Abstract
We propose the use of a context-sensitive support vector machine (csSVM) to enhance the performance of a conventional support vector machine (SVM) for identifying diffuse interstitial lung disease (DILD) in high-resolution computerized tomography (HRCT) images. Nine hundred rectangular regions of interest (ROIs), each 20 × 20 pixels in size and consisting of 150 ROIs representing six regional disease patterns (normal, ground-glass opacity, reticular opacity, honeycombing, emphysema, and consolidation), were marked by two experienced radiologists using consensus HRCT images of various DILD. Twenty-one textual and shape features were evaluated to characterize the ROIs. The csSVM classified an ROI by simultaneously using the decision value of each class and information from the neighboring ROIs, such as neighboring region feature distances and class differences. Sequential forward-selection was used to select the relevant features. To validate our results, we used 900 ROIs with fivefold cross-validation and 84 whole lung images categorized by a radiologist. The accuracy of the proposed method for ROI and whole lung classification (89.88 ± 0.02%, and 60.30 ± 13.95%, respectively) was significantly higher than that provided by the conventional SVM classifier (87.39 ± 0.02%, and 57.69 ± 13.31%, respectively; paired t test, p < 0.01, and p < 0.01, respectively). We conclude that our csSVM provides better overall quantification of DILD.
Keywords: Computed tomography, Computer-aided diagnosis, Image processing, Lung diseases
Introduction
The use of automated classification systems for the detection of diseased or abnormal tissues is becoming an important aspect of computer-aided diagnosis. Several automatic classification systems have been proposed for classifying lung disease images. Uppaluri et al. developed systems for classifying emphysema and diffuse interstitial lung disease (DILD) using 2-D textural analysis [1, 2]. Xu et al. developed a system that used 3-D textural analysis to solve the same problem [3, 4]. Prasad et al. proposed a multi-level classification method for emphysema classification [5], and Chabat et al. proposed a textural system for identifying obstructive lung disease [6]. Support vector machines (SVMs) are often used for image classification and have been shown to outperform other classifiers, such as Bayesian classifiers, artificial neural networks (ANNs), and generalized linear models [7–9]. SVM classifiers have been used to identify emphysema [10–12] and classify five regional disease patterns and normal tissues in DILD patients [13, 14].
In clinical practice, application of SVMs in selected region of interest (ROI)-based data with unambiguously classified regions is needed to extend this approach to whole lung data. Whole lung quantification allows for the evaluation and quantification of disease patterns over the entire lung, thereby allowing assessment of pathogenesis and response to therapy. Previous research has attempted to classify whole lung tissue, but these systems have been limited because they could not identify small areas of initial disease pattern due to the relatively large size of the classification pixels (i.e., 15 × 15) [3, 4] or because they had poor accuracy [14].
Furthermore, most traditional classifiers assume that regions in the image data may be independently classified into several classes. Thus, these classifiers label an ROI based on its feature properties and do not consider spatial correlations between neighboring ROIs. In the specific case of high-resolution computerized tomography (HRCT) of the lung, neighboring ROIs are highly correlated because regional disease patterns typically contain more than one ROI. Traditional classifiers do not consider the rich information that can be gleaned by considering the regions neighboring each ROI.
In the context of remote sensing image classification, researchers have proposed the use of context-sensitive classifiers that consider neighboring information in the appropriate classification of ROIs [15]. In general, context-sensitive classification methods yield excellent classification results for high-resolution images because high-resolution images contain more ROIs. For this reason, incorporation of spatial correlations and relationships provides contextual knowledge to the classifier and improves classification accuracy.
Previous image analysis approaches have addressed this issue. Remote sensing problems, such as land cover classification at the rural–urban fringe, have been analyzed by assuming a probabilistic relaxation of spatial information [16–19]. More recent research has incorporated context information by using a Markov random field (MRF), which models the spatial correlations between neighboring ROIs by modeling the joint probability of observation and the corresponding class for processing of remote sensing images [20, 21]. In the MRF framework, however, efforts have been devoted to modeling the joint probability, which incorporates modeling of intrinsic classes. The conditional random field (CRF), another random field approach, was developed to model the distribution P(Y|X) directly, where Y is the joint label and X is the observation [22]. This model is less computationally demanding than the MRF [23].
