Abstract
Specific 13C labeling of Thr methyl groups has been accomplished by growth of a standard laboratory strain of Escherichia coli on [2-13C] glycerol in the presence of deuterated isoketovalerate, Ile, and Ala. Diversion of label from the Thr biosynthetic pathway is suppressed by including Lys, Met, and Ile in the growth media. This method complements the repertoire of methyl-labeling schemes for NMR structural and dynamic studies on proteins and is particularly useful for the study of nucleic acid binding proteins due to the high propensity of Thr residues at protein-DNA and -RNA interfaces.
NMR spectroscopy is a powerful tool for probing protein dynamics over a range of time-scales(1). The combination of progress in optimized pulse sequences, better hardware, and better isotopic labeling schemes has allowed researchers to study proteins of high molecular weight(2, 3). Historically, dynamics studies using NMR spectroscopy have focused on the protein backbone (amide groups). However, the N-H group may not reflect side-chain motions and hence the need to study side-chain dynamics. Among all the groups available to study side-chains, methyls are especially suited for the purpose as they are well-distributed through the protein structure. Furthermore, the methyl HMQC (heteronuclear multiple-quantum coherence) experiments can exploit the TROSY effect resulting from cancellation of intra-methyl dipolar relaxation interactions to give better resolved and more sensitive spectra(4). This has enabled the use of methyl groups to study the dynamics of high molecular weight proteins and their complexes.
A prerequisite for 13C-based methyl dynamics studies in large proteins is that the methyl carbons should be specifically 13C labeled (adjacent to 12C), and protonated in a perdeuterated background. Initially, methyl labeling utilized 1H,13C-pyruvate (Pyr) as the sole source of carbon in deuterated media(5). More recent strategies involve the use of selectively labeled α-keto acids to specifically incorporate protonated 13C methyls in proteins(6, 7). Commercially available precursors enable incorporation of 13CH3, 13CH2D, or 13CHD2 groups at the Ile, Leu and Val methyl sites in the protein. The CH3 isotopomer allows characterization of ms-μs motions using relaxation dispersion experiments(8, 9). The CH2D and the CHD2 isotopomers are well suited for deuterium relaxation studies (10), while the CHD2 isotopomer is ideal for carbon relaxation (11). More recently, a procedure to specifically label the Ala methyls has been published (12). This protocol involves using perdeuterated α-ketoisovalerate, succinate and isoleucine to prevent the scrambling of the 13C label. A more general labeling approach uses [1-13C]- or [2-13C]- glucose(13); or [1,3-13C]- or [2-13C]- glycerol(14) as the sole carbon source to express proteins which are labeled at the methyl carbons. A disadvantage of using glucose instead of glycerol as the carbon source is that the conversion of glucose to Pyr dilutes the 13C label by 50%.
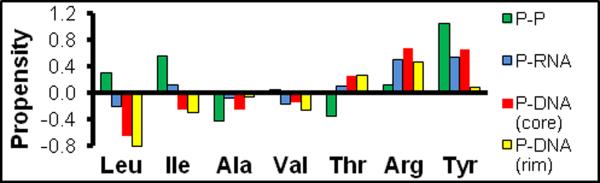
The methyl-labeling techniques developed to-date have focused on Ala, Ile, Leu and Val side-chains, all of which lack hydrogen bonding capabilities. In contrast, Thr can participate in hydrogen bonding as well as nonpolar interactions. Due to the different properties of methyl-containing amino acids, the distribution of these residues differs considerably for protein-protein, protein-RNA, and protein-DNA interfaces (Figure 1). In the case of DNA-protein interfaces, Leu (−.69), Ile (−.25), Ala (−.16), and Val (−.17) are all under-represented, showing negative propensities (given after residue name), while Ser (0.25), Thr (.27), Phe (.35), Trp (.41), Tyr (.46), and Arg (.57) are over-represented at protein-DNA interfaces. Thr residues are found with nearly equal propensity in both the core and rim of protein-DNA interfaces. Consequently, Thr residues potentially provide better probes of DNA- and RNA-protein interfaces than any other methyl containing amino acid currently utilized for NMR studies. Even though Thr methyls have a narrow chemical shift range, their proximity to nucleic acids is expected to result in a broader dispersion of shifts.
Figure 1.
