SUMMARY
The histone lysine methyltransferase NSD2 (MMSET/WHSC1) is implicated in diverse diseases and commonly overexpressed in multiple myeloma due to a recurrent t(4;14) chromosomal translocation. However, the precise catalytic activity of NSD2 is obscure, preventing progress in understanding how this enzyme influences chromatin biology and myeloma pathogenesis. Here we show that dimethylation of histone H3 at lysine 36 (H3K36me2) is the principal chromatin-regulatory activity of NSD2. Catalysis of H3K36me2 by NSD2 is sufficient for gene activation. In t(4;14)-positive myeloma cells, the normal genome-wide and gene-specific distribution of H3K36me2 is obliterated, creating a chromatin landscape that selects for a transcription profile favorable for myelomagenesis. Catalytically active NSD2 confers xenograft tumor formation upon t(4;14)-negative cells, and promotes oncogenic transformation of primary cells in an H3K36me2-dependent manner. Together our findings establish H3K36me2 as the primary product generated by NSD2, and demonstrate that genomic disorganization of this canonical chromatin mark by NSD2 initiates oncogenic programming.
INTRODUCTION
Histone lysine methylation signaling is a principal chromatin-regulatory mechanism that influences fundamental nuclear processes (Kouzarides, 2007). Lysine (K) residues can accept up to three methyl groups to form mono-, di-, and tri-methylated derivatives (Kme1, Kme2, and Kme3, respectively). Methylated histone species are sensed and linked to downstream biological functions by methyllysine-binding proteins, in a manner specified by the extent and sequence context of the methylation event (Taverna et al., 2007). The human genome encodes greater than fifty predicted protein lysine methyltransferases (PKMTs), of which several are deregulated in human disease (Kouzarides, 2007). To understand how this diverse nuclear signaling network influences chromatin biology and disease, it is essential to elucidate the precise activity of individual PKMTs on their substrates.
The vast majority of PKMT enzymes contain a conserved catalytic SET domain (Dillon et al., 2005). NSD2 (also named MMSET and WHSC1) is a SET domain-containing PKMT implicated in diverse human diseases. For example, NSD2 haploinsufficiency is implicated in the developmental disorder Wolf Hirschhorn syndrome (WHS) (Stec et al., 1998), which is characterized by growth and mental retardation, congenital heart defects, and antibody deficiencies, and NSD2-deficeint mice exhibit a spectrum of defects resembling WHS (Nimura et al., 2009).
NSD2 is also implicated in the pathogenesis of the hematologic malignancy multiple myeloma (MM) (Anderson and Carrasco, 2011). MM is the 2nd most common blood cancer, accounting for 1% of all cancers and 2% of cancer deaths in the United States (Chng et al., 2007; Palumbo and Anderson, 2011). 15–20% of MM patients carry a translocation between chromosomes 4 and 14 [t(4;14)(p16.3;q32)], which places the transcription of two genes, NSD2 and FGFR3, under the control of strong IgH intronic Eμ enhancer and 3′ enhancer, respectively, and leads to aberrant upregulation of these two genes (Chesi et al., 1998). Notably, overexpression of NSD2, and not FGFR3, is thought to be important for t(4;14)-mediated myeloma pathogenesis (Keats et al., 2003; Santra et al., 2003). Thus, NSD2 plays an important role during mammalian development and its overexpression is implicated in cancer.
The physiologic catalytic activity of NSD2 is obscure, particularly because there are several conflicting reports in the literature. Specifically, NSD2 and/or NSD2 isoforms have been proposed to generate numerous different histone marks, including trimethylation of H3 at lysines K4 (H3K4me3), K27 (H3K27me3), and K36 (H3K36me3), trimethylation of H4 at lysine K20 (H4K20me3), dimethylation of H4 at lysine 20 (H4K20me2), and dimethylation of H3 at lysine 36 (H3K36me2) (Kim et al., 2008; Li et al., 2009; Marango et al., 2008; Martinez-Garcia et al., 2011; Nimura et al., 2009; Pei et al., 2011). Biologically, NSD2 is reported to repress transcription via generation of H3K36me3 (Nimura et al., 2009), H4K20me3 (Marango et al., 2008), or H2K27me3 (Kim et al., 2008), and to mediate localization of 53BP1 to DNA damage foci through generation of H4K20me2 (Pei et al., 2011). Thus, the true enzymatic and chromatin-regulatory functions of NSD2 are unclear.
Here we employ multiple independent biochemical and cellular approaches to investigate and resolve the discrepancies regarding NSD2 enzymatic activity. We find that the principal physiologic activity of NSD2 at chromatin is dimethylation of H3K36, and in the process rule out generation of H3K36me3, H4K20me2, and several other putative methyl products of NSD2. We also elucidate the mechanism by which NSD2 – via catalysis of H3K36me2 – initiates oncogenic programming, revealing a new link between disruption of chromatin homeostasis and cancer.
RESULTS
NSD2 exclusively mono- and di-methylates nucleosomal histone H3 at lysine 36 in vitro
To understand NSD2 enzymatic specificity, we determined the methylation activity of the NSD2 SET domain (NSD2SET; see Figure S1A for schematic) using native nucleosomes as physiologically relevant substrates. This analysis revealed that NSD2SET exclusively methylates nucleosomal histone H3, similar to the positive control G9aSET, whereas SET8/PR-Set7 methylates its known substrate H4 (Figure 1A). Further, full-length NSD2 methylates nucleosomal H3, but not the other core histones including H4 (Figure 1B and S1B). Next, methylation reactions with NSD2SET on recombinant nucleosomes (which lack all modifications) were subject to mass spectrometry analysis to identify the specific histone lysine residue substrate(s) of NSD2 and the extent of methylation at modified residue(s) (Figure 1C). Employing this strategy, the only methylation events detected were mono- and di-methylation at H3K36 (Figure 1C; e.g. H4K20 methylation was not detected); these results were independently confirmed by western analysis (Figure 1D; data not shown). NSD2 was reported to repress transcription via a H3K36me3-specific methylation activity (Nimura et al., 2009). However, NSD2 did not trimethylate H3K36 on recombinant nucleosomes (Figure 1C and 1D). Furthermore, NSD2SET-mediated methylation increased levels of H3K36me1 and H3K36me2, but not H3K36me3, on native nucleosomes (Figure 1E). In contrast, H3K36me3 levels on recombinant and native nucleosomes were specifically increased by the known H3K36me3 lysine methyltransferase, SETD2 (Edmunds et al., 2008) (Figure 1F; data not shown). Full-length NSD2 and the major NSD2 variant RE-IIBP demonstrated the same activity as NSD2SET, generating H3K36me1 and H3K36me2, but not H3K36me3, on nucleosomes (Figure 1E; Figure S1C and S1D). Thus, in vitro NSD2 mono- and di-methylates, but does not trimethylate, H3K36.
