Abstract
Mt4 is a cDNA representing a phosphate-starvation-inducible gene from Medicago truncatula that is down-regulated in roots in response to inorganic phosphate (Pi) fertilization and colonization by arbuscular mycorrhizal fungi. Split-root experiments revealed that the expression of the Mt4 gene in M. truncatula roots is down-regulated systemically by both Pi fertilization and colonization by arbuscular mycorrhizal fungi. A comparison of Pi levels in these tissues suggested that this systemic down-regulation is not caused by Pi accumulation. Using a 30-bp region of the Mt4 gene as a probe, Pi-starvation-inducible Mt4-like genes were detected in Arabidopsis and soybean (Glycine max L.), but not in corn (Zea mays L.). Analysis of the expression of the Mt4-like Arabidopsis gene, At4, in wild-type Arabidopsis and pho1, a mutant unable to load Pi into the xylem, suggests that Pi must first be translocated to the shoot for down-regulation to occur. The data from the pho1 and split-root studies are consistent with the presence of a translocatable shoot factor responsible for mediating the systemic down-regulation of Mt4-like genes in roots.
Phosphate is an essential mineral nutrient for plant growth and development and is acquired from the soil by Pi transporters located in the roots (Bieleske, 1973). Translocation of phosphate within the plant is predominantly unidirectional, moving from the root to the shoot via the xylem; however, a considerable amount of phosphate is also retranslocated to the root via the phloem (Jeschke et al., 1997). The source is likely to be from both xylem-derived Pi and inorganic and organic sources from old leaves (Schachtman et al., 1998). This retranslocation process assists root growth (Smith et al., 1990) and may help reduce the buildup of toxic levels of phosphate in the shoot (Schjørring and Jensén, 1984).
The phosphate status of the shoot is known to control the root processes involved in Pi uptake and its subsequent translocation; however, the precise mechanisms of this control are unknown). The retranslocation of phosphate from the shoot to the root is likely to be involved, acting as a feedback-regulatory mechanism responsible for mediating the shoot control of phosphate levels within the plant (Drew and Saker, 1984; Schjørring and Jensén, 1984; Marschner and Cakmak, 1986). Feedback mechanisms regulating the movement of ions between the root and the shoot have also been proposed for sulfate (Jensén and König, 1982) and potassium (Pitman, 1977). Retranslocated Pi and organic derivatives of phosphate (Hall and Baker, 1972) may have dual functions both as nutrients for root processes and as signaling molecules (Drew and Saker, 1984). The precise points of regulation in the root are unclear, although some evidence implies that the loading of Pi in the xylem is central to the regulation of phosphate flux in the plant (Drew and Saker, 1984). The Arabidopsis mutant pho1 (Poirier et al., 1991) exhibits normal Pi uptake rates from the soil to the root, but is blocked in the loading of Pi into the xylem, which is genetic evidence for at least partial regulation of Pi transport by xylem loading.
Plants respond to decreasing Pi in the environment by increased root growth, increased expression of Pi transporters (Muchhal et al., 1996; Leggewie et al., 1997; Liu et al., 1998b), and alterations in metabolism, including secretion of acid phosphatases (Lefebvre et al., 1990) and RNAses (Bariola et al., 1994), which assist in the liberation of Pi from the rhizosphere. Internally, phosphate retranslocation from the shoot to the root increases (Lefebvre et al., 1990; Heuwinkel et al., 1992; Jeschke et al., 1997). Recently, however, it has been shown that in Pi-starved plants, a larger percentage of retranslocated phosphate is returned to the shoot, indicating that phosphate cycling occurs (Jeschke et al., 1997).
At the molecular level, relatively little is known about the Pi-starvation response in plants and even less about its regulation. Phosphate-starvation-inducible Pi transporters (Muchhal et al., 1996; Leggewie et al., 1997; Smith et al., 1997; Liu et al., 1998b), RNAses (Bariola et al., 1994), a β-glucosidase (Malboobi and Lefebvre, 1997), and a number of genes of unknown function (Burleigh and Harrison, 1997; Liu et al., 1997) have been cloned and provide a starting point from which to analyze the signal transduction pathways involved. The molecular mechanisms by which the shoot regulates Pi uptake and translocation are also unknown. As demonstrated by pho2, an Arabidopsis mutant that accumulates toxic levels of Pi in the shoot (Delhaize and Randall, 1995), at least one plant gene is involved in how plants sense or respond to shoot Pi levels.
One of the most widespread responses to Pi deprivation is the development of an association with AM fungi. These obligate symbionts share a symbiotic relationship with approximately 80% of angiosperm species, enhancing plant growth by transferring Pi from the soil to the plant. AM fungi have been forming associations with plants for at least 400 million years (Smith and Read, 1997). Consequently, the co-evolution of Pi-acquisition strategies by plants and the AM fungal symbiosis should be considered in the study of the Pi-starvation response in plants.
