Abstract
Myxococcus xanthus DK1622 contains inner (IM) and outer membranes (OM) separated by a peptidoglycan layer. Integral membrane, β-barrel proteins are found exclusively in the OM where they form pores allowing the passage of nutrients, waste products and signals. One porin, Oar, is required for intercellular communication of the C-signal. An oar mutant produces CsgA but is unable to ripple or stimulate csgA mutants to develop suggesting that it is the channel for C-signaling. Six prediction programs were evaluated for their ability to identify β-barrel proteins. No program was reliable unless the predicted proteins were first parsed using Signal P, Lipo P and TMHMM, after which TMBETA-SVM and TMBETADISC-RBF identified β-barrel proteins most accurately. 228 β-barrel proteins were predicted from among 7331 protein coding regions, representing 3.1% of total genes. Sucrose density gradients were used to separate vegetative cell IM and OM fractions, and LC-MS/MS of OM proteins identified 54 β-barrel proteins. Another class of membrane proteins, the lipoproteins, are anchored in the membrane via a lipid moiety at the N-terminus. 44 OM proteins identified by LC-MS/MS were predicted lipoproteins. Lipoproteins are distributed between the IM, OM and ECM according to an N-terminal sorting sequence that varies among species. Sequence analysis revealed conservation of alanine at the +7 position of mature ECM lipoproteins, lysine at the +2 position of IM lipoproteins, and no noticable conservation within the OM lipoproteins. Site directed mutagenesis and immuno transmission electron microscopy showed that alanine at the +7 position is essential for sorting of the lipoprotein FibA into the ECM. FibA appears at normal levels in the ECM even when a +2 lysine is added to the signal sequence. These results suggest that ECM proteins have a unique method of secretion. It is now possible to target lipoproteins to specific IM, OM and ECM locations by manipulating the amino acid sequence near the +1 cysteine processing site.
Introduction
The life cycle of Myxococcus xanthus involves a vegetative stage, in which cells feed on bacteria and organic detritus, and a developmental stage in which thousands of cells aggregate to form a multicellular fruiting body containing spores. Fruiting body development involves intercellular communication with at least six extracellular signals [1]. However, the receptors and sensory pathways of these signaling pathways are largely unknown. Identification of outer membrane (OM) proteins in M. xanthus may reveal components of these signaling pathways that are used to export or import signals.
The OM acts as a selective barrier that allows the passage of nutrients, water and chemical signals through pores formed by porin proteins. In porins, antiparallel β-strands are arranged to form a cylindrical β-barrel structure lined with hydrophilic residues that create a water-filled channel [2]. Some porins allow passive diffusion of small solutes with molecular weights up to 600 Da [3]. Active diffusion of specific nutrients through porins is carried out by TonB systems, which utilize energy provided by the inner membrane (IM) to mediate solute passage through the OM [4]. Some porins allow passage of specific substrates, such as fatty acids in the case of FadL [5]. Porins are synthesized as precursors with an N-terminal signal sequence that aids transport across the IM via the general secretory (Sec) pathway [6]. The signal sequences are hydrolyzed by signal peptidases present in the IM. Chaperones in the periplasm facilitate protein folding and insertion into the OM using the Omp85 machinery [7].
Databases such as Pfam can help identify OM proteins, but only if the protein contains a domain with appreciable identity to a domain of known function [8]. Unfortunately, most bacterial genomes contain hypothetical proteins that are not represented in the Pfam database. For example, the M. xanthus genome encodes 40% hypothetical proteins [9]. Thus, bioinformatic programs that can predict the OM protein β-barrel structure would be useful since this structure is unique to porins [10].
The IM and OM also contains lipoproteins that are anchored by a lipid-modified N-terminal cysteine residue. Lipoproteins are transported as precursors via the Sec pathway to the IM where they are processed at the conserved N-terminal lipobox. The lipobox consists of four amino acids (L−3-[A/S/T]−2-[G/A]−1-C+1) around the signal peptide cleavage site with the +1 cysteine serving as the site of covalent modification [11]. Lipoprotein maturation involves attachment of a diacylglycerol group to the +1 cysteine sulfhydryl group via a thioester linkage, cleavage of the signal peptide, and acylation of the +1 α-amino group. The lipid moieties anchor the N-terminus of the proteins in lipid bilayers.
In Escherichia coli, localization of lipoproteins to the IM requires aspartate at the +2 position of the mature lipoprotein [12]. Lipoproteins lacking this sorting signal are transported to the OM via the Lol pathway, which is an ABC transport system located in the IM [13]. The signal sequences directing Lol avoidance differ among bacteria. In Pseudomonas aeruginosa, lysine and serine at positions +3 and +4 lead to IM retention of lipoproteins [14]. Some bacteria secrete lipoproteins and may possess novel mechanisms to sort lipoproteins to the external environment [15]. M. xanthus secretes at least 11 lipoproteins to the extracellular matrix (ECM) whose mechanism of targeting is unknown [16].
In this paper we identified OM proteins using bioinformatic and proteomic tools. Two prediction programs TMBETA-SVM and TMBETADISC-RBF identified 228 β-barrel OM proteins in the genome, of which 54 were detected in DK1622 vegetative cells by LC-MS/MS. We show that one of these proteins, Oar, is essential for C-signal transmission during fruiting body development. Lipoprotein sorting into IM, OM, and extracellular compartments was also examined. Alanine at the +7 position mediates ECM localization, even when a signal for IM localization is also present, suggesting that there are at least two lipoprotein secretion pathways.
Results
The first goal of this study was to identify M. xanthus OM porins and lipoproteins using bioinformatic and proteomic approaches, then examine one porin, Oar, for a role in fruiting body development. The second goal of this study was to identify the trafficking signals for IM, OM and ECM lipoproteins. Site directed mutagenesis was then used to identify the ECM trafficking signal for the major ECM lipoprotein FibA.
β-barrel prediction in M. xanthus proteome
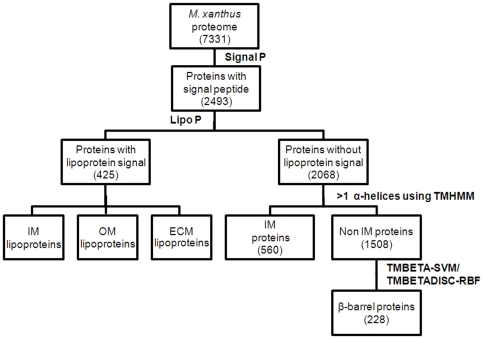
Integral OM proteins are synthesized with an N-terminal signal sequence. The signal sequence enables transport of proteins across the IM by the Sec system, and can be predicted using the program Signal P [17]. The M. xanthus proteome was examined for candidates with a signal peptide, which generated 2493/7331 candidates (Figure 1). In the next step, predicted signal peptide containing proteins were classified as lipoproteins or non-lipoproteins using the Lipo P program [13]. 425 out of 2493 signal peptide-containing proteins were predicted to be lipoproteins (Figure 1). The non-lipoproteins were further segregated into IM and non-IM proteins. Integral IM proteins have transmembrane alpha helices that are rich in hydrophobic amino acids. While some integral membrane proteins have only a single transmembrane domain, we felt that parsing out proteins with a single predicted transmembrane domain was a bit risky. Therefore, proteins with at least two putative transmembrane helices using the TMHMM program were classified as IM proteins [13]. 560/2068 (27%) proteins with putative signal peptides were identified as IM proteins.
Figure 1. Scheme to identify OM proteins utilizing bioinformatic programs.
The Signal P program identified 2493 signal peptide containing proteins among the putative 7331 member M. xanthus proteome. Of these 425 lipoproteins were identified using Lipo P. Of the 2068 proteins without a lipoprotein signal 560 were integral IM proteins identified using TMHMM. The non-IM proteins include periplasmic proteins, secreted proteins and OM proteins. Finally, integral OM proteins containing a β-barrel domain were identified using TMBETA-SVM plus TMBETADISC-RBF.