Several CRF framework methods, such as the discriminative random field and the support vector random field, have been applied to the segmentation of man-made structures [24] and brain tumors [8]. Although these approaches performed better than the MRF, they were developed for binary image segmentation and could not be applied if more than two classes were present. In the present study, we hypothesized that, in the case of diffuse lung disease, neighboring ROIs will be correlated, and hence that consideration of local contextual information for a ROI under consideration will improve the classification accuracy. To test this hypothesis, we developed a context-sensitive support vector machine (csSVM) and compared the accuracy of the csSVM with that of an existing multiclass SVM [25] by incorporating local context information into the classification of normal and regional disease patterns across whole lung images. In addition, we improved previous random field approaches using the heuristic modification to extend the binary classification problem to multiclass problems.
Materials and Methods
Automatic Quantification System
HRCT images of the whole chest were selected from a collection of images from 106 patients in the department of radiology, Asan Medical Center (Seoul, South Korea). All images were acquired with 220 mA and 140 kVp, with the patients at full inspiration. Images were reconstructed (1 mm slice thickness, 10 mm slice intervals) using an enhancing (B70f) reconstruction kernel with a 16-detector row CT (Sensation 16, Siemens Medical Solutions, Forchheim, Germany).
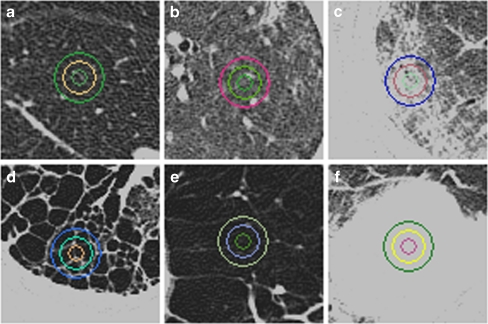
For the training set, a thoracic radiologist with 10 years of clinical experience assessed a total of 900 ROIs (20 × 20 pixels) and marked them as normal (n = 150), ground-glass opacity (n = 150), reticular opacity (n = 150), honeycombing (n = 150), emphysema (n = 150), or consolidation (n = 150; Fig. 1). The size of each ROI (20 × 20) was assessed according to previous methods [13] and expert knowledge for considering subtleties associated with the size of diffuse lung disease patterns. Only one ROI was selected in each image to minimize clustering effects. If several ROIs were selected in one patient, the ROIs were chosen from different lobes.
Fig. 1.
HRCT scans of the chest (window level, −850 HU; width, 400 HU). On each image, note the three different sizes of circular (16-, 32-, and 64-pixel diameters) highlighted regions of interest (ROI) typical of any particular condition. a Normal, b ground-glass opacity, c reticular opacity, d honeycombing, e emphysema, f consolidation
A total of 13 textural and 8 shape features were computed to characterize each ROI. The textural features were histogram features (mean, SD, skewness, and kurtosis), gradient features (mean and SD), run-length matrices (short- and long primitive emphasis), and gray-scale level co-occurrence (contrast, correlation, energy, and homogeneity) [6]. The bin size (Q) of the run-length encoding and co-occurrence were optimized to 196 and 32, respectively [26]. The shape features were from three types of white top-hat transforms (mean and SD), black top-hat transforms (mean and SD), and cluster analysis (number of clusters, cluster area mean and SD, circularity mean and SD, and aspect ratio mean and SD). Thus, each ROI was represented as a 21-dimensional feature vector with the values given in Table 1.
Table 1.
