Abstract
The use and benefits of a multimodality approach in the context of breast cancer imaging are discussed. Fusion techniques that allow multiple images to be viewed simultaneously are discussed. Many of these fusion techniques rely on the use of color tables. A genetic algorithm that generates color tables that have desired properties such as satisfying the order principle, the rows, and columns principle, have perceivable uniformity and have maximum contrast is introduced. The generated 2D color tables can be used for displaying fused datasets. The advantage the proposed method has over other techniques is the ability to consider a much larger set of possible color tables, ensuring that the best one is found. We asked radiologists to perform a set of tasks reading fused PET/MRI breast images obtained using eight different fusion techniques. This preliminary study clearly demonstrates the need and benefit of a joint display by estimating the inaccuracies incurred when using a side-by-side display. The study suggests that the color tables generated by the genetic algorithm are good choices for fusing MR and PET images. It is interesting to note that popular techniques such as the Fire/Gray and techniques based on the HSV color space, which are prevalent in the literature and clinical practice, appear to give poorer performance.
Keywords: Image fusion, Image display, Visual perception, Image perception, Image visualization, Clinical image viewing, Magnetic resonance imaging, Positron emission tomography, Breast, Genetic algorithm, Color mixing, Image enhancement, Artificial intelligence, Image analysis, Human visual system
Introduction
Application of a multimodality approach is advantageous for detection, diagnosis, and management of many ailments. Obtaining the spatial relationships between images using different modalities and conveying them to the reader maximizes the benefit that can be achieved. The process of obtaining the spatial relationships and manipulating the images so that corresponding pixels in them represent the same physical location is called image registration. Combining the registered images into a single image is called image fusion.
The advantage of a fused image lies in our inability to accurately visually judge spatial relationships between images when they are viewed side by side. Depending on background texture, mottle, shades, and colors, identical shapes and lines may appear to be different sizes [1]. This can be demonstrated by well-known simple optical illusions. The most obvious application is to combine a functional image that identifies a region of interest but lacks structural information necessary for localization, with an anatomical image providing this information.
In this paper, we examine the benefits of a multimodality approach in the context of breast cancer imaging. We then briefly discuss registration techniques before launching into possible fusion options. An overview of fusion techniques widely accepted in literature as well as a novel genetic algorithm-based one are briefly presented. The remainder of the text is devoted to a study in which radiologists were asked to perform a set of tasks reading fused positron emission tomography (PET)/magnetic resonance imaging (MRI) breast images obtained using several different fusion techniques.
In this context, F-18-FDG PET [2–5] and high-resolution and dynamic contrast-enhanced MRI [6–11] have steadily gained acceptance in addition to X-ray mammography and ultrasonography. Initial experience with combined PET (functional imaging) and MRI (anatomical localization) has yielded positive results, providing benefits compared to either modality alone [12–15]. While no clinical scanners currently exist capable of acquiring images from both modalities, prototypes have been developed [16, 17], and commercial devices are in the not too distant future.
Background
Registration
Since the breast is entirely composed of soft tissue, it easily deforms and requires non-rigid registration. Significant effort has been devoted to developing registration procedures suitable for medical images in general and more recently PET/MRI breast images [12, 18–20]. The registration procedure was selected for this work based on its efficiency, consistency, operator independence, and suitability for use in a clinical environment [18–20].
This finite element method registration procedure relies on corresponding features visible in both modalities (provided by fiducial skin markers (FSMs) taped to predetermined locations on the skin of the breast) and controlled deformations (by similar patient-prone positioning) to produce a dense displacement field describing the deformation between the two modalities. The displacement field can then be used to transform (warp) the MRI image bringing it into spatial alignment with the PET image.
Fusion for Visualization
Even when viewing the registered images side-by-side, spatial relationships may be difficult to ascertain. A combined MRI/PET image has the benefits of directly showing the spatial relationships between the two modalities, increasing the sensitivity, specificity, and accuracy of diagnosis. Most research has been dedicated to obtaining the spatial alignment between images, while comparatively little research has been devoted to finding the optimum fusion technique for presenting the images after registration. This is in part due to the secondary nature of this problem and in part due to the relative rarity of multimodal datasets until the recent clinical availability of PET/CT scanners.
Several factors need to be considered when choosing a visualization technique. These include information content, observer interaction, ease of use, observer understanding, and reader confidence. Undoubtedly, it is desirable to maximize the amount of information present in the fused image. Ideally, the registered images would be viewed as a single image that contains all of the information contained in both the MRI image and the PET image. Limitations in the dynamic range and colors visible on current display devices, as well as limitations in the human visual system make this nearly impossible.
This loss of information can be partially compensated by making the fused display interactive. Some sort of control over the fusion technique can be provided which allows the observer to change the information that is visible in the fused display.
The design of this control is an important part of the fusion process. How simple is it to use the control? How much training is required? Do the display options offered by the control aid the observer or just complicate the observation process? How responsive is the control?
The last and perhaps the most important factor relates to the observer’s understanding of the fused volume. For example, radiologists understand what they are looking at when they examine a grayscale MRI image, or PET image, i.e., variations in intensity and texture have a meaning. In the ideal case, the knowledge and experience the observer has in examining the individual modalities would be directly applicable to the fused images.
Some previous research has been devoted to discover new and optimum ways to take two images and display them as a single one. These techniques, primarily developed and used in other fields, include color overlay, color mixing, techniques based directly on color spaces, and spatial and temporal interlacing. For a review of these techniques, see Baum et al [21].
One very important fact that needs to be kept in mind when selecting the optimum fusion technique is that the choice will be both application and observer dependent.
It is well known that various vision deficiencies, such as deuteranomaly, influence how individuals perceive color. This means that in general, there will not be a fusion technique that is ideal for everyone. Even among those without any documented vision deficiencies, the optimal choice will vary with factors such as experience and training.
It is also necessary to consider environmental factors that affect how observers will perceive the displayed colors. The viewing conditions also play a role in selecting the most appropriate color table. This includes conditions such as the lighting in the room, the color of the background, and the gamma and black offset of the display device.
Throughout this paper, it is assumed that gamma correction, as well as other corrections for the viewing conditions are applied prior to deployment. The focus is on finding the optimum fusion technique for a general radiological audience, and as such, the techniques are not tuned towards any one individual. This needs to be the case for deployment in a clinical setting.