We have previously cloned a cDNA (Mt4) from Medicago truncatula, which is induced in roots in response to Pi starvation (Burleigh and Harrison, 1997). The Mt4 gene is down-regulated by both Pi fertilization and AM fungal colonization and as such may provide the means to study these two closely linked processes at the molecular level. The Mt4 cDNA is 0.5 kb (Burleigh and Harrison, 1997) and shares sequence identity with a Pi-starvation-inducible gene from tomato (Lycopersicon esculentum L.), TPSI1 (Liu et al., 1997), although identity is limited to a 30-bp sequence located in the center of both transcripts. Both genes are composed of numerous short, overlapping ORFs (Liu et al., 1997; Burleigh and Harrison, 1998), which is similar to the structure of ENOD40, a plant gene encoding a peptide signal involved in nodule development (van de Sande et al., 1996). Whereas the functions of Mt4 and TPSI1 are unknown, their sensitivity to Pi fertilization and their potential to encode small peptides make them good candidates as signaling molecules involved in the Pi-starvation response (Liu et al., 1997; Burleigh and Harrison, 1998).
Here we present evidence for the systemic down-regulation of the Mt4 gene by both Pi fertilization and AM colonization, identify Mt4-like genes in Arabidopsis and soybean, and demonstrate that At4 from Arabidopsis is not down-regulated in the Pi-accumulation mutant pho1 in response to Pi fertilization. From these results we propose that a translocatable shoot factor may be involved in the Pi-mediated systemic down-regulation of Mt4-like genes.
MATERIALS AND METHODS
Growth of Plants under Pi-Limiting and Pi-Sufficient Conditions
The plants used in the experiments were Medicago truncatula Gaertn cv Jemalong, line A17, soybean (Glycine max. L. cv Harisoy), corn (Zea mays L. cv Iochief), tomato (Lycopersicon esculentum L. cv Rutgers), Arabidopsis (Columbia ecotype), and the Arabidopsis Pi-accumulation mutant pho1 (Poirier et al., 1991). Seeds of M. truncatula, soybean, corn, and tomato were germinated on filter paper for 1 week, planted in sterile Turface (A.H. Hummert Seed, St. Louis, MO). Arabidopsis seeds were germinated and grown on sterile sand. All of the plants were fertilized with one-half-strength Hoagland solution (Arnon and Hoagland, 1940), pH 6.0, containing 0 (−Pi) or 1.0 mm (+Pi) KH2PO4 and grown for 4 weeks before harvest. Root samples were harvested for the preparation of RNA from all species, except Arabidopsis, for which whole plants were harvested. Shoot samples were harvested for Pi assays for all but the Arabidopsis samples.
To compare expression of At4 in leaves and roots of cv Columbia and the pho1 mutant, the Arabidopsis plants used in the experiment were germinated and grown in Metro Mix (Scott, Marysville, OH) for 3 weeks. Approximately 20 plants per treatment were transplanted to sterile sand in 11-cm pots, fertilized with one-half-strength Hoagland solution (Arnon and Hoagland, 1940), pH 6.0, containing 0 (−Pi) KH2PO4 and grown for an additional 3 weeks before harvest. Root and shoot samples were harvested for the preparation of RNA.
Split-Root Experiments
For the Pi split-root experiments, seedlings were prepared and grown in Turface for 10 d as described above. The roots of individual plants were separated into two equal parts, placed into separate containers, fertilized weekly with nutrient solution containing either 0.02 mm (low Pi) or 1.0 mm (high Pi) KH2PO4 (see Fig. 1), and grown for an additional 4 weeks. The controls included a split-root treatment in which both halves of the root system received low-Pi fertilizer and a split-root treatment in which both halves received high-Pi fertilizer. At harvest, roots were frozen in liquid nitrogen for RNA extraction. Root and shoot samples were also harvested for Pi assays.
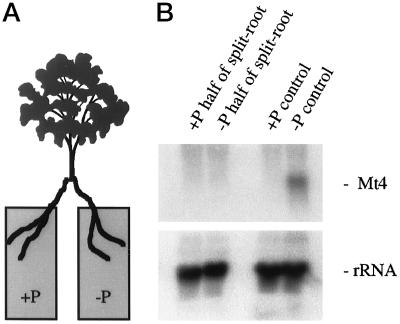
Figure 1.
The systemic down-regulation of Mt4 by Pi fertilization. A, Illustration of a split-root plant. B, Northern blot of total RNA isolated from the roots of M. truncatula grown in a split-root design with one-half of the root system receiving high-Pi (+Pi half of split-root) and one-half receiving low-Pi (−Pi half of split-root) fertilizer. In split-root controls both halves received high-Pi (+Pi control) or low-Pi (−Pi control) fertilizer. The blot was probed with a 32P-labeled Mt4 cDNA (top panel) and a 32P-labeled pSR1-2B3 (18S rRNA, bottom panel).