Summary of 13 textural features and 8 shape features that represent each ROI
| Category | Descriptor | Dimension |
|---|---|---|
| Textual feature | Histogram | Mean |
| S.D. | ||
| Skewness | ||
| Kurtosis | ||
| Gradient | Mean | |
| S.D. | ||
| Run-length matrix | Short primitive emphasis | |
| Long primitive emphasis | ||
| Co-occurrence matrix | Angular second moment | |
| Contrast | ||
| Correlation | ||
| Inverse difference moment | ||
| Entropy | ||
| Shape feature | Top-hat transform | White top-hat mean |
| White top-hat S.D. | ||
| Black top-hat mean | ||
| Black top-hat S.D. | ||
| Cluster | LAA Area | |
| Number of cluster | ||
| Mean | ||
| S.D. |
The validity of these descriptors for characterizing the textural and shape information during classification of lung parenchyma has been demonstrated previously [7, 10–12, 14]. Applying all 21 features listed in Table 1, we conducted sequential forward feature selection (SFS), which showed superior performance in previous studies [6, 10, 12], to remove irrelevant features that might have led to misclassification.
A near-optimal feature set of 900 ROIs, calculated from SFS, was employed to build the classifier. Subsequently, 84 whole lung images were quantified by the previously trained classifier. In addition, a radiologist was asked to draw an area map of 84 whole lung images, using dedicated in-house software, to validate the accuracy of the classification by the ROI comparison. The basic diagram for the automatic quantification system is shown in Fig. 2.
Fig. 2.
Automatic quantification system procedure diagram used for the detection of diffuse interstitial lung disease
Proposed Method
Overview
Our method employs a multiclass SVM [25]. The two components, observation potential and local context potential, capture the relationship between observation and labeling using multiclass SVM, and between a classified ROI and its neighboring ROIs.
Let the observed data from an input image be given as X = {xi} i ∈ S and the corresponding labels as Y = {yi} i ∈ S, where xi is the data from ith ROI, and S is the total set of ROIs. In this work, yi can have n classes, i.e., yi ∈ {1, 2,…, n}. Thus, the joint distribution over the labels yi, given the observation Xi, can be written as:
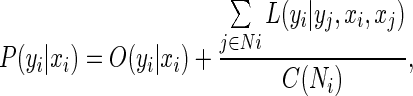 |
1 |
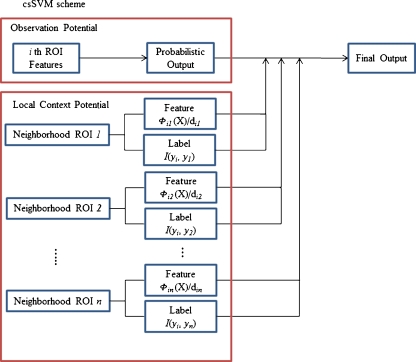
where O(.) and L(.) are the observation and local context potential, respectively, and Ni is a neighboring ROI, the ith ROI, and C(Ni) is the number of ith neighborhoods. A schematic diagram of this process is illustrated in Fig. 3.
Fig. 3.
Schematic diagram of the csSVM, consisting of the observation potential and the local context potential, for classifying a current ROI using the current ROI’s features along with the neighborhood ROIs’ features and labels
Observation Potential
The observation potential measures the likelihood that the ith ROI will have the label yi ∈ {1, 2,…, n} in a given image xi, ignoring information about other ROIs. In this paper, we used a multiclass SVM to calculate the observation potential. Multiclass probability outputs were required to calculate the observation potential. We used the method of Wu et al. [27], who introduced methods that transfer the support vector into multiclass probability estimates, to obtain multiclass probability estimates for the observation potential.
