Abstract
Background
The competence of the tsetse fly Glossina pallidipes (Diptera; Glossinidae) to acquire salivary gland hypertrophy virus (SGHV), to support virus replication and successfully transmit the virus depends on complex interactions between Glossina and SGHV macromolecules. Critical requisites to SGHV transmission are its replication and secretion of mature virions into the fly's salivary gland (SG) lumen. However, secretion of host proteins is of equal importance for successful transmission and requires cataloging of G. pallidipes secretome proteins from hypertrophied and non-hypertrophied SGs.
Methodology/Principal Findings
After electrophoretic profiling and in-gel trypsin digestion, saliva proteins were analyzed by nano-LC-MS/MS. MaxQuant/Andromeda search of the MS data against the non-redundant (nr) GenBank database and a G. morsitans morsitans SG EST database, yielded a total of 521 hits, 31 of which were SGHV-encoded. On a false discovery rate limit of 1% and detection threshold of least 2 unique peptides per protein, the analysis resulted in 292 Glossina and 25 SGHV MS-supported proteins. When annotated by the Blast2GO suite, at least one gene ontology (GO) term could be assigned to 89.9% (285/317) of the detected proteins. Five (∼1.8%) Glossina and three (∼12%) SGHV proteins remained without a predicted function after blast searches against the nr database. Sixty-five of the 292 detected Glossina proteins contained an N-terminal signal/secretion peptide sequence. Eight of the SGHV proteins were predicted to be non-structural (NS), and fourteen are known structural (VP) proteins.
Conclusions/Significance
SGHV alters the protein expression pattern in Glossina. The G. pallidipes SG secretome encompasses a spectrum of proteins that may be required during the SGHV infection cycle. These detected proteins have putative interactions with at least 21 of the 25 SGHV-encoded proteins. Our findings opens venues for developing novel SGHV mitigation strategies to block SGHV infections in tsetse production facilities such as using SGHV-specific antibodies and phage display-selected gut epithelia-binding peptides.
Author Summary
Tsetse fly (Diptera; Glossinidae) transmits two devastating diseases to farmers (human African Trypanosomiasis; HAT) and their livestock (Animal African Trypanosomiasis; AAT) in 37 sub-Saharan African countries. During the rainy seasons, vast areas of fertile, arable land remain uncultivated as farmers flee their homes due to the presence of tsetse. Available drugs against trypanosomiasis are ineffective and difficult to administer. Control of the tsetse vector by Sterile Insect Technique (SIT) has been effective. This method involves repeated release of sterilized males into wild tsetse populations, which compete with wild type males for females. Upon mating, there is no offspring, leading to reduction in tsetse populations and thus relief from trypanosomiasis. The SIT method requires large-scale tsetse rearing to produce sterile males. However, tsetse colony productivity is hampered by infections with the salivary gland hypertrophy virus, which is transmitted via saliva as flies take blood meals during membrane feeding and often leads to colony collapse. Here, we investigated the salivary gland secretome proteins of virus-infected tsetse to broaden our understanding of virus infection, transmission and pathology. By this approach, we obtain insight in tsetse-hytrosavirus interactions and identified potential candidate proteins as targets for developing biotechnological strategies to control viral infections in tsetse colonies.
Introduction
Tsetse flies (Glossina sp.) are found exclusively in sub-Saharan Africa and are efficient vectors of African trypanosomes, causative agents of sleeping sickness in humans and nagana in domesticated animals [1]–[3]. Sleeping sickness is invariably fatal if untreated and, until now, the available drugs for sleeping sickness have been unsatisfactory, some being toxic and all difficult to administer [4], and resistance to drugs is increasing [5]. Hence, the search for novel strategies must continue among which are vector-based strategies [6]. Tsetse control remains the most feasible management technique to combat trypanosomiasis and the application of the sterile insect technique (SIT) within the concept of area-wide integrated insect management (AW-IPM), has had promising successes [7], [8]. This strategy relies heavily on colony mass rearing of flies in contained production facilities. The problem is that the production of some species of tsetse such as Glossina pallidipes colonies are vulnerable to infections by a salivary gland hypertrophy virus (SGHV) [9]–[12]; which in a proportion of infected flies leads to hypertrophy (hyperplasia) of the salivary glands (hereafter referred to as SGs) and gonadal lesions. Consequently, fly productivity and fecundity drastically drops, often leading to colony collapse, making colony rearing and SIT applications difficult to implement.
A critical step during SGHV infection of tsetse is the viral replication following ingestion of virus-contaminated blood meals [13]. Although it is yet to be established how long after infection the virus is transmitted, it is likely that a requisite to the transmission of the virus is replication and secretion of the virus into the SG lumen. It is currently unknown whether virus transmission is modified by tsetse saliva that is also deposited at the feeding site to enable the blood feeding process [14], [15]. Further, it is currently unknown how SGHV influences fly gene expression in the SGs or how exactly tsetse immune system defends the fly from the injurious consequences of SGHV infection. To date, the non-redundant (nr) protein database of GeneBank has 156 Glossina proteins, 17 of which are annotated as found in the fly's SGs [16]. This is in addition to 8 proteins from a previous limited transcriptome analysis of G. morsitans morsitans saliva [17]–[20]. Given that knowledge on the mechanisms behind virus replication and transmission processes remains very limited, further studies are required to characterize the molecular interactions between Glossina and its SGHV.
The hypothesis of this study is that the competence of Glossina sp. to acquire SGHV and to successfully transmit mature virions to its offspring and to other flies in the colony depends on interactions between Glossina and SGHV macromolecules. Hence, the Glossina SG secretome must encompass a spectrum of proteins required for all the different facets of the SGHV infection cycle. In this study, we investigated the secretome of hypertrophied and non-hypertrophied SGs of the tsetse fly Glossina pallidipes to gain a deeper understanding of the composition and putative roles of the fly's saliva in the Glossina-SGHV interactions, aiming particularly at the identification of potential targets for development of virus mitigation strategies in mass rearing facilities of G. pallidipes.
Materials and Methods
Tsetse fly strain and SGHV detection
Two groups of G. pallidipes flies, teneral (i.e. within 24 h post emergence) and ten-day old were obtained from the IAEA Insect Pest Control Laboratory Seibersdorf, Austria. To detect the presence of Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) infections in teneral flies, total DNA was extracted from one intermediate excised leg of individual flies using the ZR Genomic DNA kit (Zymo Research, USA). The DNA was amplified by PCR using primers and conditions described previously [21], [22]. Flies with negative PCR results were considered as non-infected (hereafter referred to as non-hypertrophied), while flies with positive PCR results were considered as symptomatically infected. The symptomatically SGHV-infected flies (hereafter referred to as hypertrophied) used in this study were naturally infected during the fly mass-rearing, i.e. they acquired SGHV from their mothers. These naturally-infected flies were used in this study because artificial infection of Glossina using SGHV preparations from hypertrophied salivary glands either orally or by injection does not lead to hyperplasia in the same generation (unpublished data). The hyperplasia of the SGs was confirmed microscopically during subsequent dissections. The ten-day old experimental flies were divided into eight groups based on hours post feeding (hereafter referred to as hpf) and whether they were hypertrophied or not. These were groups 1A and B at 0 hpf (non-fed = teneral, non-hypertrophied or hypertrophied), and groups of ten-day old flies at 48 (groups 2A and B), 72 (groups 3A and B) and 96 (groups 4A and B) hpf. Flies in groups 1A and B were dissected immediately after PCR results. The flies in the other groups were given a blood meal and maintained in the insectaria in standard rearing conditions [23] until the dissections of the SGs at the respective hpf.
Harvesting of fly saliva
Harvesting of saliva was performed by adaptation of previously described methods [13], [17], [24], [25]. Briefly, flies were anesthetized by a cold shock (10 min; 4°C), and dissected. For each group, 10 and 40 pairs of intact SGs were aseptically collected from hypertrophied and non-hypertrophied flies, respectively, in 500 µl ice-cold, sterile PBS (pH 7.4) supplemented with EDTA-free protease inhibitor cocktail (Roche Applied Sciences, Germany). Saliva fluid was allowed to diffuse out of the glands into the buffer for 2.5 h on ice and the buffer was subsequently separated from the SGs by brief centrifugation (500 rpm; 2 min; 4°C). The supernatants (i.e., saliva = diffusate from intact SGs) were filtered (0.45-µm filter) and immediately frozen in 100 µl aliquots at −80°C until further analysis.
Gel electrophoresis and in-gel digestion
To determine the tsetse SGs protein profiles, the saliva samples were separated using SDS-PAGE (12% acrylamide) [26] for 1.1 cm. The gel was stained with Coomassie Brilliant Blue using the Colloidal Staining Kit (Invitrogen). To prepare the proteins for each of the eight groups described above, equal portions of the central third of each entire gel lane, i.e. the middle section spanning the complete gel lane, and containing the saliva proteins was excised. We selected this middle region of the lanes to avoid contaminations from neighboring lanes. The resultant gel sections were cut into approximately 1 mm3 cubes and in-gel protein digestions performed as previously described [27]. Supernatants were transferred to fresh Eppendorf microtubes, and the remaining peptides were extracted by incubating the gel pieces with 5% trifluoroacetic acid (TFA)/H2O, followed by 15% acetonitrile (ACN)/1% TFA. The extracts were combined, reduced in volume in a speed vacuum and dissolved in 20 µl of 0.1% formic acid/H2O. The peptides resulting from this digestion were analyzed by LTQ-Orbitrap Nano liquid chromatography coupled to electrospray and tandem mass spectrometry (nanoLC-MS/MS) as previously described [28].
Identification and quantitation of the salivary gland proteins
Raw MS/MS files from the LTQ-Orbitrap were generated by MaxQuant software version 1.1.1.36, supported by Andromeda as the database search engine for peptide identification [28]–[31]. MS/MS spectra were searched against a concatenated G. m. morsitans decoy database generated by reversing the protein sequences. The database used for peptide/protein searches was derived from the SGs expressed sequence tag (EST) library available from the International Glossina Genome Initiative (IGGI) (http://old.genedb.org/genedb/glossina/). Protein sequences of common contaminants, e.g. trypsin and keratins were used in MaxQuant's “contaminants.fasta” database. MaxQuant was used with a peptide tolerance of 10 parts per million while all other settings were kept as default with one extra addition of asparagine or glutamine de-amidation as variable modification [32] to allow de-amidated peptides to be used for quantification. Bioinformatic analysis of the MaxQuant/Andromeda Work flow output and the analysis of the abundances of the identified proteins were performed with the Perseus module (available at the MaxQuant suite). We accepted peptides and proteins with a false discovery rate (FDR) of less than 1% and proteins with at least 2 unique peptides.
Signal peptide prediction and gene ontology term mapping of G. pallidipes SG proteins
To suggest putative functions for individual tsetse SGs secretome components, the accepted proteins were inputted into Blast2GO v.2.4.8 (http://www.blast2go.org/) [33] and categorized by molecular function (MF), biological process (BP) and cellular component (CC). Gene Ontology (GO) term mapping was based on sequence similarity to previous GO mapped sequences available in the Uniprot database and by merging GOs identified after the InterProScan searches. Signal secretion peptides were predicted from the results arising from InterProScan searches [34], while potential secretion signal peptide sequences and determination of cleavage site positions predicted using SignalP v. 3.0. (http://www.cbs.dtu.dk/services/SignalP/).
Structural and functional annotation of SGHV-encoded proteins
To predict the conserved domains of SGHV-encoded proteins, the viral protein sequences were inputted into InterPro suite (http://www.ebi.ac.uk/Tools/pfa/iprscan/). Structural and functional annotations were determined by pasting the single-letter amino acid codes of the proteins into the Sequence Annotated by Structure (SAS) interface (http://www.ebi.ac.uk/thornton-srv/databases/sas/) [35]. Further annotations of the information obtained from the PDB output were performed at the pfam site (http://pfam.sanger.ac.uk/).