The remaining 1508 signal peptide containing proteins, comprising periplasmic, secreted, and OM proteins, were subjected to β-barrel prediction programs to identify integral OM proteins. Six prediction programs were evaluated, TMBETA-NET, PRED-TMββ, TMBETA-SVM, TMBETADISC-RBF, BOMP, and TMB-HUNT, using 40 M. xanthus protein sequences with predicted β-barrel domains obtained from the Pfam database (Table 1). These proteins are homologous to well-studied OM proteins from other organisms as demonstrated by BLASTP. The 40 proteins represent 10 protein families including TonB dependent receptors, Omp85 and OmpH, secretins, efflux proteins, and organic solvent tolerance proteins [18], [19], [20], [21]. Except for BOMP (10%) and TMB-HUNT (63%) all prediction programs identified >77% of the 40 OM proteins (Table 1). PRED-TMββ identified the most β-barrel proteins (38/40) (95%). TMBETADISC-RBF, TMBETA-SVM and TMBETA-NET identified 34/40 (85%), 33/40 (83%) and 31/40 (78%) β-barrel proteins, respectively.
Table 1. M. xanthus β-barrel domain proteins obtained from the Pfam database.
| MXAN Number | Function | 1a | 2b | 3c | 4d | 5e | 6f |
| AN0272 | TonB dependent receptor | + | + | + | + | + | + |
| MXAN0518 | TonB dependent receptor | + | + | + | − | − | − |
| MXAN0562 | Phosphate selective porin (PhoE) | + | + | + | + | − | + |
| MXAN0990 | Outer membrane efflux protein | + | − | + | − | − | − |
| MXAN1316 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN1450 | TonB dependent receptor (Oar) | + | + | + | + | + | − |
| MXAN2514 | Secretin (GspD) | + | + | + | + | − | − |
| MXAN2708 | Organic solvent tolerance protein (OstA) | + | + | + | + | + | − |
| MXAN3106 | Secretin (GspE) | + | + | + | + | + | − |
| MXAN3431 | Outer membrane efflux protein | + | + | + | − | − | − |
| MXAN3883 | Fimbrial usher protein (FUP) | + | + | + | + | + | − |
| MXAN3905 | Outer membrane efflux protein | + | + | + | + | + | − |
| MXAN4176 | Outer membrane efflux protein | + | + | + | + | + | − |
| MXAN4198 | Outer membrane efflux protein | + | + | + | + | + | − |
| MXAN4559 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN4727 | Structural protein (OmpH) | + | + | − | − | + | − |
| MXAN4728 | Omp85 protein | + | + | + | − | − | − |
| MXAN4746 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN4772 | OmpA protein | − | + | − | − | + | − |
| MXAN5023 | TonB dependent receptor | + | + | + | + | − | − |
| MXAN5030 | Outer membrane efflux transporter | + | + | + | + | + | − |
| MXAN5042 | OmpA protein | + | − | − | − | − | − |
| MXAN5069 | Aquaporin Z (ApqZ) | + | − | − | − | + | − |
| MXAN5772 | Secretin (PilQ) | + | + | + | + | + | − |
| MXAN5956 | Major intrinsic protein | + | − | + | − | − | − |
| MXAN6044 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN6176 | Outer membrane efflux protein | + | + | + | + | + | − |
| MXAN6246 | OmpA | + | − | − | − | − | − |
| MXAN6487 | Outer membrane efflux protein | + | + | + | + | + | − |
| MXAN6547 | TonB dependent receptor | + | + | + | + | − | − |
| MXAN6579 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN6716 | TonB dependent receptor | + | + | + | + | − | − |
| MXAN6845 | TonB dependent receptor | + | + | + | + | + | − |
| MXAN6911 | TonB dependent receptor | + | + | + | + | + | + |
| MXAN7037 | OmpA | + | + | + | + | + | − |
| MXAN7040 | Fatty acid transport (FadL) | + | + | + | + | + | + |
| MXAN7238 | Outer membrane efflux protein | + | + | − | + | + | − |
| MXAN7331 | TonB dependent receptor | + | + | + | + | − | − |
| MXAN7397 | OmpA | − | − | − | − | + | − |
| MXAN7436 | Outer membrane efflux protein | + | + | + | + | − | − |
Programs that correctly predicted a β-barrel protein are indicated by a positive sign (+) while failing to do so is indicated by a negative sign (−).
PRED-TMββ.
TMBETADISC-RBF.
TMBETA-SVM.
TMBETA-NET.
TMB-HUNT.
BOMP.
TMBETA-NET, TMBETA-SVM, TMBETADISC-RBF and PRED-TMββ were tested on 21 M. xanthus IM, periplasmic, and ECM proteins to eliminate programs that generate false positives (Table 2). The IM and periplasmic proteins were obtained from the Pfam database while the ECM proteins were previously identified from proteomic studies [16]. PRED-TMββ produced 11/20 (55%) false positives, TMBETA-NET and TMBETADISC-RBF generated 2/20 (9.5%) false positives, and TMBETA-SVM produced no false positives.
Table 2. M. xanthus IM, periplasmic and ECM proteins1.
| MXAN Number | Function | Predicted localization | 1a | 2b | 3c | 4d |
| MXAN0468 | peptidylprolyl cis-trans isomerase | Periplasm | + | − | − | − |
| MXAN0977 | di-haem cytochrome-c peroxidase | Periplasm | − | − | − | − |
| MXAN1066 | PTS system, IIA component | Periplasm | + | − | − | − |
| MXAN1389 | alkaline phosphatase | Periplasm | + | − | − | − |
| MXAN2832 | permease | Periplasm | − | − | − | − |
| MXAN2951 | ABC transporter, periplasmic substrate binding protein | Periplasm | − | − | − | − |
| MXAN3420 | multicopper oxidase (CumA) | Periplasm | − | − | − | − |
| MXAN0274 | biopolymer transport protein, ExbD/TolR family | IM | − | − | − | − |
| MXAN0559 | ABC transporter,ATP-binding protein (Mac1) | IM | + | − | − | − |
| MXAN2505 | general secretory pathway protein K (GspK) | IM | − | − | − | + |
| MXAN2570 | acetate–CoA ligase | IM | − | − | − | − |
| MXAN3182 | Serine threonine kinase | IM | + | − | − | − |
| MXAN4829 | isoquinoline 1-oxidoreductase, beta subunit (IorB) | IM | + | − | − | − |
| MXAN5123 | sensor histidine kinase MrpA (MrpA) | IM | + | + | − | − |
| MXAN0075 | amidohydrolase | ECM | − | − | − | − |
| MXAN1424 | unknown | ECM | + | + | − | − |
| MXAN1493 | unknown | ECM | + | − | − | − |
| MXAN2375 | unknown | ECM | − | − | − | − |
| MXAN3885 | Spore coat U | ECM | + | − | − | + |
| MXAN5686 | unknown | ECM | + | − | − | − |
IM and periplasmic proteins were obtained from the Pfam database while ECM proteins were previously identified by Curtis et al [16].
PRED-TMββ.
TMBETADISC-RBF.
TMBETA-SVM.
TMBETA-NET.
In the absence of a stand-alone program for TMBETA-NET, β-barrel proteins were identified using TMBETA-SVM and TMBETADISC-RBF, which predicted 240/1508 and 414/1508 proteins respectively. 228 proteins were identified by both programs equivalent to ∼3.1% of the genome (Table S1). Analyses of several Gram-negative bacteria suggests that 2–3% of the genome encodes porins [22].
Identification of OM proteins in M. Xanthus
The OM fraction was purified, then subjected to LC-MS/MS [23]. Using the bioinformatic scheme shown in figure 1, 54 β-barrel proteins (Table 3) were identified along with 44 lipoproteins (Table 4). The bioinformatic scheme enabled the identification of cytoplasmic, IM, and periplasmic contaminants in the OM preparation (Table S2).