Figure 1. NSD2 mono- and di-methylates H3K36 in vitro.
(A) NSD2SET exclusively methylates H3 on native nucleosomes. Autoradiogram of methylation assays on HeLa-purified nucleosomes using the indicated recombinant enzymes. Coomassie stain of histones is shown below.
(B) Full-length NSD2 exclusively methylates H3 on native nucleosomes. Autoradiogram and Coomassie of methylation assays as in (A) using full-length NSD2 purified from Sf9 cells. Lane 1: Nucleosome (Nu) only, lane 2: control reaction with IPed material from non-transduced Sf9 cells. Left panel: Coomassie stain of purified full-length NSD2 protein.
(C and D) Mono- and di-methylation of H3K36 on recombinant nucleosomes by NSD2. (C) Quantitative mass spectrometry of NSD2SET methylation assays on recombinant nucleosomes. Control reaction lacks NSD2SET. Mass spectrum shows H3K36me0 (Left) peptide levels decrease whereas H3K36me1 (middle) and H3K36me2 (right) peptides are detected upon NSD2SET methylation (see extended methods). No other methylation events (e.g. H3K36me3, H4K20me2) were detected. Arrows: locations where modified peptides are expected in the -NSD2SET reaction. (D) Western analysis of NSD2SET methylation assays as in (C) using the indicated antibodies.
(E) NSD2 generates H3K36me1 and H3K36me2 on native nucleosomes. Western analysis with the indicated antibodies of methylation assays with NSD2SET (left panel) and full-length NSD2 (right) on HeLa nucleosomes. Control reaction lacks NSD2.
(F) Trimethylation of H3K36 on native nucleosomes by SETD2SET. Western analysis of methylation assays of NSD2SET and SETD2SET on HeLa nucleosomes. Control reaction has no enzyme.
(G) NSD2 does not methylate at H4K20. Western analysis of methylation assays of NSD2SET on recombinant H4. Control reaction lacks NSD2SET. Total H4 is shown as a loading control.
NSD2 was recently proposed to regulate localization of 53BP1 to DNA damage foci via dimethylation of H4K20 (Pei et al., 2011). However, our data reveal that NSD2 (full-length, NSD2SET, and RE-IIBP) does not methylate H4K20, in contrast to the positive control enzyme SET8, as determined by immunoblot analysis and mass spectrometry on multiple substrates including H4 peptides, recombinant H4, purified H4, recombinant nucleosomes, and native nucleosomes (Figure 1C–E, 1G, S1B–E; data not shown). In addition, NSD2 and RE-IIBP do not methylate the previously implicated substrate sites H3K4 (Marango et al., 2008) and H3K27 (Kim et al., 2008) (Figure 1D; 1E, S1C; S1D; data not shown). We note that peptide arrays were used to test the epitope and methyl-state specificity of antibodies used in our study (Bua et al., 2009) (Figure S1F; data not shown). Thus, under our experimental conditions, our results do not support previous claims that NSD2 can methylate H3K4, H3K27, or H4K20 substrates. Taken together, we conclude that in vitro NSD2 is principally an H3K36 mono- and di-methylase.
Dimethylation of H3K36 is the principal catalytic function of NSD2 on histones in cells
Overexpression of NSD2 in 293T cells resulted in increased global H3K36me2 levels, but had no effect on the levels of H3K36me3, H4K20me2, and several other marks (Figure 2A). Next, NSD2 activity was investigated in KMS11 cells, a model t(4;14)+ myeloma cell line in which the IgH enhancer drives overexpression of NSD2 (Lauring et al., 2008). RNA interference (RNAi)-mediated knockdown of NSD2 expression in KMS11 cells, using two independent shRNAs (Figure S1A), led to a specific decrease in levels of H3K36me2, but not other histone methyl marks including H4K20me2 and H3K36me3 (Figure 2B). In contrast, global levels of H3K36me3, but not H3K36me2, were decreased in SETD2-depleted KMS11 cells (Figure 2C). In addition, NSD2 depletion in HT1080 and U2OS cancer cell lines decreased H3K36me2 levels without changing H3K36me3 levels (Figure 2D).
Figure 2. NSD2 generates H3K36me2 in cells.
(A) NSD2 overexpression increases global H3K36me2 levels in cells. Western analysis with the indicated antibodies of whole cell extract (WCE) from 293T cells transfected with full-length NSD2 or control vector. Total H3 is shown as a loading control.
(B) Depletion of NSD2 by RNAi specifically decreases global H3K36me2 level in t(4;14)+ myeloma cells. Western analysis as in (A) of WCE from t(4;14)+ KMS11 myeloma cells stably expressing control shRNA or two independent shRNAs targeting NSD2.
(C) Depletion of SETD2 reduces H3K36me3 levels in myeloma cells. Western analysis of WCE as in (B) from KMS11 cells expressing the indicated shRNAs.
(D) Depletion of NSD2 specifically decreases global H3K36me2 levels in non-myeloma cancer cell lines. Western analysis with the indicated antibodies of WCE from HT1080 (top panel) and U2OS (bottom panel) cells transfected with control siRNA or two independent siRNAs targeting NSD2.
(E) Schematic of KMS11, TKO, and NTKO cell lines (see Lauring et al., 2008). Homologous recombination in t(4;14)+ KMS11 cells targeting either inactivation of the t(4;14)-associated NSD2 allele (TKO) or the wild-type NSD2 allele (NTKO).
(F), (G) and (H) Loss of t(4;14)-associated NSD2 expression in myeloma cells leads to depletion of H3K36me2, but not other histone methylation marks. (F) Western analysis of WCE from KMS11, TKO and NTKO cells using the indicated antibodies. (G) Long exposure and quantitation of NSD2 western analysis in the indicated cell lines. (H) Quantitative mass spectrometry of the relative amounts of the indicated methylation events in KMS11 cells versus two independent TKO cell lines. Fold Δ: ratio of KMS11 over TKO samples. ± standard deviation from 2 replicas analyzed in triplicate.