Results
Strategy
For a comprehensive analysis of the G. pallidipes SG proteins, we adopted a four-step strategy. (1) Electrophoretic profiling and identification of tsetse saliva proteins by nanoLC-MS/MS, (2) cataloguing of the MS-supported proteins by gene ontology mapping using Blas2GO suite, (3) confirmation of the presence of N-terminal signal peptide sequences in these proteins by InterProScan and SignalP suites, and (4) prediction of the potential Glossina-SGHV interactions by analyzing the proteins expressed in hypertrophied SGs.
Electrophoretic profiling and proteomic analysis
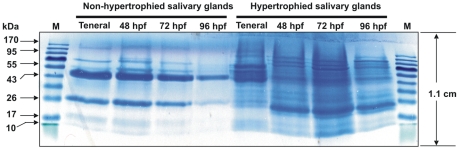
The rationale of using 10 and 40 pairs of intact glands from hypertrophied and non-hypertrophied salivary glands respectively, follows from documented reports that hypertrophied salivary glands are enlarged at least four times their normal thickness [36], [37]. The saliva was harvested from hypertrophied and non-hypertrophied SGs dissected at various time points after feeding as described in the materials and methods and electrophoretic profiles were made (Figure 1). In general, the electrophoretic protein expression profiles detected in the non-hypertrophied and hypertrophied glands correlated well with the viral loads that we have reported in our previous study for these glands [36], [37]. The proteins ranged from <10 kDa to >170 kDa. Visual observation of the SDS-PAGE gel revealed not only clear similarities, but also quantitative differences between the protein profiles of hypertrophied and non-hypertrophied SGs in terms of both proteic band intensities and the presence or absence of several protein bands. Three trends are demonstrated from the protein profiles: (1) A fairly constant protein quantity, albeit a slight decrease at 96 hpf for non-hypertrophied SGs and a maximal quantity at 72 hpf for hypertrophied SGs, (2) A (multiple) high intensity protein band (26 kDa region) in the profile of non-hypertrophied SGs relative to the hypertrophied SGs, and (3) at all the time points, the majority of protein bands in the 19–25 kDa, 29–43 kDa, 55–70 kDa and >95 kDa range present in the secretome of hypertrophied SGs have low abundance or are not detectable in the non-hypertrophied SGs.
Figure 1. SDS-PAGE profiling of G. pallidipes salivary glands proteins.
Shown in the Colloidal Blue Stained SDS-PAGE gel are saliva proteins from non-hypertrophied (lanes 2–5) and hypertrophied salivary glands (lanes 6–9) of G. pallidipes dissected at 0 hpf (Teneral), 48 hpf, 72 hpf and 96 hpf. Lanes 1 and 10 were loaded with Pre-stained protein marker (Fermentas), for which the sizes are indicated in kDa.
A MaxQuant/Andromeda search of LC-MS/MS data against the nr NCBI database was performed for all eight saliva protein samples in the gel lanes (Figure 1). The saliva samples at 72 hpf were arbitrarily chosen for further analysis are they seemed to contain maximum amount of saliva proteins and may also contain the maximum amount of SGHV proteins. This selection criterion is in agreement with previous studies indicating that tsetse saliva production is at peak production 3 days after taking a blood meal [17]. Analysis of these samples by the Perseus module of the MaxQuant suite yielded 521 protein hits, 31 of which were SGHV-encoded based on the known genome and predicted proteome of the virus [28], [38]. Based on an FDR limit of <1% and detection of at least 2 unique peptides per protein (see methods), a further search of G. m. morsitans SG EST dataset identified 317 protein hits. Twenty-five of these proteins were SGHV-encoded (Table S1).
Differential protein expression in hypertrophied and non-hypertrophied SGs
Based on the MS/MS data and the electrophoretic protein profile in Figure 1, a comparison of the protein expression patterns in the non-hypertrophied and hypertrophied SGs within and between the different time points revealed noteworthy findings. Firstly, the relative composition of saliva proteins in the non-hypertrophied SGs remained fairly constant from 0 hpf to 72 h pf, followed by a slight drop at 96 hpf, unlike in the case of the non-hypertrophied SGs (Figure 1). This is in agreement with a previous report by Van Den Abbeele and his group [17]. Secondly, at all the time points, at least 32 Glossina proteins are highly up regulated i.e. they are expressed in high abundance in the hypertrophied SGs but were either detected in very low abundance or not detectable at all in the non-hypertrophied SGs (Table 1). Thirdly, some Glossina proteins are down regulated in the hypertrophied SGs relative to the non-hypertrophied SGs. Nine of the most down-regulated proteins at all the time points are indicated in Table 2. Taken together, these differential protein expressions between hypertrophied and non-hypertrophied SG implies that infection of Glossina by SGHV greatly alters the protein expression pattern in flies with hypertrophied SGs. Lastly, the maximal expression of proteins was found in the hypertrophied SGs at 72 hpf (Figure 1). This agrees well with previous reports that saliva production in Glossina reaches maximum 2–3 days after a blood meal [17].
Table 1. Thirty-two G. pallidipes secretome proteins uniquely expressed in hypertrophied salivary glands.
| Mol. wt range | Description of the proteins expressed in high abundance in hypertrophied salivary glands | ||
| Protein Name | Accession No. | Functional annotation | |
| <26 kDa | Bis-(5′-nucleosyl)-tetraphosphatase | XP_001969373.1 | DNA replication, metabolic stress and apoptosis |
| Niemann-Pick TypeC-2 | ADD20212.1 | Mesoderm development | |
| RNA polymerase II - BTF3 | ADD19794.1 | Transcription | |
| deoxyUTP-pyrophosphatase | ADD20717.1 | Chromosomal integrity | |
| Transitional ER ATPase | XP_002016299.1 | Molecular chaperones | |
| ADP-ribosylation factor 1 | ADE42873.1 | Vesicle coat protein assembly | |
| DNA replication factor/protein phosphatase inhibitor SET/SPR-2 | ADD19576.1 | Movement of histones; nucleasome assembly; chromatin fluidity | |
| GTPase -binding nuclear protein Ran/TC4/gSP1 | ADD18812.1 | Nuclear transport and DNA replication | |
| Cu/Zn-superoide dismutase | ADD19264.1 | REDOX processes | |
| 26–43 kDa | Ras-like GTP-binding protein Rho1 | ADD18925.1 | Control of cytoskeletal changes |
| Proteasome activator complex subunit 3 | ADD19370.1 | Binds to 20S proteasome | |
| Aldo/keto reductase | ADD18539.1 | Oxidoreductase activities | |
| 26S proteasome non-ATPase regulatory complex | ADD18827.1 | Integrity of 26S proteasome complex | |
| Quiescin-sulfhydryl oxidase4 | XP_002048727.1 | Localized in high concentrations in cells with heavy secretory load | |
| 43–55 kDa | Serine protease inhibitor4 | ABC25075.1 | Chaperoning; Toll signalling |
| 60s acidic ribosomal protein P0 | ADD19996.1 | Proteins synthesis | |
| S-adenosylmethionine synthetase | ADD19751.1 | Supplies metabolic methyl groups by catalyzing synthesis of AdoMet | |
| ATP-dependent RNA helicase | XP_001968815.1 | Splicing and ribosome biogenesis | |
| Vacuolar ATPase-S1 Ac45 | ADD18511.1 | REDOX; NAD/NADH-binding | |
| Imaginal growth factor-3 | ABC25095.1 | Chitinase-related GH18; interaction with surface glycoproteins | |
| Hsp-cognate70Cb | CAB38172.2 | Protein folding | |
| Yellow protein precursor | ADD19747.1 | Controls adult pigmentation | |
| >55 kDa | Catalase | ADD20421.1 | Protection from peroxides |
| Hsp70/Hsp90 organizing protein | ADD19147.1 | Protein-protein interactions; chaperoning; transcription; protein transport complexes | |
| Juvenile hormone esterase | ADD18773.1 | Hydrolase | |
| Hsp60 (GroEL chaperonin) | ADD20133.1 | Productive protein folding | |
| Angiotensin-converting-enzyme | XP_002002462.1 | Membrane-located metallopeptidase | |
| Amidophosphoribosyltransferase | XP_002083899.1 | Purine biosynthesis | |
| Lysosomal-α-manosidase | ADD18519.1 | Carbohydrate metabolism | |
| C1-Tetrahydrofolate synthase | ADD18346.1 | Carbon metabolism | |
| Endocytosis/signalling protein EHD1 | ADD19069.1 | Endocytosis; vesicle transport; signal transduction | |
| Gamma-glutamyl phosphate reductase | ADD19821.1 | L-proline biosynthetic pathway | |
The thirty-two G. pallidipes proteins expressed in high abundance in the hypertrophied SGs at all the time points (0, 48, 72 and 96 hpf) but were either detected in very low abundance or not detectable at all in the non-hypertrophied salivary glands.
Table 2. Nine G. pallidipes proteins that are down-regulated in hypertrophied salivary glands.
| Protein Name | Mol. Wt (kDa) | Accession No. | Description of functional domains |
| Tsal2 protein precursor | 17.541 | ADD19043.1 | DNA/RNA non-specific endonuclease |
| 5′-nucleotidase family salivary protein | 18.229 | ADD20435.1 | Cellular energy metabolism |
| Tsal1 protein precursor | 22.509 | ADD20565.1 | DNA/RNA non-specific endonuclease |
| Larval serum protein 2 | 23.716 | ADD18255.1 | store of amino acid for synthesis of adult proteins |
| Salivary antigen 5 precursor | 28.901 | ADD18879.1 | CAP family protein with unknown function |
| 20S proteasome regulatory-β-type | 30.592 | ADD19908.1 | Central enzyme of non-lysosomal protein degradation |
| Salivary secreted adenosine | 47.831 | ADD20094.1 | A metallo-dependent hydrolase |
| Salivary gland growth factor-1 | 5.388 | ADD18584.1 | Adenosine deaminase-related growth factor |
| Apyrase-related protein | 53.977 | ADD18451.1 | Cellular energy metabolism |
Summary of nine of the most down regulated proteins in the hypertrophied salivary glands. The proteins are arranged in the order of molecular weights.