Local Context Potential
The local context potential measures how neighboring ROIs i and j are affected by each other and compensates for the error derived from the observation potential. In the MRF framework, the Ising model is a commonly used local context potential [28]. This method penalizes discontinuities in the labeling of neighboring ROIs i and j by a cost β [29]. However, it captures the dissimilarities only in the labels of adjacent ROIs and does not consider neighborhood-to-neighborhood discontinuities in the feature vectors. In the present study, we treated label and feature vector dissimilarities, to model the local context potential. Thus, the local context potential is modeled as:
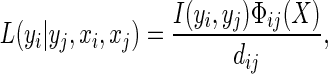 |
2 |
where the function I(yi, yj) is the interaction vector, dij is the Euclidean distance between the ith and jth ROI, Фij(X) is a function of the computed feature vector distance between the ith and jth ROI based on observation X. The function I(yi, yj) is a 1 × k vector that contains 1 in the yjth column, −1 in the yith column, and 0 in the other columns when yi = yj; if yi ≠ yj, I(yi, yj) contains 0 on all columns. Thus, it may be represented as:
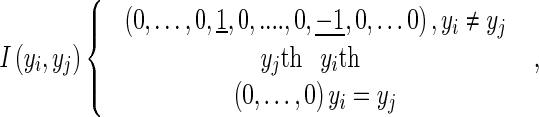 |
3 |
Since the yi have categorical values, {1, 2, …., n}, the interaction vector adjusts the observation potential’s posterior probability by the amount Фij(X)/dijC(Ni). The function Фij(X) can be modeled as:
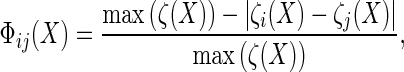 |
4 |
where the function ζk maps the kth ROI into a feature vector. This function yields larger values because neighboring elements are more similar. Thus, a neighboring ROI that has a feature vector similar to that of the currently classified ROI has more of an influence on classification of the current ROI than an ROI with a dissimilar feature vector.
Experimental Setting
Previous kernel function tests for the SVM indicated that a radial basis function performed better than other kernel functions [12]. Thus, we applied a kernel function of the radial basis function against a polynomial, and the associated parameters (e.g., cost C and gamma value) were optimized by a grid search. The purpose of inference is to identify an optimal joint label, given an image X. Inference was performed iteratively unless the improvement in the probability of a joint label dropped below the predetermined threshold. The conventional SVM and observation potential in our proposed method employed a classifier that used the LIBSVM [30]. Computation time and accuracy were measured by running the MATLAB (The MathWorks, Natick, MA, USA) routine on a PC with an i5 Core, 2.67 GHz, and 2 GB RAM.
Statistical Analysis
The conventional SVM and our csSVM were initially tested on 900 ROIs using 20 applications of fivefold cross-validation. The fivefold cross-validation was performed as follows: the 900 ROIs of each class were divided uniformly and randomly into five exclusive stratified subsets. Each fold was in turn held aside as a test set, and the other exclusive four subsets were used to train the classifiers. The average of the fivefold cross-validation results yielded the overall result. Eighty-four whole lung images were tested by the conventional SVM and our csSVM to evaluate image classification accuracy and computational time. Computational times were compared by measuring the testing time (the time required to classify each whole lung image) as calculated by the conventional SVM or our csSVM. All testing results were compared with a t test, and an α-value of 0.01 was considered statistically significant.
Experiment Results
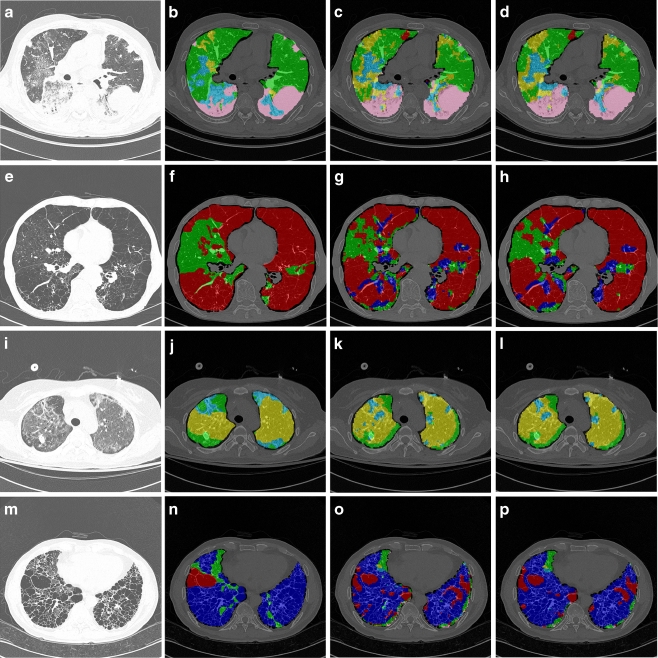
Table 2 shows the accuracy of our proposed method and the conventional multiclass SVM for whole lung images and ROI-based data (paired t test, p < 0.01). We also measured computational times required for whole lung quantification using either classifier. In this task, we used features that had been extracted prior to conducting the actual quantification. Thus, the times in Table 2 indicate the time elapsed during quantification by each classifier. Figure 4 illustrates an example of regional lung disease pattern classification by conventional SVM and by our proposed csSVM.