KMS11 cells in which the t(4;14)-driven NSD2 allele has been inactivated by homologous recombination (translocation knock-out (TKO) cells) have attenuated tumorigenicity compared to parental KMS11 cells or cells with targeted inactivation of only the wild-type allele (non-translocation knock-out (NTKO); see Figure 2E) (Lauring et al., 2008). In two independent TKO cell lines, which lack the t(4;14) NSD2 allele, global H3K36me2 levels were lower than in KMS11 and NTKO cells, and no changes in H3K36me3 or H4K20me2 levels were observed (Figure 2F). Accordingly, NSD2 protein abundance was 6 to 7 fold higher in KMS11 cells than in TKO cell lines (Figure 2G and S1G).
An unbiased quantitative mass spectrometry screen, comparing levels of histone methyl marks between KMS11 and TKO cells, revealed that H3K36me2 levels are 3-fold higher in KMS11 cells versus TKO cells (Figure 2H). Conversely, levels of unmethylated H3K36 and H3K36me1 are lower by a factor of two in KMS11 cells relative to TKO cells, suggesting conversion of these marks to H3K36me2 in KMS11 cells (Figure 2H). No other significant differences in histone methylation were observed (Figure 2H). Thus, we conclude that NSD2 does not trimethylate H3K36 or dimethylate H4K20, and NSD2 is required for the majority of H3K36me2 present in several cancer cell types, including t(4;14)+ myeloma cells.
t(4;14)+-driven NSD2 overexpression triggers genome-wide reprogramming of H3K36me2
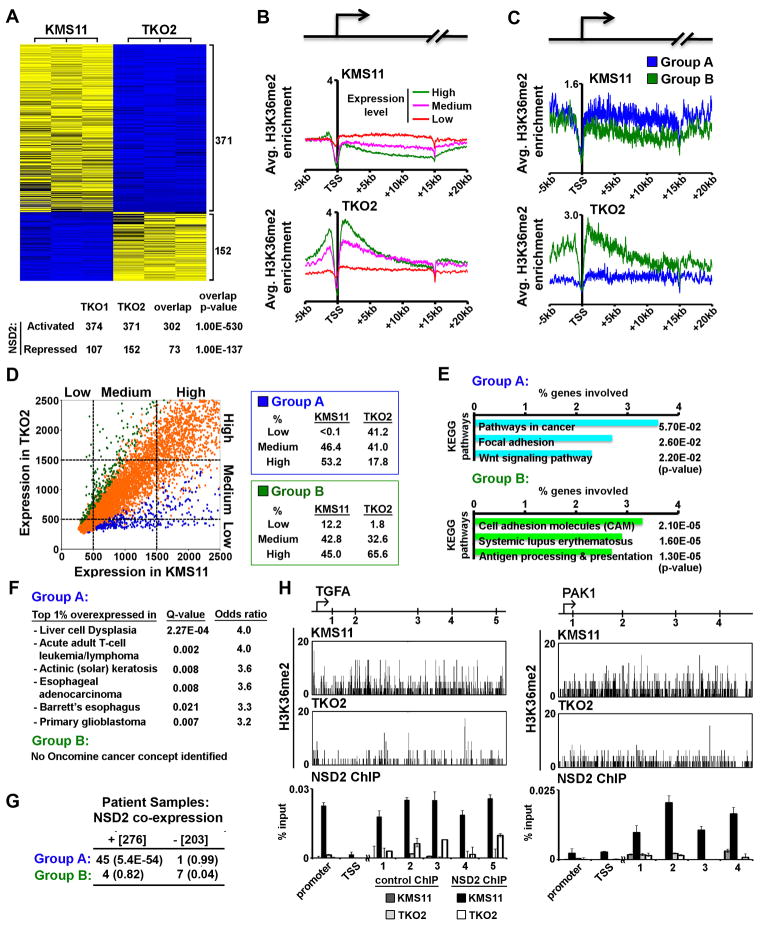
We next used ChIP-seq (chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing) to determine the genomic distribution of H3K36me2 in the absence or presence of the t(4;14) translocation (TKO2 and KMS11 cells, respectively). In TKO2 cells, which express NSD2 only from one wild-type allele (see Figure 2E, 2G, S1G), H3K36me2 signal was preferentially enriched in intragenic versus intergenic regions on a representative chromosome (Figure 3A, bottom panel for higher resolution image). In contrast, in t(4;14)+ KMS11 cells the H3K36me2 signal does not map to gene bodies but rather is dispersed throughout the chromosome (Figure 3A). On a genome-wide scale, in two independent H3K36me2 ChIP-seq experiments, the majority of H3K36me2 peaks in TKO2 cells map to intragenic regions, whereas in KMS11 cells, H3K36me2 peaks are largely found within intergenic regions (Figure 3B; Figure S2A; independent biological ChIP-seq replica are shown in Supplementary Figures). Despite the high global levels of H3K36me2 in KMS11 cells, peak-calling programs identified relatively few peaks due to the widespread enrichment of H3K36me2 throughout the KMS11 genome (e.g. on chromosome 21, 35 peaks in KMS11 cells versus 236 peaks in TKO cells (Figure 3A)). To circumvent potential computational bias in calling peaks, the genomic distribution of the raw number of H3K36me2-associated reads per nucleotide was determined. As shown in Figure 3C, independent of read numbers, H3K36me2-bound nucleotides in KMS11 cells are found dispersed throughout the genome, with an intergenic/intragenic ratio paralleling the intergenic/intragenic composition of the whole genome (Figure 3C; Figure S2B). In contrast, in TKO2 cells, H3K36me2-bound nucleotides, like H3K36me2 peaks, preferentially map within gene bodies (Figure 3C; Figure S2B).
Figure 3. t(4;14)-driven NSD2 overexpression obliterates the normal genome-wide and gene-specific distribution of H3K36me2.
(A), (B) and (C) t(4;14)-associated NSD2 overexpression obliterates the normal genome-wide distribution of H3K36me2. (A) H3K36me2 ChIP-seq signals from KMS11 and TKO2 cells across a representative chromosome are shown. Top two insets: H3K36me2 distribution on chromosome 21 in the indicated cell lines. H3K36me2 peak #s are indicated. Bottom two insets: Higher resolution track of H3K36me2 ChIP signal from 23–29 megabasepairs (mb) on chromosome 21. Genes are marked below the track. (B) Percent of H3K36me2-enriched peaks localized to either intergenic (yellow) or intragenic (purple) regions throughout the genome in KMS11 and TKO2 cells. Number of H3K36me2 peaks in each cell line is shown. (C) Distribution of nucleotides associated with the indicated number of H3K36me2 ChIP-seq reads mapping to intergenic (yellow) and intragenic (purple) regions in KMS11 and TKO2 cells. p-values are calculated for the indicated read thresholds. The intergenic versus intragenic composition of the whole genome is shown.