It is a necessity, however, to consider the task that the images produced by the application of the color table are going to be used for. There have been demonstrations of how a properly designed fusion technique can aid a task while an improperly designed one can complicate matters [1]. Because of this, the discussion is constrained to joint display of MRI/PET images.
An interesting fact is that a 2D color lookup table (LUT) can be used to implement all of the previous mentioned fusion techniques with the exception of interlacing. Even when using interactive techniques, the fusion procedure being used at a given instance in time can generally be described using 2D color tables. This work takes advantage of this fact when presenting the fusion techniques studied to the reader.
Two-Dimensional Color Table Basics
Color tables, also known as color maps, are simple yet powerful tools. The grayscale values of the two source images for a given pixel serve as indices in the color map. Looking up the indices in the color map gives the color that the same pixel in the fused image should have.
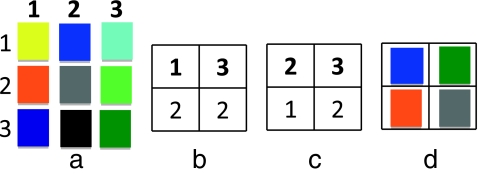
An example of a 3 × 3 color table is shown in Figure 1a. The vertical numbering represents intensity values in the first source image, while the horizontal numbers represent intensity values in the second source image. If the first source image is given by Figure 1b and the second source by Figure 1c, then using this color table to fuse the two images will result in Figure 1d.
Fig. 1.
An example of using a 3 × 3 color table (a) to fuse images represented by (b) and (c), and the result of the operation (d)
Materials and Methods
Fusion Software
Fusion was performed using the Fusion Viewer1 software package [22, 23]. Fusion Viewer was developed in-house and available for free. It has been designed and implemented in C# with a modular object oriented design for increased extensibility and compatibility, along with simplified distribution.
Fusion Viewer provides both traditional and novel options for displaying and fusing 3D datasets. A simple plug-in interface allows rapid development of novel fusion techniques. In addition to fusion capabilities, several options are provided for mapping 16-bit datasets onto an 8-bit display, and both traditional maximum intensity projections (MIP) and MIPs of fused volumes are supported.
Genetic Algorithm Automated Generation of Multivariate Color Tables
A genetic algorithm for automatic generation of color tables that have desired properties is presented here. The advantage the proposed method has over other color table generation techniques is its ability to consider a much larger set of possible color tables, ensuring that the best one is found.
The algorithm searches for 2D color tables satisfying predetermined criteria. First, the underlying definition for the color tables will be discussed and then the criteria, which determine the genetic algorithm’s fitness function, will be presented.
Color Mixing Color Table Definition
Color mixing is a technique that can be used to take any number of one-channel images (N) and create a fused RGB image [21] It is performed using Eq. 1. Here, R, G, B represent the red, green, and blue channels in the displayed image, respectively, Si represents the intensity in the ith source image, Ri, Gi, Bi are the weighting factors for the red channel, green channel, and blue channel, respectively. They determine the contribution of source i to each of the output channels.
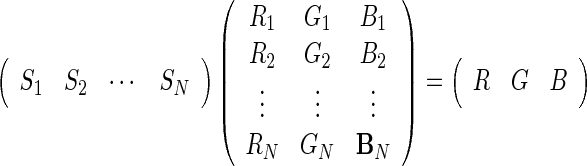 |
1 |
Let the source intensities be normalized from zero to one. Applying Eq. 1 is then equivalent to taking the intensity axis of source i and lying it along the line segment formed by connecting (0,0,0) to (Ri, Gi, Bi) in the RGB color space. The output image is then formed by summing the projections of each of these onto the red, green, and blue axes.
The technique can be extended by allowing the vectors (Ri, Gi, Bi) to point in any direction. For example, as the source intensity increases, the red in the fused image decreases. The technique can also be extended by using an offset so that the vectors (Ri, Gi, Bi) do not need to be located at the origin. After making these extensions, the color mixing technique can be represented by Eq. 2. Where OXi represents the offset from the origin along the X axis for the contribution from source i.
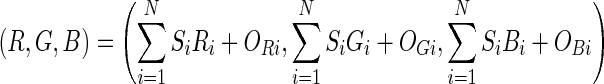 |
2 |
Color Difference
Before an algorithm which generates color tables can be created, there needs to be a way to quantitatively define guidelines or requirements to be used when generating the color tables. The result of evaluating a color table with these guidelines will be the fitness factor used to determine the reproduction of the color tables within the genetic algorithm.
To aid in defining these guidelines, a way to determine the difference between two colors is first introduced. Traditionally, this difference is defined as the Euclidean distance, ∆E, between the two colors in the CIE L*a*b* space and is given by:
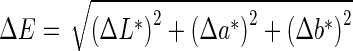 |
3 |
Where L* is related to intensity values (lightness), and a* and b* correspond to chromaticity values; a* and b* represent color-opponent dimensions with a* running roughly from red to green and b* running from yellow to blue. The validity of Eq. 3 comes from the assumption that the color space is perceptually uniform and orthogonal. In order for the color space to be perceptually uniform, the Euclidean distance between two colors within the space needs to be directly related to the perceived closeness of the colors. It has been shown that this assumption is not quite true for the CIE L*a*b* color space [24], since unacceptable errors arise for certain regions of the color gamut [25]. For this reason and for explicit definition in the RGB color space, which is required for display on current hardware, we choose to use the more recently developed computationally efficient measurement used by CompuPhase in their PaletteMaker application [25, 26].
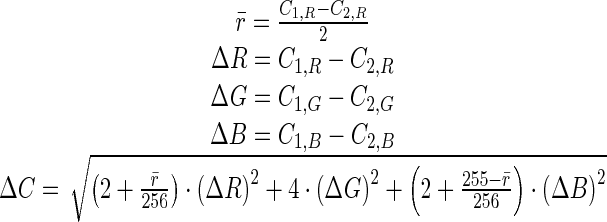 |
4 |
Here, ΔC is the perceived difference between the two colors defined in nonlinear RGB space (sRGB) as (C1,R,C1,G,C1,B) and (C2,R,C2,G,C2,B), where CX,Y is the value for the Yth channel for the Xth color, and has values from 0 to 255. The color difference assumes display on a standard computer monitor (CRT, LCD) with a gamma of approximately 2.5 in a typical office viewing environment.