Propensity for selected residues found within protein-protein (P-P), protein-RNA (P-RNA), or protein-DNA (P-DNA) interfacial regions. The P-DNA interface is divided into a core (solvent inaccessible) and a rim region (partially solvent accessible). Propensity is defined as ln(fi/fi°) where fi is the area of ith residue in the interfacial region and fi° is the area occupied by the residue on the remaining surface of the protein. Total P-DNA propensities for each residue are given in the text. Data from (15–17).
Presented here is a technique to uniquely label the methyl group of threonine residues of a protein expressed using a standard Escherichia coli T7 expression host(18) without genetic modification. The technique works in H2O as well as ~100% D2O media. We show that there is no 13C-13C coupling to the adjacent carbon and that 13C-H cross-peaks in the deuterated sample arise from the -13CHD2 isotopomer.
We use [2-13C]-glycerol as the primary carbon source in the bacterial growth medium. The metabolism of glycerol in E. coli is well understood (see Figure S1). Glycerol is converted to Pyr in the glycolytic pathway and then enters the TCA cycle as acetyl-CoA by condensation with oxaloacetate (OAA) to form citrate. Following the 2-C atom from glycerol through the TCA cycle, the labeled carbon becomes either C1 or C4 of OAA. The OAA can then either get converted to Asp or can re-enter the TCA cycle by reacting with another molecule of acetyl-CoA. On re-entering the TCA cycle, both C1 and C4 of OAA will be lost as CO2. Therefore, the 13C label on glycerol either goes to Asp and subsequently to Thr or is lost as CO2. The side-chain carboxyl group of Asp ultimately forms the methyl group on Thr, with the methyl protons arising from the media as well as from the carbon source. In 100% D2O containing media it has been shown that 45% of the Thr methyls are labeled as 13CHD2 while 81% of the Thr Cβ are deuterated(19). The high level of deuteration on the Cβ position leads to favorable relaxation properties in larger proteins.
A number of biosynthetic pathways can lead to a dilution of the 13C label (see Figure S1). Pyruvate can be converted to Ala, Leu, Val and Ile while Asp can be converted to Met and Lys. Thr itself can be converted to Ile. Therefore, inhibiting these pathways is important to prevent dilution of the label. This was accomplished by adding deuterated Ala, α-ketoisovalerate, Ile, and protonated Met and Lys to the media before inducing protein expression (see supporting information for more detail). Since the carbon lost from the TCA cycle due to biosynthetic needs is obtained from CO2, NaH13CO3 was added to the media to enhance labeling (see below). The labeling scheme was tested on the 130 residue RNA binding domain of the transcriptional termination factor rho (rho130) (20). To estimate the 13C incorporation at the methyl group of Thr, Met was labeled by adding 13C methyl labeled Met to the media. The Thr labeling efficiency was tested using the following sources of 13C: a) [2-13C] glycerol b) [2-13C] glycerol and NaH13CO3 (2g/L) c) [2-13C] glycerol and NaH13CO3 (10g/L) and d) NaH13CO3 (10g/L). Media containing 50% and 100% D2O was used to test the contribution of the solvent to protonation of the Thr methyl.
The methyl region of the C-H correlated spectra of uniformly 13C labeled rho130 and the specifically labeled sample (only the Thr and Met methyls labeled) is shown in Figure 2 (panels A and B). The Thr labeling protocol is extremely specific and reduces the intensity of the other methyl peaks by ~95%, significantly reducing the crowding in this region of the spectrum. Figure 2 B also shows the 1-D slice along the carbon dimension of a threonine peak. The lack of any splitting shows that the carbon adjacent to the methyl 13C carbon is not labeled. Figure 2 C shows the spectrum of the deuterated specifically labeled sample acquired using a refocused-INEPT experiment which selects for 13CHD2 signals(11). A C-H HMQC experiment showed that that 13CHD2 isotopomer is the predominant protonated species (approx. 90% 13CHD2 and 10% 13CH2D) when the cells are grown in 100% D2O. In the case of expression in 50% D2O, the relative proportion of 13CH3, 13CH2D and 13CHD2 is approx. 40:40:20, thus some fraction of the hydrogens on the Thr methyl group are supplied directly from the solvent as well as from the protonated carbon source.
Figure 2.
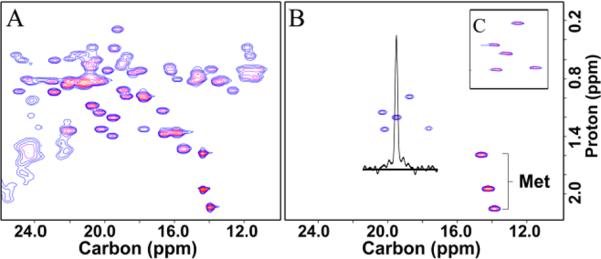
2D 1H-13C spectra of rho130. A) HSQC of uniformly 13C labeled protein, B) HSQC of rho130 specifically labeled at Thr and Met methyl groups, and C) refocused-INEPT experiment on deuterated rho130 sample showing the Thr CHD2 peaks.