Table 3. OM β-barrel proteins identified by LC-MS/MS.
| MXAN Number1 | Function | No. of peptides |
| MXAN0219a | Hypothetical protein | 1 |
| MXAN0518a | TonB-dependent receptor | 1 |
| MXAN0659b | Putative lipoprotein | 5 |
| MXAN0662b | Hypothetical protein | 3 |
| MXAN0751b | Conserved domain protein | 4 |
| MXAN0855b | Putative chemotaxis MotB protein | 5 |
| MXAN0924a | Hypothetical protein | 2 |
| MXAN1426b | Hypothetical protein | 2 |
| MXAN1450a | TonB-dependent receptor (Oar) | 40 |
| MXAN1689 | Conserved hypothetical protein | 1 |
| MXAN2417b | Conserved hypothetical protein | 1 |
| MXAN2462b | Hypothetical protein | 1 |
| MXAN2514a | General secretion pathway protein D | 4 |
| MXAN2536 | Putative long-chain-fatty-acid-CoA ligase | 6 |
| MXAN2659a | Hypothetical protein | 17 |
| MXAN2906b | Penicillin acylase family protein | 8 |
| MXAN3106a | Bacterial membrane secretin (secretin) family | 10 |
| MXAN3160b | Peptidase, M13 (neprilysin) family | 22 |
| MXAN3774b | Conserved Hypothetical protein | 11 |
| MXAN3780 | Patatin-like phospholipase family protein | 3 |
| MXAN3953b | Hypothetical protein | 2 |
| MXAN4085 | Peptidylprolyl cis-trans isomerase, FKBP-type | 1 |
| MXAN4293b | Hypothetical protein | 4 |
| MXAN4295 | Patatin-like phospholipase family protein | 6 |
| MXAN4365 | Outer membrane receptor family | 1 |
| MXAN4652 | Putative Flp pilus assembly protein CpaB | 1 |
| MXAN4728a | OMP85 family protein | 4 |
| MXAN4746a | TonB-dependent receptor | 5 |
| MXAN5023a | TonB dependent receptor | 5 |
| MXAN5152b | OmpA family protein | 3 |
| MXAN5194 | OmpA domain protein | 2 |
| MXAN5453b | Hypothetical protein | 6 |
| MXAN5685b | Hypothetical protein | 2 |
| MXAN5743b | Hypothetical protein | 11 |
| MXAN5756b | TolB protein | 2 |
| MXAN5931a | Hypothetical protein | 7 |
| MXAN6079b | Putative molybdopterin oxidoreductase, iron-sulfur binding subunit | 15 |
| MXAN6090b | Hypothetical protein | 10 |
| MXAN6196a | Hypothetical protein | 3 |
| MXAN6487a | Outer membrane efflux protein domain protein | 11 |
| MXAN6521b | Putative lipoprotein | 1 |
| MXAN6829 | Hypothetical protein | 4 |
| MXAN6891b | Hypothetical protein | 2 |
| MXAN6911a | TonB-dependent receptor | 8 |
| MXAN7037 | Putative chemotaxis MotB protein | 1 |
| MXAN7039b | Putative lipoprotein | 34 |
| MXAN7040a | FadL | 13 |
| MXAN7104b | M3 (thimet oligopeptidase) family peptidase | 10 |
| MXAN7112b | Conserved Hypothetical protein | 3 |
| MXAN7196b | Hypothetical protein | 1 |
| MXAN7203a | Putative 28 kDa outer membrane protein | 5 |
| MXAN7407a | Hypothetical protein | 6 |
| MXAN7436a | Outer membrane efflux protein | 4 |
| MXAN7513 | Hypothetical protein | 1 |
All proteins labeled with an ‘a’ or ‘b’, were identified in both this study and the Kahnt et al. study using proteomic approaches. Proteins identified by proteomics and classified as β-barrel proteins in the Kahnt et al. study, are identified with an ‘a’ superscript. Proteins identified by proteomics in the Kahnt et al. study and likely misclassified by them, are denoted with a ‘b’. Proteins with no superscript are β-barrel proteins unique to this study.
Table 4. OM lipoproteins identified by LC-MS/MS.
| MXAN Number | Function | No. of peptides |
| MXAN0283 | Putative lipoprotein | 1 |
| MXAN0522 | Putative lipoprotein | 2 |
| MXAN0533 | NAD dependent epimerase/dehydratase family | 1 |
| MXAN0662 | Hypothetical protein | 3 |
| MXAN0751 | Conserved domain protein | 4 |
| MXAN0934 | Protease DO family protein | 14 |
| MXAN1063 | Putative lipoprotein | 1 |
| MXAN1162 | Putative lipoprotein | 5 |
| MXAN1176 | Peptidylprolyl cis-trans isomerase, cyclophilin-type | 1 |
| MXAN1342 | Putative lipoprotein | 1 |
| MXAN1451 | Putative lipoprotein MlpA | 4 |
| MXAN1623 | peptidase, M16 (pitrilysin) family | 8 |
| MXAN1689 | Conserved hypothetical protein | 1 |
| MXAN2091 | Peptidase, M16 (pitrilysin) family | 3 |
| MXAN2286 | Peptidyl-dipeptidase Dcp | 4 |
| MXAN2417 | Conserved hypothetical protein | 1 |
| MXAN2470 | 5′-nucleotidase family protein | 1 |
| MXAN2660 | Putative lipoprotein | 6 |
| MXAN2968 | Efflux transporter, RND family, MFP subunit | 4 |
| MXAN3060 | Adventurous gliding motility protein CglB | 4 |
| MXAN3084 | Social gliding motility protein Tgl | 2 |
| MXAN3103 | Putative lipoprotein | 3 |
| MXAN3440 | Peptidase, M13 (neprilysin) family | 4 |
| MXAN3581 | Peptidyl-dipeptidase A | 4 |
| MXAN4641 | Hypothetical protein | 1 |
| MXAN4747 | Putative lipoprotein | 1 |
| MXAN4900 | Putative lipoprotein | 15 |
| MXAN4966 | Putative lipoprotein | 10 |
| MXAN5331 | Putative lipoprotein | 2 |
| MXAN5361 | Putative 5′-nucleotidase | 1 |
| MXAN5390 | Putative lipoprotein | 1 |
| MXAN5684 | Putative lipoprotein | 5 |
| MXAN5933 | Peptidase, M48 (Ste24 endopeptidase) family | 5 |
| MXAN6381 | Hypothetical protein | 1 |
| MXAN6521 | Putative lipoprotein | 1 |
| MXAN6660 | Hypothetical protein | 3 |
| MXAN6720 | Putative lipoprotein | 2 |
| MXAN6978 | Putative lipoprotein | 2 |
| MXAN6985 | Hypothetical protein | 1 |
| MXAN7108 | Putative lipoprotein | 1 |
| MXAN7110 | Peptidyl-prolyl cis-trans isomerase, FKBP-type | 8 |
| MXAN7220 | Putative lipoprotein | 1 |
| MXAN7333 | Putative lipoprotein | 4 |
| MXAN7438 | Putative cobalt-zinc-cadmium resistance protein | 3 |
Oar is required for C-signaling
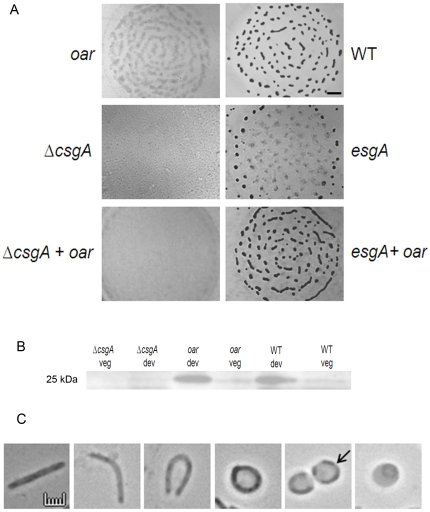
Porins form hydrophilic channels through which extracellular signals may pass. oar mutants exhibit delayed aggregation and are unable to sporulate [24]. Oar appears to be a TonB dependent receptor. To determine whether an essential developmental signal passes through the Oar pore, oar cells were mixed pair wise with mutants unable to produce each of the essential developmental signals A, B, C, D, E and S [1]. If the oar mutant is proficient in producing an extracellular signal, it would be expected to rescue development of a mutant unable to produce such a signal. oar cells were mixed in 1∶1 ratio with mutants from each signal-producing class, and rescued development of all but csgA mutants (Figure 2A).
Figure 2. Role of oar in cell signaling.
(A) Extracellular complementation of oar (LS2453) cells with ΔcsgA (LS2441) and esg (JD300). WT (DK1622) cells were used as a control. Bar is 1 mm (B) Western blot analysis of vegetative cells and 24 h developing cells using anti-CsgA primary antibody. (C) Morphology of oar cells during development. The first panel represents 24 h developing WT cells while the subsequent panels represent various oar cell shapes as they ultimately transform into spheroplast (extreme right panel). Bar is 1 µm.