This definition of color difference is aided in defining the requirements for the color tables which follow.
Color Table Requirements
RGB Color Space It was decided that the color tables produced need to be defined in the 8-bit per channel RGB color space supported by most applications. This is a nonlinear gamma corrected RGB color space, so that colors will appear properly on a typical CRT or LCD display. This is required in order to facilitate easy use and guaranteed compatibility of the color tables produced. This was taken into consideration when selecting the formula for color differences.
Order Principle Trumbo defines several desirable properties of color tables [27]. One of these is the order principle. Basically, if a color table satisfies the order principle, then the colors chosen to represent the data values should be perceived as ordered in the same order as the data values. Spectral color tables where large variations of hue occur do not satisfy this principle. This is important because the pixel values in the original medical data represent physical quantities, such as the standardized uptake values (SUV) in the PET images. This is the information radiologists need to have, if one pixel is shown as blue and another red, the readers will be unable to determine which pixel has a higher SUV without referring to the color table. While the color table will not be a secret from the radiologist evaluating the fused data, the less they need to refer to the color table, the more efficiently and confidently they can read the images.Further, a side effect of a color table not satisfying the order principle is that the color table often creates false segmentation when applied to the image. The color contours created in the image emphasize particular pixel values.To guarantee that the order principle is satisfied, a representation of the color table based on the color mixing technique is used by the algorithm.
Rows and Columns Principle The rows and columns principle is also defined by Trumbo [27]. It states that the colors in the color table should be chosen so that two source images do not obscure one another.This is particularly important in this situation. Each of the input images and their gray levels mean something to the radiologist. This meaning needs to be preserved in the fused images. The reader needs be able to tell the intensity of each of the source images by examining the fused image.This is ensured by making the color’s 1D color tables corresponding to each source as different as possible from each other. In other words, the first row of the color table should consist of colors as different as possible from the first column. This can quantitatively be measured by maximizing ΔC, in Eq. 4, for the average color in the first row of the table and the average color for the first column in the table. This color difference will be referred to as ΔCsources. Due to the linear model of the color mixing technique, this property will then be distributed throughout the rest of the color table.
Perceivably Uniform The ideal color table should be perceivably uniform. In other words, the ΔC between neighboring entries in the color table should be a constant throughout the table. This can be measured by finding ΔC for all neighbors and then examining its variance. The smaller the variance, the better. We will refer to this variance as var(ΔCtable).This is an important factor because it minimizes the reliance on the color table, due to the fact that the radiologist’s intuition about the location of the color in the color table is more likely to be correct.
Maximized Contrast The contribution from each source should have as much contrast as possible. As contrast increases for a source, it gets easier to see the variations in the fused image due to that source. Due to using the color mixing model, we need only to examine the endpoints of the first row and column of the color table to know the range of colors available for each of the sources to use.Maximizing ΔC between the first entry in the first column and the last entry in the first column of the color table will maximize the contrast for the first source. Similarly, maximizing ΔC between the first entry in the first row and the last entry in the first row of the color table will maximize the contrast for the second source.In addition, to ensure good contrast throughout the color table, it is desirable to have the contrast along the diagonal of the color table maximized. This is done by maximizing ΔC for the first entry in the first row and column of the color table and the last entry in the last row and column of the color table. Contrast throughout the remainder of the color table is also evaluated by maximizing the mean ΔC for all neighboring pixels in the color table.In summary, we will evaluate contrast using four factors: the contrast for the first source (ΔCs1), the contrast for the second source (ΔCs2), the contrast along the diagonal (ΔCdiag), and the mean contrast between neighboring pixels mean (ΔCtable).
Desirable Properties Not Considered It should be noted that in the current implementation, the algorithm does not consider all of the desired properties of a color table. For example, no preference is given to any particular color. Humans may find some colors easier to look at and examine for long periods of time than others.Well-known simultaneous contrast and chromatic contrast effects are not considered [28]. These effects describe how the appearance of a particular color may change based on the surrounding colors in the image.Another effect that the human visual system has on images that is usually ignored is that the color of an object influences its perceived size [29, 30]. For example, if a lesion is colored red-purple, it would appear larger than if it had been colored green.
Algorithm Execution
A relatively simple and standard genetic algorithm is used for the generation of the color tables. Each color table is defined by 12 real numbers that have a range from −1 to 1. These numbers represent the following variables from Eq. 2: R1, OR1, G1, OG1, B1, OB1, R2, OR2, G2, OR2, B2, OB2. These coefficients when used with the color mixing equation completely define a color table.
To start, an initial population of color tables is randomly generated. An iterative loop is then entered. Each member of the population is then evaluated and ranked based on the requirements of the desired color table. A new population is then generated, where the contribution from each member of the previous generation to the new generation is based upon its ranking. This process is repeated for a large number of iterations.
Evaluation of a population of color tables involves testing them for each requirement as previously described. The numeric results of the evaluation of a given color table can then be weighted and summed to give the fitness score for that member of the population. This process is shown as Eq. 5. Each of the three desirable properties included in the fitness factor are given equal weight. Prior to creating the fitness factors, each term is normalized by the mean value of that term for the entire population and threshold to a maximum absolute value of 2. This prevents any single term from dominating and insures improvements in any term can influence the fitness factor.
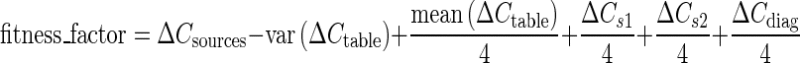 |
5 |
The members of the current generation with the highest fitness scores are automatically included in the next generation. The rest of the members in the next generation are created by splicing or mutating the members in the current population.
When creating a population member by mutation, a member of the previous generation is chosen randomly with a probability proportional to its fitness score. The new population member is then created from the old one by making one or two random changes to its defining coefficients.
When creating a population member by splicing, two members of the previous generation are chosen at random with a probability proportional to their fitness scores. The new population member is generated by taking the first X coefficients of it from the first chosen member and remaining 12-X coefficients from the second member. The point of splicing, X, which determines the amount of each of the chosen color tables that gets transferred to the new color table, is chosen at random.