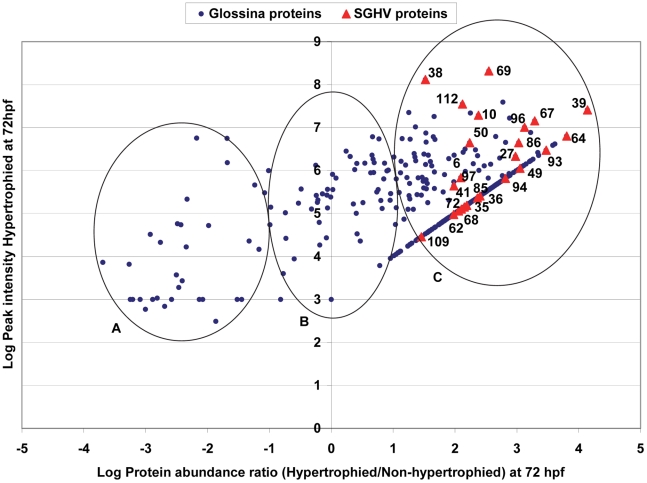
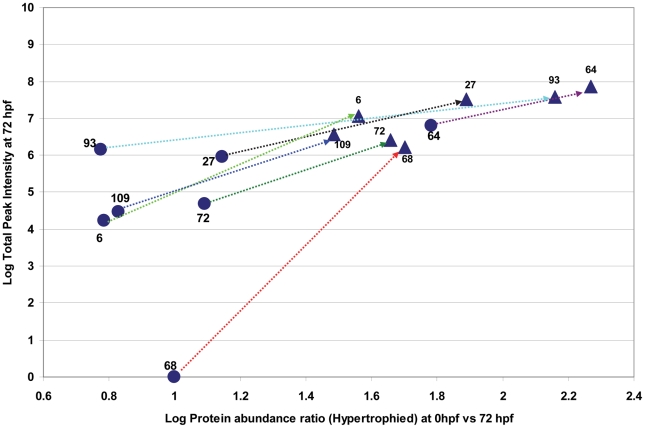
Based on the analysis of the differential protein expression patterns, we focused on the abundances of saliva proteins from hypertrophied and non-hypertrophied SGs at 72 hpf relative to 0 hpf in order to investigate perturbations of the protein expression in the fly SGs. We first compared Glossina and SGHV protein abundance ratios between hypertrophied and non-hypertrophied SGs (Figure 2) to find the really significance differences between the two samples. Three abundance patterns could be deduced. First, 39.4% (115/292) of Glossina and 52% (13/25) SGHV proteins were abundantly expressed in hypertrophied flies relative to non-hypertrophied flies (Figure 2A). Second, 14.4% (42/292) of Glossina proteins showed relatively low abundance regardless of SGHV infection (Figure 2B). Lastly, 46.2% (135/292) and 48% (12/25) of Glossina and SGHV proteins were specifically expressed in the hypertrophied SGs, respectively (Figure 2C). A plot of the shift in the abundance of a selection of seven SGHV proteins uniquely expressed in hypertrophied flies at 72 hpf relative to teneral flies is shown in Figure 3. The host proteins abundantly expressed in hypertrophied SGs at 72 hpf showed a similar trend (data not show). These uniquely expressed proteins at 72 hpf have implications in the Glossina-SGHV molecular interactions (see discussion).
Figure 2. Plot of abundance of G. pallidipes (dots) and SGHV (triangles) proteins.
The figure shows protein abundance ratios between hypertrophied and non-hypertrophied salivary glands collected at 72 hpf. G. pallidipes and SGHV proteins are indicated in blue dots and red triangles respectively. Shown are the most abundantly expressed proteins (group A), the least abundant (group B) and the proteins detected in the hypertrophied salivary glands but were not detectable in the non-hypertrophied salivary glands (group C). To determine the ratio for group C the value for the non-detectable proteins was set to 1000. This value is just below the lowest value obtained for the least abundant protein (cn8877 Salivary gland growth factor-2).
Figure 3. The shift in abundance of SGHV proteins from 0 hpf to 72 hpf.
The figure shows the shift observed in the expression of 7 SGHV proteins detected in the secretome of hypertrophied salivary glands of G. pallidipes dissected at 72 hpf (circles) relative to 0 hpf (triangles).
Gene ontology and signal peptide sequence mapping
Secondly, we conducted gene ontology (GO) mapping on the saliva proteins. At least one GO term could be assigned to 285 of the 317 detected saliva proteins. Five (1.8%) Glossina proteins were deemed of unknown function after blast searches against the nr databases and annotation augmentation. The GO terms assigned to individual Glossina proteins are shown in Table S1, while the terms assigned to SGHV proteins are shown in Table S2.
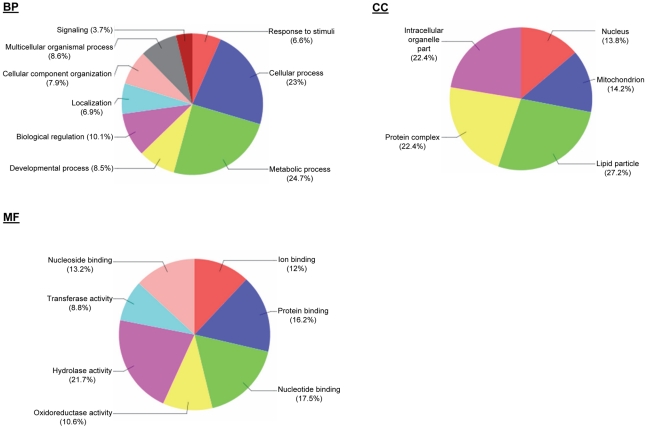
Based on the GO annotations, the MS/MS-supported proteins were grouped into three categories: (1) the broad biological processes (BP) the proteins are involved in, (2) the predicted molecular functions (MF) they perform, and (3) the sub-cellular structures/locations/macromolecular complexes or components (CC) these proteins associate with (Figure 4). The three most common BPs were metabolic processes (24.7%), cellular processes (23%) and biological regulation (10.1%) whereas the three most common MFs were nucleotide/nucleoside-binding (combined percentage of 30.7%), hydrolase activity (21.7%), protein binding (16.2%) and transferase activity (13.2%). The multilevel CC analysis returned five sub-categories, of which the highest proportion (27.2%) of the proteins showed association with lipid-related metabolism (sub-categories of CC, Figure 4). These observations are not entirely unexpected. Other studies have shown considerable changes in cellular organization and metabolism in diseased insects. For instance, during the infection of mosquitoes by densonucleosis (DNV) and crane fly by Tipula iridescent (TIV) viruses, an enlarged nucleus in target cells was noted to be accompanied by a large increase in nuclear DNA synthesis and a massive enlargement of fat body cells [39]. We also noted that a relatively high proportion (23%) of the identified tsetse SG proteins is associated with cellular processes (BP). Although a full explanation for this observation remains to be confirmed, cellular modifications would be expected in SGHV-infected Glossina SG cells, in terms of the formation of vesicles and/or multivesicular bodies associated with various organelles such as the endoplasmic reticulum. Indeed, Sang et al., showed that the cytoplasm of SGHV-infected cells of G. m. centralis are heavily vacuolated, show complete disintegration of cytoplasmic organelles, including the smooth and rough ER and the mitochondria, leaving the nuclei scattered around [40]. These membrane and organelle changes could also be involved in processes such as viral mRNA translation, assembly of protein complexes of both SGHV and Glossina origin, as well as cell-to-cell transport.
Figure 4. Annotation of G. pallidipes salivary glands secretome proteins.
Gene Ontology (GO) terms for categorization of G. pallidipes salivary gland secretome proteins by molecular function (MF), biological process (BP) and cellular component (CC) to suggest the functional components of the secretome.
Lastly, we used the InterProScan suite to determine which of the identified saliva proteins contained predicted secretion signals. This analysis revealed that 96.5% (282/292) of Glossina and (6/25) and 24% of SGHV MS/MS-supported proteins contained predicted secretion signals, respectively. Further, SignalP analysis of these proteins confirmed that 23% (65/282) of these Glossina proteins and the 6 SGHV proteins contained N-terminal signal peptide sequences. Table S1 shows the 65 G. pallidipes saliva proteins with signal peptides predicted by SignalP (hereafter referred to as the SG secretome proteins). Structural and functional annotation of all detected SGHV-encoded proteins revealed that 14 were non-structural (NS) an 11 were structural/capsid (VP) proteins, respectively (Table S2).
It is noteworthy that some of the detected Glossina proteins may not be synthesized in the SGs. For this, alternative mechanisms for the translocation of proteins into SGs should be taken into account. We expected Glossina to have a large number of saliva effector macromolecules with complex effects due to their unique blood-sucking nature. Perhaps some of the identified proteins and macromolecules are synthesized in other organs and then transported into the SGs through hemolymph, by yet-to-be identified mechanisms. This is likely because SGs are capable of sequestering proteins from the hemolymph and then secreting them [41]–[43].
Discussion
Differential protein expression in hypertrophied and non-hypertrophied SGs
Functional analysis of the Glossina proteins that are abundantly expressed in the hypertrophied SGs but are either not detectable or are expressed in very low abundance in the non-hypertrophied SGs revealed that approximately 31% (10/32) may be involved in DNA replication (see Table 1). These uniquely expressed proteins also include molecular chaperones (transitional ER ATPase), and proteins involved in regulation of signalling (serine protease inhibitor 4), and protein-protein interactions (Hsp70/Hsp90 organizing protein) productive protein folding (Hsp60 or GroEL_like chaperonin protein). Other noteworthy proteins include quiescin-sulfylhydryl oxidase 4, a protein usually localized in high concentrations in cells with heavy secretory loads, Ras-like GTP binding protein (Rho1) which is involved in the control of cytoskeletal changes, and imaginal disc growth factor 3, a chitinase-related GH18 protein involved in the interactions with surface glycoproteins.
Some of the down-regulated proteins such as tsetse salivary gland proteins 1 and 2 (Tsal1/2) and tsetse antigen 5 protein are of unknown function. It is possible that the need for bulk synthesis of proteins for maturation of the virions in hypertrophied SGs could have led to the down regulation of the larval serum protein-2, which serves as a store of amino acid for synthesis of adult proteins. Additionally, the down regulation of salivary apyrase, which has been postulated to facilitate blood location and blood feeding [44], is understandable because flies with hypertrophied SGs almost stop feeding 10–15 days post emergence (unpublished data). Overall, the up-regulation of these proteins, coupled to the down-regulation of the proteins described in Table 2 signifies the alteration of protein expression in hypertrophied Glossina SGs as SGHV hijacks key molecular processes as discussed later in this article.
Our approach in establishing the Glossina SG secretome was based on the assumption that the SG proteins would have N-terminal signal sequences. Two clear trends were inferred from the cataloguing of saliva proteins in this study: (1) a high proportion of Glossina SG secretome proteins indeed have signal peptide sequences, and (2) only a low proportion of the secreted proteins associated with Glossina saliva (∼4.9%) are of unknown function. The percentage of non-annotatable Glossina SG secretome proteins is substantially lower than the average of non-annotatable proteins across other systems such as the pea aphid Acyrthosiphon pisum genome (30%) [45]. This high percentage of annotable proteins in tsetse SGs is probably due to the highly specific function of Glossina saliva in blood feeding.
It is likely that our stringent analysis and assumption that the secretome proteins must have N-terminal sequences to substantiate their inclusion into the Glossina secretome may have excluded some important proteins from our secretome pool. These proteins do not necessarily need to be excluded from being part of the Glossina secretome. As this is the first study of the secretome of the Glossina SGs, there is insufficient data available to provide insights into the roles of a majority of the identified proteins. However, potential effector roles of many of these candidate proteins, particularly in the interactions between Glossina and SGHV macromolecules, can be predicted based on their homology or similarity to other proteins involved in vector-virus interactions. In this regard, we discuss a selection of the secretome proteins in the context of six categories of functional proteins, and in relation to SGHV infection.
Protein folding machinery
A high abundance of heat shock proteins (HSPs) was identified in the Glossina hypertrophied SG secretome. HSP induction by viruses is not the result of a meager “canonical” HSP-mediated heat shock response (HSR), but rather the effect of virus-controlled transcriptional/translational switches, sometimes involving individual viral products [46]. Our data present strong indication of hijacking of the host cell chaperon machinery for correct folding of abundant SGHV proteins rapidly synthesized in bulk, and for their correct assembly into viral components during the different phases of viral replication. These observations have been documented in previous studies (see [47]–[49] for review). For instance, we detected Torsin-like-protein-precursor in hypertrophied SGs as opposed to the non-hypertrophied SGs. This protein is localized in the endoplasmic reticulum (ER) lumen and is involved in unfolded protein binding as well as chaperone-mediated protein folding [50], [51].