The design of the mycorrhizal split-root experiment was identical to that of the Pi split-root experiment, except that mycorrhizal spore inoculum was added to one-half of the split-root system at the initiation of the experiment, all treatments received fertilizer containing 0.02 mm KH2PO4, and plants were grown for 5 weeks before harvest. Details of plant inoculation and quantitation of fungal colonization were described previously (Harrison and Dixon, 1993). Glomus versiforme spores were collected from colonized leek plants, surface-sterilized, washed three times in sterile distilled water, quantified, and placed on the roots of treatment plants at the initiation of the experiment. Included were two replications of the mycorrhizal split-root treatment, a nonmycorrhizal control, and a control in which one-half of the split-root system was mock inoculated with the water used to prepare the fungal spore inoculum. At harvest, roots were frozen in liquid nitrogen for RNA extraction. Shoot samples were harvested for Pi assays. Root samples were harvested and colonization assessed by the modified gridline-intersect method, as described previously (Harrison and Dixon, 1993).
Southern- and Northern-Blot Analyses
The isolation of total RNA and genomic DNA was as described previously (Burleigh and Harrison, 1997). The preparation of probes and both Southern and northern blotting and hybridization were carried out as described by Sambrook et al. (1989) using 1 and 10 μg of DNA and RNA per treatment, respectively. Blots were hybridized at 42°C in 50% formamide, 4× SSPE, 1% SDS, 0.5× Denhardt's solution, and 100 μg mL−1 salmon-sperm DNA. The final washing conditions were 2× SSPE, 1% SDS at 65°C. Northern blots were hybridized with a labeled 18S rRNA probe (pSR1-2B3) (Eckenrode et al., 1985) to provide a control for loading and transfer and to allow for the comparison of expression levels between treatments of the same blot.
RT-PCR
The RT-PCR technique used to identify At4 transcripts in RNA preparations from roots, leaves, or whole plants of wild-type Arabidopsis and the mutant pho1 was carried out according to the method of Kawasaki (1990). One microgram of total RNA was reverse transcribed using Superscript reverse transcriptase (GIBCO-BRL) and oligonucleotide-(dT)15 primers. These RT reactions were then standardized based on their ribosomal cDNA content. To assess ribosomal content, RT reactions were amplified by PCR using the ribosomal primer pair NS1/NS21 (Simon et al., 1992) under nonlimiting reaction conditions and the products separated by gel electrophoresis, blotted, and probed with a 32P-labeled pSR1-2B3 cDNA (18S rRNA) (Eckenrode et al., 1985). Band intensities were quantified using a phosphor imager (Molecular Dynamics, Sunnyvale, CA). The standardized RT reactions were then amplified by PCR using the Arabidopsis primer pair (see Fig. 4) and probed using either the 32P-labeled At4 cDNA or a 32P-end-labeled oligonucleotide based on the conserved sequence (GGAAAGGGCAACTTCGATCCTTTGGCATTT) of the Mt4 cDNA sequence. The PCR reactions consisted of an initial denaturation step at 94°C for 5 min followed by 26 (ribosomal cDNA) or 28 cycles (At4) of melting at 94°C for 30 s, annealing at 48°C (ribosomal cDNA) or 63°C (At4) for 30 s, and extension at 72°C for 30 s using a Geneamp 2400 thermal cycler (Perkin-Elmer). For a comparison of At4 levels in roots and shoots, the PCR reactions with the At4 primers were taken to 35 cycles. End labeling of the oligonucleotide was carried out according to the method of Sambrook et al. (1989). Hybridization with the end-labeled oligonucleotide was carried out at 55°C in 6× SSPE, 10 mm NaH2PO4, 1 mm EDTA, 0.5% SDS, 100 μg/mL denatured salmon-sperm DNA, and 0.1% nonfat dry milk. The final washing conditions were 2× SSPE, 1% SDS at 42°C.
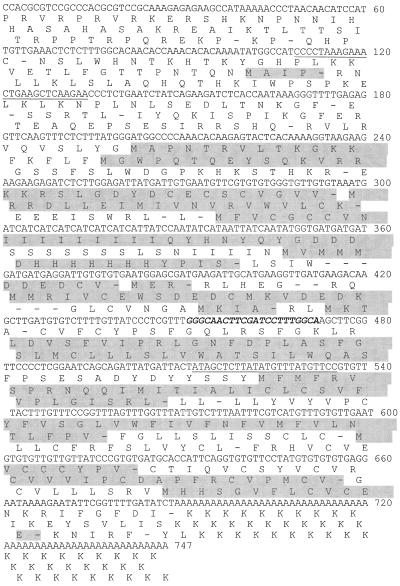
Figure 4.
The nucleotide and translated amino acid sequences of At4. Bases are numbered at right. Predicted ORFs are shaded. The region of nucleotide identity with Mt4 from M. truncatula and TPSI1 from tomato is in bold italics. The primer pair used in PCR experiments is underlined.
Pi Content Determinations
The Pi content of the roots and shoots was determined using a phosphomolybdate colorimetric reaction according to the method of Chen et al. (1956). Tissue previously frozen in liquid nitrogen was powdered using a mortar and pestle, air dried at 50°C, rehydrated in a solution containing 1.4% ascorbic acid, 0.36% ammonium molybdate, and 4.2% H2SO4, incubated at 50°C for 30 min, and assayed at A800. The Pi content was expressed as micrograms of Pi per milligram of dry weight and three replicate samples were assayed for each treatment. Although this method measures Pi, it should be noted that there is some breakdown of labile phosphate esters during this assay (Ames, 1966).