(D) Normal H3K36me2 distribution within genes is abolished in t(4;14)+ myeloma cells. Average H3K36me2 ChIP signal across 20,910 annotated genes in KMS11 (left; blue) and TKO2 (right; red) cells. The transcribed bodies of all genes were normalized to 15 kilobasepairs (kb). The arrow in the schematic illustrates the TSS.
To specifically investigate H3K36me2 distribution within gene bodies, the average signal of H3K36me2 across 20,910 annotated genes was determined in TKO2 and KMS11 cells. In TKO2 cells, H3K36me2 signal is modest in the promoter regions, peaks proximal to the transcription start site (TSS), and gradually decays downstream into the transcribed body away from the TSS (Figure 3D, right inset; Figure S2C). In contrast, in KMS11 cells, there is no prominent TSS-proximal area of H3K36me2 enrichment and little overall variance in H3K36me2 signal intensity across the average gene unit (Figure 3D, left inset; Figure S2C). The results in TKO2 cells are consistent with a ChIP-chip-based genome-wide binding study in Drosophila of H3K36me2 distribution (Bell et al., 2007). Thus, in mammalian cells we propose that H3K36me2 normally maps to gene bodies, and t(4;14)-driven overexpression of NSD2 disrupts the physiologic genomic organization of H3K36me2.
NSD2 promotes transcription
Previous studies have implicated NSD2 as a negative regulator of transcription (Kim et al., 2008; Marango et al., 2008; Nimura et al., 2009), though H3K36me2 is generally associated with gene activation (Bannister et al., 2005; Kizer et al., 2005; Krogan et al., 2003). To investigate the relationship between NSD2 and transcription, comparative gene expression analysis between KMS11 cells and TKO cells and between KMS11 cells stably expressing control shRNA or two independent NSD2-targeting shRNAs were determined (Figure 4A and S3A). Employing a linear model statistical analysis (Smyth et al., 2003), NSD2 expression correlated with upregulation for the large majority of genes displaying statistically significant expression changes (Figure 4A and S3A). Real-time PCR independently confirmed the differential expression of a number of genes identified in the microarray analyses (Figure S3B). Based on these data we conclude that NSD2 expression is linked to transcription activation.
Figure 4. NSD2 activates transcription and promotes an oncogenic program in KMS11 cells.
(A) Depletion of NSD2 by targeted inactivation of t(4;14)-associated NSD2 allele in KMS11 cells correlates with global downregulation of transcription. Genome-wide expression profiling of KMS11 cells in comparison to the two TKO cell lines. Top: heatmap representation of differentially expressed genes in TKO2 versus KMS11 cells (n=3). Numerical expression values for each gene were median centered and scaled to range from −1.0 (blue; low expression) to 1.0 (yellow; high expression). Bottom: overlap of differentially expressed genes between the two TKO cell lines. Activated: upregulated in KMS11/TKO cells; repressed: downregulated in KMS11/TKO cells. p-values: statistical significance of the overlap.
(B) Positive correlation between H3K36me2 enrichment and gene expression is abolished by NSD2 overexpression. Average H3K36me2 profile in the indicated cell lines across the 20,910 annotated genes separated into three groups based on absolute expression units from the datasets described in (A).
(C) In KMS11 cells, H3K36me2 levels are elevated at NSD2-dependent upregulated genes. The average H3K36me2 profile in the indicated cell lines across the top 500 genes upregulated in KMS11/TKO2 cells (Group A: blue) and the top 500 genes downregulated in KMS11/TKO2 cells (Group B: green).
(D) NSD2 promotes expression of normally silent genes. Top: scatter plot of absolute gene transcript levels for 20,910 annotated genes in KMS11 versus TKO2 cells. The percent of Group A (blue) and Group B (green) genes that fall within the 3 expression quantile categories (low, medium, and high) is shown at right.
(E) t(4;14)-driven NSD2 overexpression promotes expression of genes involved in cancer-signaling pathways. KEGG pathway analysis of group A and group B genes using p-value cutoff=0.1 and sorted by the percentage of genes within a functional group.
(F) Genes upregulated in t(4;14)+ myeloma cells constitute a common signature in diverse cancers. Oncomine cancer concept analysis of group A and B genes with genes overexpressed in different cancers using Q-value cutoff=0.05 and odds ratio cutoff=3. Significant cancer concepts sorted by the odds ratio.
(G) Genes upregulated in t(4;14)+ KMS11 cells are co-expressed with NSD2 in myeloma patient samples. Table: number of genes that positively (276) or negatively (203) correlate with NSD2 expression in the Multiple Myeloma Research Consortium patient sample database and the overlap with group A and B genes. p-values: statistical significance of the overlap.
(H) NSD2 directly binds at target genes. ChIP analysis of NSD2 occupancy across the TGFA and PAK1 genes in KMS11 and TKO2 cells. Top: schematics of the two genes. Arrow indicates the TSS and numbers the location of ChIP primer pairs. Middle: snapshot of H3K36me2 ChIP-seq signal at TGFA and PAK1 in the indicated cell lines. Bottom: NSD2 ChIP signals at the indicated primer pairs. Protein A beads used in control ChIP. Error bars indicate the standard error of the mean (s.e.m.) from 3 experiments.
We next determined the relationship between NSD2-mediated H3K36me2 enrichment and gene expression level. Using the expression profiles obtained from KMS11 and TKO2 cells, the 20,910 annotated genes present in the data sets were partitioned into high, medium, and low expression groups based upon absolute expression level and the average H3K36me2 distribution for each group was determined. In TKO2 cells, a clear correlation was observed between H3K36me2 and gene expression: the most highly expressed genes have the highest H3K36me2 signal and the lowest expressed genes have little H3K36me2 enrichment (Figure 4B, bottom panel; Figure S3C). These results provide genome-wide evidence that H3K36me2 localization at gene bodies positively correlates with transcription levels in mammalian cells.