The csgA gene is required for the production of the C-signal [25]. ΔcsgA cells can be rescued for development by mixing with csgA + cells even if those cells are also developmentally defective [26]. 1∶1 mixtures containing oar and ΔcsgA cells failed to undergo fruiting body development suggesting that oar cells are unable to provide C-signal to ΔcsgA cells. In contrast, the esg gene product is required for the synthesis of a branched chain fatty acid required for production of the E-signal [27]. esg mutations do not completely eliminate synthesis of branched chain fatty acids because there is a second pathway that can also produce them, albeit at smaller concentrations, causing some fruiting body development and sporulation as observed in Figure 2A. When oar cells were mixed with esg mutant cells, fruiting body development and sporulation was restored to wild type levels (Figure 2A).
oar cells produce CsgA at levels comparable to WT cells (Figure 2B), suggesting that oar has difficulty transmitting the C-signal. Another property suggestive of a defect in C-signaling is the inability to ripple (data not shown). During early development cells move in traveling waves known as ripples. Rippling requires C-signaling to regulate the cellular reversal period [28], [29]. The absence of both rippling and extracellular complementation of csgA mutants suggests a defect in C-signal transmission. However, the oar phenotype does not entirely phenocopy csgA. When the oar mutant was mixed 1∶1 with WT cells, 99% of the spores that germinated were of WT origin suggesting that the oar sporulation defect cannot be bypassed with an extracellular signal. csgA mutants, in contrast, typically form 30–50% of the spores when mixed with WT.
Examination of oar cells revealed a striking and novel defect. While 100% of 24 h developing WT cells show rod-shaped morphology, 62% of the oar cells were bent into horseshoe shapes that eventually circularize. The outer membrane appears to pull away from the rest of the cell to form spheroplasts (Figure 2c). These defects were not observed in oar vegetative cells (data not shown). In a csgA oar double mutant, only 8% of cells had oar-like, morphology. These results suggested that C-signal accumulation causes deformation of the cell envelop with lethal consequences.
ECM and IM lipoproteins utilize different sorting signals in M. xanthus
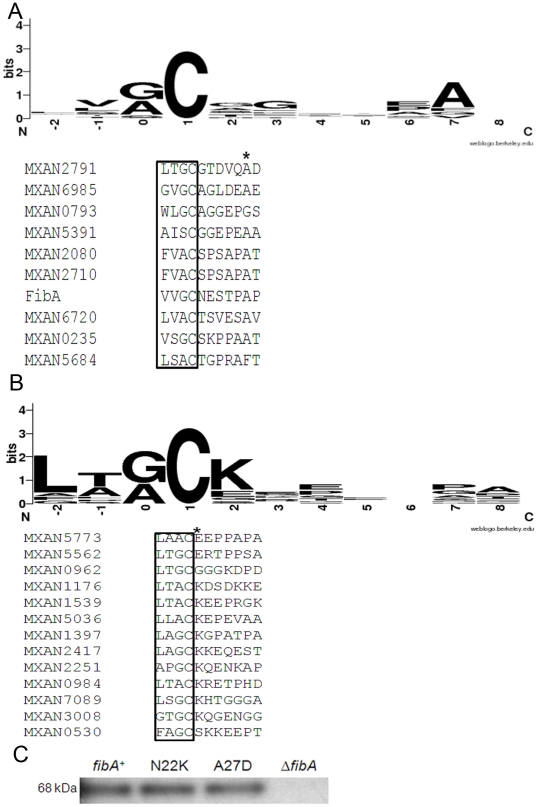
In other Proteobacteria, lipoproteins are sorted into the IM and OM using amino acid residues near the lipid modification site. Since E. coli does not secrete lipoproteins, the mechansim by which they make their way to the ECM are largely unknown. To investigate lipoprotein targeting to the ECM, the lipobox and the first eight amino acids of the mature ECM and IM lipoproteins were examined by multiple sequence alignment using WebLogo [30]. 7/10 ECM proteins, including by far the most abundant ECM protein FibA, possess alanine at the +7 position (Figure 3A). No amino acid conservation was observed in OM lipoproteins identified by LC-MS/MS (data not shown). This result suggests that +7 alanine may have a role in targeting lipoproteins to the ECM.
Figure 3. Bioinformatic analysis of first eight amino acids of N-terminus of M. xanthus lipoproteins.
(A) Multiple sequence alignment of ECM proteins using WebLogo. The lipobox (highlighted by a box made of dashed lines) and the following seven amino acids of the N-terminal region of mature lipoproteins were aligned using WebLogo. Seven ECM lipoproteins have alanine at the 7th position (highlighted by a solid box). (B) 8/12 predicted IM lipoproteins have lysine at the 2nd position (highlighted by a solid box). (C) Western blot analysis of 18 h developing cells using Mab2105 primary antibody. Strains used include LS2760 (WT FibA), LS2208 (ΔfibA), LS2761 (N22K FibA), LS2764 (A27D FibA).
Because the mechanism of inner membrane targeting is also unknown in M. xanthus, and highly variable among the Proteobacteria, 12 putative IM lipoproteins were identified by LC-MS/MS from purified inner membranes (Table 5). 8/12 IM lipoproteins have lysine at the second position suggesting that lysine at the +2 position may be be essential for inner membrane targeting, as it is in E. coli (Figure 3B). The signal sequence of MXAN1176, a lipoprotein with lysine at the +2 position, localized mCherry in the IM [31].
Table 5. Putative lipoproteins identified by LC-MS/MS from IM fraction.
| MXAN | Function | No. of peptides |
| MXAN 0530 | Putative lipoprotein | 1 |
| MXAN 0962 | Putative lipoprotein | 2 |
| MXAN 0984 | Heavy metal efflux transporter, RND family, MFP subunit | 3 |
| MXAN 1176 | Peptidylprolyl cis-trans isomerase, cyclophilin-type | 5 |
| MXAN 1397 | PBS lyase HEAT-like repeat protein | 3 |
| MXAN 1539 | Putative lipoprotein | 6 |
| MXAN 2417 | Conserved hypothetical protein | 8 |
| MXAN 3008 | Adventurous gliding motility protein AglU | 5 |
| MXAN 5036 | Conserved domain protein | 4 |
| MXAN 5562 | Putative lipoprotein | 3 |
| MXAN 5773 | Putative lipoprotein | 3 |
| MXAN 7089 | Putative lipoprotein | 1 |
Site directed mutagenesis of fibA was carried out to determine whether alanine at the +7 position is essential for ECM localization. Since no amino acid conservation was observed in the N-terminus of OM lipoproteins, alanine (GCC) was changed to aspartate (GAC) at the 27th position (+7 in mature FibA) as this substitution involved minimal nucleotide modification. The modified fibA gene was expressed in plasmid pZJY156 under control of the constitutive pilA promoter then introduced into ΔfibA strain LS2208. As a positive control, WT fibA, was introduced into LS2208 with the same vector system. Both strains produced comparable amounts of FibA as revealed by Western analysis of whole cells (Figure 3C). Immuno transmission electron microscopy was carried out to quantify FibA localization in the ECM. Cells were allowed to form biofilms on formvar coated nickel grids in submerged culture [32]. FibA secretion was then induced by incubating the grids in cohesion buffer. The cells were probed with anti-FibA (Mab2105) followed by anti-mouse antibody conjugated to 10 nm gold particles, and then examined by transmission electron microscopy. Gold particles on or around 10–12 cells were enumerated.
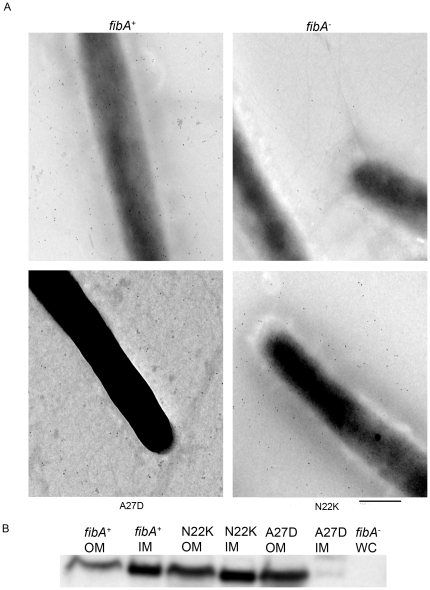
Approximately 70 gold particles cell−1 are associated with the ECM and the cell surface of strain LS2760, which produces WT FibA (Figure 4A). In contrast only a few gold particles were attached to the surfaces of LS2208 (ΔfibA) or LS2764 (A27D) (3.1 and 6.1 particles cell−1, respectively). Clearly, the +7 alanine is essential for localizing FibA to the ECM. Membrane separation of 7 h developing cells was performed in order to examine the localization of FibA to IM and OM locations. WT FibA is distributed almost equally in IM and OM during starvation suggesting that transport of FibA to the ECM occurs stepwise (Figure 4B). Conversely, the A27D change led to exclusively OM localization.