RESULTS
Systemic Down-Regulation of Mt4 in Response to Pi Fertilization and to Colonization by an AM Fungus
We initiated a series of split-root experiments to determine whether the down-regulation of Mt4 by Pi fertilization occurs systemically. A M. truncatula plant was grown with its root system divided equally between two containers and these received either low-Pi or high-Pi fertilizer (Fig. 1A). The two halves of the root system were harvested separately and Mt4 transcript levels were assessed by northern-blot analyses. We were unable to detect transcripts in either the high-Pi or the low-Pi half of the split-root system (Fig. 1B). Mt4 transcripts were abundant in roots in which both halves of the root system received low-Pi fertilizer and were absent in roots in which both halves received high-Pi fertilizer. This indicates that Pi fertilization of one-half of the root results in the systemic down-regulation of Mt4 expression throughout the whole root system.
The levels of Pi in the leaves reflected the two fertilization regimes (Table I, experiment A). Pi levels were relatively low in roots in which both halves of the root system received low-Pi fertilizer and were relatively high in roots in which both halves received high-Pi fertilizer. However, the treatment in which one-half of the split roots received low-Pi fertilizer and the other half received high-Pi fertilizer had a differential response; the half receiving low-Pi fertilizer did not accumulate high levels of Pi, despite the accumulation of Pi in the fertilized half of the root system and in the leaves. These results suggest that Pi accumulation was not responsible for the systemic down-regulation of Mt4 gene expression.
Table I.
Pi levels in roots and shoots from the homolog (A) and split-root experiments (B)
| Experiment | Species | Tissue | Treatment | Average Pi Content |
|---|---|---|---|---|
| μg Pi mg−1dry wt | ||||
| A | M. truncatula | Root | +Pi control | 3.76 (0.04) |
| Root | −Pi control | 1.23 (0.07) | ||
| Shoot | +Pi control | 5.13 (0.00) | ||
| Shoot | −Pi control | 2.47 (0.15) | ||
| Root | +Pi half | 3.51 (0.05) | ||
| Root | −Pi half | 1.66 (0.02) | ||
| Shoot | ±Pi | 6.11 (0.08) | ||
| Shoot | ±Fungus | 2.93 (0.09) | ||
| B | Soybean | Shoot | +P | 4.52 (0.80) |
| Shoot | −P | 1.29 (0.17) | ||
| Corn | Shoot | +P | 2.18 (0.28) | |
| Shoot | −P | 0.66 (0.10) | ||
| Tomato | Shoot | +P | 1.67 (0.27) | |
| Shoot | −P | 1.06 (0.34) |
The Pi content was determined using a phosphomolybdate assay and is the average of three determinations (n = 3). Numbers in parentheses are sd values.
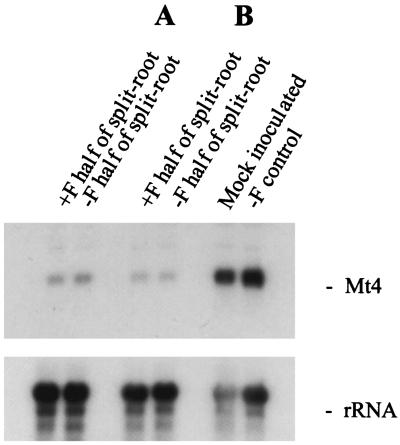
A similar experimental design was used to determine whether the down-regulation of Mt4 by mycorrhizal colonization occurs systemically. Transcript levels were high in split roots from an uncolonized plant, as well as from a plant in which one-half of the root system was inoculated with the spore wash (Fig. 2). Split roots in which one-half of the root system was colonized by the AM fungus had low levels of Mt4 transcripts in both halves of the root system, indicating that down-regulation of Mt4 transcripts by mycorrhizal fungi also occurs systemically. This systemic down-regulation of Mt4 expression was observed in two split-root plants (Fig. 2). The level of fungal colonization in these two plants was 54% and 50% of the root length.
Figure 2.
The systemic down-regulation of Mt4 by mycorrhizal colonization. Northern blots of total RNA isolated from the roots of M. truncatula grown in a split-root design with one-half of the root system grown with (+F half of split-root) and one-half grown without (−F half of split-root) G. versiforme colonization for two plants (A and B). In split-root controls one-half of the root system was mock inoculated or both halves were uncolonized (−F control). The blot was probed with a 32P-labeled Mt4 cDNA (top panel) and a 32P-labeled pSR1-2B3 (18S rRNA, bottom panel).
The level of Pi in the leaves of a split-root plant in which one-half of the root system was colonized by an AM fungus was approximately equal to that of a plant in which both halves remained uncolonized (−Pi control) (Table I, experiment A). These results suggest that Pi accumulation was not likely to be responsible for the observed systemic down-regulation of Mt4 gene expression in mycorrhizal split roots. Also, no visible growth differences were observed between the treatments (data not shown). Enhanced Pi nutrition and growth effects in mycorrhizal plants may not be detectable in experiments of short duration or when growth conditions limit the ability of AM fungi to adequately accumulate trace levels of Pi present in the substrate.