Table 2.
Overall accuracy and computational time for whole lung images and ROI-based data for the conventional SVM method and the proposed method
| Classifier | Accuracy (%) | Time (s) | |
|---|---|---|---|
| Whole lung (p < 0.01) | ROI (p < 0.01) | Whole lung (p < 0.01) | |
| Conventional SVM | 57.69 ± 13.31 | 87.39 ± 0.02 | 2.25 ± 0.41 |
| Proposed method | 60.30 ± 13.95 | 89.88 ± 0.02 | 3.05 ± 0.91 |
Fig. 4.
Whole lung quantification results for comparing the conventional SVM to the proposed method. Each pixel was coded by the classification result, indicated by a semi-transparent color (normal, green; ground-glass opacity, yellow; reticular opacity, cyan; honeycombing, blue; emphysema, red; and consolidation, pink). a Original HRCT image (case 1), b drawing by an expert (case 1), c quantification results from the conventional SVM (case 1), d quantification results from the csSVM (case 1), e original HRCT image (case 2), f drawing by an expert (case 2), g quantification results from the conventional SVM (case 2), h quantification results from the csSVM (case 2), i original HRCT image (case 3), j drawing by an expert (case 3), k quantification results from the conventional SVM (case 3), l quantification results from the csSVM (case 3), m original HRCT image (case 4), n drawing by an expert (case 4), o quantification results from the conventional SVM (case 4), p quantification results from the csSVM (case 4)
Discussion and Conclusion
In this study, we developed a csSVM that incorporates neighborhood information, including neighboring ROI features and labels. Our method can be applied to multiclass classification problems that cannot be addressed by previously proposed methods employing conditional random fields [9, 24]. Although the overall computational time of our csSVM was 35% greater than that of the conventional SVM, the overall accuracy of our csSVM was significantly better, by 2.61% for the whole lung and by 2.49% for ROI-based data.
In medical imaging analysis, our multiclass csSVM may be applied to classification-based image quantification to estimate texture-based emphysema indices in patients with chronic obstructive pulmonary disease and to assess the pathogenesis of DILD. Furthermore, our proposed csSVM may be applicable to other areas, such as remote sensing and computer vision, in which spatial relationships are important. Other classifiers, such as ANNs and Bayesian classifiers, could be extended to context-sensitive classifiers to obtain more accurate results.
The present study has several limitations. First, application of the proposed method to the whole lung yielded lower accuracy classification results (60.30 ± 13.95%) than did the ROI-based classification system (89.88 ± 0.02%). ROIs in the ROI-based data were selected in regions that were representative of the disease, whereas the ROIs classified throughout an entire lung would include transitional regions, which could contribute to disagreements in disease pattern. This problem may be overcome by using adaptive ROIs on borderlines. Another possible explanation is that different quantification methods were used by the expert radiologist and the classifier. The expert radiologist divided the whole lung into six classes using in-house software (a line-based drawing program), whereas the classifier quantified the whole lung using a pixel-based method. This discrepancy in quantification should be further studied in experiments in which experts classify whole images using a pixel-based rather than a line-based program.