In KMS11 cells, high H3K36me2 signal did not correlate with gene expression, indicating that H3K36me2 is not strictly required for transcription (Figure 4B, top panel; Figure S3C). However, the observation of t(4;14)-dependent changes in cellular transcription profiles suggests that at a subset of genes, localized H3K36me2 enrichment is sufficient to activate expression. We therefore analyzed H3K36me2 distribution within genes that are differentially expressed in KMS11 cells relative to TKO2 cells, grouped into the top 500 upregulated genes (group A) or the top 500 downregulated genes (group B). As shown in Figure 4C, within KMS11 cells the average H3K36me2 signal is higher in group A than group B, and within TKO2 cells the average H3K36me2 signal is higher in group B than group A (also see Figure S3D). These findings indicate that in KMS11 cells, H3K36me2 enrichment at a distinct set of genes positively correlates with transcription (i.e. group A versus group B). Further, a scatter plot comparing absolute expression levels for all 20,910 genes in KMS11 versus TKO2 cells revealed that a large fraction of group A genes are expressed at low levels in TKO2 cells (Figure 4D). Together, these results argue that (i) H3K36me2 is sufficient but not necessary to induce transcription, and (ii) in KMS11 cells, NSD2-mediated localized elevation of H3K36me2 induces transcription at normally inert genes.
NSD2 promotes an oncogenic gene expression program
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was employed to characterize the biological signaling pathways associated with t(4;14)-driven NSD2 overexpression. The top functional categories of group A genes included cancer-associated and cell migration signaling pathways (Figure 4E). In contrast, group B genes are associated with immunologic-related B cell functions (Figure 4E). Moreover, the signature of upregulated genes comprising group A, but not group B genes, overlaps with the transcriptional signature of several cancers present in the Oncomine Concept Database (Figure 4F). Finally, group A genes show significant overlap with genes that are co-upregulated with NSD2 in myeloma patient samples (Figure 4G and Figure S3E). Thus, NSD2 overexpression in KMS11 cells might alter the balance of transcriptional programs away from normal plasma cell gene expression pathways to activation of pathways that promote myelomagenesis.
Next, a systematic quantitative direct ChIP strategy was used to determine if group A genes are direct targets of NSD2. In all regions spanning four oncogenes (TGFA, PAK1, MET, and RRAS2) that have greater H3K36me2 signal in KMS11 cells than in TKO2 cells, NSD2 binding was significantly higher in KMS11 cells than in TKO2 cells (Figure 4H; Figure S3F). Together, these data argue for a model in which t(4;14)-driven NSD2 overexpression leads to aberrant enrichment of H3K36me2 at normally silent cancer-associated genes, triggering increased expression of these oncogenes.
NSD2 catalytic activity promotes transcriptional activation at oncogenic loci
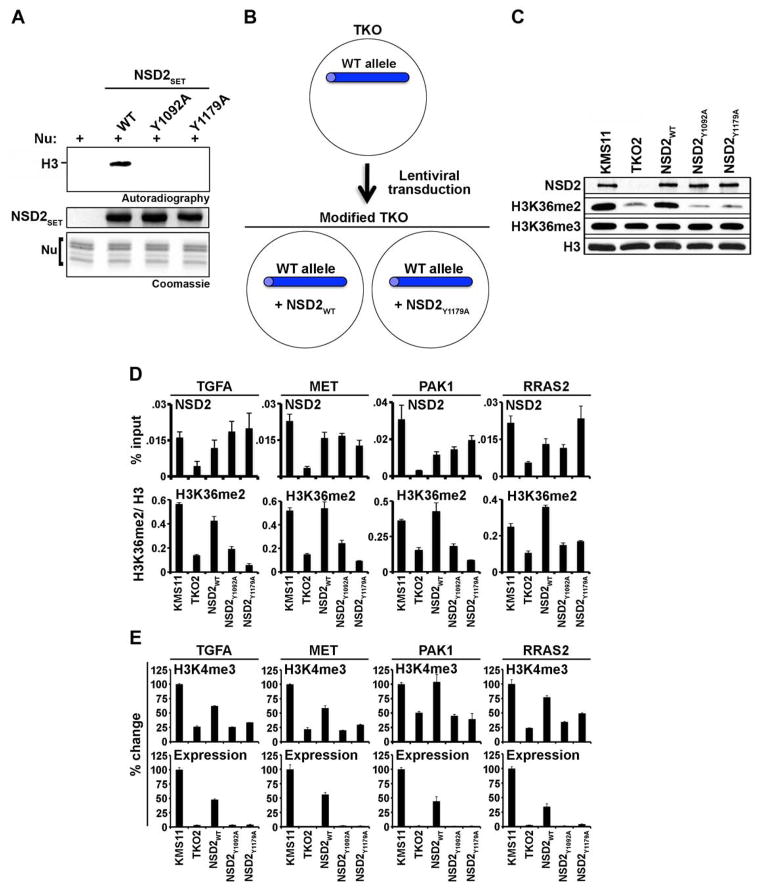
To test the specific role of NSD2 methylation activity in aberrant oncogene induction, we screened for NSD2 catalytic mutants based on sequence homology with other SET domain-containing proteins (Dillon et al., 2005)(Figure S1A). NSD2 proteins harboring several amino acid substitutions reported in the literature to inactivate NSD2 (Marango et al., 2008; Martinez-Garcia et al., 2011; Pei et al., 2011) retained robust methylation activity (Figure S4A; data not shown). Instead, a tyrosine to alanine substitution at amino acid Y1092 or Y1179 (Y1092A and Y1179A, respectively) abolished NSD2 methylation of nucleosomes in vitro (Figure 5A). To model t(4;14)-driven expression of NSD2, a reconstitution system was established in which TKO2 cells were complemented with NSD2WT, NSD2Y1092A, or NSD2Y1179A (cell lines named NSD2WT, NSD2Y1092A, or NSD2Y1179A, respectively; see schematic Figure 5B) to restore NSD2 amounts to those detected in KMS11 cells (Figure 5C). Only NSD2WT, but not the NSD2 catalytic mutants, reconstituted global H3K36me2 signal to the amounts detected in KMS11 cells (Figure 5C).
Figure 5. NSD2 catalytic activity is required for transcriptional activation at oncogenic loci.
(A) Identification of catalytically inactive NSD2 mutants. Top: autoradiogram of in vitro methylation assay with NSD2SET and the two mutants (Y1092A and Y1179A) on native nucleosomes. Coomassie stain of NSD2SET proteins (middle panel) and nucleosomes (bottom panel) is shown.