Mt4-Like Genes in Arabidopsis and Soybean
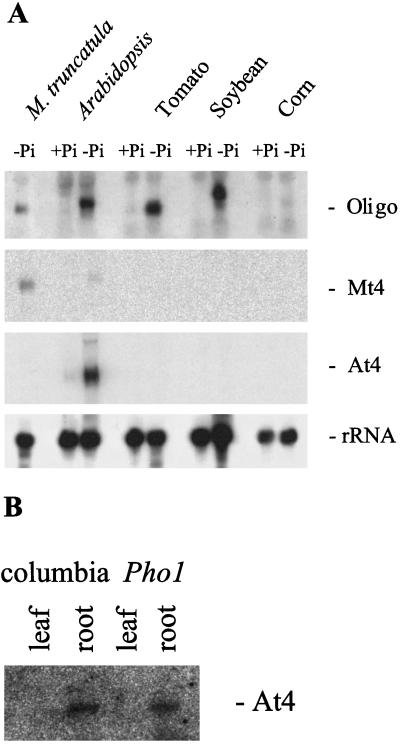
Because a Pi-starvation-inducible homolog of Mt4 has been described in tomato (Liu et al., 1997), we searched for Mt4-like genes in other plant species to determine whether they may be a widely conserved component of the Pi-deprivation responses. RNA was prepared from whole Arabidopsis plants and from the roots of soybean, corn, and tomato grown with either low-Pi or high-Pi fertilizer. Initial northern-blot analyses, in which the whole Mt4 sequence was used as a probe, detected only Mt4 transcripts in RNA from Pi-starved M. truncatula (Fig. 3A). However, the use of a 30-bp oligonucleotide probe, representing the sequence conserved between Mt4 and the tomato homolog TPSI1, enabled the detection of Pi-starvation-inducible transcripts in Arabidopsis and soybean. These transcripts were approximately 0.7 and 1.0 kb in length, respectively. The 0.5-kb transcript identified in tomato corresponded to the size of the TPSI1 transcript (Liu et al., 1997). Transcripts were not detected in corn receiving either low-Pi or high-Pi fertilizer.
Figure 3.
Detection of Mt4-like genes in Arabidopsis, tomato, and soybean. A, Northern blot of total RNA isolated from plants receiving either low-Pi (−Pi) or high-Pi (+Pi) fertilizer. The blot was probed with the conserved sequence (Oligo), the Mt4 cDNA, the At4 cDNA, or a 32P-labeled pSR1-2B3 (rRNA). B, Expression of At4 in Arabidopsis roots. RT-PCR products from RNA extracted from leaves and roots of Pi-deprived Arabidopsis plants (wild type and pho1 mutant) using At4-specific primers. The blot was hybridized with 32P-labeled At4 cDNA.
The Pi levels of the soybean and corn leaves were consistent with the two fertilization regimes: plants receiving low-Pi fertilizer had relatively low Pi levels, whereas plants receiving high-Pi fertilizer had relatively high Pi levels (Table I, experiment B). However, the amount of Pi in the leaf tissue from the two tomato treatments was similar, although plants receiving low-Pi fertilizer were stunted in growth and the leaves were purple in color, which are classic symptoms of Pi starvation. The Arabidopsis plants receiving low-Pi fertilizer were also stunted relative to the plants receiving high-Pi fertilizer (data not shown).
The Arabidopsis At4 Gene
To initiate studies of the Mt4-like gene in Arabidopsis in which access to a number of mutants could provide the means to further characterize Mt4-like gene expression, we decided to isolate the Arabidopsis cDNA identified in the previous northern-blot analyses. Using the 30-bp sequence conserved in Mt4 and TPSI1 as the target sequence, we searched an Arabidopsis expressed sequence tag database and identified a cDNA, which we named At4 (accession no. AF055372). Sequence analysis revealed that At4 was 747 bp in length (Fig. 4). This corresponds to the size of the transcript identified in RNA isolated from Pi-starved Arabidopsis plants using the conserved oligonucleotide probe (Fig. 3A). This northern blot was stripped and reprobed with the At4 cDNA and it hybridized to the same Pi-starvation-inducible transcript from Arabidopsis (Fig. 3A). As observed with the Mt4 cDNA, the At4 cDNA did not hybridize well to transcripts from other species. The Arabidopsis RNA used on this northern blot was prepared from whole Arabidopsis plants and, therefore, to determine whether At4 transcripts are present in both the leaves and roots of Pi-deprived plants, RNA was prepared from these tissues individually and the presence of the At4 transcript was assessed by RT-PCR followed by hybridization with the At4 probe. The At4 PCR product was detected in RNA samples from Arabidopsis root tissue but not in RNA samples from leaves (Fig. 3B).