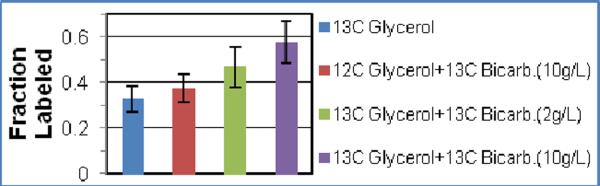
The efficiency of 13C labeling of the Thr methyls using glycerol alone is 32% (Figure 3). If all of the 13C from the 2-C of glycerol was used for Thr synthesis then the γ-methyl would be labeled to a level of 50% (see Figure S1). Numerical simulations show that with 32% labeling, approximately 40% of the carbon flux in the TCA cycle is diverted to biosynthetic needs. Because this carbon is replenished by carboxylation of phosphoenolpyruvate with 12CO2 to produce OAA, the observed labeling efficiency is less than 50%. Since the incorporated CO2 becomes the C4 position in OAA, the methyl labeling efficiency can be increased by adding NaH13CO3 to the media prior to induction. Figure 3 shows that an 80% increase in signal intensity, relative to that obtained with [2-13C] glycerol alone, was obtained on adding 10g/L of NaH13CO3 to the media. Use of NaH13CO3(10g/L) alone is as efficient as using [2-13C] glycerol.
Figure 3.
Mean normalized peak intensities when using different 13C sources. Individual peaks were first normalized to the corresponding peaks in a uniform 13C labeled sample to account for intensity differences due to polarization transfers. Each set was then normalized to the most intense Met resonance. The bars indicates the range of values observed for the 5 Thr residues.
When [2-13C] glycerol is used as the carbon source the carbonyls of Arg, Asp, Asn, Glu, Gln, Leu, Pro, and Thr residues will be selectively labeled. The 13C carbonyl can be used to filter the 2D 1H-15N spectrum (Figure S2), showing only peaks from NH groups preceding the labeled carbonyl. The intensity of the peaks in the filtered spectrum is consistent with the expected labeling pattern reported in earlier studies (13, 14, 21) (Figure S3). Since carbonyl editing allows the identification of the preceding residue type for NH, this information can be used in the mainchain resonance assignment process (22, 23). Note that the residues that are labeled at the carbonyl position with [2-
13C] glycerol are complementary to the set of residues (I, L, Y, V, A, P) identified as being useful for assignment purposes using individual 13C carbonyl labeled amino acids (23).
In addition to selective labeling at the carbonyl position, the use of [2-13C] glycerol also leads to selective labeling of the Cα position (13, 14, 21). An analysis of peak intensities in the Cα filtered 1H-15N spectra of rho130 (Figure S4) shows that this labeling pattern is preserved with the Thr labeling scheme. The simplification of the C-H spectrum facilitates Cα relaxation studies, an important contribution to the characterization of backbone dynamics.
To summarize, the Thr labeling protocol can be easily incorporated into standard protein expression protocols. The labeled methyl carbon is not coupled to the adjacent carbon, and when using 100% deuterated media, 13CHD2 is the predominant species. Also, due to the use of deuterated α-ketoisovalerate, Ala, Met, and Ile, even proteins expressed in H2O based media would have a significantly deuterated methyl background. These characteristics make this protocol ideal for relaxation studies. We note that although the labeling efficiency of the Thr methyl with respect to 13C exceeds 50%, the fraction of 13CHD2 methyls is estimated to be ~25% when 100% D2O is used, reducing the overall efficiency relative to other methyl-labeling schemes. However, the fraction of the 13CHD2 isotopomer can be increased by raising the H2O content of the media. The protocol also selectively enriches the labeling of the carbonyl carbons, providing useful information for resonance assignments. This protocol complements the set of methyl labeling schemes available to the NMR community and should be particularly useful in the study of protein-DNA interactions.
Supplementary Material
ACKNOWLEDGMENTs
We would like to thank Virgil Simplaceanu and Roberto Gil for support of the NMR spectrometers and for help in data collection.
Funding This work was supported by the Mellon College of Science at CMU and an NIH Merit grant 5R37-GM029207 to L.J-J.