Many viruses interact with HSPs at different infection stages. In Epstein-Barr Virus (EBV), virus attachment at cell membrane receptors activates signal transduction pathways interfering with the heat shock response (HR) [52]. Surface exposed hsc70 and hsp70 proteins are involved in virus entry into cells [53]–[55] and hsc70 may play an active role in virus entry into the host cell as well as at a post-attachment step [56]. In addition, hsp70 chaperones are involved in disassembly of oligomeric protein structures and viral internalization into host cells (see review in [57]–[63]). Viral proteins, including E1A of Adenovirus [64], large T antigen (T ag) of Simian vacuolating virus 40 [65], [66], ICP4 of Human simplex virus (HSV-1), IE2 of human cytomegalovirus, and nuclear antigen 3 (EBNA3) of EBV [67]–[72], modulate hsp70 by direct interaction with different components of the basal transcription apparatus. Additionally HSP70-cognate 4 (HSP70-4) plays an important role in the homeostasis and suppression of O'nyong-nyong virus (ONNV) replication and in the establishment of latent infections in the mosquito Anopheles gambiae [73]. It is noteworthy that HSP70-4 was detected in the secretome of hypertrophied SG, as opposed to the non-hypertrophied SGs. Although it remains to be established in the case of SGHV infection of Glossina, this protein may play roles in the establishment of asymptomatic SGHV infections in Glossina [13].
Pathogen recognition and defense response proteins
We also identified some members of inducible secreted polypeptides, including C-type lectins (CTLs) (Table S1). CTLs play important roles in insect defense by recognizing pathogen-associated molecular patterns (PAMPS) [74], [75] in invading pathogens [76], [77]. The expression of lectins during virus infection would be expected because they have been shown to mediate immune functions [78], including activation of the lectin complement pathway that also exists in arthropods, thus binding to carbohydrates expressed on viral glycoproteins. In this regard, we detected members of thioester-containing proteins (TEPs) (Table S1) which have been described in the complement system of Drosophila melanogaster and An. gambiae [79]. It is therefore likely that the hypertrophied SGs express CTLs to block glycoprotein-mediated attachment of SGHV to non-infected SG cells. Although CTLs generally diffuse during SGHV infection, their binding to circulating virus could effectively reduce viral infection of lectin-expressing SG cells. Binding of CTLs to virus and subsequent deposition of complement components on the virus membrane can also lead to enhanced infection of cells that express complement receptors.
Protein export machinery
ADP-ribosylation factor (ARF) is an abundant protein that reversibly associates with Golgi membranes, and is implicated in the regulation of membrane traffic through the secretory pathway [80]–[82]. This pathway is important for processing of viral contents into complexes capable of nuclear penetration. ARFs have been shown to be up-regulated and involved in virus infection [83]–[85], and possibly explains the detection of ARF in the hypertrophied SGs at 48, 72 and 96 hpf, as opposed to non-hypertrophied and teneral-hypertrophied SGs at 0 hpf. The data presented here indicate the recruitment/hijacking of ARFs to membranes by SGHV and may provide clues for future identification of the pathways utilized by the virus in the replication process in hypertrophied SG.
Proteases and protease inhibitors
Six types of serine proteases were detected in the secretome of hypertrophied SG but not in the non-hypertrophied SG. Additionally, a precursor of phosphenol oxidase activating factor of the prophenol-oxidase-activating system (proPO-AS), an important component of the innate immune response in insects [86]–[88], was also detected. These two proteins are involved in initiating a signal cascade that eventually leads to melanization reaction which includes the formation of toxic intermediary compounds to kill invading viruses [87]. Studies have shown that the baculovirus P74 is a viral attachment protein [89]–[91]. Cleavage of P74 by trypsin is crucial for infection, probably by to exposing a receptor binding domain [92], enabling interaction with host receptors. SGHV P74 was not detected in this study, which is probably due to its low abundance as demonstrated in our previous study [28]. The expression of the serine proteases in hypertrophied SG is probably required for the interaction of SGHV with the host SG cells receptors as has been reported for several entomopathogens [93], [94].
Housekeeping genes
RNA polymerase II general transcription factor (BTF3), the translation elongation factor EF-1 gamma (EF1γ), and the ATP-dependent RNA helicase were among the factors detected in the secretome of hypertrophied SG. BTF3 is a general transcription factor necessary for activation of a number of viral promoters by RNAP II [95], while EF1γ is involved in the regulation of protein assembly and folding [96], [97]. The detection of these proteins in hypertrophied SGs, coupled to the presence of RNA helicase is desirable for the expression of replication- and maturation-related genes. In addition, proteins involved in signal transduction were also detected (in SGs dissected 48, 72 and 96 hpf) including GTPase-activating protein (GAP), cAMP-dependent protein kinase, and Ras-related small GTPase (Rho type). GAP is known to be necessary for efficient virus infection and replication [98], and is implicated in the regulation of anterograde traffic between the ER and the Golgi complex, while cAMP-dependent protein kinase is implicated in the regulation of virus infection and virus-induced cell-cell fusion.
Cytoskeletal proteins
Most viruses use components of the host cytoskeleton to move within cells. Upon virus infection, virions or sub-viral nucleoprotein complexes are transported from the cell surface to the site of viral transcription and replication. During viral escape, particles containing proteins and nucleic acids move again from the site of their synthesis to that of virus assembly and further to the plasma membrane [99], [100]. Viral (sub) particles, particularly in members of herpesviridae, adenoviridae, parvoviridae, poxviridae and baculoviridae use the microtubule and the actin cytoskeleton. In this study, actin 5C, actin 87E and actin depolymerizing factor were detected in all saliva samples except in the non-hypertrophied SG collected 96 hpf. F-actin capping protein was detected in the saliva of hypertrophied SGs 48 hpf, while actin 57B was detected, albeit in low abundance, in hypertrophied SGs 72 hpf. Myosin heavy chain, which drives transport along actin filaments [99] was detected in the saliva of hypertrophied SGs except at 96 hpf, which probably indicates reduced active transport of SGHV virions at this time point. While all the 6 cytoskeletal proteins were detected in the SGs 96 hpf, none were detected in the secretome of non-hypertrophied SGs 96 hpf. It is possible that this could be due to the hyperplasia of the SGs by SGHV, which could potentially lead to lysis of SG cells during advanced stages of viral infection. Further studies are required to investigate these observations.
Functional annotation of SGHV- proteins detected in the saliva of G. pallidipes
The movement and/or replication of viruses in insect vectors require specific interactions between viruses and host components. DNA viruses have evolved mechanisms to evade the host restrictions at entry, cytoplasmic transport, replication, protein synthesis, innate (and for mammalian viruses, adaptive immune) recognition, and egress from the infected cells. The SGHV genome is a circular double-stranded (ds) DNA molecule [38]. Nuclear-replicating viruses with ds DNA genomes such as herpesvirus engage in all aspects of cellular metabolism [101]. Although it has yet to be established for SGHV, DNA viruses adopt the host transcriptional apparatus and all cellular pathways required for processing and transport of their mRNAs. The host cellular mechanisms translate and turn over viral proteins, while transport of viral macromolecules takes place through cellular organelles and structures. In this study, structural and functional annotation of the identified SGHV proteins also indicate their engagement with Glossina cellular metabolism. In the following sections, we briefly discuss potential roles played by these SGHV proteins and their possible interactions with Glossina SG secretome proteins during the different facets of the viral infection cycle. Inferences below are drawn from other nuclear-replicating DNA viruses, including adenoviruses, hepadnaviruses, herpesviruses, papillomaviruses, and polyomaviruses.
SGHV entry into host cells
The stepwise entry of DNA viruses into host cells requires viral attachment to cell surface receptors and lateral movements of the virus-receptor complex to specialized sites on the plasma membrane [102]–[105]. In closely-related baculoviruses, per os infectivity factor proteins (PIFs) have been shown to be involved in viral attachment to the host cells. [38], [106]–[110]. In the current study, PIFs had very low abundance as was also noted in our previous proteomics study of GpSGHV [28]. In this study, SGHV085 was annotated to be a tyrosine kinase-dependent signaling structural protein, and is probably involved in transport of SGHV polypeptide into the nucleus (see next section) [111]. Additionally, viruses in general elicit signals following attachment to the host cell membrane to circumvent the host defense mechanism. In this regard, annotation revealed SGHV046 as a glutathione S-transferase-like protein, thus pointing to its involvement in this type of signaling.
Bidirectional cytoplasmic transport of SGHV particles to the nucleus
DNA viruses that replicate their genomes in the nucleus use microtubule motors for trafficking towards the nucleus and the periphery during egress after replication [111]–[113]. Bidirectional transport allows precise delivery of capsids to ensure nuclear targeting, and has been demonstrated in HSV-1) [114], [115] and in human adenovirus 2/5 (Ad2/5) [116]–[118]. Incoming DNA viruses expose proteins on the capsid that preferentially recruit microtubule motor complexes [112], and may release tegument proteins before they traffic to the nucleus [119]. To regulate capsid transport, protein phosphorylation by viral and/or host cellular kinases modulate tegument protein composition as in the case of vaccinia virus [120]. In this study, cAMP-dependent protein kinase detected in the SG secretome is probably involved in anterograde trafficking of SGHV. Additionally, the SGHV041 protein detected here is a Casein kinase (isoform-δ), which is likely to be involved in phosphorylating cytoskeletal components both in anterograde and egress [121], [122]. Early during infection, some viruses such as δ-2 human herpes virus 8 and hepatitis C virus induce Rho GTPases [98], [123], which alter the dynamics by increasing the acetylation of actin microfilaments thereby enhancing viral capsid trafficking transport to the nucleus and establishment of successful infection. Again, Ras-related small GTPase (Rho type) as well as GTPase-activating protein (GAP) were detected in the Glossina hypertrophied SGs, and suggest their participation in viral trafficking towards the nucleus. Finally, although the role of spectrins in cytoplasmic transport is not clear, this study identified SGHV010 to have spectrin repeat domains, indicating its potential involvement in SGHV anterograde trafficking.
Docking, uncoating/disassembly and release of SGHV DNA into nucleus
Cytoplasmic transport is followed by viral genome docking and uncoating at the nuclear pore complex (NPC), a stepwise programme involving partial proteome degradation of incoming capsid or tegument proteins [124], [125]. Although it is not clear how uncoating at the NPC occurs, experiments with some viruses such as herpes B virus have indicated that capsids are transported to the nuclear membrane where they bind to NPCs and release their genome into the nucleus [126]. Additionally, cytoplasmic processing of incoming capsids makes them competent for docking to the NPC [114], and probably prevents the naked viral chromatin from traveling through the cytoplasm, which could trigger DNA-sensing host innate immune responses as has been demonstrated in adenovirus [127]. The SGHV006 protein detected in this study was determined to have α/β-hydrolase catalytic domain, a signature domain for lecithin∶cholesterol acyltransferase (LACT) which is involved in membrane docking of viruses to NPC, as well as in nucleocytoplasmic transport of capsids (see [114], [128]–[131] for review).