The At4 cDNA is predicted to contain numerous short, overlapping ORFs, which is similar to Mt4 and TPSI1 (Fig. 4). However, none of the At4 ORFs share any amino acid identity with the ORFs of Mt4 or TPSI1. The conserved nucleic acid sequence located between bp 451 and 472 of the At4 transcript is potentially translated in two frames, but not the conserved amino acid sequence, WKGQLR/LSFGI, found in both Mt4 and TPSI1 (Fig. 5).
Figure 5.
Comparison of the conserved nucleotide sequences of Mt4 from M. truncatula, TPSI1 from tomato, and At4 from Arabidopsis. Perfectly conserved nucleotides are represented by asterisks and well-conserved nucleotides are represented by dots.
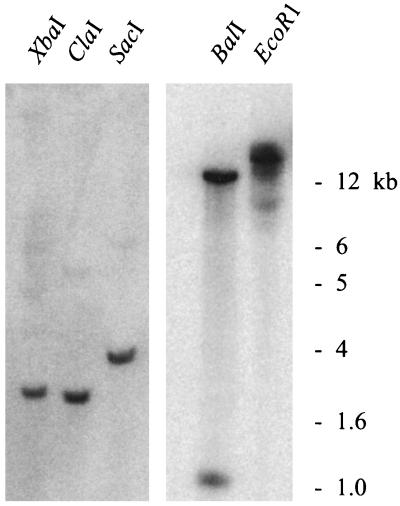
A Southern blot of Arabidopsis genomic DNA revealed one copy of At4 in the genome (Fig. 6). BalI digestion resulted in two bands, as predicted by the presence of a BalI site within the cDNA.
Figure 6.
Southern blot of Arabidopsis genomic DNA. Hybridization of a 32P-labeled At4 cDNA to a gel blot of DNA digested with five different restriction enzymes. Size markers are on the right.
Expression of At4 in the Pi-Accumulation Mutant pho1
The Arabidopsis pho1 mutant is impaired in its ability to load Pi into the xylem and therefore to translocate Pi to the shoot (Poirier et al., 1991). To determine whether the down-regulation of Mt4-like genes by Pi is dependent on Pi translocation to the shoot, we examined the expression of the At4 gene in the pho1 mutant. Because of the limited amount of pho1 tissue available, we used RNA extracted from whole plants coupled with RT-PCR and hybridization with the At4 or conserved oligonucleotide probe. As shown in Figure 3B, At4 is expressed in roots and not in leaves, and therefore this approach measures expression in the roots.
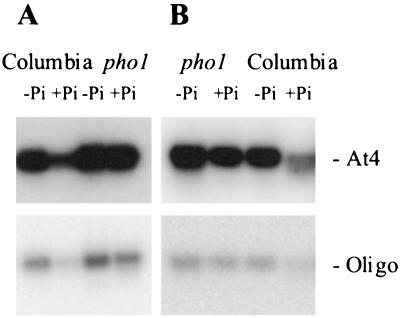
At4 transcripts were abundant in wild-type plants receiving low-Pi fertilizer, but not in plants receiving high-Pi fertilizer, as determined using either the At4 cDNA or the oligonucleotide based on the conserved region as the probes (Fig. 7). In contrast, At4 transcripts were abundant in pho1 plants receiving both low-Pi and high-Pi fertilizer. These results were observed in two independent experiments (Fig. 7). Thus, the At4 transcript is not down-regulated in response to Pi in the pho1 mutant, suggesting that down-regulation is dependent on the translocation of Pi to the shoot.
Figure 7.
Southern blots of RT-PCR products derived from wild-type Arabidopsis and the Pi-accumulation mutant pho1. Total RNA was extracted from plants grown with (+Pi) or without (−Pi) phosphate fertilization, reverse transcribed, standardized, amplified by PCR using a primer pair based on the At4 sequence, blotted, and probed with either a 32P-labeled At4 cDNA (At4) or an end-labeled oligonucleotide identical to the conserved sequence (Oligo) for two independent experiments (A and B).
DISCUSSION
The identification of Pi-starvation-inducible Mt4-like genes in members of the Fabaceae, Solanaceae, and Brassicaceae families indicates that these genes are relatively widespread among dicots and supports the hypothesis that these genes are a common component of the Pi-starvation response. Surprisingly, these genes could only be identified using a 30-bp probe, based on the region of identity between the Mt4 and TPSI1 cDNAs, and were not detected when full-length cDNAs were used as the probes. In contrast, we previously identified a homolog of Mt4 in M. sativa by northern-blot analysis using the entire Mt4 cDNA as a probe (Burleigh and Harrison, 1997). Likewise, Liu et al. (1997) identified homologs of TPSI1 in other solanaceous plants using the entire TPSI1 cDNA as a probe. As might be expected, the homologs from closely related species share greater amino acid identity than homologs from distantly related species. The inability to detect an Mt4-like gene in corn, a representative monocot, implies that these genes may have a relatively recent evolutionary history. These results may reflect subtle differences in the mechanisms that monocots and dicots use in their response to Pi starvation.