For the stopping criteria, the algorithm can be halted when the member with the highest fitness score does not change for a number of generations. There is no fear of running the algorithm for too many generations due to the nature of the problem.
After the algorithm has finished executing, the member of the final population with the highest fitness score represents the “best” color table that the algorithm could come up with. The algorithm can either be run several times or the top members of the final population can be considered, providing a set of color tables that can then be evaluated by human observers.
Algorithm Advantages
The genetic algorithm proposed here provides several advantages currently not available elsewhere. It searches the entire color gamut for linear mapping models that maximize properties that are believed to allow accurate interpretation of the images being fused. Maximized contrast allows larger differences and consequently easier differentiation between colors. Adherence to the order principle and uniformity allows interpretation of the colors in a natural intuitive manner. Adherence to the rows and columns principle allows a more accurate differentiation of the information from the images fused.
As will be shown in the following section, numerous fusion techniques already exist and are commonly used. Existing methods have been selected mainly because they may produce esthetically pleasing images or because they are believed to adhere to one of these desired properties. The color tables produced using this genetic algorithm are the first ones to be quantitatively evaluated for their ability to meet all of these properties simultaneously.
Fusion Techniques Investigated
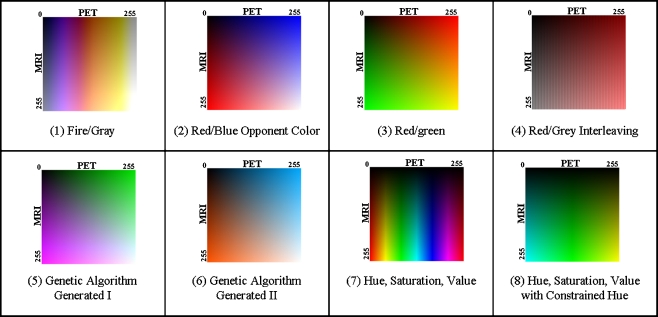
In addition to two color tables produced by the genetic algorithm, six other promising fusion for visualization techniques were selected by the authors to be investigated further. Each of these techniques is shown in Figure 2 and discussed below:
Fig. 2.
Color tables for the different fusion techniques investigated in the study. The vertical direction corresponds to the MRI intensity and the horizontal to the PET intensity. The origin is located in the upper left
Fire/Gray
This technique was created using color overlay. The fire color table from ImageJ [31] was applied to the PET image and a grayscale color table to the MRI image. Color was used for PET and variations in intensity for MRI, since MRI is a higher resolution modality, and the human visual system is more sensitive to small changes in intensity than to small changes in color [32–34]. The technique was selected for study due to its use in other research applications.
Red/Blue
This technique is based on the CIE L*a*b* color space. It is performed by assigning shades of blue to the PET image and shades of red to the MRI image. These colors are selected based on opponent color theory [35]. The basic assertion of opponent color theory is that red and blue are perceived separately and changes to one will not affect the perceived amount of the other. This assertion does not hold for most other color pairs.
Red/Green
This is another color overlay technique. A red color table is applied to the PET image and a green color table to the MRI image. It was selected for inclusion in the study because of its use in the Mayo Clinic’s Analyze [36] software package.
Gray/Red Interlace
This fusion technique is based on spatial interlacing. Fused images were created by taking alternating columns from each of the source images. For example, the odd columns in the fused image are taken from odd columns in the PET image while the even columns are taken from even columns in the MRI image. The PET image was first pseudo-colored using a red color table. This technique was selected for the study because of its popularity in the research literature [1, 37, 38].
Hue, Saturation, Value
The hue, saturation, value (HSV) color space is a model designed to describe how humans perceive color. Hue is the color, saturation is the amount of the color, and value is the intensity of the color. For this technique, hue was used to present the PET image, and value was used to present the MRI image. The saturation was kept constant. Like the Fire/Gray technique, the MRI was used for the intensity because of the human visual system’s higher sensitivity to changes in intensity over changes in color. This color table was selected because of its popularity throughout the literature.
HSV, Constrained Hue
This technique is similar to the previously discussed hue, saturation, value technique except that it uses a constrained hue angle. Instead of the color varying over the entire spectrum, it was only varied within the cyan to green to yellow region [33]. Constraining the hue angle helps prevent false segmentation due to changes in color and provides colors that appear to have a natural ordering to a human observer.
The Study
To test the validity of fusion-based visualization, the eight techniques previously discussed and shown in Figure 2 were used to conduct a study with four radiologists from the Department of Radiology at SUNY Upstate Medical University. The study was designed to answer the following questions.
Are the spatial relationships between images better conveyed when viewed side-by-side or as a fused image?
Which fusion techniques do the radiologists prefer?
Which fusion techniques do the radiologists think they can use the best?
Which fusion techniques are easiest for the radiologists to use?
Which fusion techniques most accurately allow the original MRI and PET information to be recovered by the radiologist?
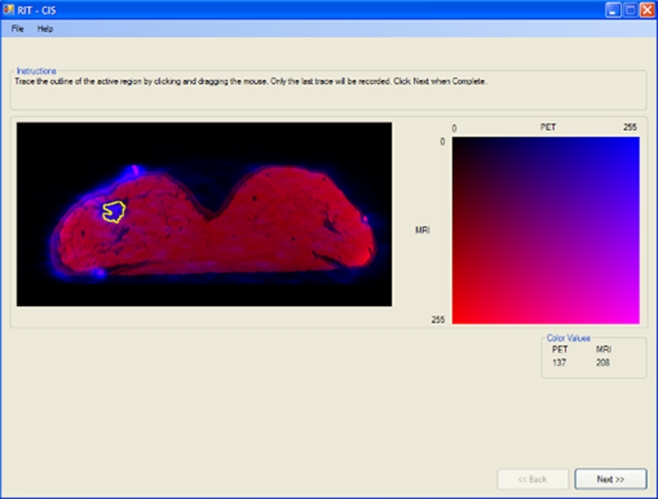
Nine different images with no malignant lesions were presented using each of the eight techniques to four experienced nuclear medicine physicians. Each time, the nine images were presented in a random order. For each physician, the techniques were also presented in a random order. The nuclear medicine physicians were asked to perform the following tasks using the interface shown in Figure 3.
