SUMMARY
To prevent ATR activation, telomeres deploy the single-stranded DNA binding activity of TPP1/POT1a. POT1a blocks the binding of RPA to telomeres, suggesting that ATR is repressed through RPA exclusion. However, comparison on the DNA binding affinities and abundance of TPP1/POT1a and RPA indicates that TPP1/POT1a by itself is unlikely to exclude RPA. We therefore analyzed the central shelterin protein TIN2, which links TPP1/POT1a (and POT1b) to TRF1 and TRF2 on the double-stranded telomeric DNA. Upon TIN2 deletion, telomeres lost TPP1/POT1a, accumulated RPA, elicited an ATR signal, and showed all other phenotypes of POT1a/b deletion. TIN2 also affected the TRF2-dependent repression of ATM kinase signaling but not to TRF2-mediated inhibition of telomere fusions. Thus, while TIN2 has a minor contribution to the repression of ATM by TRF2, its major role is to stabilize TPP1/POT1a on the ss telomeric DNA, thereby allowing effective exclusion of RPA and repression of ATR signaling.
INTRODUCTION
Mammalian telomeres are comprised of numerous copies of shelterin assembled on the telomeric TTAGGG repeat DNA (Palm and de Lange, 2008). Shelterin recognizes telomeres primarily with two duplex telomeric DNA binding factors (TRF1 and TRF2). It also contains one (in human) or two (in rodents) POT1 proteins that bind to single-stranded TTAGGG repeats. The POT1 proteins connect to TRF1 and TRF2 through protein interactions involving TPP1 and TIN2 (Liu et al., 2004b; Houghtaling et al., 2004; Ye et al., 2004b), forming a multi-subunit complex that can be isolated intact from human cells (Ye et al., 2004a; O'Connor et al., 2006).
TIN2 is a central component of shelterin that not only connects TPP1/POT1 to the other shelterin components but also stabilizes TRF1 and TRF2 on the duplex telomeric repeat array (Liu et al., 2004a; Ye et al., 2004a; Kim et al., 2004). TIN2 contributes to telomere length regulation but its precise role in telomere protection has not been established (Abreu et al., 2010; Ye and de Lange, 2004; Kim et al., 1999). The function of TIN2 is of particular interest since it is mutated in a subset of Dyskeratosis Congenita (DC) patients (Savage et al., 2008; Walne et al., 2008; Tsangaris et al., 2008).
The role of TIN2 in connecting TPP1/POT1 to TRF1/2 is relevant to the regulation of telomerase-mediated telomere maintenance (Loayza and de Lange, 2003; Kim et al., 1999; Abreu et al., 2010). However, the significance of this link to telomere protection has not been established and several authors have suggested that the TPP1/POT1 heterodimer protects telomeres in a manner that does not require its association with TIN2/TRF1/TRF2 (e.g. (Giraud-Panis et al., 2010; Baumann and Price, 2010; Flynn et al., 2011)). Here we present evidence indicating that TIN2 is critical for the protective functions of TPP1/POT1.
The essential function of TPP1/POT1 is to prevent the ATR kinase dependent DNA damage response. The risk of inappropriate activation of the ATR kinase is inherent to the structure of mammalian telomeres, which contain single-stranded TTAGGG repeats owing to the greater length of the 3’ ended strand of the telomeric repeat array. This 3’ overhang can either be single-stranded or loop back and invade the duplex telomeric TTAGGG repeats. In the latter configuration, called the t-loop, single-stranded TTAGGG repeats are exposed in a displacement loop (D loop) at the base of the t-loop (McElligott and Wellinger, 1997; Makarov et al., 1997; Chai et al., 2006; Griffith et al., 1999). The length of the single-stranded telomeric DNA is estimated to be 40–400 nt (Chai et al., 2006; Zhao et al., 2008), which is sufficient for the binding of RPA - the sensor in the ATR pathway ((Zou and Elledge, 2003); reviewed in (Cimprich and Cortez, 2008)).
RPA recruits ATR to ss DNA through an interaction with ATRIP (Ball et al., 2005; Zou and Elledge, 2003; Ball et al., 2007; Kumagai et al., 2004; Namiki and Zou, 2006; Xu et al., 2008). Its three subunits (RPA70, RPA32, and RPA14) form a complex that binds ss DNA with a Kd of 10−9-10−11 M (Wold, 1997). The optimal RPA binding site is 30 nt whereas RPA forms an unstable complex on substrates of 8–10 nt (Lavrik et al., 1999; Kim et al., 1992; Sibenaller et al., 1998; Blackwell and Borowiec, 1994; Blackwell et al., 1996). In vitro, two RPA units bound to ~75 nt of DNA are sufficient to recruit ATRIP (Zou and Elledge, 2003) and in a Xenopus egg extract system, a 35 nt ss DNA gap can activate the ATR kinase, presumably by binding a single RPA (MacDougall et al., 2007). Based on this data, the telomeric 3’ overhang is of sufficient length to meet the minimal requirements for RPA-mediated ATR activation. It was therefore proposed that telomeres might repress ATR signaling by excluding RPA from their single-stranded moiety.