The At4 cDNA from Arabidopsis shares similarities with both Mt4 from M. truncatula and TPSI1 from tomato: (a) all of these genes are induced by Pi starvation, (b) they are single-copy genes, (c) their transcripts range in size from approximately 0.5 to 0.7 kb, (d) they are composed of numerous short, overlapping ORFs (8 for Mt4, 6 for TPSI1, and 12 for At4), and (e) they share a short region of nucleic acid identity located approximately in the middle of the transcript. Although initially hypothesized to encode the conserved polypeptide sequence WKGQLR/LSFGI, the absence of a corresponding ORF in At4 now suggests that the function of this region may not be at the protein level. Alternatively, the other nonconserved ORFs may be important. In mammalian systems short, overlapping ORFs are sometimes present in the 5′ untranslated regions of genes, where they play a role in the translational regulation of a larger, translated ORF located at the 3′ end of the transcript (Reynolds et al., 1996). ENOD40, a plant gene encoding a growth regulator involved in nodule development (van de Sande et al., 1996), is also composed of numerous short, overlapping ORFs. It is interesting that the plant genes ENOD40, Mt4, At4, and TPSI1 do not contain any obvious large ORFs at their 3′ ends.
The pho1 mutant of Arabidopsis is unable to load Pi into the xylem and therefore Pi translocation to the shoot is severely reduced (Poirier et al., 1991). Examination of At4 expression in wild-type plants and the pho1 mutant revealed that whereas Pi fertilization resulted in the reduction of At4 transcripts in wild-type Arabidopsis, Pi fertilization did not reduce At4 gene expression in the pho1 mutant. These analyses suggest that the down-regulation of At4 in response to Pi fertilization is dependent on the translocation of Pi to the shoot. Thus, our results not only provide further support for the shoot control of root responses involved in phosphate nutrition (Drew and Saker, 1984; Jeschke et al., 1997), but also provide the first example, to our knowledge, of this control operating at the level of gene expression.
Using a split-root technique we demonstrated that the down-regulation of the Mt4 gene by mycorrhizal colonization was a systemic effect: colonization of one-half of a split-root system resulted in the down-regulation of Mt4 expression in those roots and in the other half of the roots not directly exposed to the fungus. However, in these experiments, colonization did not significantly enhance the Pi status of the plant, suggesting that Pi most likely was not involved in this down-regulation. These results support our previous suggestion that the down-regulation of the Mt4 gene in the roots of M. truncatula is controlled by two pathways that are at least initially independent: one dependent on Pi fertilization and the other dependent on root contact with AM fungi (Burleigh and Harrison, 1997). The function of Mt4 in these two closely linked processes remains to be determined.
Mt4 gene expression was also systemically down-regulated by Pi fertilization. Liu et al. (1998a) have also recently reported that Pi fertilization can systemically down-regulate TPSI1 and two Pi transporters in tomato. We found that Pi did not accumulate in the unfertilized half of the split roots, despite the accumulation of Pi in the fertilized half of the root system and in the shoot. This is similar to the findings of Drew and Saker (1984), although in their case it was a transitory response. Because of the absence of Pi accumulation, we conclude that this nutrient was not directly responsible for the observed systemic down-regulation of Mt4 expression in the unfertilized half of the split roots. However, it is possible that the recycling of phosphate between the shoot and the root (Drew and Saker, 1984; Jeschke et al., 1997), although not accumulating as Pi in the Pi-starved half of the split-root system, may nonetheless have caused the down-regulation of Mt4 gene expression in these roots. Alternatively, based on the results of both the pho1 and Pi split-root experiments, we suggest that the translocation of Pi to the shoot results in the production or activation of a shoot factor that is subsequently translocated to the root, where it down-regulates Mt4-like genes. Whereas Pi itself is not likely the signal, organic forms of phosphate, such as ATP (Ziegler, 1975), may act as signaling molecules. The proposed shoot-derived signal molecule could be a translocating activator of Mt4-like genes, whereby its absence or inactivation as a result of high shoot phosphate levels results in Mt4 down-regulation in the root. Several organic acids such as malate, succinate, shikimate, and oxalate increase in phloem sap during phosphate starvation (Dinkelacker et al., 1989; Hoffland et al., 1992; Jeschke et al., 1997) and could be candidates for this proposed factor. Finally, other shoot-derived molecules could be influential, as is the case for certain amino acids that act as signaling molecules in nitrogen uptake (Muller and Touraine, 1992).
The hypothesis that a shoot signal is responsible for the down-regulation of Mt4 gene expression will be further tested.
ACKNOWLEDGMENTS
The authors thank Drs. Melina Lopez-Meyer and Christian Dammann for helpful discussions and for critical reading of the manuscript. We thank Laura Blaylock for growing some of the Arabidopsis plants.
Abbreviations:
- AM
arbuscular mycorrhizal
- ORF
open reading frame
- RT
reverse transcription
Footnotes
This work was supported by The Samuel Roberts Noble Foundation.