Figure 4. Identification of M. xanthus lipoprotein sorting signals.
(A) Immuno transmission electron microscopy of developing cells using monoclonal antibody Mab2105. M. xanthus cells were allowed to form a biofilm on a formvar-carbon-coated nickel grid for 3 h. The cells were probed with Mab2105, which reacts primarily with FibA followed by anti-mouse antibodies conjugated with 10 nm colloidal gold particles. Bar is 500 nm. (B) Western blot analysis of membrane fractions purified from 7–8 h developing cells.
Because +2 lysine mediates retention of IM proteins (Figure 3B, [31]) we wondered whether FibA with +2 lysine would be retained in the IM. Site directed mutagenesis was used to replace an asparagine (AAC) codon with a lysine (AAG) codon at the 22nd position (+2 in mature FibA). The modified fibA gene was also expressed in pZJY156 under control of the constitutive pilA promoter and introduced into LS2208. The strain produced comparable amounts of FibA to the WT (Figure 4A). Immuno transmission electron microscopy of this strain revealed large numbers of gold particles associated with the ECM (112 particles cell−1) suggesting that the N22K substitution does not abort ECM localization of FibA. Furthermore, N22K FibA showed similar distribution to the WT in IM and OM further confirming that lysine at the +2 position does not exclusively target FibA to the IM (Figure 4B). These results suggest that ECM lipoproteins are sorted differently than those retained in the IM.
Discussion
A stepwise approach was used to identify OM porin proteins from the 7331 member M. xanthus proteome because no single program was successful. Our approach involved parsing the proteome with existing software, Signal P, Lipo P and TMHMM, to generate a smaller pool of candidates. Those proteins that possessed a type I signal sequence, but were devoid of transmembrane helices and a lipobox were examined using TMBETA-SVM and TMBETADISC-RBF. This approach identified 228 putative β-barrel proteins and dramatically reduced the number of false positives. When TMBETADISC-RBF was used on the whole M. xanthus proteome, it identified 915 β-barrel proteins (12% of the genome), which is far greater than is typical of Gram-negative bacteria.
Out of 228 putative β-barrel proteins, 54 were identified by LC-MS/MS in the vegetative cell OM fraction. In a complementary approach, Kahnt et al used biotinlyation of M. xanthus whole cells and OM vesicles to identify β-barrel proteins [9]. Kahnt et al used PRED-TMββ to identify β-barrel proteins from among the biotinylated proteins, which we found to generate over 50% false positives. Furthermore, 22/298 biotinylated proteins that were not predicted to be β-barrel proteins in the Kahnt et al study are predicted by our work to be β-barrel proteins. All in all, 43 proteins were identified by LC-MS/MS in both studies (Table 3). Because most membrane protein enrichment methods cannot avoid protein contamination from other cellular compartments, use of a robust bioinformatic approach can help accurately identify integral OM proteins from a pool of enriched candidates.
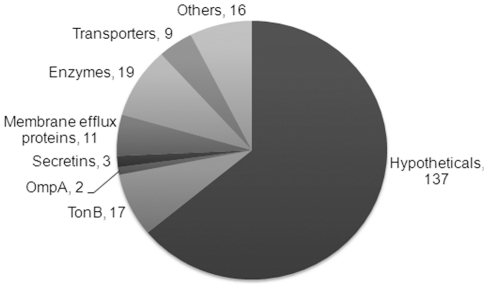
The majority (∼60%) of the predicted OM proteins have no known functions (Figure 5). Many putative β-barrel proteins are predicted to be involved in transport. M. xanthus potentially encodes 17 TonB dependent receptors of which only five were detected in vegetative cells. TonB dependent receptors are involved in energy dependent uptake of specific substrates, such as iron, which may be poorly permeable across the membrane or may be present in very low concentrations in the environment. The energy for transport is derived from the proton motive force across the IM and is delivered by a protein complex consisting of TonB, ExbB and ExbD [33], [34].
Figure 5. Pie chart classifying 228 OM β-barrel proteins according to function.
Most M. xanthus OM proteins have no known function. The second major class of proteins includes TonBs, which are required for transport of a specific substrate. Transport of small molecules are carried out by transporters and OmpA, membrane efflux proteins are required for export of toxins and secretins form a large OM pore that allow export by Type II secretion system.
The C-signal is a developmental signal produced by the csgA gene. One M. xanthus TonB dependent receptor, Oar, appears to be essential for C-signal transmission since an oar mutant fails to rescue sporulation of csgA cells in mixture. Contact and cell alignment is essential for C-signaling [35], [36]. While the inability of oar cells to rescue ΔcsgA development could arise due to improper alignment of cells caused by their abnormal shapes, another possibility is that oar is the OM channel used to export the C-signal. The latter possibility is suggested by the fact that oar is also required for the periodic movement of cells in traveling waves, sometimes referred to as ripples because of their visual similarity with ripples on the surface of water. The C-signal is the only signal known to be required for rippling. Rippling begins early in development, long before oar cells begin to bend. Thus it remains an intriguing possibility that oar is the porin for the C-signal.
Unlike csgA mutants, which retain their long, thin shape during their defective attempt at development, oar mutants become deformed in a novel manner. oar cells begin to bend in a central location, eventually forming circular cells whose outer membrane begins to pull away from the cell. At 24 hours, nearly 2/3 of oar cells are deformed. Loss of csgA restores normal cell morphology to oar cells suggesting that the morphological defect is due to accumulation of the C-signal. Curiously, the morphological problem appears to begin at the center of the cells and might suggest that C-signaling is mediated by side-to-side contact rather than polar contact as previously hypothesized [35]. While some evidence suggests that the C-signal is the CsgA protein [37], indirect evidence suggests that CsgA is an enzyme that acts on lipid molecules [38]. It is possible that the product of this enzyme reaction may destabilize the cell membrane.
The most abundant ECM lipoprotein is FibA [16], [39]. Alanine at the +7 position appears to be conserved in most ECM proteins [16]. An amino acid change from alanine to aspartate at the +7 position (A27D) in FibA leads to OM retention, suggesting that the +7 alanine is crucial for export to the ECM. In WT cells FibA is found in equal amounts in IM and OM suggesting stepwise passage through the cell envelope. If secretion occurs in a stepwise manner substitution of asparagine with lysine at the +2 position might be expected to cause FibA to accumulate in the IM, the first transit point in a temporal sequence. However, this substitution did not block export to the OM and ECM. These results suggest the M. xanthus secretion system for ECM proteins differs from the one for membrane proteins.
Klebsiella oxytoca also possesses two mechanisms for lipoprotein transport, a Lol system that moves proteins to the OM, and a type II secretion system for the export of the cell surface exposed lipoprotein PulA [40], [41]. K. oxytoca uses +2 Asp as a LolA avoidance tag such that proteins with +2 Asp are retained in the inner membrane while those without it move to the OM. The work presented in this paper suggests that M. xanthus has a similar system. However, the PulA secretion pathway requires +2 Asp to retain PulA in the IM temporarily until the type II secretion system transports it outside the cell. FibA does not have a +2 Asp so the mechanism of secretion is fundamentally different from K. oxytoca. Whether FibA secretion depends on a type II secretion system remains unknown. The M. xanthus genome predicts the presence of two type II secretion systems and future studies should reveal whether these are utilized for lipoprotein export. While, the mechanism of ECM lipoprotein trafficking remains unknown, our study provides a valuable tool to identify subcellular location of various lipoproteins based on sequence information, and a method to target specific proteins to IM, OM or ECM.