(B) Schematic of NSD2 cellular reconstitution system. Full-length NSD2 or catalytically inactive NSD2 (NSD2Y1092A, and NSD2Y1179A) were introduced into TKO2 cells by lentiviral transduction.
(C) Complementation of TKO2 cells with NSD2WT but not catalytically inactive NSD2 increases global H3K36me2 to KMS11 levels. Western analysis of WCE from KMS11 cells, TKO2 cells, and TKO2 cells stably transduced with wild-type or catalytically inactive NSD2.
(D and E) Complementation of TKO2 cells with NSD2WT but not with catalytic mutants increases H3K36me2 levels at target oncogenes and activates expression of these genes. (D) ChIP analyses of NSD2 occupancy (top; y axis: %input) and H3K36me2 signal (bottom; y axis: H3K36me2/H3) at four genes at the location indicated in Figures 4H and S3F: TGFA (#4), MET (#2), PAK1 (#2) and RRAS2 (#5). Error bars indicate s.e.m. from 3 experiments. (E) ChIP analyses of H3K4me3 levels at promoter regions (top) and quantitative RT-PCR analysis of mRNA transcripts (bottom) of the genes in (D).
At chromatin, NSD2 occupancy at t(4;14)-associated NSD2-target genes TGFA, MET, PAK1 and RRAS2 was partially to fully reconstituted in NSD2WT, NSD2Y1092A, or NSD2Y1092A cell lines (Figure 5D). However, only NSD2WT, but not the catalytic mutants, reconstituted H3K36me2 ChIP signal at the four genes to levels approaching those observed in KMS11 cells (Figure 5D). Moreover, co-occurrence of NSD2 binding and H3K36me2 elevation was associated with H3K4me3 promoter enrichment and transcription of the NSD2-target genes (Figure 5E). These data further support a direct role for NSD2 dimethylation of H3K36 in activating transcription and argue that NSD2 overexpression can change the state of chromatin from an inert to active form that promotes transcription.
Catalytically active NSD2 restores tumorigenicity of t(4;14)-negative myeloma cells
KMS11 cells proliferate more rapidly and grow more aptly in an anchorage-independent environment relative to TKO2 cells (Lauring et al., 2008). Moreover, proliferation of RNAi-mediated NSD2-depleted KMS11 cells is attenuated relative to control cells, suggesting NSD2 dosage regulates cancer cell phenotypes (Figure S4B). Consistent with the RNAi results, complementation of TKO2 cells with NSD2WT, but not the NSD2 catalytic mutants, increased the cellular proliferation rate to match that of KMS11 cells (Figure 6A). In addition, NSD2WT cells, but not TKO2 and NSD2Y1179A cells, form colonies in methylcellulose (a measure of anchorage-independent growth) with efficiency comparable to KMS11 cells (Figure 6B).
Figure 6. Catalytically active NSD2 confers tumorigenicity upon t(4;14)− negative cells.
(A) Complementation with NSD2WT but not catalytically inactive NSD2 accelerates the proliferation rate of TKO2 cells. Cell numbers in the indicated lines were determined over 9 days. Error bars indicate s.e.m. from 3 independent experiments.
(B) Complementation of TKO2 cells with NSD2WT but not with NSD2Y1179A promotes anchorage-independent growth. The ability of the indicated cells lines to grow colonies in methylcellulose is shown. Bar graph indicates # of colonies/per field 21 days after seeding. Error bars indicate s.e.m. from 3 independent experiments.
(C and D) Catalytically active NSD2 confers xenograft tumor formation and invasion capacity upon t(4;14)-negative cells. (C) KMS11, TKO2, NSD2, and NSD2Y1179A cell lines stably expressing GFP-luciferase biomarkers were introduced into SCID/Beige mice via intraperitoneal injection. Top: western analysis of WCE using anti-GFP antibodies. Total H3 is shown as a loading control. Middle: # of tumor-bearing mice at day 28 from 2 independent experiments. Bottom: representative bioluminescent images of mice 4 weeks after injection. (D) Cell lines as in (C) were introduced into non-irradiated SCID/Beige mice via intravenous tail vein injection. Top: western analysis of WCE as in (C). Middle: # of tumor-bearing mice at day 28. Bottom: representative bioluminescent images of mice 4 weeks after injection.
A mouse xenograft model system was used to assess the role of NSD2 methylation activity in tumor formation in vivo. SCID/Beige mice were injected intraperitoneally with KMS11, TKO2, NSD2WT, and NSD2Y1179A cells stably expressing GFP-luciferase biomarkers to allow for in vivo live-cell imaging (Creusot et al., 2008). 28 days after injection – in two independent experiments and in multiple mice – KMS11 and NSD2WT cells formed tumors in the peritoneal cavity, whereas TKO2 and NSD2Y1179A cells failed to grow (Figure 6C). In myeloma disease, proliferation of malignant plasma cells within the bone marrow (BM) compartment eventually overwhelms and disrupts normal BM function (Anderson and Carrasco, 2011). The role of NSD2 in BM seeding and invasion was determined in non-irradiated SCID/Beige mice injected intravenously with the biomarked cell lines. In multiple mice, the signal from both TKO2 and NSD2Y1179A cells was not detected one week after injection (Figure 6D and S4C). In contrast, in 5/5 injections with KMS11 cells and 4/5 injections with NSD2WT cells, the signal grew stronger over time and was consistent with the cells seeding in the BM of major bones such as femur and pelvis (Figure 6D; Figure S4C). Immunohistochemical staining of bone marrow sections confirmed the presence of KMS11 cells in the femur medullary cavities (data not shown). Thus, we conclude that catalytically active NSD2 directly promotes neoplastic growth in vivo.
NSD2 is a general oncoprotein
Analysis of the Oncomine database comparing normal to cancer gene expression data sets revealed preferential and significant upregulation of NSD2 transcript in numerous cancers (Figures 7A and 7B). Moreover, RNAi-mediated NSD2 depletion reduced the proliferation rate of HT1080 fibrosarcoma and U2OS osteosarcoma cell lines (Figure S4D). Thus, in addition to t(4;14)+ myeloma, NSD2 may contribute to the development of other cancer types. Indeed, in the t(4;14)− KMS12-PE and U266 myeloma cell lines, which have lower amounts of NSD2 and H3K36me2 relative to t(4;14)+ cells (Figure S4E), the expression of NSD2WT, but not NSD2Y1179A, increased (i) global H3K36me2 levels (Figure 7C), (ii) cell proliferation rates (Figure 7D), and (iii) the ability of cells to grow in anchorage-independent conditions, relative to the parental cell lines (Figure 7E). Thus, increased NSD2 expression in t(4;14)− cell lines promotes cancer-associated cellular phenotypes.