Development of SGHV transcription and replication
Upon infection, some viruses such as Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) establish centers for transcription, DNA replication and progeny nucleocapsid assembly [101], and others express at least one regulatory protein that interacts directly with similar domains such as the promyelocytic leukemia protein nuclear bodies (PML-NBs) [101]. In this study, SGHV112 annotation revealed the presence of the helix-turn-helix characteristic domains of regulatory proteins [132] involved in DNA-protein interactions. Gamma-interferons were detected in this study in the hypertrophied SG dissected as early as 48 hpf, as well as a 19.3 kDa Glossina protein encoded by GMsg-6444 (a SUMMO-binding protein). The ubiquitin-like protein SUMO is a partner protein to viral replication center and are dramatically enhanced by interferons [133]. Viral proteins associating with these centers have the ability to stimulate lytic infection and induction of reaction from quiescence [134]. This supports observations that majority of SGHV infections in tsetse colonies are in fact asymptomatic [13], and that there is virus induction in infected flies after an initial blood meal (unpublished data). Also detected in this study were SGHV035 and SGHV036, homologs of thymidylate synthase and deoxycytidylate hedroxymethylase, respectively. The former is involved in regulating a balanced supply of dNTPs during DNA replication [135], [136], while the latter is involved in pyrimidine metabolism [137], [138]. Further, our annotation predicted that SGHV039 (a HSP90-like ATPase) is possibly involved in regulation of unwinding of DNA supercoil strands. Additionally, SGHV062, a p53 transcription factor-like protein containing β-sandwich domain of the sec23/24 superfamily was detected. Proteins of this family are involved in chromosomal segregation and are induced early during the S-phase of the cell cycle [139]–[142], and hence have direct roles in viral DNA transcription and replication. Further, detected in this study was an ABC ATPase-like family protein (SGHV064). Studies have implicated members of this protein family to be involved in translation initiation, ribosome biosynthesis and virus capsid assembly of other viruses such as HIV [143], [144]. Taken together, the presence of these Glossina and SGHV proteins in the hypertrophied SGs are an indication that SGHV genome associates with the periphery of PML-NBs, and that viral replication compartments would develop from these sites, as has been observed in other viral systems for instance in HSV-1 [145], [146].
SGHV maturation and nuclear egress
SGHV049 detected in this study was predicted to be a pre-mRNA splicing factor-9-like protein. The WD40/G-β-repeats represent in this protein is a signature domain for proteins that associate with the spliceosome [147]). Other detected proteins that have potential roles in SGHV maturation were: (1) SGHV072, a FAD-dependent sulfhydryl oxidase (with a late promoter motif and hence likely to be involved in virion maturation, [148]); (2) SGHV093, an uncharacterized endonuclease type-IIe-like protein with DNA-binding and cleavage activities [149]; (3) SGHV027, a chitinase-II (O-glycosyl hydrolase) protein, which like the chitinases family-18 proteins may be involved in virus maturation (see [150]–[152] for review); (4) SGHV038, a protein containing α-β-barrel active site, thus likely to be involved in the expression of receptor proteins for membrane transport (egress) [153] and (5) SGHV096, a metal (Mn2+) and ion (S2O4 2−)-binding protein with multiple TMs, thus likely to be part of SGHV ion-channel proteins. Newly assembled enveloped viruses recruit periphery directed motors, and are transported to the plasma membrane on the microtubules upon binding of the outer membrane [113] proteins and fuse with plasma membrane. With involvement of host tyrosine kinases [112], the virus may eventually be launched away from the infected cell and spread the viral progeny. Although it is unclear whether SGHV travel in vesicles or as capsid as baculoviruses do, SGHV097 was predicted to be a vesicle-associated membrane protein and could be involved in targeting and/or fusion of virus-containing vesicles to the target membranes [119], [154].
Conclusions
The Glossina pallidipes species is found in several African countries and rearing facilities have been established in Kenya, Ethiopia and Tanzania with the aim of strengthening the fly eradication campaigns in African. Salivary gland hypertrophy virus infection leads to drastic negative effects on the productivity and stability of the G. pallidipes colonies and in certain cases to colony collapse. In addition, a recent study [155] has demonstrated that this virus is widely distributed in the wild populations of G. pallidipes which further complicates the tsetse eradication campaigns because the mass-reared colonies are normally established from flies collected from the target wild populations. Due to this negative effect, it is necessary to develop virus management strategies to enable sustenance of a healthy and productive colony size for the fly eradication campaigns. Designing a strategy that would interrupt the replication/transmission cycle of the virus in the colonies requires a comprehensive understanding of the mechanism involving the vector-virus interactions. The aim of our study was to establish an extensive protein map of G. pallidipes salivary gland secretome proteins and SGHV proteins in hypertrophied salivary glands, using stringent mass spectrometry criteria to validate the potential proteins, and to establish possible Glossina-SGHV interactions. The substantial differences between the protein profiles of the secretomes of hypertrophied and non-hypertrophied salivary glands, and the analysis of the nanoLC-MS/MS-supported secretome proteins data obtained demonstrate that a large proportion of the proteins identified are indeed secreted.
The data presented in this study demonstrates that the G. pallidipes salivary gland secretome encompasses a wide spectrum of proteins that may be required for the different facets of the SGHV infection cycle from viral attachment to egress of the virions from infected Glossina cells. On the basis of our previous proteomic analysis of GpSGHV virions [28] and the data presented in this study, some interactions between Glossina and SGHV proteins can be predicted (Table 3). It is to be noted that the per os infectivity factors (PIFs) were only detected in very low abundance, and although they are not included in our final pool of the salivary proteins, they probably play a vital role in the oral infection of Glossina sp. One other SGHV protein that was noted to be present in very low abundance was the baculovirus ODV-E66 (PIF4) homolog. This protein is known to be involved in the initial attachment of the baculovirus in the mid-gut epithelia of infected insects, and it is likely to play a similar role in the case of Glossina.
Table 3. Putative Glossina- SGHV protein interactions.
| Glossina secretome proteins | SGHV proteins(ORFs in brackets) | Putative Glossina-SGHV protein Interactions |
| C-Lectins, hsc70, hsp70 & 90, TEPs | Per os infectivity factors (1, 102, 53, 76), glutathione-S-transferase (46) | Viral entry & signalling |
| Actins, Rho GTPase, GAP, molecular chaperones, ARFs | Lecithin-cholesterol acyltransferase (6), casein kinase I-δ (41), spectrin (10) | Birectional cytoplasmic transport & docking at nuclear pore complex (NPC) |
| HSPs, Translation initiation factors, RNA-helicase | Thymidylate synthase (35), dihydrofolate reductase thymidylate synthase (36), HSP90-like ATPase (39), p53-transcription factor-like (62), ABC-ATPase (64) | Viral DNA transcription,replication and translation |
| NTPase-/Torsin-like, DnaJ/Hsp40, molecular chaperones, hsp70-4, ER-PDI | Pre-mRNA splicing factor (49), FAD-sulfhydryl oxidase (72), type-IIe restriction enzyme (93), chitinase-II (27), maltodextrin glycosyltransferase (38), transport-channel proteins (85, 96), vesicle-associated membrane protein (97) | Maturation & nuclear egress of mature virions from infected salivary gland cells |
TEPs = thioester-containing proteins; hsc = heat shock cognate; hsp = heat shock protein; GAP = GTPase-activating protein; ARFs = ADP-ribosylation factor; ER-PDI = endoplasmic reticulum protein disulphide isomerase.
Summary of the putative interactions between Glossina and SGHV proteins during the different facets of the virus replication cycle. The SGHV ORFs encoding the respective viral proteins are indicated (bold and in brackets).
Taken together, the identification of putative host-viral protein interactions opens novel avenues for the development of mitigation strategies against GpSGHV infections. Such strategies could include immune-interventions whereby virus-specific antibodies against PIF proteins could be supplemented in the blood meals used in the membrane-feeding in tsetse production facilities. This would lead to immuno-complexion of SGHV virions in the blood meals, which would block the horizontal transmission of SGHV from fly to fly. In addition, phage display-selected gut epithelia-binding peptides such as derived from the ODV-e66 protein (PIF-4) homolog could be designed to impede the attachment of SGHV to the Glossina mid-gut and subsequent movement of the virus into the fly hemocoel. Such chemically-synthesized oligopeptides or the phage display library expressing the active peptides could be supplemented in the flies' blood meals and, thereby, upon ingestion of the meal, the peptides would out-compete the viral homologs (ODV-e66) in the attachment to the mid-gut receptors [156]. By this approach, vertical transmission of the virus from mother-to-offspring would be interrupted. Finally, SGHV-specific genes could also be targets for RNA interference (RNAi).
Future perspectives
The roles of Glossina and SGHV proteins identified in this study need to be experimentally established. Still to be investigated are questions particularly with regard to Glossina specificity and how SGHV overcomes various transmission barriers in the tsetse fly. Also to be resolved are the roles played in Glossina specificity by SGHV proteins, Glossina proteins, virus receptors, Glossina symbionts, as well as the role of hemolymph and other tissues in the viral transmission process. In addition, the barriers to transovarial (vertical) transmission of SGHV in tsetse remain grossly under-investigated. We hope that with the cataloguing of Glossina salivary glands secretome proteins, the detection of SGHV proteins in hypertrophied salivary glands and the advent of new molecular technologies, that such roles can be elucidated further and eventually exploited to initiate novel strategies for controlling SGHV infections in tsetse mass rearing facilities.
Supporting Information
Sixty-five salivary gland secretome proteins of G. pallidipes . Sixty-five G. pallidipes salivary gland secretome proteins supported by nanoLC-MS/MS, confirmed by gene ontology (GO) annotation, presence of signal peptide sequences and by Blasts on the NCBI and G. m. morsitans databases.
(DOC)
Twenty-five SGHV-encoded proteins detected in the G. pallidipes hypertrophied salivary glands.
(DOC)
Acknowledgments
The authors acknowledge Carmen Marin and A. Hasim Mohammed for tsetse rearing. LC-MS/MS measurements were done at Biqualys, Wageningen (www.biqualys.nl).