Materials and Methods
Bioinformatic analysis
M. xanthus protein sequences were obtained from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Myxococcus_xanthus_DK_1622/NC_008095.faa). Prediction of signal peptide, lipoprotein, and transmembrane helices in protein sequences were made using Signal P (http://www.cbs.dtu.dk/services/SignalP/), Lipo P (http://www.cbs.dtu.dk/services/LipoP/) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) respectively[10], [17], [42]. M. xanthus β-barrel domain proteins were obtained from the Pfam database (http://pfam.janelia.org/). The majority of the β-barrel domain proteins belong to the OM β-barrel protein (MBB) superfamily.
Six β-barrel prediction methods were evaluated for their ability to discriminate M. xanthus β-barrel proteins including TMB-HUNT (http://bmbpcu36.leeds.ac.uk/~andy/betaBarrel/AACompPred/aaTMB_Hunt.cgi), TMBETA-SVM (http://tmbeta-svm.cbrc.jp/), TMBETA-NET (http://psfs.cbrc.jp/tmbeta-net/), TMBETADISC-RBF (http://rbf.bioinfo.tw/~sachen/OMPpredict/TMBETADISC-RBF.php), PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB/input.jsp) and BOMP (http://services.cbu.uib.no/tools/bomp) [42], [43], [44], [45], [46], [47].
Bacterial strains and growth condition
Table 6 lists the bacterial strains, plasmids and primers used in this study. M. xanthus DK1622 cells were grown in CYE broth [1% Bacto casitone (Difco), 0.5% yeast extract (Difco), 10 mM 4-morpholinepropanesulfonic acid (MOPS) buffer (pH 7.6), and 0.1% MgSO4)] at 32°C with vigorous shaking. To solidify the media, Bacto agar (Difco) was added at a concentration of 1.5%. E. coli cells were grown in Luria-Bertani (LB) medium. Kanamycin was added to CYE or LB media at a final concentration of 50 µg ml−1.
Table 6. Bacterial strains, plasmids and primers used in this study.
| M. xanthus strains | Genotype | Reference or source |
| DK1622 | Wild type | [56] |
| LS2208 | ΔfibA | Lawrence Shimkets |
| LS2441 | ΔcsgA | Lawrence Shimkets |
| LS2453 | oar, Kmr | Lawrence Shimkets |
| LS2456 | oar csgA, Kmr | Lawrence Shimkets |
| LS2760 | LS2208 containing plasmid pSTB31, Kmr | This study |
| LS2761 | LS2208 containing plasmid pSTB27, Kmr | This study |
| LS2764 | LS2208 containing plasmid pSTB28.1, Kmr | This study |
| JD300 | esg, Kmr | [57] |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pZJY156 | Shuttle vector | [52] |
| pUC19 | Cloning vector | [58] |
| pSTB20 | pCR2.1-TOPO carrying pilA promoter and fibA gene | This study |
| pSTB21 | pCR2.1-TOPO carrying full length fibA | This study |
| pSTB22 | pUC19 carrying 500 bp, XbaI-SalI fragment from pSTB20 | This study |
| pSTB23 | pSTB22 with N22K substitution in the FibA | This study |
| pSTB24.1 | pSTB22 with A27D substitution in the FibA | This study |
| pSTB25 | 500 bp, XbaI-SalI from pSTB23 cloned into pSTB21 | This study |
| pSTB26.1 | 500 bp, XbaI-SalI from pSTB24.1 cloned into pSTB21 | This study |
| pSTB27 | pZJY156 carrying pilA promoter and modified fibA from pSTB25 | This study |
| pSTB28.1 | pZJY156 carrying pilA promoter and modified fibA from pSTB26.1 | This study |
| pSTB31 | pZJY156 carrying pilA promoter and WT fibA from pSTB20 | This study |
| Primers 1 | ||
| A | 5′TCTAGAGGGAGCGCTTCGGATGCGTAGGCTGATCG 3′ | |
| B | 5′CTTCTGCACGAGCATGGGGGTCCTCAGAGAAGGTTGCAACGG 3′ | |
| C | 5′ ACCCCCATGCTCGTGCAGAAGAGAGTTCGCGGAGCG 3′ | |
| D | 5′ GGTACCCCTCGAGCCGCTGCCCAAGTAG 3′ | |
| FibA2DF | 5′ GAGTCCACCCCTGACCCCGAGGCCGAC 3′ | |
| FibA2DR | 5′ GTCGGCCTCGGGGTCAGGGGTGGACTC3′ | |
| FibAKF | 5′ GTTGTCGGTTGCAAGGAGTCCACCCCTGCC 3′ | |
| FibAKR | 5′ GGCAGGGGTGGACTCCTTGCAACCGACAAC 3′ |
Underline indicates an overlap of 21 nucleotides.
Membrane separation
Membrane separation was carried out as described by Simunovic et al with a few modifications [48]. A 1 L culture of M. xanthus DK1622 cells was grown to a density of 2×108 cells ml−1. Cells were harvested by centrifugation, washed with chilled distilled water, and resuspended in 40 to 50 ml of 23.5% sucrose in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.6. Freshly prepared chicken egg white lysozyme (300 µg ml−1) (Sigma Chemical Co., St. Louis, Mo.) and EDTA (pH 7.6) (1 mM) were added, and incubated overnight at 4°C with gentle stirring. The cells were resuspended in 6 ml of ice-cold double-distilled water with vigorous pipetting to induce spheroplast formation, and stirred for 30 min at 4°C. Spheroplasts were collected by centrifugation at 12,000× g for 10 min at 4°C. Following centrifugation the supernatant was collected and saved while the pellet was resuspended in 3 volumes of 5 mM EDTA (pH 7.6). One tablet of complete EDTA-free protease inhibitor (Roche, Indianapolis, Ind.) was added, and the suspension stirred for 1 h. The supernatant was added back to the suspension and stirred for an additional 30 min. 1 ml of RNase A (10 mg ml−1, Sigma Chemical Co.) and 1 ml of DNase type II (10 mg ml−1, Sigma Chemical Co.) were added, and stirred for 30 min. Rod-shaped cells were pelleted by centrifugation at 5000× g for 10 min at 4°C. Spheroplasts were collected by ultracentrifugation at 100,000× g for 3 h at 4°C in a 70.1 Ti rotor (Beckman Coulter). Membrane pellets were resuspended in 23.5% sucrose in 20 mM HEPES, 5 mM EDTA, pH 7.6 using a Dual 21 tissue homogenizer (Kimble Kontes, Vineland, N.J.), and incubated overnight with gentle stirring at 4°C. The membrane suspension was loaded on top of a three-step gradient consisting of 10 ml of 60% sucrose, 10 ml of 48% sucrose, 10 ml of 35% sucrose in 20 mM HEPES, 5 mM EDTA, pH 7.6. The membrane fractions were separated by ultracentrifugation at 120,000× g for 4 h at 4°C in a Beckman swinging bucket rotor (SW28). The OM fraction migrated to the middle of the 35% sucrose layer. It was collected, diluted with HE0.1 buffer [20 mM HEPES, 0.1 mM EDTA, pH 7.6], concentrated by ultracentrifugation at 120,000× g for 3 h at 4°C in a 70.1 Ti rotor, then stored at −20°C.
The membrane enrichments were collected as described by Simunovic et al [23]. The membrane enrichment that migrated at the 48/60% sucrose layer interface consisted of IM and hybrid membrane (HM) fraction, which were collected and concentrated by ultracentrifugation using a SW28 rotor at 120,000× g for 4 h at 4°C. Concentrated enrichments were layered on top of a discontinuous sucrose gradient consisting of 4 ml 70% sucrose, 4 ml 60% sucrose, 15 ml 55% sucrose, 3 ml 40% sucrose, and 3 ml 30% sucrose in 20 mM HEPES, 5 mM EDTA, pH 7.6. The gradients were centrifuged using SW28 rotor at 70,000× g for 20 h. The membrane enrichment that migrated between the 60 and 70% sucrose layers consisted of IM fraction. This fraction was collected, concentrated by ultracentrifugation, then stored at −20°C for proteomic analysis.
Membrane separation of developing cells was carried out by growing a 500 ml culture to a final density of 5×108 cell ml−1. The cells were harvested by centrifugation and resuspended in 10 ml water. The cells were spread on two 33- by 22-cm trays containing TPM agar [10 mM Tris HCl, pH 7.6, 1 mM KH(H2)PO4, pH 7.6, 10 mM MgSO4, 1.5% agar (Difco)] and incubated at 32°C for 7 h. Developing cells were harvested with a razor blade and resuspended in 40 to 50 ml of 23.5% sucrose in 20 mM HEPES, pH 7.6. The membrane separation was carried out as described above.