On a PET monochrome image, click on the region of maximum metabolic activity (glandular tissue).
On a grayscale LUT, select the gray level that corresponds to the gray level value of the region of maximum metabolic activity in the PET image.
On an MRI image, click on the morphological region that corresponds to the region of maximum metabolic activity (the same location clicked on the PET image).
On a grayscale LUT, select the gray level that corresponds to the gray level value of the region of maximum activity in the MRI.
Trace the region of maximum metabolic activity on both the PET and MRIs.
On a fused image, click on the region of maximum metabolic activity.
Click on the corresponding color on the LUT to identify the associated PET and MRI values.
Trace the region of maximum metabolic activity on the fused image.
Evaluate degree of difficulty while performing the task.
Evaluate understanding of the fusion technique used.
Indicate preference for the fusion technique.
Fig. 3.
Interface used by radiologists to evaluate fusion techniques
Images used for the study were acquired using a dedicated PET/CT scanner (GE Discovery ST with BGO detector and 4-slice CT) and a 1.5T MRI System (Philips Intera). PET images were obtained with patient prone and breasts freely suspended, immediately after intravenous administration of 10 mCi of F-18-FDG with nine FSMs taped on the skin of each breast. They were reconstructed in a 128 × 128 × 47 matrix, with 4.25 mm voxel size. To assure that the stress conditions in the imaged breast are similar in different modalities, a replica of the MRI breast coil made of plastic with very low absorption for 511 keV photons was used in PET scans.
In MRI scans, the patient was prone with both breasts suspended into a single-well housing a standard Philips clinical breast RF receiver coil. A high-resolution 3D fast field echo technique with TR/TE = 14/3 was applied to obtain MRI breast images. An image matrix of 512 × 512 × 120 was used in reconstruction with 0.7 × 0.7 × 1.4 mm3 voxel size. The field of view (360 × 360 mm) was centered over the breasts.
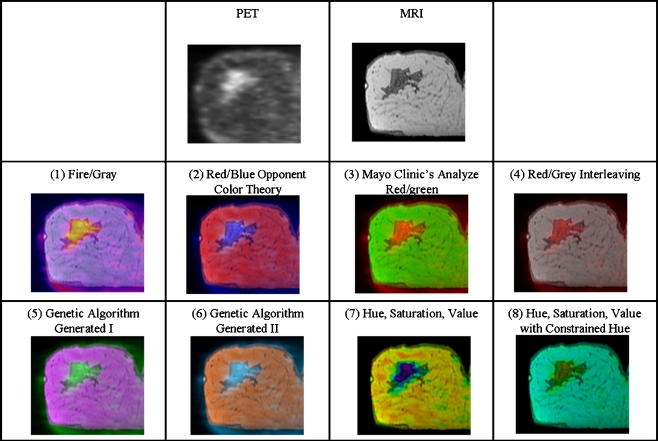
The registration was performed using the previously mentioned procedure. After registration, slices through regions of interest were selected from each dataset for use in the study. The slices were extracted and fused using the Fusion Viewer application and correspond to the standard views, coronal, sagittal, and axial. The regions of interest were selected to be regions containing above average metabolic activity. Figure 4 contains examples of images used during the study.
Fig. 4.
Fused images created using each technique. The PET and MRI source images used are shown in the top row
Results and Discussions
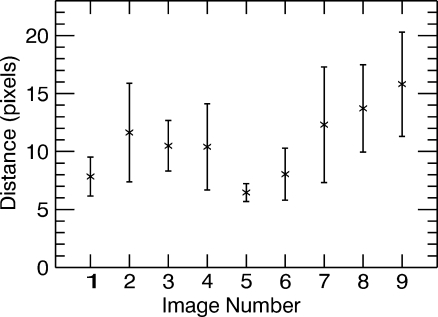
The results presented in Figures 5, 6, 7, 8, 9, and 10 answer the five questions the study was set out to address. Figure 5 shows a plot of the average distance, in pixels, over all participants, between the location of the area of maximum metabolic activity selected on the MRI and the PET grayscale images when viewed side-by-side. One pixel corresponds to area of 0.7 × 0.7 mm. All error bars shown here represent the standard error. It can be seen that the average distance, over all nine images, is approximately 10.7 pixels; in other words, there is up to approximately a 1-cm difference between the two regions selected when viewing PET and MRI images side by side. This discrepancy might increase the risk of performing a biopsy in the wrong location, misdiagnosis, and justifies the need for a fused display.
Fig. 5.
Distance in pixels between corresponding locations chosen on PET and MRI images when viewed side by side
Fig. 6.
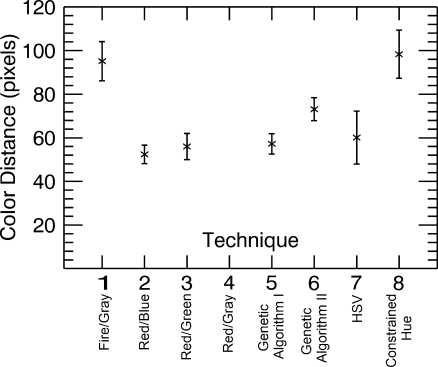
Distance in pixels between corresponding locations chosen on PET and fused images
Fig. 7.
Difficulty ratings assigned when performing tasks for each technique. 1 corresponds to “hardest” and 5 to “easiest”
Fig. 8.
Ability level assigned by observers for each technique. 1 means they do not think they are capable of using it, and 5 means they believe they are an expert at using the technique
Fig. 9.
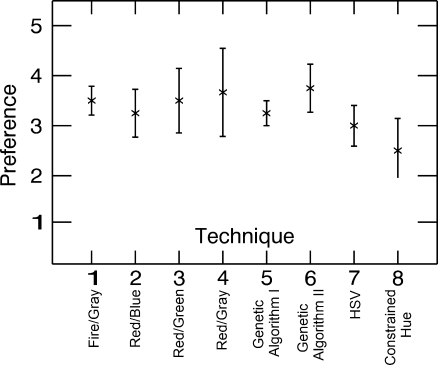
Preference level assigned by observers for each technique. 1 corresponds to no preference and 5 corresponds to a preferred technique
Fig. 10.