Second, our proposed method was applied to 2D images, which prevented us from removing systematic errors, such as those introduced by an airway or vessel [3]. Use of 3D images would provide better results for whole lung quantification, although the computational cost would be significantly greater. Finally, csSVM was only applied to the quantification of DILD patient lungs. To examine the general effectiveness of the model, further evaluations should be performed using datasets from different organs or tissues.
In our proposed csSVM method, the accuracy of DILD quantification was greater than that provided by traditional SVM, with only a slight increase in computational load. We propose that csSVM be considered for the quantification of DILD.
Acknowledgment
This study was supported by a Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund; KRF-2007-313-D00980).
References
- 1.Uppaluri R, Mitsa T, Sonka M, Hoffman EA, McLennan G. Quantification of pulmonary emphysema from lung computed tomography images. Am J Respir Crit Care. 1997;156:248–254. doi: 10.1164/ajrccm.156.1.9606093. [DOI] [PubMed] [Google Scholar]
- 2.Uppaluri R, Hoffman EA, Sonka M, Hunninghake GW, McLennan G. Interstitial lung disease—A quantitative study using the adaptive multiple feature method. Am J Respir Crit Care. 1999;159:519–525. doi: 10.1164/ajrccm.159.2.9707145. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Sonka M, McLennan G, Guo JF, Hoffman EA. MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE Trans Med Imag. 2006;25:464–475. doi: 10.1109/TMI.2006.870889. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Beek EJR, Yu HJ, Guo JF, McLennan G, Hoffman EA. Computer-aided classification of interstitial lung diseases via MDCT: 3D adaptive multiple feature method (3D AMFM) Acad Radiol. 2006;13:969–978. doi: 10.1016/j.acra.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Prasad M, Sowmya A, Wilson P. Multi-level classification of emphysema in HRCT lung images. Pattern Anal Appl. 2009;12:9–20. doi: 10.1007/s10044-007-0093-7. [DOI] [Google Scholar]
- 6.Chabat F, Yang GZ, Hansell DM. Obstructive lung diseases: Texture classification for differentiation at CT. Radiology. 2003;228:871–877. doi: 10.1148/radiol.2283020505. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Seo JB, Lee JG, Kim SS, Kim N, Kang SH. Performance testing of several classifiers for differentiating obstructive lung diseases based on texture analysis at high-resolution computerized tomography (HRCT) Comput Methods Programs Biomed. 2009;93:206–215. doi: 10.1016/j.cmpb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Lee CH, Schmidt M, Murtha A, Bistritz A, Sander M, Greiner R. Segmenting brain tumors with conditional random fields and support vector machines. Proc Comput Vis Biomed Image Appl. 2005;3765:469–478. doi: 10.1007/11569541_47. [DOI] [Google Scholar]
- 9.Lee CH, Greiner R, Schmidt M: Support vector random fields for spatial classification. Knowledge discovery in databases: Pkdd 2005 3721:121-132, 2005
- 10.Lee Y, Kim N, Seo JB, Lee J, Kang SH. The performance improvement of automatic classification among obstructive lung diseases on the basis of the features of shape analysis, in addition to texture analysis at HRCT. Proc. SPIE (Medical Imaging) 2007;6512:65124F. [Google Scholar]
- 11.Park YS, Seo JB, Kim N, Chae EJ, Oh YM, Lee SD, Lee Y, Kang SH. Texture-based quantification of pulmonary emphysema on high-resolution computed tomography: Comparison with density-based quantification and correlation with pulmonary function test. Investig Radiol. 2008;43:395–402. doi: 10.1097/RLI.0b013e31816901c7. [DOI] [PubMed] [Google Scholar]