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Wageningen University Phd Sandwich Program (www.wur.nl), The Netherlands Fellowship Program Grant Award CF7548/2011 (www.nuffic.nl), and the FAO/IAE Programme (www.iaea.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Geiger A, Ravel S, Mateille T, Janelle J, Patrel D, et al. Vector competence of Glossina palpalis gambiensis for Trypanosoma brucei s.l. and genetic diversity of the symbiont Sodalis glossinidius. Mol Biol Evol. 2007;24:102–109. doi: 10.1093/molbev/msl135. [DOI] [PubMed] [Google Scholar]
- 2.Gooding RH, Krafsur ES. Tsetse genetics: Contributions to biology, systematics, and control of tsetse flies. Annu Rev Entomol. 2005;50:101–123. doi: 10.1146/annurev.ento.50.071803.130443. http://arjournals.annualreviews.org/loi/ento. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steelman CD. Effects of external and internal arthropod parasites on domestic livestock production. Annu Rev Entomol. 1976;21:155–178. doi: 10.1146/annurev.en.21.010176.001103. [DOI] [PubMed] [Google Scholar]
- 4.Barrett MP. The rise and fall of sleeping sickness. The Lancet. 2006;367:1377–1378. doi: 10.1016/S0140-6736(06)68591-7. [DOI] [PubMed] [Google Scholar]
- 5.Matovu E, Seebeck T, Enyaru JCK, Kaminsky R. Drug resistance in Trypanosoma brucei spp., the causative agents of sleeping sickness in man and nagana in cattle. Microbes and Infection. 2001;3:763–770. doi: 10.1016/s1286-4579(01)01432-0. [DOI] [PubMed] [Google Scholar]
- 6.Rio RV, Hu Y, Aksoy S. Strategies of the home-team: symbioses exploited for vector-borne disease control. Trends Microbiol. 2004;12:325–336. doi: 10.1016/j.tim.2004.05.001. PM:15223060. [DOI] [PubMed] [Google Scholar]
- 7.Hendrichs JP, Kenmore P, Robinson AS, Vreysen MJB. Area-wide integrated pest management (AW-IPM): Principles, practice and prospects. In: Vreysen MJB, Robinson AS, Hendrichs J, editors. Area-wide Control of Insect Pests: From Research to Field Implementation. Dordrecht, The Netherlands: Springer; 2007. pp. 3–33. [Google Scholar]
- 8.Msangi AR, Saleh KM, Kiwia N, Malele II, Mussa WA, et al. Success in Zanzibar: Eradication of tsetse. In: Tan K-H, editor. Area-wide Control of Fruit Flies and other Insect Pests. Penang (Malaysia): Penerbit Universiti Sains Malaysia; 2000. pp. 57–66. [Google Scholar]
- 9.Ellis DS, Maudlin I. Salivary gland hyperplasia in wild caught tsetse from Zimbabwe. Entomol Exp Appl. 1987;45:167–173. [Google Scholar]
- 10.Jaenson TGT. Virus-like rods associated with salivary gland hyperplasia in tsetse, Glossina pallidipes. Trans R Soc Trop Med Hyg. 1978;72:234–238. doi: 10.1016/0035-9203(78)90200-6. [DOI] [PubMed] [Google Scholar]
- 11.Jura WGZO, Odhiambo TR, Otieno LH, Tabu NO. Gonadal lesions in virus-infected male and female tsetse, Glossina pallidipes (Diptera: Glossinidae). J Invertebr Pathol. 1988;52:1–8. doi: 10.1016/0022-2011(88)90095-x. [DOI] [PubMed] [Google Scholar]
- 12.Jura WGZO, Otieno LH, Chimtawi MMB. Ultrastructural evidence for trans-ovum transmission of the DNA virus of tsetse, Glossina pallidipes (Diptera: Glossinidae). Curr Microbiol. 1989;18:1–4. [Google Scholar]
- 13.Abd-Alla AMM, Kariithi H, Parker AG, Robinson AS, Kiflom M, et al. Dynamics of the salivary gland hypertrophy virus in laboratory colonies of Glossina pallidipes (Diptera: Glossinidae). Virus Res. 2010;150:103–110. doi: 10.1016/j.virusres.2010.03.001. doi: 10.1016/j.virusres.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Caljon G, Van Den AJ, Sternberg JM, Coosemans M, De Baetselier P, et al. Tsetse fly saliva biases the immune response to Th2 and induces anti-vector antibodies that are a useful tool for exposure assessment. Int J Parasitol. 2006;36:1025–1035. doi: 10.1016/j.ijpara.2006.05.002. PM:16777113. [DOI] [PubMed] [Google Scholar]
- 15.Caljon G, De Ridder K, De Baetselier P, Coosemans M, Van Den AJ. Identification of a tsetse fly salivary protein with dual inhibitory action on human platelet aggregation. PLoS One. 2010;5:e9671. doi: 10.1371/journal.pone.0009671. PM:20351782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alves-Silva J, Ribeiro JMC, Abbeele JJ-D, Attardo G, Hao Z, et al. An insight into the sialome of Glossina morsitans morsitans. BMC Genomics. 2010;11:213. doi: 10.1186/1471-2164-11-213. http://www.biomedcentral.com/1471-2164/11/213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Den Abbeele J, Caljon G, Dierick JF, Moens L, De Ridder K, et al. The Glossina morsitans tsetse fly saliva: General characteristics and identification of novel salivary proteins. Insect Biochem Mol Biol. 2007;37:1075–1085. doi: 10.1016/j.ibmb.2007.06.006. http://www.sciencedirect.com/science/article/B6T79-4P12J32-1/2/ca95a2da52c60c548b7611d864c5a521. [DOI] [PubMed] [Google Scholar]
- 18.Cappello M, Li S, Chen X, Li C-B, Harrison L, et al. Tsetse thrombin inhibitor: bloodmeal-induced expression of an anticoagulant in salivary glands and gut tissue of Glossina morsitans morsitans. Proc Natl Acad Sci U S A. 1998;95:14290–14295. doi: 10.1073/pnas.95.24.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Aksoy S. A family of genes with growth factor and adenosine deaminase similarity are preferentially expressed in the salivary glands of Glossina m. morsitans. Gene. 2000;252:83–03. doi: 10.1016/s0378-1119(00)00226-2. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Kwon J, Aksoy S. Characterization of genes expressed in the salivary glands of the tsetse fly, Glossina morsitans morsitans. Insect Mol Biol. 2001;10:69–76. doi: 10.1046/j.1365-2583.2001.00240.x. [DOI] [PubMed] [Google Scholar]
- 21.Abd-Alla A, Bossin H, Cousserans F, Parker A, Bergoin M, et al. Development of a non-destructive PCR method for detection of the salivary gland hypertrophy virus (SGHV) in tsetse flies. J Virol Methods. 2007;139:143–149. doi: 10.1016/j.jviromet.2006.09.018. http://dx.doi.org/10.1016/j.jviromet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Abd-Alla AMM, Salem TZ, Parker AG, Wang Y, Jehle JA, et al. Universal primers for rapid detection of Hytrosaviruses. J Virol Methods. 2011;171:280–283. doi: 10.1016/j.jviromet.2010.09.025. http://dx.doi.org/10.1016/j.jviromet.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Feldmann U, Luger D, Barnor H, Dengwat L, Ajagbonna B, et al. Tsetse fly mass rearing: Colony management, deployment of sterile flies, related research and development. 1992. pp. 167–180. Tsetse control, diagnosis and chemotherapy using nuclear techniques. Proceedings of a seminar jointly organized by the International Atomic Energy Agency and the Food and Agriculture Organization of the United Nations and held in Muguga, Kenya, 11–15 February 1991. Vienna, Austria: IAEA.
- 24.Caljon G, Van Den AJ, Stijlemans B, Coosemans M, De Baetselier P, et al. Tsetse fly saliva accelerates the onset of Trypanosoma brucei infection in a mouse model associated with a reduced host inflammatory response. Infect Immun. 2006;74:6324–6330. doi: 10.1128/IAI.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo E, deBianchi AG, James AA, Marinotti O. The major acid soluble proteins of adult female Anopheles darlingi salivary glands include a member of the D7-related family of proteins. Insect Biochem Mol Biol. 2002;32:1419–1427. doi: 10.1016/s0965-1748(02)00062-0. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavge of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. http://dx.doi.org/10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Ince AI, Boeren SA, van Oers MM, Vervoort J, Vlak JM. Proteomic analysis of Chilo iridescent virus. Virol. 2010;405:253–258. doi: 10.1016/j.virol.2010.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kariithi HM, Ince AI, Boeren S, Vervoort J, Bergoin M, et al. Proteomic analysis of Glossina pallidipes Salivary Gland Hypertrophy Virus virions for immune intervention in tsetse fly colonies. J Gen Virol. 2010;91:3065–3074. doi: 10.1099/vir.0.023671-0. doi: 10.1099/vir.0.023671-0. [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. PM:19029910. [DOI] [PubMed] [Google Scholar]
- 30.Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. PM:19373234. [DOI] [PubMed] [Google Scholar]
- 31.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. DOI: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 32.Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. http://bioinformatics.oxfordjournals.org/content/21/18/3674.abstract. [DOI] [PubMed] [Google Scholar]
- 34.Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. http://bioinformatics.oxfordjournals.org/content/17/9/847.abstract. [DOI] [PubMed] [Google Scholar]
- 35.Milburn D, Laskowski RA, Thornton JM. Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng. 1998;11:855–859. doi: 10.1093/protein/11.10.855. PM:9862203. [DOI] [PubMed] [Google Scholar]
- 36.Whitnall ABM. The trypanosome infections of Glossina pallidipes in the Umfolosi game reserve, Zululand. Onderstepoort J Vet Sci Anim Ind. 1934;2:7–21. [Google Scholar]
- 37.Abd-Alla A, Cousserans F, Parker A, Bergoin M, Chiraz J, et al. Quantitative PCR analysis of the salivary gland hypertrophy virus (GpSGHV) in a laboratory colony of Glossina pallidipes. Virus Res. 2009;139:48–53. doi: 10.1016/j.virusres.2008.10.006. doi: 10.1016/j.virusres.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Abd-Alla AMM, Cousserans F, Parker AG, Jehle JA, Parker NJ, et al. Genome analysis of a Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) reveals a novel large double-stranded circular DNA virus. J Virol. 2008;82:4595–4611. doi: 10.1128/JVI.02588-07. doi: 10.1128/JVI.02588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris ON. Metabolic changes in diseased insects. 3. Nucleic acid metabolism in Lepidoptera infected by densonucleosis and Tipula iridescent viruses. J Invertebr Pathol. 1970;16:180–186. doi: 10.1016/0022-2011(70)90058-3. PM:5482777. [DOI] [PubMed] [Google Scholar]
- 40.Sang RC, Jura WGZO, Otieno LH, Ogaja P. Ultrastructural changes in the milk gland of tsetse Glossina morsitans centralis (Diptera; Glissinidae) female infected by a DNA virus. J Invertebr Pathol. 1996;68:253–259. doi: 10.1006/jipa.1996.0093. [DOI] [PubMed] [Google Scholar]
- 41.Doyle D, Laufer H. Sources of larval salivary gland secretion in the dipteran Chironomus tentans. J Cell Biol. 1969;40:61–78. doi: 10.1083/jcb.40.1.61. PM:5782452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar D. On the contribution of haemolymph to the salivary proteins of the red cotton bug, Dysdercus koenigii F. (Heteroptera, Pyrrhocoridae). Cell Mol Life Sci. 1979;35:765–767. http://dx.doi.org/10.1007/BF01968233. [Google Scholar]
- 43.Carolan JC, Fitzroy CIJ, Ashton PD, Douglas AE, Wilkinson TL. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics. 2009;9:2457–2467. doi: 10.1002/pmic.200800692. DOl 10.100Vpmic.200800692. [DOI] [PubMed] [Google Scholar]
- 44.Ribeiro JMC, Vaughan JA, Azad AF. Characterization of salivary apyrase activity of three rodent flea species. Comp Biochem Physiol B Comp Biochem. 1990;95:215–218. doi: 10.1016/0305-0491(90)90067-4. [DOI] [PubMed] [Google Scholar]
- 45.The International Aphid Genomics Consortium. Genome Sequence of the Pea Aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. http://dx.doi.org/10.1371%2Fjournal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoro MG. Heat shock factors and the control of the stress response. Biochem Pharmacol. 2000;59:55–63. doi: 10.1016/s0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 47.Morimoto RI, Santoro MG. Stress-inducible responses and heat shock proteins: New pharmacologic targets for cytoprotection. Nat Biotech. 1998;16:833–838. doi: 10.1038/nbt0998-833. [DOI] [PubMed] [Google Scholar]
- 48.Young JC, Hartl FU. Chaperones and transcriptional regulation by nuclear receptors. Nat Struct Mol Biol. 2002;9:640–642. doi: 10.1038/nsb0902-640. http://dx.doi.org/10.1038/nsb0902-640. [DOI] [PubMed] [Google Scholar]