Distance in pixels on the color table between the color observers thought they chose from the fused image and color they actually chose. Maximum distance is approximately 362 pixels
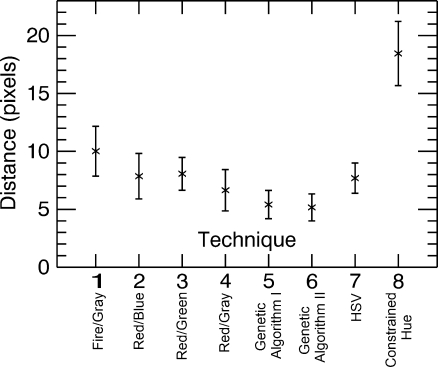
Figure 6, on the other hand, shows the average distance, over all observers and images for each technique, in pixels between the location of the area of maximum metabolic activity selected on the fused image and the corresponding location selected on the grayscale PET image. It can be seen that the average distance is reduced particularly with techniques 5 (Genetic Algorithm I) and 6 (Genetic Algorithm II), the two color tables generated by the genetic algorithm. Performance is worst with technique 8 (Constrained Hue), possibly because as the range of colors gets smaller they become increasingly difficult to differentiate. An analysis of variance (ANOVA) test was performed to statistically quantify the differences between the techniques. Technique 5 (Genetic Algorithm I) shows a significant improvement over techniques 1 (Fire/Gray) and 8 (Constrained Hue; t < 0.05, i.e. difference in the two means is significant), and technique 6 (Genetic Algorithm II) shows a significant improvement over techniques 1 (Fire/Gray), 3 (Red/Green), and 8 (Constrained Hue; t < 0.05).
Note that popular techniques such as the Fire/Gray (technique 1) and techniques based on the HSV color space (techniques 7 and 8), which are prevalent in the literature and clinical practice, appear to give poorer performance. One common misconception is that a larger range of colors will allow smaller variations to be seen, when in reality, sharp changes in color might attract the focus making variations within the individual colors difficult to discern. Also, radiologists do not necessarily have the same skills at interpreting colors and identifying specific characteristics (e.g., hue and intensity) as artists or color specialists might have.
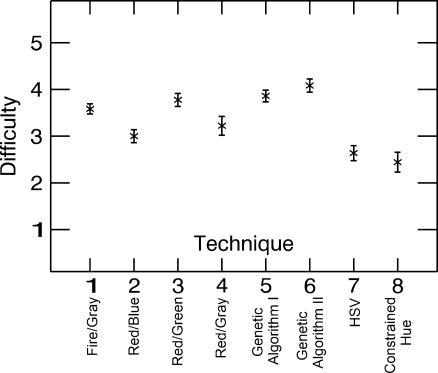
Figure 7 shows results of the difficulty rating assigned by the observer when performing the tasks with each technique, where 5 represents easiest and 1 represents hardest. An ANOVA test was performed to statistically quantify the differences between the techniques. It can be seen that techniques 5 (Genetic Algorithm I) and 6 (Genetic Algorithm II) are ranked significantly easier to use than techniques 1 (Fire/Gray), 2 (Red/Blue), 4 (Red/Gray), 7 (HSV), and 8 (Constrained Hue; t < 0.05). Techniques 1 (Fire/Gray) and 3 (Red/Green) also have high rankings, which may reflect past experience using those techniques.
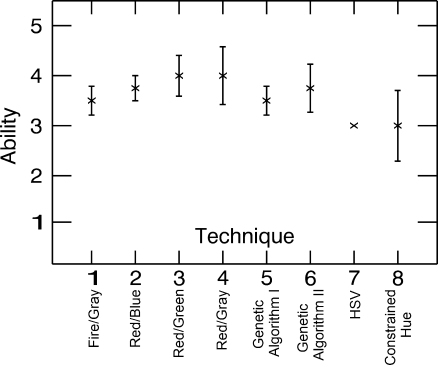
Figure 8 shows the level of ability each observer believes he or she has for using each of the techniques, where 1 means they do not think they are capable of using it and 5 means they believe they are experts at using the technique. The rankings are fairly even with techniques 3 (Red/Green) and 4 (Red/Gray) having the highest and techniques 7 (HSV) and 8 (Constrained Hue) the lowest. It is not surprising that techniques 7 and 8 rank the lowest, as it is unlikely that radiologists have significant experience describing colors using HSV color space.
Figure 9 shows the preferences assigned to each technique by the observers. No technique is given significantly higher preference than any other. The higher preferences are assigned to techniques 1 (Fire/Gray), 3 (Red/Green), 4 (Red/Gray), and 6 (Genetic Algorithm II). It is interesting to note that the techniques the observers claimed to prefer and understand the best were not necessarily the ones with which they were best able to complete the tasks. This suggests that thorough testing should be performed before selecting a fusion technique rather than choosing the technique most radiologists seem to prefer.
Figure 10 shows the distance on the color tables between the color at a point selected in the fused images and that color as identified on the color table by the observer. This distance represents the observer’s ability to decode the coloring used for the fusion. It represents the ability of the observer to recover the original MRI and PET value at a given point in the image. Observers achieved the smallest distances with techniques 1 (Fire/Gray), 3 (Red/Green), and 5 (Genetic Algorithm I). Technique 4 (Red/Gray) could not be evaluated with this metric since colors could appear at more than one location in the color table.
This study consisted of multiple tasks and a large amount of collected data. A subset of the results, those that most directly answer the proposed study questions, has been presented here. Further details on the study, including public access to the tools used, data used, and data collected (both raw and analyzed) can be found in an electronic archive of the study.[39] This includes the results not presented here such as those from tracing the region of higher metabolic activity.
Conclusions
This initial limited study clearly demonstrates the need and benefit of a joint display because of the inaccuracy when using a side-by-side display. In many cases, the differences between the techniques are qualitatively and quantitatively significant. The study suggests that the color tables generated by the genetic algorithm, particularly technique 5, are good choices for fusing MRI and PET images. This is best illustrated in Figure 6, which demonstrates the spatial accuracy of the technique. This property is hardest to obtain through other tools, as compared, for example, to the MRI and PET values for a point in the image, which are usually accessible to the observer through other means.
