.1204435. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaz CM, Hu Y, Daniel R, Gaddis S, Kittrell F, Medina D. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene. 2002;21:6366–6376. doi: 10.1038/sj.onc.1205816. [DOI] [PubMed] [Google Scholar]
- Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- Barbour AP, Reeder JA, Walsh MD, Fawcett J, Antalis TM, Gotley DC. Expression of the CD44v2–10 isoform confers a metastatic phenotype: importance of the heparan sulfate attachment site CD44v3. Cancer Res. 2003;63:887–892. [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Coates PJ, Gu X, Nylander K. DeltaNp63 isoforms regulate CD44 and keratins 4, 6, 14 and 19 in squamous cell carcinoma of head and neck. J Pathol. 2007;213:384–391. doi: 10.1002/path.2237. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Zhu H, Chu A, Iida N, Zhang L, Hung MC. Interaction between the adhesion receptor, CD44, and the oncogene product, p185HER2, promotes human ovarian tumor cell activation. J Biol Chem. 1997;272:27913–27918. doi: 10.1074/jbc.272.44.27913. [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Dou J, Pan M, Wen P, Li Y, Tang Q, Chu L, Zhao F, Jiang C, Hu W, Hu K, Gu N. Isolation and identification of cancer stem-like cells from murine melanoma cell lines. Cell Mol Immunol. 2007;4:467–472. [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol. 2000;32:78–84. doi: 10.1016/s0168-8278(00)80192-0. [DOI] [PubMed] [Google Scholar]
- Gao AC, Lou W, Dong JT, Isaacs JT. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res. 1997;57:846–849. [PubMed] [Google Scholar]
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44(+)CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R, Gatter K, Bartek J, Lane D, Harris AL. Increased expression of mutant forms of p53 oncogene in primary lung cancer. Lancet. 1990;335:675–679. doi: 10.1016/0140-6736(90)90801-b. [DOI] [PubMed] [Google Scholar]
- Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem. 2001;276:27716–27720. doi: 10.1074/jbc.C100121200. [DOI] [PubMed] [Google Scholar]
- Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Piltti K, Kerosuo L, Hakanen J, Eriksson M, Angers-Loustau A, Leppa S, Salminen M, Sariola H, Wartiovaara K. E6/E7 oncogenes increase and tumor suppressors decrease the proportion of self-renewing neural progenitor cells. Oncogene. 2006;25:4880–4889. doi: 10.1038/sj.onc.1209492. [DOI] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, Wakeham A, Shahinian A, Catzavelos C, Rak J, et al. CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity. Blood. 1997;90:2217–2233. [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Bishop JM. Expression of CD44 is repressed in neuroblastoma cells. Mol Cell Biol. 1991;11:5446–5453. doi: 10.1128/mcb.11.11.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren S, Inganas M, Norberg T, Lindgren A, Nordgren H, Holmberg L, Bergh J. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Schonthal AH, Galante J, Stark GR. p130/E2F4 binds to and represses the cdc2 promoter in response to p53. J Biol Chem. 2001;276:1998–2006. doi: 10.1074/jbc.M005101200. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Weber GF, Bronson RT, Ilagan J, Cantor H, Schmits R, Mak TW. Absence of the CD44 gene prevents sarcoma metastasis. Cancer Res. 2002;62:2281–2286. [PubMed] [Google Scholar]
- Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90(+) Cancer Stem Cells in Human Liver Cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61(Suppl 2):1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- Yu Q, Toole BP, Stamenkovic I. Induction of apoptosis of metastatic mammary carcinoma cells in vivo by disruption of tumor cell surface CD44 function. J Exp Med. 1997;186:1985–1996. doi: 10.1084/jem.186.12.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- Zolota V, Tsamandas AC, Melachrinou M, Batistatou A, Scopa C. Expression of CD44 protein in renal cell carcinomas: association with p53 expression. Urol Oncol. 2002;7:13–17. doi: 10.1016/s1078-1439(01)00129-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.