- 49.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. The FASEB Journal. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. http://www.fasebj.org/content/15/7/1118.abstract. [DOI] [PubMed] [Google Scholar]
- 50.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. PM:8637592. [DOI] [PubMed] [Google Scholar]
- 51.Hartl FU, Martin J, Neupert W. Protein folding in the cell: the role of molecular chaperones Hsp70 and Hsp60. Annu Rev Biophys Biomol Struct. 1992;21:293–322. doi: 10.1146/annurev.bb.21.060192.001453. PM:1525471. [DOI] [PubMed] [Google Scholar]
- 52.Cheung RK, Dosch HM. The Growth Transformation of Human B Cells Involves Superinduction of hsp70 and hsp90. Virology. 1993;193:700–708. doi: 10.1006/viro.1993.1178. http://www.sciencedirect.com/science/article/B6WXR-45P69M0-81/2/73d30c4a76f9934949374dc99d5b2770. [DOI] [PubMed] [Google Scholar]
- 53.Lopez S, Arias CF. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 2004;12:271–278. doi: 10.1016/j.tim.2004.04.003. http://www.sciencedirect.com/science/article/B6TD0-4CB0JS2-2/2/1c51e523457fb3bf4f0c5780e8c1089d. [DOI] [PubMed] [Google Scholar]
- 54.Zarate S, Cuadras MA, Espinosa R, Romero P, Juarez KO, et al. Interaction of Rotaviruses with Hsc70 during Cell Entry Is Mediated by VP5. J Virol. 2003;77:7254–7260. doi: 10.1128/JVI.77.13.7254-7260.2003. http://jvi.asm.org/cgi/content/abstract/77/13/7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Vargas J, Romero P, Lopez S, Arias CF. The Peptide-Binding and ATPase Domains of Recombinant hsc70 Are Required To Interact with Rotavirus and Reduce Its Infectivity. J Virol. 2006;80:3322–3331. doi: 10.1128/JVI.80.7.3322-3331.2006. http://jvi.asm.org/cgi/content/abstract/80/7/3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerrero CA, Bouyssounade D, Zarate S, Isa P, Lopez T, et al. Heat Shock Cognate Protein 70 Is Involved in Rotavirus Cell Entry. J Virol. 2002;76:4096–4102. doi: 10.1128/JVI.76.8.4096-4102.2002. http://jvi.asm.org/cgi/content/abstract/76/8/4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niewiarowska J, D'Halluin JC, Belin MT. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Experimental Cell Research. 1992;201:408–416. doi: 10.1016/0014-4827(92)90290-o. http://www.sciencedirect.com/science/article/B6WFC-4DKKG5M-95/2/6540548439e4f5dc957c1fb838424a61. [DOI] [PubMed] [Google Scholar]
- 58.Chroboczek J, Gout E, Favier AL, Galinier R. Novel partner proteins of adenovirus penton. Curr Top Microbiol Immunol. 2003;272:37–55. doi: 10.1007/978-3-662-05597-7_2. [DOI] [PubMed] [Google Scholar]
- 59.Ungewickell E. The 70-kd mammalian heat shock proteins are structurally and functionally related to the uncoating protein that releases clathrin triskelia from coated vesicles. EMBO Journal. 1985;4:3385–3391. doi: 10.1002/j.1460-2075.1985.tb04094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saphire ACS, Guan T, Schirmer EC, Nemerow GR, Gerace L. Nuclear Import of Adenovirus DNA in Vitro Involves the Nuclear Protein Import Pathway and hsc70. Journal of Biological Chemistry. 2000;275:4298–4304. doi: 10.1074/jbc.275.6.4298. http://www.jbc.org/content/275/6/4298.abstract. [DOI] [PubMed] [Google Scholar]
- 61.Rapoport I, Boll W, Yu A, Bocking T, Kirchhausen T. A Motif in the Clathrin Heavy Chain Required for the Hsc70/Auxilin Uncoating Reaction. Mol Biol Cell. 2008;19:405–413. doi: 10.1091/mbc.E07-09-0870. http://www.molbiolcell.org/cgi/content/abstract/19/1/405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chromy LR, Oltman A, Estes PA, Garcea RL. Chaperone-Mediated In Vitro Disassembly of Polyoma- and Papillomaviruses. J Virol. 2006;80:5086–5091. doi: 10.1128/JVI.80.10.5086-5091.2006. http://jvi.asm.org/cgi/content/abstract/80/10/5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greene LE, Eisenberg E. Dissociation of clathrin from coated vesicles by the uncoating ATPase. J Biol Chem. 1990;265:6682–6687. http://www.jbc.org/content/265/12/6682.abstract. [PubMed] [Google Scholar]
- 64.Phillips B, Abravaya K, Morimoto RI. Analysis of the specificity and mechanism of transcriptional activation of the human hsp70 gene during infection by DNA viruses. J Virol. 1991;65:5680–5692. doi: 10.1128/jvi.65.11.5680-5692.1991. http://jvi.asm.org/cgi/content/abstract/65/11/5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon MC, Fisch TM, Benecke BJ, Nevins JR, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 66.Kingston RE, Cowie A, Morimoto RI, Gwinn KA. Binding of polyomavirus large T antigen to the human hsp70 promoter is not required for trans activation. Mol Cell Biol. 1986;6:3180–3190. doi: 10.1128/mcb.6.9.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caswell R, Hagemeier C, Chiou CJ, Hayward G, Kouzarides T, et al. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74(Pt 12):2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 68.Caswell R, Bryant L, Sinclair J. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J Virol. 1996;70:4028–4037. doi: 10.1128/jvi.70.6.4028-4037.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colberg-Poley AM, Santomenna LD, Harlow PP, Benfield PA, Tenney DJ. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furnari BA, Poma E, Kowalik TF, Huong SM, Huang ES. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young P, Anderton E, Paschos K, White R, Allday MJ. Epstein-Barr virus nuclear antigen (EBNA) 3A induces the expression of and interacts with a subset of chaperones and co-chaperones. J Gen Virol. 2008;89:866–877. doi: 10.1099/vir.0.83414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sim C, Hong Y, Tsetsarkin K, Vanlandingham D, Higgs S, et al. Anopheles gambiae heat shock protein cognate 70B impedes o'nyong-nyong virus replication. BMC Genomics. 2007;8:231. doi: 10.1186/1471-2164-8-231. http://www.biomedcentral.com/1471-2164/8/231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunological Reviews. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. http://dx.doi.org/10.1111/j.1600-065X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 75.Watanabe A, Miyazawa S, Kitami M, Tabunoki H, Ueda K, et al. Characterization of a Novel C-Type Lectin, Bombyx mori Multibinding Protein, from the B. mori Hemolymph: Mechanism of Wide-Range Microorganism Recognition and Role in Immunity. J Immunol. 2006;177:4594–4604. doi: 10.4049/jimmunol.177.7.4594. http://jimmunol.org/content/177/7/4594.abstract. [DOI] [PubMed] [Google Scholar]
- 76.Medzhitov R, Janeway CA. Innate immunity: impact on the adaptive immune response. Current Opinion in Immunology. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. http://www.sciencedirect.com/science/article/B6VS1-4547CRX-B7/2/2efe6e6e7e9e1eeddd2b258f435fd187. [DOI] [PubMed] [Google Scholar]
- 77.Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology. 1989;54 Pt 1:1–13. doi: 10.1101/sqb.1989.054.01.003. http://dx.doi.org/10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. PM:11287977. [DOI] [PubMed] [Google Scholar]
- 79.Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40:903–908. doi: 10.1016/j.molimm.2003.10.010. PM:14698229. [DOI] [PubMed] [Google Scholar]
- 80.D'Orsogna MR, Chou T. Optimal Cytoplasmic Transport in Viral Infections. PLoS ONE. 2009;4:e8165. doi: 10.1371/journal.pone.0008165. http://dx.doi.org/10.1371%2Fjournal.pone.0008165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. http://www.sciencemag.org/content/249/4969/635.abstract. [DOI] [PubMed] [Google Scholar]
- 82.Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci U S A. 1990;87:1238–1242. doi: 10.1073/pnas.87.3.1238. http://www.pnas.org/content/87/3/1238.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belov GA, tan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, et al. Hijacking Components of the Cellular Secretory Pathway for Replication of Poliovirus RNA. J Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. http://jvi.asm.org/cgi/content/abstract/81/2/558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma J, Zhang M, Ruan L, Shi H, Xu X. Characterization of two novel ADP ribosylation factors from the shrimp Marsupenaeus japonicus. Fish & Shellfish Immunology. 2010;29:956–962. doi: 10.1016/j.fsi.2010.08.003. http://www.sciencedirect.com/science/article/B6WFN-50SGPDS-2/2/1c79dc46f5e9ecfe9ee32bfc3086538e. [DOI] [PubMed] [Google Scholar]
- 85.Zhang M, Ma J, Lei K, Xu X. Molecular cloning and characterization of a class II ADP ribosylation factor from the shrimp Marsupenaeus japonicus. Fish & Shellfish Immunology. 2010;28:128–133. doi: 10.1016/j.fsi.2009.10.008. http://www.sciencedirect.com/science/article/B6WFN-4XG3SBG-1/2/995c4c2826b2b231f0a1a818699556c7. [DOI] [PubMed] [Google Scholar]
- 86.Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. DOI: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 87.Franssens V, Simonet G, Breugelmans B, Van Soest S, Van Hoef V, et al. 2008 doi: 10.1016/j.peptides.2007.07.032. The role of hemocytes, serine protease inhibitors and pathogen-associated patterns in prophenoloxidase activation in the desert locust, Schistocerca gregaria. Peptides 29: 235–241. http://dx.doi.org/10.1016/j.peptides.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 88.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 89.Haas-Stapleton EJ, Washburn JO, Volkman LE. P74 Mediates Specific Binding of Autographa californica M Nucleopolyhedrovirus Occlusion-Derived Virus to Primary Cellular Targets in the Midgut Epithelia of Heliothis virescens Larvae. J Virol. 2004;78:6786–6791. doi: 10.1128/JVI.78.13.6786-6791.2004. http://jvi.asm.org/cgi/content/abstract/78/13/6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao L, Zhou W, Xu H, Zheng Y, Qi Y. The Heliothis armigera single nucleocapsid nucleopolyhedrovirus envelope protein P74 is required for infection of the host midgut. Virus Research. 2004;104:111–121. doi: 10.1016/j.virusres.2004.03.005. http://www.sciencedirect.com/science/article/B6T32-4CBW7SC-1/2/06f1c1f8d51e8fd409db57b6fa72bc12. [DOI] [PubMed] [Google Scholar]
- 91.Faulkner P, Kuzio J, Williams GV, Wilson JA. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J Gen Virol. 1997;78:3091–3100. doi: 10.1099/0022-1317-78-12-3091. PM:9400957. [DOI] [PubMed] [Google Scholar]
- 92.Slack JM, Lawrence SD. Evidence for proteolytic cleavage of the baculovirus occlusion-derived virion envelope protein P74. J Gen Virol. 2005;86:1637–1643. doi: 10.1099/vir.0.80832-0. PM:15914841. [DOI] [PubMed] [Google Scholar]
- 93.Rukmini V, Reddy CY, Venkateswerlu G. Bacillus thuringiensis crystal delta-endotoxin: role of proteases in the conversion of protoxin to toxin. Biochimie. 2000;82:109–116. doi: 10.1016/s0300-9084(00)00355-2. PM:10727765. [DOI] [PubMed] [Google Scholar]
- 94.Westenberg M, Veenman F, Roode EC, Goldbach RW, Vlak JM, et al. Functional analysis of the putative fusion domain of the baculovirus envelope fusion protein F. J Virol. 2004;78:6946–6954. doi: 10.1128/JVI.78.13.6946-6954.2004. PM:15194771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moncollin V, Fischer L, Cavallini B, Egly JM, Chambon P. Class II (B) general transcription factor (TFIIB) that binds to the template-committed preinitiation complex is different from general transcription factor BTF3. Proc Natl Acad Sci U S A. 1992;89:397–401. doi: 10.1073/pnas.89.1.397. PM:1729710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koonin EV, Mushegian AR, Tatusov RL, Altschul SF, Bryant SH, et al. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain–study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. PM:7703850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caldas T, Laalami S, Richarme G. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J Biol Chem. 2000;275:855–860. doi: 10.1074/jbc.275.2.855. PM:10625618. [DOI] [PubMed] [Google Scholar]