Figure 7. NSD2 is an oncoprotein that contributes to the tumorigenicity of various cancers.
(A) and (B) NSD2 upregulation in diverse cancers present in the Oncomine database. (A) # of Oncomine datasets in which NSD2 is differentially expressed relative to controls w/cutoffs of >3-fold change and p-value<0.05. Red: upregulated; blue: downregulated. (B) Boxplots indicate (from top to bottom) the maximum, 75th percentile, median, 25th percentile and minimum of 15 representative datasets in which NSD2 is upregulated in cancer (C) tissue (grey) compared to normal (N) tissue. A, cutaneous melanoma; B, ductal breast carcinoma; C, skin squamous cell carcinoma; D, infiltrating bladder urothelial carcinoma; E, lung adenocarcinoma; F, skin squamous melanoma; G, gastric intestinal type adenocarcinoma; H, gastric mixed adenocarcinoma; I, esophageal adenocarcinoma; J, ovarian serous cystadenocarcinoma; K, acute myeloid leukemia; L, fibrosarcoma; M, malignant fibrous histiocytoma; N, synovial sarcoma; O, leiomyosarcoma.
(C) Complementation with NSD2WT but not catalytically inactive NSD2 increases global H3K36me2 levels in t(4;14)− myeloma cells. Western analysis of WCE from the t(4;14)− KMS12-PE (left) and U266 (right) cell lines stably transduced with NSD2WT, NSD2Y1179A or empty virus.
(D) and (E) NSD2 but not NSD2Y1179A increases proliferation and anchorage independent growth of t(4;14)− myeloma cells. Growth curves of the cell lines in (C) were determined as in Figure 6A. (E) Methylcellulose colony formation assay of cell lines as in Figure 6B. Error bars for (D) and (E) indicate s.e.m. from at least 3 independent experiments.
(F) NSD2 but not NSD2Y1179A promotes oncogenic transformation of p19ARF−/− MEFs. Left: western analysis of WCE from p19ARF−/− MEFs stably transduced with NSD2WT, NSD2Y1179A or empty virus. Right: numbers of colonies formed in methylcellulose for each cell line. Error bars indicate s.e.m. from 3 independent experiments.
(G and H) NSD2 increases H3K36me2 at and expression of Fos, Igf2, Figf, and Mdk in p19ARF−/− MEFs. (G) Quantitative RT-PCR analysis of the transcript levels of the indicated genes. (H) H3K36me2 ChIP in the transcribed bodies of the indicated genes. Error bars indicate s.e.m. from 3 experiments.
Primary mouse cells can be transformed by two genetic “hits”: either loss of a tumor suppressor coupled with activation of an oncogene or activation of two oncogenes (Land et al., 1983). The transformation activity of candidate oncogenes can therefore be tested in primary cells that have already been subject to a first hit, for example loss of the p19ARF tumor suppressor. We therefore tested the ability of NSD2WT and NSD2Y1179A to transform p19ARF knockout mouse embryonic fibroblasts (p19ARF−/− MEFs). Expression of NSD2WT resulted in higher global levels of H3K36me2 relative to control and NSD2Y1179A-expressing cells (Figure 7F and Figure S4F). Strikingly, NSD2WT-expressing p19ARF−/− MEFs efficiently formed colonies in methylcellulose, whereas control and NSD2Y1179A-expressing cells did not (Figure 7F; Figure S4F). To our knowledge, NSD2 is the only PKMT shown to behave like a classic oncogene in this prototypical transformation assay.
The genome-wide gene expression profile of NSD2WT-expressing p19ARF−/− MEFs revealed higher expression of several cancer-linked genes relative to control and NSD2Y1179A-expressing p19ARF−/− MEFs (Figure S4G), which was confirmed by real time PCR for Fos, Igf2, Figf, and Mdk genes (Figure 7G). The H3K36me2 signal within the transcribed body of Fos, Igf2, Figf, and Mdk mirrors the expression pattern, being higher in NSD2WT-expressing cells than in control- and NSD2Y1179A-expressing cells (Figure 7H). Together, our results argue that in p19ARF−/− MEFs, as in myeloma cells, NSD2-driven H3K36me2 elevation regulates transcriptional programs that promote cellular transformation.
DISCUSSION
Elucidation of NSD2’s physiologic enzymatic activity at chromatin
Despite clinical evidence supporting a role for NSD2 in the etiology of t(4;14)+ multiple myeloma, the enzymatic activity of NSD2 and how it relates to cancer development has been obscure (Chng et al., 2007). The correct assignment of catalytic specificity for enzymes like NSD2 is crucial for understanding how histone modifications regulate chromatin and for the efforts to develop epigenetic-based therapeutics for diseases like MM. Indeed, erroneous conclusions about the enzymatic and chromatin-regulatory functions of NSD2 could stymie progress in identifying drugs for the treatment of t(4;14)+ myeloma. Here we demonstrated that the principal methylation activity of NSD2 at chromatin is generation of H3K36me2, and that this activity drives NSD2-associated oncogenic programming.
Our findings are in agreement with a study reporting in vitro dimethylation activity at H3K36 by the three NSD family members NSD1, NSD2, and NSD3/WHSC1L1 (Li et al., 2009). However, our results conflict with a recent study concluding, based on immunoblot analysis, that NSD2 trimethylates H3K36 (Nimura et al., 2009), an activity we do not detect. As antibodies against Kme3 epitopes frequently crossreact with the dimethyl species (Egelhofer et al., 2011; Fuchs et al., 2011), misinterpretation of immunoblot specificity may have led to the incorrect conclusion that NSD2 is an H3K36 trimethylase (Nimura et al., 2009). Indeed, the difference in function between H3K36me2 and H3K36me3 has important clinical implication. We find that NSD2 is an oncoprotein and that it generates the bulk of H3K36me2 in a number of cell types (Figure 2B and 2D). In contrast, SETD2, which synthesizes the bulk of H3K36me3 in diverse cells (Bell et al., 2007; Edmunds et al., 2008), acts like a tumor suppressor in clear cell renal cell carcinoma (Dalgliesh et al., 2010; Duns et al., 2010). Thus, the state of methylation at H3K36 – me2 versus me3 – may link modification of this residue to dramatically different functional consequences, and highlights the exquisite level of biological regulation that can be achieved by subtle changes in histone methylation.