LITERATURE CITED
- Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- Arnon DI, Hoagland DR. Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci. 1940;50:463–483. [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bieleske RL. Phosphate pools, phosphate transport and phosphate availability. Annu Rev Plant Physiol. 1973;24:225–252. [Google Scholar]
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. A cDNA from the arbuscular mycorrhizal fungus Glomus versiforme with homology to a cruciform DNA-binding protein from Ustilago maydis. Mycorrhiza. 1998;7:301–306. [Google Scholar]
- Chen PS, Toribara TY, Warner H. Microdeterminations of phosphorus. Anal Biochem. 1956;28:1756–1758. [Google Scholar]
- Delhaize E, Randall PJ. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;107:207–213. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelacker B, Römheld V, Marschner H. Citric acid excretion and precipitation of calcium citrate in the rhizosphere of white lupin (Lupinus albus L.) Plant Cell Environ. 1989;12:285–292. [Google Scholar]
- Drew MC, Saker LR. Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: evidence of non-allosteric regulation. Planta. 1984;160:500–507. doi: 10.1007/BF00411137. [DOI] [PubMed] [Google Scholar]
- Eckenrode VK, Arnold J, Meagher RB. Comparison of the nucleotide sequence of soybean 18S rRNA with the sequence of other small subunit rRNAs. J Mol Evol. 1985;21:259–269. doi: 10.1007/BF02102358. [DOI] [PubMed] [Google Scholar]
- Hall SM, Baker DA. The chemical composition of Ricinus phloem exudate. Planta. 1972;106:131–140. doi: 10.1007/BF00383992. [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dixon RA. Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microbe Interact. 1993;6:643–654. [Google Scholar]
- Heuwinkel H, Kirkby EA, Le Bot J, Marschner H. Phosphorus deficiency enhances molybdenum uptake by tomato plants. J Plant Nutr. 1992;15:549–558. [Google Scholar]
- Hoffland E, van den Boogaard R, Nelemans JA, Findenegg GR. Biosynthesis and root exudation of citric and malic acid in phosphate-starved rape. New Phytol. 1992;122:675–680. [Google Scholar]
- Jensén P, König T. Development of regulation mechanisms for SO24− influx in spring wheat roots. Physiol Plant. 1982;55:459–464. [Google Scholar]
- Jeschke WD, Kirkby EA, Peuke AD, Pate JS, Hartung W. Effects of P deficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L.) J Exp Bot. 1997;48:75–91. [Google Scholar]
- Kawasaki ES. Amplification of RNA. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 21–27. [Google Scholar]
- Lefebvre DD, Duff SMG, Fife CA, Julien-Inalsingh C, Plaxton WC. Response to phosphate deprivation in Brassica nigra suspension cells. Plant Physiol. 1990;93:504–511. doi: 10.1104/pp.93.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie G, Willmitzer L, Riesmeier JW. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Raghothama KG. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998a;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact. 1998b;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Malboobi MA, Lefebvre DD. A phosphate-starvation inducible β-glucosidase gene (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol. 1997;34:57–68. doi: 10.1023/a:1005865406382. [DOI] [PubMed] [Google Scholar]
- Marschner H, Cakmak I. Mechanism of phosphorus-induced zinc deficiency in cotton. II. Evidence for impaired shoot control of phosphorus uptake and translocation under zinc deficiency. Physiol Plant. 1986;68:491–496. [Google Scholar]
- Muchhal US, Pardo JM, Raghathama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed]
- Muller B, Touraine B. Inhibition of NO3− uptake by various phloem-translocated amino acids in soybean seedlings. J Exp Bot. 1992;43:617–623. [Google Scholar]
- Pitman MG. Ion transport in the xylem. Annu Rev Plant Physiol. 1977;28:71–88. [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K, Zimmer AM, Zimmer A. Regulation of RARβ2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J Cell Biol. 1996;134:827–835. doi: 10.1083/jcb.134.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants: from soil to cell. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjørring JK, Jensén P. Phosphorus nutrition of barley, buckwheat and rape seedlings. I. Influence of seed-borne P and external P levels on growth, P content and 32P/31P-fractionation in shoots and roots. Physiol Plant. 1984;61:577–583. [Google Scholar]
- Simon L, Lalonde M, Bruns TD. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhia fungi colonizing roots. Appl Environ Microbiol. 1992;58:291–295. doi: 10.1128/aem.58.1.291-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Dong B, Delhaize E. The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 1997;11:83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Smith FW, Jackson WA, van den Berg PJ. Internal phosphorus flows during development of phosphorus stress. Aust J Plant Physiol. 1990;17:451–464. [Google Scholar]
- Smith SE, Read DJ, eds (1997) Mycorrhizal Symbiosis. Academic Press, San Diego, CA
- van de Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A and others. Modification of phytohormone response by a peptide encoded by ENOD40 of legumes and a nonlegume. Science. 1996;273:370–373. doi: 10.1126/science.273.5273.370. [DOI] [PubMed] [Google Scholar]
- Ziegler H. Nature of transported substances. In: Zimmermann HH, Milburn JA, editors. Transport in Plants. I. Phloem Transport. Encyclopedia of Plant Physiology, Vol 1. New York: Springer-Verlag; 1975. pp. 59–100. [Google Scholar]