- 98.Sklan EH, Staschke K, Oakes TM, Elazar M, Winters M, et al. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J Virol. 2007;81:11096–11105. doi: 10.1128/JVI.01249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campbell EM, Hope TJ. Gene therapy progress and prospects: viral trafficking during infection. Gene Ther. 2005;12:1353–1359. doi: 10.1038/sj.gt.3302585. [DOI] [PubMed] [Google Scholar]
- 100.Dohner K, Sodeik B. The role of the cytoskeleton during viral infection. Curr Top Microbiol Immunol. 2005;285:67–108. doi: 10.1007/3-540-26764-6_3. DOI: 10.1007/3-540-26764-6_3. [DOI] [PubMed] [Google Scholar]
- 101.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 102.Greber UF, Gastaldelli M. Junctional gating: the achilles' heel of epithelial cells in pathogen infection. Cell Host Microbe. 2007;2:143–146. doi: 10.1016/j.chom.2007.08.004. PM:18005729. [DOI] [PubMed] [Google Scholar]
- 103.Greber UF. Signalling in viral entry. Cell Mol Life Sci. 2002;59:608–626. doi: 10.1007/s00018-002-8453-3. PM:12022470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. PM:8221887. [DOI] [PubMed] [Google Scholar]
- 105.Dugan AS, Eash S, Atwood WJ. Update on BK virus entry and intracellular trafficking. Transpl Infect Dis. 2006;8:62–67. doi: 10.1111/j.1399-3062.2006.00153.x. PM:16734628. [DOI] [PubMed] [Google Scholar]
- 106.Braunagel SC, Summers MD. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr Drug Targets. 2007;8:1084–1095. doi: 10.2174/138945007782151315. PM:17979668. [DOI] [PubMed] [Google Scholar]
- 107.Slack JM, Dougherty EM, Lawrence SD. A study of the Autographa californica multiple nucleopolyhedrovirus ODV envelope protein p74 using a GFP tag. J Gen Virol. 2001;82:2279–2287. doi: 10.1099/0022-1317-82-9-2279. PM:11514740. [DOI] [PubMed] [Google Scholar]
- 108.Slack JM, Lawrence SD, Krell PJ, Arif BM. A soluble form of p74 can act as a per os infectiviry factor to the Autographa californica multiple nucleopolyhedrovirus. J Gen Virol. 2010;91:915–918. doi: 10.1099/vir.0.017145-0. [DOI] [PubMed] [Google Scholar]
- 109.Sparks WO, Harrison RL, Bonning BC. Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology. 2011;409:69–76. doi: 10.1016/j.virol.2010.09.027. PM:20970820. [DOI] [PubMed] [Google Scholar]
- 110.Peng K, van Oers MM, Hu Z, van Lent JW, Vlak JM. Baculovirus per os infectivity factors form a complex on the surface of occlusion-derived virus. J Virol. 2010;84:9497–9504. doi: 10.1128/JVI.00812-10. PM:20610731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lin W, Hata T, Kasamatsu H. Subcellular distribution of viral structural proteins during simian virus 40 infection. J Virol. 1984;50:363–371. doi: 10.1128/jvi.50.2.363-371.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Greber UF. Viral trafficking violations in axons: the herpesvirus case. Proc Natl Acad Sci U S A. 2005;102:5639–5640. doi: 10.1073/pnas.0501696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marek M, Merten OW, Galibert L, Vlak JM, van Oers MM. Baculovirus VP80 Protein and the F-Actin Cytoskeleton Interact and Connect the Viral Replication Factory with the Nuclear Periphery. J Virol. 2011;85:5350–5362. doi: 10.1128/JVI.00035-11. doi: l0.1128JM.OOO15-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mabit H, Nakano MY, Prank U, Saam B, Dohner K, et al. Intact microtubules support adenovirus and herpes simplex virus infections. J Virol. 2002;76:9962–9971. doi: 10.1128/JVI.76.19.9962-9971.2002. DOI: 10.1128/JVI.76.19.9962–9971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohkawa T, Volkman LE, Welch MD. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol. 2010;190:187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suomalainen M, Nakano MY, Boucke K, Keller S, Greber UF. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001;20:1310–1319. doi: 10.1093/emboj/20.6.1310. PM:11250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP, et al. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. PM:10037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kelkar SA, Pfister KK, Crystal RG, Leopold PL. Cytoplasmic dynein mediates adenovirus binding to microtubules. J Virol. 2004;78:10122–10132. doi: 10.1128/JVI.78.18.10122-10132.2004. PM:15331745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luxton GWG, Haverlock S, Coller KE, Antinone SE, Pincetic A, et al. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci U S A. 2005;102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Newsome TP, Scaplehorn N, Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. PM:15297625. [DOI] [PubMed] [Google Scholar]
- 121.Granzow H, Klupp BG, Mettenleiter TC. The pseudorabies virus US3 protein is a component of primary and of mature virions. J Virol. 2004;78:1314–1323. doi: 10.1128/JVI.78.3.1314-1323.2004. PM:14722286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith GA, Enquist LW. Break ins and break outs: viral interactions with the cytoskeleton of Mammalian cells. Annu Rev Cell Dev Biol. 2002;18:135–161. doi: 10.1146/annurev.cellbio.18.012502.105920. PM:12142276. [DOI] [PubMed] [Google Scholar]
- 123.Naranatt PP, Krishnan HH, Smith MS, Chandran B. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol. 2005;79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, et al. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. PM:16420530. [DOI] [PubMed] [Google Scholar]
- 125.Delboy MG, Roller DG, Nicola AV. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J Virol. 2008;82:3381–3390. doi: 10.1128/JVI.02296-07. PM:18234803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rabe B, Glebe D, Kann M. Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J Virol. 2006;80:5465–5473. doi: 10.1128/JVI.02303-05. PM:16699026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. PM:18288107. [DOI] [PubMed] [Google Scholar]
- 128.Chizmadzhev YA. The mechanisms of lipid-protein rearrangements during viral infection. Bioelectrochemistry. 2004;63:129–136. doi: 10.1016/j.bioelechem.2003.10.016. doi: IO.lOl6lj.bioelechem.2003.IO.OI6. [DOI] [PubMed] [Google Scholar]
- 129.Greber UF, Puntener D. DNA-tumor virus entry–from plasma membrane to the nucleus. Semin Cell Dev Biol. 2009;20:631–642. doi: 10.1016/j.semcdb.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 130.Jovasevic V, Liang L, Roizman B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol. 2008;82:3311–3319. doi: 10.1128/JVI.01919-07. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smede M, Hussain M, Asgari S. A lipase-like gene from Heliothis virescens ascovirus (HvAV-3e) is essential for virus replication and cell cleavage. Virus Genes. 2009;39:409–417. doi: 10.1007/s11262-009-0407-5. [DOI] [PubMed] [Google Scholar]
- 132.Wintjens R, Rooman M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. PM:8831795. [DOI] [PubMed] [Google Scholar]
- 133.Lallemand-Breitenbach V, de TH. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. PM:20452955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Everett RD, Orr A, Preston CM. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. PM:9857173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gribaudo G, Riera L, Lembo D, De AM, Gariglio M, et al. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J Virol. 2000;74:4979–4987. doi: 10.1128/jvi.74.11.4979-4987.2000. PM:10799571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gribaudo G, Riera L, Rudge TL, Caposio P, Johnson LF, et al. Human cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent fibroblasts. J Gen Virol. 2002;83:2983–2993. doi: 10.1099/0022-1317-83-12-2983. PM:12466474. [DOI] [PubMed] [Google Scholar]
- 137.Cohen PS. Translational regulation of deoxycytidylate hydroxymethylase and deoxynucleotide kinase synthesis in T4-infected Escherichia coli. Virology. 1972;47:780–786. doi: 10.1016/0042-6822(72)90569-7. PM:4551997. [DOI] [PubMed] [Google Scholar]
- 138.Lewis IP, Cohen SS. Deoxycytidylate hydroxymethylase from virus-infected E. coli. Methods in Enzymology. 1963:131–136. [Google Scholar]
- 139.Dahl J, You J, Benjamin TL. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol. 2005;79:13007–13017. doi: 10.1128/JVI.79.20.13007-13017.2005. PM:16189003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 141.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci U S A. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Turpin E, Luke K, Jones J, Tumpey T, Konan K, et al. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J Virol. 2005;79:8802–8811. doi: 10.1128/JVI.79.14.8802-8811.2005. PM:15994774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. DOI 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rodnina MV. Protein synthesis meets ABC ATPases: new roles for Rli1/ABCE1. EMBO Rep. 2010;11:143–144. doi: 10.1038/embor.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maul GG, Everett RD. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75(Pt 6):1223–1233. doi: 10.1099/0022-1317-75-6-1223. PM:8207389. [DOI] [PubMed] [Google Scholar]
- 146.Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. PM:8599237. [DOI] [PubMed] [Google Scholar]
- 147.Shih SR, Krug RM. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 1996;15:5415–5427. [PMC free article] [PubMed] [Google Scholar]
- 148.Hakim M, Fass D. Dimer interface migration in a viral sulfhydryl oxidase. J Mol Biol. 2009;391:758–768. doi: 10.1016/j.jmb.2009.06.070. 001:1 0.1 01 l:Vj.jmb.2009.06.070. [DOI] [PubMed] [Google Scholar]
- 149.Xia YN, Burbank DE, Uher L, Rabussay D, Van Etten JL. Restriction endonuclease activity induced by PBCV-1 virus infection of a Chlorella-like green alga. Mol Cell Biol. 1986;6:1430–1439. doi: 10.1128/mcb.6.5.1430. PM:3023890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Basaillon M, Senechal S, Bernier L, Lemay G. A glycosyl hydrolase activity of mammalian reovirus s1 protein can contribute to viral infection through a mucus layer. J Mol Biol. 1999;286:759–773. doi: 10.1006/jmbi.1998.2495. [DOI] [PubMed] [Google Scholar]
- 151.Hiramatsu S, Ishihara M, Fujie M, Usami S, Yamada T. Expression of a chitinase gene and lysis of the host cell wall during Chlorella virus CVK2 infection. Virology. 1999;260:308–315. doi: 10.1006/viro.1999.9824. PM:10417265. [DOI] [PubMed] [Google Scholar]
- 152.Saville GP, Patmanidi AL, Possee RD, King LA. Deletion of the Autographa californica nucleopolyhedrovirus chitinase KDEL motif and in vitro and in vivo analysis of the modified virus. J Gen Virol. 2004;85:821–831. doi: 10.1099/vir.0.19732-0. PM:15039525. [DOI] [PubMed] [Google Scholar]
- 153.Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, et al. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution. 2010;64:3024–3034. doi: 10.1111/j.1558-5646.2010.01049.x. PM:20550574. [DOI] [PubMed] [Google Scholar]
- 154.Nagel CH, Dohner K, Fathollahy M, Strive T, Borst EM, et al. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J Virol. 2008;82:3109–3124. doi: 10.1128/JVI.02124-07. doi: 10.1128/JVI.02124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kariithi HM, Ahmadi, M, Parker AG, Elashry AM, Franz G, et al. Prevalence and genetic diversity of salivary gland hypertrophy virus in the wild populations of Glossina pallidipes (Diptera: Glossinidae) from southern and eastern Africa. Applied Environmental Microbiology. 2011 in press. [Google Scholar]
- 156.Sparks WO, Rohlfing A, Bonning BC. A peptide with similarity to baculovirus ODV-E66 binds the gut epithelium of Heliothis virescens and impedes infection with Autographa californica multiple nucleopolyhedrovirus. J Gen Virol. 2011;92:1051–1060. doi: 10.1099/vir.0.028118-0. doi: 10.1099/vir.0.028118-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sixty-five salivary gland secretome proteins of G. pallidipes . Sixty-five G. pallidipes salivary gland secretome proteins supported by nanoLC-MS/MS, confirmed by gene ontology (GO) annotation, presence of signal peptide sequences and by Blasts on the NCBI and G. m. morsitans databases.
(DOC)
Twenty-five SGHV-encoded proteins detected in the G. pallidipes hypertrophied salivary glands.
(DOC)