NSD2 was recently reported to function within the tumor suppressor ATM kinase DNA damage response pathway (Pei et al., 2011). The authors claimed that NSD2 dimethylates H4K20 in vitro, and that in ATM-mediated cellular responses to DNA damage, this activity mediates 53BP1 localization and function at DNA damage foci. However, it is counterintuitive for an oncoprotein like NSD2 to be a key component in the canonical ATM tumor suppressor pathway. Moreover, utilizing multiple approaches, we observe no methylation activity for NSD2 at H4K20 (Figure 1A–E, 1G, 2A, 2B, S1B, S1C, S1E; data not shown). Thus, while it is intriguing to postulate a role for NSD2 in DNA repair pathways, it does not occur through direct H4K20 methylation.
NSD2 is far more active on nucleosomes than free histones (data not shown), however, two methylation activities – at H3K18 and H4K44 (which has high sequence similarity to H3K36 (Fig S5A; data not shown)) – are catalyzed by NSD2 on free histones but not on nucleosomes (data not shown; (Li et al., 2009)). The functional relevance of these two auxiliary activities is unclear and requires future investigation. Finally, the NSD2 isoform RE-IIBP was reported to methylate H3K27 based upon mass spectrometry analysis (Kim et al., 2008). However, the mass shifts attributed to methylation events in the study do not correspond to the correct mass of methyl moieties. Our analysis showed that NSD2 and RE-IIBP both dimethylate H3K36, but do not use H3K27 as a substrate (Figure 1C, 1D, 1E, S1C, S1D). Together, by employing biochemical, proteomic, and cellular methods, our results do not support previous claims that H3K4, H3K27, and H4K20 are substrate sites of NSD2, providing compelling evidence that therapeutics targeting NSD2 inhibition should focus on H3K36 dimethylation as the exclusive activity of this enzyme on nucleosomes.
Mechanism of oncogenic programming by NSD2
Monoclonal gammopathy of undetermined significance (MGUS) is an asymptomatic premalignant condition detected in 3% of the population over fifty and carries a 1% average annual risk for progression to MM (Chng et al., 2007). A subset of MGUS patients are t(4;14)+, suggesting that NSD2 overexpression might initiate myelomagenesis and that these patients may be at high risk of transitioning to myeloma (Chng et al., 2007). We propose that pathologic overexpression of NSD2 – by altering the genome-wide profile of H3K36me2 – results in localized chromatin relaxation at normally silent genes, which in turn selects for a gene expression program favorable for plasma cell transformation (Figure S5B). For example, cancer-associated genes like PAK1 are normally silent in t(4;14)− cells, but have higher expression in KMS11 cells. Our model argues that increased NSD2 levels result in stochastic NSD2 binding and H3K36me2 generation at the PAK1 gene, which is sufficient to promote transcription by a mechanism that remains to be determined. In addition to genes like PAK1, NSD2 will also stimulate transcription at genes not linked to oncogenesis. However, expression of proteins that provide an advantage for cells to proliferate and survive will be selected over time and thus constitute the majority of NSD2-associated upregulated genes in myeloma cells.
The ability of t(4;14)− cells to form xenograft tumors in mice is conferred by expression of catalytically active NSD2 (Figure 6). In addition, NSD2 expression is elevated in diverse cancer types (Figure 7A and 7B), and to our knowledge is the only PKMT that can act as an oncogene in a classic primary MEF transformation assay (Figure 7F). Like its role in bestowing tumorigenicity to TKO2 cells, the mechanism of NSD2-mediated MEF transformation is H3K36me2-dependent. Together, our results argue that NSD2-mediated disruption of H3K36me2 organization may promote oncogenic programming in many cell types, and that inhibition of this enzyme may have broad therapeutic efficacy for diverse cancers.
EXPERIMENTAL PROCEDURES
Materials
Recombinant enzymes (NSD2SET, G9ASET, SET8, SETD2SET) were expressed in E. coli and full-length NSD2 protein in Sf9 cells. H3 amino acids 1–42 were clones into pGEX-6P1 with the indicated substitutions. NSD2 antibodies were described (Lauring et al., 2008; Li et al., 2009). Methylation assays, quantitative mass spectrometry, microarray gene expression, and ChIP-seq protocol and data analysis are described in detail in the Extended Experimental Procedures.
Xenograft mouse model
10^6 cells stably expressing GFP-luciferase fusion protein were directly injected into the peritoneal cavities or tail veins of SCID/Beige mice. Tumor growth was measured weekly by bioluminescence imaging. All mice were treated in accordance with AAALAC approved guidelines at Stanford University (protocol number: 15066).
Data Access
Raw data has been submitted to NCBI Gene Expression Omnibus (GEO) under the accession number GSE29305. Genome browser tracks and additional data related to this paper are available at http://dldcc-web.brc.bcm.edu/lilab/NSD2/
Supplementary Material
Highlights.
Dimethylation of H3K36 is the principal chromatin-regulatory activity of NSD2
NSD2, via H3K36me2 catalysis, promotes transcription and cell transformation
NSD2 links genomic disorganization of H3K36me2 to oncogenic programming
NSD2 catalytic activity is required for t(4;14)+ myeloma cell tumorigenecity.
Acknowledgments
We thank D. Reinberg for NSD2 and SETD2 antibody. This work was supported in part by grants from the NIH to O.G. (R01 GM079641), W.L. (U01DA025956) and B.A.G. (DP2OD007447), GlaxoSmithKline to OG, the DOD to W.L. (PC094421), an ASMS Research Award, and NJCCR SEED grant to B.A.G, a Genentech Foundation Predoctoral Fellowship for A.J.K. and a NSF Fellowship to B.M.Z.. W.L. is a recipient of a Duncan Scholar Award. O.G. is a recipient of an Ellison Senior Scholar in Aging Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. Embo J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20:571–596. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot RJ, Yaghoubi SS, Kodama K, Dang DN, Dang VH, Breckpot K, Thielemans K, Gambhir SS, Fathman CG. Tissue-targeted therapy of autoimmune diabetes using dendritic cells transduced to express IL-4 in NOD mice. Clin Immunol. 2008;127:176–187. doi: 10.1016/j.clim.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, Belch AR, Pilarski LM. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kee HJ, Choe NW, Kim SM, Eom GH, Baek HJ, Kook H, Kook H, Seo SB. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol. 2008;28:2023–2034. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.