- 12.Kim N, Seo JB, Lee Y, Lee JG, Kim SS, Kang SH. Development of an automatic classification system for differentiation of obstructive lung disease using HRCT. J Digit Imaging. 2009;22:136–148. doi: 10.1007/s10278-008-9147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim N, Seo JB, Sung YS, Park BW, Lee Y, Park SH, Lee YK, Kang SH. Effect of various binning methods and ROI sizes on the accuracy of the automatic classification system for differentiation between diffuse infiltrative lung diseases on the basis of texture features at HRCT. Proc SPIE (Medical Imaging) 2008;6914:69743N. [Google Scholar]
- 14.Park SO, Seo JB, Kim N, Park SH, Lee YK, Sung YS, Park BW, Lee Y, Lee J, Kang SH, et al. Feasibility of automated quantification of regional disease patterns depicted on high-resolution computed tomography in patients with various diffuse lung diseases. Korean J Radiol. 2009;10:455–463. doi: 10.3348/kjr.2009.10.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards JA, Jia X. Remote sensing digital image analysis: An introduction. Berlin: Springer; 2006. [Google Scholar]
- 16.Eklundh JO, Yamamoto H, Rosenfeld A. A relaxation method for multispectral pixel classification. Pattern analysis and machine intelligence. IEEE Trans PAMI. 1980;2:72–75. doi: 10.1109/TPAMI.1980.4766973. [DOI] [PubMed] [Google Scholar]
- 17.Rosenfeld A, Hummel RA, Zucker SW. Scene labeling by relaxation operations. IEEE T Syst Man Cybern. 1976;6:420–433. doi: 10.1109/TSMC.1976.4309519. [DOI] [Google Scholar]
- 18.Lee T, Richards JA: Pixel relaxation labelling using a diminishing neighbourhood effect. Proc. IGARSS’89 and Canadian Symposium on Remote Sensing 12th:634–637, 1989
- 19.Kalayeh HM, Landgrebe DA. Adaptive relaxation labeling. IEEE Trans Pattern Anal. 1984;6:369–372. doi: 10.1109/TPAMI.1984.4767530. [DOI] [PubMed] [Google Scholar]
- 20.Bruzzone L, Prieto DF. Adaptive relaxation labeling context-based approach to unsupervised change detection in multitemporal remote-sensing images. IEEE T Image Process. 2002;11:452–466. doi: 10.1109/TIP.2002.999678. [DOI] [PubMed] [Google Scholar]
- 21.Melgani F, Serpico SB. A statistical approach to the fusion of spectral and spatio-temporal contextual information for the classification of remote-sensing images. Pattern Recogn Lett. 2002;23:1053–1061. doi: 10.1016/S0167-8655(02)00052-1. [DOI] [Google Scholar]
- 22.Lafferty JFP, McCallum A: Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proc. Intl. Conf. on Machine Learning:282–289, 2001
- 23.Feng XJ, Williams CKI, Felderhof SN. Combining belief networks and neural networks for scene segmentation. IEEE Ttrans Pattern Anal. 2002;24:467–483. doi: 10.1109/34.993555. [DOI] [Google Scholar]
- 24.Kumar S, Hebert M. Discriminative random fields. Int J Comput Vision. 2006;68:179–201. doi: 10.1007/s11263-006-7007-9. [DOI] [Google Scholar]
- 25.Hsu CW, Lin CJ. A comparison of methods for multi-class support vector machines. IEEE T Neural Netw. 2002;13(2):415–425. doi: 10.1109/72.991427. [DOI] [PubMed] [Google Scholar]
- 26.Haralick R, Shanmugam K, Dinstein IH. Textural features for image classification. Systems, man and cybernetics. IEEE Trans. 1973;3:610–621. [Google Scholar]
- 27.Wu TF, Lin CJ, Weng RC. Probability estimates for multi-class classification by pairwise coupling. J Mach Learn Res. 2004;5:975–1005. [Google Scholar]
- 28.Kumar S, Hebert M: Discriminative random fields: A discriminative framework for contextual interaction in classification: IEEE Comput Soc, 2003
- 29.Ising E. Beitrag zur Theorie des Ferromagnetismus. Z Phys A: Hadrons Nucl. 1925;31:253–258. [Google Scholar]
- 30.Chang CC, Lin CJ, LIBSVM : A library for support vector machines, 2001. Software available at http://www.csie.ntu.edu.tw/∼cjlin/libsvm