Phenol extraction of OM proteins
Phenol extraction of OM proteins was carried out as described in Hancock and Nikaido [49]. An equal volume of 88% phenol, pH 6.8, was added to the OM protein sample that was prepared and frozen as mentioned above. The OM sample was incubated at 70°C for 10 min. The mixture was immediately cooled on ice for 10 min, and then centrifuged for 10 min at 5000× g. The upper, aqueous layer was discarded. To the interface and the phenol phase an equal volume of distilled water was added, incubated at 70°C for 10 min, cooled on ice, and centrifuged at 5000× g for 10 min. After the aqueous phase was discarded, protein was extracted from the phenol phase using two volumes of acetone each time. The acetone fractions were combined and the acetone was removed by air-drying. The pellet was resuspended in 100 µl of 1× PBS buffer [137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4].
In-gel trypsin digestion
150 µg protein was boiled in loading buffer [52.5 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue, 100 mM dithiothreitol (DTT)] for 10 min and cooled on ice for 10 min. The sample was loaded on a 4–20% gradient polyacrylamide gel. The gel was run at 70 V until the dye entered the gel. Protein detection was performed using Bio-safe Coomassie Stain (Bio-Rad). The portion of the gel containing protein was cut into small pieces, and destained with 100 µl of water for 15 min. The gel pieces were washed sequentially for 15 minutes each with 50% acetonitrile, 100% acetonitrile, and 100 mM ammonium bicarbonate containing 50% acetonitrile (vol/vol). The gel pieces were dried under vacuum, treated with 100 µl of 10 mM DTT in 40 mM ammonium bicarbonate at 56°C for 45 min, alkylated with 100 µl of 55 mM iodoacetamide, 40 mM ammonium bicarbonate, and incubated for 30 min at room temperature in the dark. The gel pieces were washed with acetonitrile for 15 min, and then dried under vacuum. The gel pieces were rehydrated with 2 µg µl−1 proteomics-grade trypsin (Promega) in 40 mM ammonium bicarbonate and incubated at 37°C overnight. Solutions from multiple trypsin digestions were pooled. The gel slices were washed once with 50% acetonitrile in 25 mM ammonium bicarbonate, twice with 5% formic acid, and twice with acetonitrile for 15 min each. The washes were combined with the solutions from the previous step and dried under vacuum.
Identification of OM proteins by LC/MS-MS
The peptides obtained from trypsin digestion were loaded on a PicoFrit C18 column (8-cm by 50-µm) (New Objective, Woburn, MA), and separated on an Agilent 1100 capillary LC (Palo Alto, CA), which interfaced directly to a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). The mobile phases consisted of A (H2O and 0.1% formic acid) and B (acetonitrile and 0.1% formic acid). The peptides were eluted from the column during a 90 min linear gradient from 5 to 60% of total solution composed of mobile phase B into the mass spectrometer at a flow rate of 200 ηl min−1. MS/MS spectra were acquired on the nine most abundant precursor ions from each MS scan with a repeat count and duration of 1 and 5 s each. Dynamic exclusion was enabled for 200 s. The MS/MS spectra were converted into peak lists by mzMXL2Other and ReAdW software [50].
Database searches were performed using Mascot 1.9 software (Matrix Science, Boston, MA) against a M. xanthus protein database obtained from NCBI. The search parameters included full tryptic enzymatic cleavage, up to three missed cleavages, peptide tolerance of 1000 ppm, fragment ion tolerance of 0.6 Da. Fixed modification was set as carbamidomethyl due to carboxyamidomethylation of cysteine residues (+57 Da) while variable modification was set as oxidation of methionine residues (+16 Da) and deamidation of asparagines residues (+1 Da). The proteins identified were statistically validated using ProValT algorithm as implemented in ProteoIQ (BioInquire, Athens, GA) [51]. Only proteins with a false-discovery rate of less than 1% were considered to be statistically significant.
Extracellular complementation
M. xanthus cells were grown to a cell density of 5×108 cells ml−1, harvested and resuspended to a final concentration of 5×109 cells ml−1. oar cells were mixed with various strains in 1∶1 ratio and 10 µl of the cell mixture was spotted on TPM agar plates. The plates were incubated at 32°C and after five days digital images were acquired.
Microscopic analysis
5×108 M. xanthus cells were spotted on TPM agar plates and incubated for 24 h. Cells were resuspended in a drop of TPMF buffer [TPM containing 10% ficoll], and examined with a phase contrast microscope (Leica Microsystems, DM55008). Digital images were obtained at 1000× magnification using a QIQICAM FAST 1394 camera (Compix Inc).
Site directed mutagenesis and cloning of fibA
The fibA gene was expressed from pZJY156 [52]. The fibA gene was fused with the pilA promoter using the gene splicing by overlap extension (SOEing PCR) method [53]. The primers are listed in Table 6. Primers A and B were used for amplification of the pilA promoter including the ribosomal binding site and the start codon [54]. Primers C and D were used for the amplification of full length fibA. Primers B and C were designed to have an overlap of 21 nucleotides (underlined region). M. xanthus DK1622 genomic DNA was used as a template. The two PCR products and primer pair A and D were then used for SOEing PCR. PCR products were separated on 0.8% agarose, excised, extracted using the Gel Extraction kit (Qiagen), and cloned into pCR2.1-TOPO (Invitrogen) to create pSTB20. Full length fibA was cloned into pCR2.1-TOPO to create pSTB21. A 500 bp, XbaI-SalI fragment containing the pilA promoter and the N-terminal region of FibA from pSTB20 was cloned into pUC19 to create pSTB22, which was used as the template for site directed mutagenesis. The primers used for the site directed mutagenesis are listed in Table 6. Primers FibA2DF and FibA2DR were used for replacing alanine with aspartate at 27th position while FibAKF and FibAKR were used for replacing asparagine with lysine at the 22nd position (Table 6). PCR was carried out using a high fidelity DNA polymerase I (Expand High Fidelity PCR system, Roche). The PCR products obtained were treated with DpnI to eliminate the methylated template, and then transformed into E. coli Top10 cells. Plasmids were isolated from transformants and sequenced. The plasmids encoding FibA with N22K or A27D amino acid substitutions were called pSTB23 and pSTB24.1 respectively. The 500 bp, XbaI-SalI from pSTB23 and pSTB24.1 were cloned at the same site in pSTB21 to create pSTB25 and pSTB26.1, respectively. The new plasmids contained full length fibA encoding the N22K or A27D substitutions expressed from the pilA promoter. The plasmids, pSTB25 and pSTB26.1 were digested with XbaI and EcoRI, and the 2800 bp fragment containing the pilA promoter and fibA gene was cloned into pZJY156 to create pSTB27 and pSTB28.1 plasmids. These plasmids were then transformed into M. xanthus LS2208 to create LS2761 and LS2764. Full length fibA expressed from the pilA promoter was cloned into pZJY156 to create pSTB31 and also transformed into M. xanthus LS2208 to create LS2760. Expression of fibA from the pilA promoter was verified by western blotting using monoclonal antibody Mab2105 [55].
Western blot analysis
5 µg of cell lysate or 10 µg of membrane fractions, were separated on a 4–20% SDS-PAGE gradient gel (Bio Rad). Proteins were transferred to an Immobilin-P, PVDF membrane (Milipore). The membrane was blocked with 3% bovine serum albumin (BSA) in PBST (1× PBS containing 0.1% Tween 20). The proteins were probed with the Mab2105 (1∶500 dilution) or anti-CsgA (1∶5000) that was prepared in PBST containing 0.1% BSA [55]. This was followed by washing three times with PBST. The membrane blot was then probed with horseradish peroxidase-conjugated goat anti-mouse IgG or anti-rabbit IgG, which were diluted to 1∶10,000 in PBST containing 0.1% BSA. The membrane was washed three times with PBST and developed with the ECL luminescence detection kit (Amersham).