The genetic algorithm shows promising results, although it needs to be refined to consider additional desired properties. The test for the compliance of the rows and columns principle should consider the implications of opponent color theory. Further, the algorithm should be expanded to consider the set of all 2D color tables and not just those that can be represented using the extended color mixing model. The most complex part of this expansion will be the redefinition of the tests for the desired properties. The current results can be made interactive by simple scaling of the source axes, but attempts to generate fully interactive fusion operators may provide interesting results and useful insights.
The fusion techniques used during this study were applied to 2D datasets (actually a single cross-section of 3D datasets). Since the operators are applied locally at the voxel level, they can be applied to full 3D datasets without any modification. The fused images can then be viewed as a series of slices (or orthogonal reconstructions) as is most commonly done in practice today.
The results presented here are not translatable if the desire is the fused 3D datasets that have been rendered using projection based or surface-based techniques. In these cases, fusion is no longer occurring at the voxel level, and other fusion methods will be more appropriate for handling the information provided in the renderings.
It is clear that a larger study including a greater number of readers should be performed to confirm and support the results of this study. Observer performance studies ought to be performed, and interactive versions of the fusion techniques need to be investigated due to their ability to present significantly more information from the underlying images, as compared to static techniques. The most appropriate technique will most likely depend on the specific tasks, and future studies should focus on evaluation while the radiologist is performing specific clinical duties.
The study does make clear the need for fused displays and encourages fusion capabilities to be introduced into PACS and RIS systems. It stresses the need for manufacturers of multimodal systems, such as PET/CT, to investigate ways to improve the presentation of the acquired data.
Acknowledgements
We wish to thank Drs. M. Lisi, M. McGrath, J. Tam, and M. Khan for their participation in this study. The authors wish to thank the reviewers who helped improve this manuscript with their comments.
This research was partially supported by Carol M. Baldwin Breast Cancer Research Award and by the Department of Radiology at SUNY Upstate Medical University.
Footnotes
References
- 1.Rehm K, Strother SC, Anderson JR, Schaper KA, Rottenberg DA. Display of merged multimodality brain images using interleaved pixels with independent color scales. J Nucl Med. 1994;35:1815–1821. [PubMed] [Google Scholar]
- 2.Bombardieri E, Crippa F. PET imaging in breast cancer. Q J Nucl Med. 2001;45:245–256. [PubMed] [Google Scholar]
- 3.Scheidhauer K, Walter C, Seemann MD. FDG PET and other imaging modalities in the primary diagnosis of suspicious breast lesions. Eur J Nucl Med Mol Imaging. 2004;31(Suppl. 1):S70–S79. doi: 10.1007/s00259-004-1528-7. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu S, Kinahan P, Tseng J, Dunnwald LK, Schubert EK, Pham P, Lewellen B, Mankoff DA. SUV varies with time after injection in 18F-FDG PET of breast cancer: characterization and method to adjust for time differences. J Nucl Med. 2003;44(7):1044–1050. [PubMed] [Google Scholar]
- 5.Eubank WB, Mankoff D, Bhattacharya M, Gralow J, Linden H, Ellis G, Lindsley S, Austin-Seymour M, Livingston R. Impact of FDG PET on defining the extent of disease and on the treatment of patients with recurrent or metastatic breast cancer. AJR Am J Roentgenol. 2004;183(2):479–486. doi: 10.2214/ajr.183.2.1830479. [DOI] [PubMed] [Google Scholar]
- 6.Eliat PA, Dedieu V, Bertino C, Boute V, Lacroix J, Constans JM, Korvin B, Vincent C, Bailly C, Joffre F, Certaines J, Vincensini D. Magnetic resonance imaging contrast-enhanced relaxometry of breast tumors: an MRI multicenter investigation concerning 100 patients. Magn Reson Imaging. 2004;22:475–481. doi: 10.1016/j.mri.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs P, Liney GP, Lowry M, Kneeshaw PJ, Turnbull LW. Differentiation of benign and malignant sub-1 cm breast lesions using dynamic contrast enhanced MRI. Breast. 2004;13:115–121. doi: 10.1016/j.breast.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Stoutjesdijk MJ, Boetes C, Jager GJ, Beex L, Bult P, Hendriks JH, Laheij RJ, Massuger L, Die LE, Wobbes T, Barentsz JO. Magnetic resonance imaging and mammography in women with a hereditary risk of breast cancer. J Natl Cancer Inst. 2001;93:1095–1102. doi: 10.1093/jnci/93.14.1095. [DOI] [PubMed] [Google Scholar]
- 9.Warner E, Plewes DB, Shumak RS, Catzavelos GC, Prospero LS, Yaffe MJ, Goel V, Ramsay E, Chart PL, Cole DE, Taylor GA, Cutrara M, Samuels TH, Murphy JP, Murphy JM, Narod SA. Comparison of breast MRI, mammography, and ultrasound for surveillance of women at high risk for hereditary breast cancer. J Clin Oncol. 2001;19:3524–3531. doi: 10.1200/JCO.2001.19.15.3524. [DOI] [PubMed] [Google Scholar]
- 10.Wright H, Listinsky J, Rim A, Chellman-Jeffers M, Patrick R, Rybicki L, Kim J, Crowe J. Magnetic resonance imaging as a diagnostic tool for breast cancer in premenopausal women. Am J Surg. 2005;190:572–575. doi: 10.1016/j.amjsurg.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Baum F, Fischer U, Vosshenrich R, Grabbe E. Classification of hypervascularized lesions in CE MR imaging of the breast. Eur Radiol. 2002;12(5):1087–1092. doi: 10.1007/s00330-001-1213-1. [DOI] [PubMed] [Google Scholar]