Footnotes
ASSOCIATED CONTENT Summary of relevant metabolic pathways and additional detail regarding the labeling procedure is available. In addition, 2D-15N-1H HSQC spectra of rho130, without editing and edited by N-Cα and N-CO coupling, and peak intensities in the edited spectra, are given in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 2.Kay LE. NMR studies of protein structure and dynamics. J Magn Reson. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Tzakos AG, Grace CR, Lukavsky PJ, Riek R. NMR techniques for very large proteins and RNAs in solution. Annu Rev Biophys Biomol Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 4.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H-13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 5.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. J Mol Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 6.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 7.Ruschak AM, Velyvis A, Kay LE. A simple strategy for 13C,1H labeling at the Ile-g2 methyl position in highly deuterated proteins. J Biomol NMR. 2010;48:129–135. doi: 10.1007/s10858-010-9449-1. [DOI] [PubMed] [Google Scholar]
- 8.Tugarinov V, Kay LE. Stereospecific NMR assignments of prochiral methyls, rotameric states and dynamics of valine residues in malate synthase G. J Am Chem Soc. 2004;126:9827–9836. doi: 10.1021/ja048738u. [DOI] [PubMed] [Google Scholar]
- 9.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: Application to a 723-residue enzyme. J Am Chem Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 10.Tugarinov V, Ollerenshaw JE, Kay LE. Probing side-chain dynamics in high molecular weight proteins by deuterium NMR spin relaxation: An application to an 82-kDa enzyme. J Am Chem Soc. 2005;127:8214–8225. doi: 10.1021/ja0508830. [DOI] [PubMed] [Google Scholar]
- 11.Ishima R, Louis JM, Torchia DA. Transverse 13C Relaxation of CHD2 Methyl Isotopmers To Detect Slow Conformational Changes of Protein Side Chains. J Am Chem Soc. 1999;121:11589–11590. [Google Scholar]
- 12.Ayala I, Sounier R, Use N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR. 2009;43:111–119. doi: 10.1007/s10858-008-9294-7. [DOI] [PubMed] [Google Scholar]
- 13.Lundstrom P, Teilum K, Carstensen T, Bezsonova I, Wiesner S, Hansen DF, Religa TL, Akke M, Kay LE. Fractional 13C enrichment of isolated carbons using [1-13C]- or [2- 13C]-glucose facilitates the accurate measurement of dynamics at backbone Ca and side-chain methyl positions in proteins. J Biomol NMR. 2007;38:199–212. doi: 10.1007/s10858-007-9158-6. [DOI] [PubMed] [Google Scholar]
- 14.Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H. Assigning large proteins in the solid state: a MAS NMR resonance assignment strategy using selectively and extensively 13C-labelled proteins. J Biomol NMR. 2009;44:245–260. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- 15.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 16.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas S, Guharoy M, Chakrabarti P. Dissection, residue conservation, and structural classification of protein-DNA interfaces. Proteins. 2009;74:643–654. doi: 10.1002/prot.22180. [DOI] [PubMed] [Google Scholar]
- 18.Studier FW, Moffatt BA. Use of bacterophage-T7 RNA-polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 19.Otten R, Chu B, Krewulak KD, Vogel HJ, Mulder FA. Comprehensive and cost-effective NMR spectroscopy of methyl groups in large proteins. J Am Chem Soc. 2010;132:2952–2960. doi: 10.1021/ja907706a. [DOI] [PubMed] [Google Scholar]
- 20.Briercheck DM, Wood TC, Allison TJ, Richardson JP, Rule GS. The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions. Nat Struct Biol. 1998;5:393–399. doi: 10.1038/nsb0598-393. [DOI] [PubMed] [Google Scholar]
- 21.LeMaster DM, Kushlan DM. Dynamical Mapping of E. coli Thioredoxin via 13C NMR Relaxation Analysis. J Am Chem Soc. 1996;118:9255–9264. [Google Scholar]
- 22.McCallum SA, Hitchens TK, Rule GS. Solution structure of the carboxyl terminus of a human class Mu glutathione S-transferase: NMR assignment strategies in large proteins. J Mol Biol. 1999;285:2119–2132. doi: 10.1006/jmbi.1998.2428. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi K, Ng E, Malia TJ, Wagner G. 1-13C amino acid selective labeling in a (HN)-2H-15N background for NMR studies of large proteins. J Biomol NMR. 2007;38:89–98. doi: 10.1007/s10858-007-9152-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.