Electron microscopy
M. xanthus cells were grown to a density of 5×108 cells ml−1 and diluted to 3.3×106 cells ml−1in CYE or CYEK (CYE containing 50 µg ml−1 kanamycin) broth. 4 ml of the cell suspension and a formvar-carbon-coated nickel grid (Electron Microscope Sciences) was transferred to a petri plate (60×15 mm). The plate was incubated at 32°C for 12 h. A thin biofilm on the surface of the grid was allowed to form. Starvation was induced by replacing the CYE or CYEK broth with cohesion buffer [10 mM MOPS, pH 6.8, 1 mM MgCl2, 1 mM CaCl2] and incubated for 3 h at 32°C. The grid was treated with 2% glutaraldehyde for 15 min at room temperature followed by washing five times with the cohesion buffer. The grid was blocked with 5% bovine serum albumin (BSA) in cohesion buffer for 45 min at room temperature. The grid was treated with Mab2105 antibody (1∶20 dilution) prepared in cohesion buffer containing 5% BSA for 45 min at room temperature followed by washing three times with cohesion buffer [55]. The grid was then treated with anti-mouse antibody (1∶100 dilutions) conjugated to 10 nm colloidal gold particles (Sigma-Aldrich), and incubated for 30 min at room temperature. The grid was washed three times with the cohesion buffer. The grid was washed three times with water, allowed to air dry, and observed under a FEI Technal transmission electron microscope operated at 200 kV.
Supporting Information
228 OM proteins identified by bothTMBETA-SVM and TMBETADISC-RBF.
(DOC)
List of non-outer membrane integral proteins identified by LC-MS/MS.
(DOC)
Acknowledgments
We would like to thank Jan Mrazek for helpful discussions, Krishna Bayyareddy for technical help, and Dr. John P. Shields and Dr. Jianguo Fan of the Center for Ultrastructural Research for their assistance with TEM.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by research grant MCB 0742976 from the National Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shimkets LJ, Gill RE, Kaiser D. Intercellular signaling during fruiting body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 2.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postle K. TonB system, in vivo assays and characterization. Methods Enzymol. 2007;422:245–269. doi: 10.1016/S0076-6879(06)22012-3. [DOI] [PubMed] [Google Scholar]
- 5.van den Berg B. The FadL family: unusual transporters for unusual substrates. Curr Opin Struct Biol. 2005;15:401–407. doi: 10.1016/j.sbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 7.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahnt J, Aguiluz K, Koch J, Treuner-Lange A, Konovalova A, et al. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J Proteome Res. 2010;9:5197–5208. doi: 10.1021/pr1004983. [DOI] [PubMed] [Google Scholar]
- 10.Punta M, Forrest LR, Bigelow H, Kernytsky A, Liu J, et al. Membrane protein prediction methods. Methods. 2007;41:460–474. doi: 10.1016/j.ymeth.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 2009;17:13–21. doi: 10.1016/j.tim.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Seydel A, Gounon P, Pugsley AP. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol. 1999;34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 13.Masuda K, Matsuyama S, Tokuda H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc Natl Acad Sci U S A. 2002;99:7390–7395. doi: 10.1073/pnas.112085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka SY, Narita S, Tokuda H. Characterization of the Pseudomonas aeruginosa Lol system as a lipoprotein sorting mechanism. J Biol Chem. 2007;282 doi: 10.1074/jbc.M611840200. [DOI] [PubMed] [Google Scholar]
- 15.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Curtis PD, Atwood J, 3rd, Orlando R, Shimkets LJ. Proteins associated with the Myxococcus xanthus extracellular matrix. J Bacteriol. 2007;189:7634–7642. doi: 10.1128/JB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Koski P, Hirvas L, Vaara M. Complete sequence of the ompH gene encoding the 16-kDa cationic outer membrane protein of Salmonella typhimurium. Gene. 1990;88:117–120. doi: 10.1016/0378-1119(90)90068-3. [DOI] [PubMed] [Google Scholar]
- 19.Reizer J, Reizer A, Saier MH., Jr The MIP family of integral membrane channel proteins: sequence comparisons, evolutionary relationships, reconstructed pathway of evolution, and proposed functional differentiation of the two repeated halves of the proteins. Crit Rev Biochem Mol Biol. 1993;28:235–257. doi: 10.3109/10409239309086796. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 21.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punta M, Forrest LR, Bigelow H, Kernytsky A, Liu J, et al. Membrane protein prediction methods. Methods. 2007;41:460–474. doi: 10.1016/j.ymeth.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simunovic V, Gherardini FC, Shimkets LJ. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J Bacteriol. 2003;185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Canamero M, Munoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S. Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J Bacteriol. 1993;175:4756–4763. doi: 10.1128/jb.175.15.4756-4763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimkets LJ, Gill RE, Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen DC, Bretscher AP, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 27.Toal DR, Clifton SW, Roe BA, Downard J. The esg locus of Myxococcus xanthus encodes the E1 alpha and E1 beta subunits of a branched-chain keto acid dehydrogenase. Mol Microbiol. 1995;16:177–189. doi: 10.1111/j.1365-2958.1995.tb02291.x. [DOI] [PubMed] [Google Scholar]
- 28.Shimkets LJ, Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Igoshin OA, Mogilner A, Welch RD, Kaiser D, Oster G. Pattern formation and traveling waves in myxobacteria: theory and modeling. Proc Natl Acad Sci U S A. 2001;98:14913–14918. doi: 10.1073/pnas.221579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 32.Kuner JM, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990a;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 36.Kim SK, Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990b;4:896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 37.Kruse T, Lobedanz S, Berthelsen NM, Sogaard-Andersen L. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 38.Avadhani M, Geyer R, White DC, Shimkets LJ. Lysophosphatidylethanolamine is a substrate for the short-chain alcohol dehydrogenase SocA from Myxococcus xanthus. J Bacteriol. 2006;188:8543–8550. doi: 10.1128/JB.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearns DB, Bonner PJ, Smith DR, Shimkets LJ. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J Bacteriol. 2002;184:1678–1684. doi: 10.1128/JB.184.6.1678-1684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poquet I, Kornacker MG, Pugsley AP. The role of the lipoprotein sorting signal (aspartate +2) in pullulanase secretion. Mol Microbiol. 1993;9:1061–1069. doi: 10.1111/j.1365-2958.1993.tb01235.x. [DOI] [PubMed] [Google Scholar]
- 42.Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrow AG, Agnew A, Westhead DR. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005;33:W188–192. doi: 10.1093/nar/gki384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gromiha MM, Ahmad S, Suwa M. TMBETA-NET: discrimination and prediction of membrane spanning beta-strands in outer membrane protein. Nucleic Acids Res. 2005;33:W164–167. doi: 10.1093/nar/gki367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou YY, Gromiha MM, Chen SA, Suwa M. TMBETADISC-RBF: Discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Comput Biol Chem. 2008;32:227–231. doi: 10.1016/j.compbiolchem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park KJ, Gromiha MM, Horton P, Suwa M. Discrimination of outer membrane proteins using support vector machines. Bioinformatics. 2005;21:4223–4229. doi: 10.1093/bioinformatics/bti697. [DOI] [PubMed] [Google Scholar]
- 48.Simunovic V, Gherardini FC, Shimkets LJ. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J Bacteriol. 2003;185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hancock RE, Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978;136:381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, Deutsch EW, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotech. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 51.Graham RL, Pollock CE, O'Loughlin SN, Ternan NG, Weatherly DB, et al. Multidimensional proteomic analysis of the soluble subproteome of the emerging nosocomial pathogen Ochrobactrum anthropi. J Proteome Res. 2006;5:3145–3153. doi: 10.1021/pr060293g. [DOI] [PubMed] [Google Scholar]
- 52.Zhao JY, Zhong L, Shen MJ, Xia ZJ, Cheng QX, et al. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl Environ Microbiol. 2008;74:1980–1987. doi: 10.1128/AEM.02143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horton R. PCR-mediated recombination and mutagenesis. Molecular Biotechnology. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 54.Wu SS, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behmlander RM, Dworkin M. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J Bacteriol. 1991;173:7810–7820. doi: 10.1128/jb.173.24.7810-7820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Downard J, Ramaswamy SV, Kil KS. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
228 OM proteins identified by bothTMBETA-SVM and TMBETADISC-RBF.
(DOC)
List of non-outer membrane integral proteins identified by LC-MS/MS.
(DOC)