- 12.Moy L, Ponzo F, Noz ME, Maguire GQ, Jr, Murphy-Walcott AD, Deans AE, Kitazono MT, Travascio L, Kramer EL. Improving specificity of breast MRI using prone PET and fused MRI and PET 3D volume datasets. J Nucl Med. 2007;48(4):528–537. doi: 10.2967/jnumed.106.036780. [DOI] [PubMed] [Google Scholar]
- 13.Noz ME, Maguire GQ, Jr, Moy L, Ponzo F, Kramer EL. Can the specificity of MRI breast imaging be improved by fusing 3D MRI volume data sets with FDG PET? IEEE Int Symp Biomed Imaging. 2005 [Google Scholar]
- 14.Goerres GW, Michel SC, Fehr MK, Kaim AH, Steinert HC, Seifert B, Schulthess GK, Kubik-Huch RA. Follow-up of women with breast cancer: comparison between MRI and FDG PET. Eur Radiol. 2003;13:1635–1644. doi: 10.1007/s00330-002-1720-8. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Moore MO, Lehman CD, Mankoff DA, Lawton TJ, Peacock S, Schubert EK, Livingston RB. Combined use of MRI and PET to monitor response and assess residual disease for locally advanced breast cancer treated with neoadjuvant chemotherapy. Acad Radiol. 2004;11:1115–1124. doi: 10.1016/j.acra.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Pichler BJ, Judenhofer MS, Catana C, Walton JH, Kneilling M, Nutt RE, Siegel SB, Claussen CD, Cherry SR. Performance test of an LSO-APD detector in a 7-T MRI scanner for simultaneous PET/MRI. J Nucl Med. 2006;47:639–647. [PubMed] [Google Scholar]
- 17.Catana C, Wu Y, Judenhofer MS, Qi J, Pichler BJ, Cherry SR. Simultaneous acquisition of multislice PET and MR images: initial results with a MR-compatible PET scanner. J Nucl Med. 2006;47:1968–1976. [PubMed] [Google Scholar]
- 18.Coman I, Krol A, Feiglin D, Lipson E, Mandel J, Baum KG, Unlu M, Wei L. Intermodality nonrigid breast-image registration. IEEE Int Symp Biomed Imaging. 2005 [Google Scholar]
- 19.Krol A, Unlu MZ, Baum KG, Mandel JA, Lee W, Coman IL, Lipson ED, Feiglin DH. MRI/PET nonrigid breast-image registration using skin fiducial markers. Phys Med. 2006;21(Suppl. 1):39–43. doi: 10.1016/S1120-1797(06)80022-0. [DOI] [PubMed] [Google Scholar]
- 20.Baum KG: Multimodal Breast Image Registration, Visualization, and Image Synthesis, PhD Dissertation, Rochester, NY: Rochester Institute of Technology, Chester F. Carlson Center for Imaging Science, 2008
- 21.Baum KG, Helguera M, Hornak JP, Kerekes JP, Montag ED, Unlu MZ, Feiglin DH, Krol A. Techniques for fusion of multimodal images: application to breast imaging. Proc IEEE Int Conf Image Process. 2007 [Google Scholar]
- 22.Baum KG, Helguera M, Krol A. A new application for displaying and fusing multimodal data sets. Proc SPIE Multimodal Biomed Imaging II. 2007 [Google Scholar]
- 23.Baum KG, Helguera M, Krol A. Fusion viewer: a new tool for fusion and visualization of multimodal medical data sets. J Digit Imaging. 2008;21(Suppl. 1):S59–S68. doi: 10.1007/s10278-007-9082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granger EM: “Is CIE L*a*b* good enough for desktop publishing?”. Technical report, Light Source Inc., 1994
- 25.Rimersma T, ITB CompuPhase: Colour metric. Available at http://www.compuphase.com/cmetric.htm. Accessed 17 August 2010
- 26.CompuPhase: PaletteMaker. Available at http://www.compuphase.com/software_palettemaker.htm. Accessed 17 August 2010
- 27.Trumbo BE. Theory for coloring bivariate statistical maps. Am Stat. 1981;35(4):220–226. doi: 10.2307/2683294. [DOI] [Google Scholar]
- 28.Ware C. Color sequences for univariate maps: theory, experiments, and principles. IEEE Comput Graph Appl. 2002 [Google Scholar]
- 29.Tedford WH, Jr, Bergquist SL, Flynn WE. The size-color illusion. J Gen Psychol. 1997;97:145–149. doi: 10.1080/00221309.1977.9918511. [DOI] [PubMed] [Google Scholar]
- 30.Rheingans P: Task-based Color Scale Design. SPIE Applied Image and Pattern Recognition, 35–43, 2000. doi: 10.1117/12.384882
- 31.NIH: ImageJ. Available at http://rsb.info.nih.gov/ij/. Accessed 17 August 2010
- 32.Russ JC. The Image Processing Handbook. 4. Boca Raton: CRC; 2002. [Google Scholar]
- 33.Agoston AT, Daniel BL, Herfkens RJ, Ikeda DM, Birdwell RL, Heiss SG, Sawyer-Glover AM. Intensity-modulated parametric mapping for simultaneous display of rapid dynamic and high-spatial-resolution breast MR imaging data. Radiographics. 2001;21:217–226. doi: 10.1148/radiographics.21.1.g01ja22217. [DOI] [PubMed] [Google Scholar]
- 34.Sammi MK, Felder CA, Fowler JS, Lee JH, Levy AV, Li X, Logan J, Pályka I, Rooney WD, Volkow ND, Wang GJ, Springer CS., Jr Intimate combination of low- and high-resolution image data: I. Real-space PET and H2O MRI, PETAMRI. Magn Reson Med. 1999;42:345–360. doi: 10.1002/(SICI)1522-2594(199908)42:2<345::AID-MRM17>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 35.Buchsbaum G, Gottschalk A. Trichromacy, opponent colours coding and optimum colour information transmission in the retina. Proc R Soc Lond B Biol Sci. 1983;220(1218):89–113. doi: 10.1098/rspb.1983.0090. [DOI] [PubMed] [Google Scholar]
- 36.Mayo Biomedical Imaging Resource: Analyze Software System. Available at http://www.mayo.edu/bir/Software/Analyze/Analyze.html. Accessed 17 August 2010
- 37.Spetsieris PG, Dhawan V, Ishikawa T, Eidelberg D. Interactive visualization of coregistered tomographic images. Proc Biomed Vis. 1995;86:58–63. [Google Scholar]
- 38.Lee JS, Kim B, Chee Y, Kwark C, Lee MC, Park KS. Fusion of coregistered cross-modality images using a temporally alternating display method. Med Biol Eng Comput. 2000;38:127–132. doi: 10.1007/BF02344766. [DOI] [PubMed] [Google Scholar]
- 39.Rafferty K, Baum KG, Schmidt E, Krol A, Helguera M: Multimodal display techniques with application to breast imaging. RIT digital media library, http://hdl.handle.net/1850/4648, 2007