The single-stranded DNA binding factor in shelterin, the TPP1/POT1 heterodimer, is the most likely candidate repressor of RPA. POT1 binds to 5’-(T)TAGGGTTAG-3’ with sub-nanomolar affinity (Lei et al., 2004; Loayza et al., 2004; Nandakumar et al., 2010). POT1 recognizes this site at 3’ end, a ds-ss junction, and internally, predicting that POT1 can bind along the 3’ overhang as well as to the ss DNA in the D loop (Loayza et al., 2004; Wang et al., 2007; He et al., 2006; Lei et al., 2004; Palm et al., 2009). The two mouse POT1 proteins (POT1a and POT1b) and human POT1 have similar sequence specificity and DNA affinity (He et al., 2006; Palm et al., 2009). DNA binding by human POT1 is enhanced by the N-terminus of TPP1, although TPP1 has no discernable DNA binding activity by itself (Wang et al., 2007; Xin et al., 2007). The recruitment of POT1 to telomeres critically depends on its interaction with TPP1. Deletion of TPP1, or interference with the TPP1-POT1 interaction, leads to a lack of POT1 binding to telomeres (Liu et al., 2004b; Ye et al., 2004b; Hockemeyer et al., 2007; Kibe et al., 2010). Thus, POT1 is likely to bind to ss telomeric DNA and function there as a TPP1/POT1 heterodimer.
Consistent with the RPA exclusion model, removal of POT1a/b from mouse telomeres results in rapid accumulation of RPA, induction of Telomere dysfunction induced Foci (TIFs), and phosphorylation of Chk1 (Hockemeyer et al., 2006; Lazzerini Denchi and de Lange, 2007; Gong and de Lange, 2010). This DNA damage response occurs in G1 as well as in S/G2 and is dependent on the ATR kinase and TopBP1. Additional phenotypes of POT1a/b DKO cells are deregulation of the 3’ overhang length (a phenotype specific to POT1b deletion), polyploidization due to endoreduplication, metaphases with diplochromosomes, and a low frequency of sister telomere fusion (Wu et al., 2006; Hockemeyer et al., 2006; Hockemeyer et al., 2008; Davoli et al., 2010). When TPP1 is compromised, the same phenotypes arise, confirming that TPP1 is required for POT1 function (Hockemeyer et al., 2007; Kibe et al., 2010).
We envisaged at least three, not mutually exclusive, ways in which TPP1/POT1 could exclude RPA from telomeres. First, TPP1/POT1 could be more abundant than RPA; second, TPP1/POT1 could have a higher affinity for telomeric DNA than RPA, as was suggested recently (Arnoult et al., 2009); and third, TPP1/POT1 might be locally enriched and stabilized at telomeres through TIN2-mediated tethering to the other shelterin components. Here we show that POT1 is significantly less abundant than RPA and does not have a greater affinity for telomeric DNA, even when bound to TPP1. Through conditional deletion of TIN2, we provide direct evidence that TPP1/POT1 require a connection to the TIN2/TRF1/TRF2 complex in order to repress ATR signaling. A second function of TIN2 is to promote the repression of ATM signaling (but not NHEJ) by TRF2.
RESULTS
RPA is significantly more abundant than the POT1 proteins
The abundance of RPA and POT1a/b were determined through quantitative immunoblotting of whole cell lysates using known quantities of purified trimeric RPA and mouse POT1a and POT1b as standards (Fig. S1) (Takai et al., 2010). The result indicated that the HeLa1.3 and HTC75 tumor cell lines contained 3–5 million RPA molecules per cell (Fig. 1A). This estimate is an order of magnitude higher than the previous estimates from DNA binding and DNA replication assays using cytosolic and freeze/thaw lysates (Wold and Kelly, 1988; Seroussi and Lavi, 1993; Kenny et al., 1990). In contrast, POT1a and POT1b were each expressed at only 2–7 thousand molecules per mouse embryo fibroblast (MEF) cell (Fig. 1B and Fig. S1). This is consistent with the estimate of ~2 × 104 copies of the single human POT1 in HeLa and HTC75 (Takai et al., 2010). Assuming that RPA is expressed at the same level in mouse and human cells, these data suggest that the mouse and human POT1 proteins are considerably less abundant than RPA.
Figure 1. Abundance and DNA binding features of POT1, TPP1/POT1a and RPA.
(A) and (B) Quantitative immunoblotting for RPA32, POT1a, and POT1b in whole cell lysates and comparison to recombinant standards (Fig. S1A). Right: abundance of human RPA and mouse POT1a/b based on data from three experiments as shown on the left. (C) Gel-shift reactions with the indicated probes and increasing amounts (0.16–40 nM) of human POT1 or RPA. (D) Binding of mouse TPP1/POT1a and human RPA to Tel34. Protein amounts as in (C). (E) Summary of the apparent Kd of human RPA, human POT1, and mouse TPP1/POT1a derived from gel-shift experiments as shown in (C) and (D) (average values from three experiments represented and standard deviations). Kd values were derived from mathematical curve fitting (GraphPad Prism) of PhosphorImager data on RPA and POT1 analyzed in parallel.
RPA and TPP1/POT1 have similar affinities for telomeric DNA
To determine whether POT1 has a higher affinity for telomeric DNA than RPA, we first used baculovirus-derived human POT1 and human RPA purified from E. coli. The substrate was a telomeric repeat array of 34 nt (Tel34), which is sufficiently long to accommodate RPA and contains five overlapping POT1 recognition sites including the optimal POT1 binding site at the 3’ end (TTAGGGTTAG-3’). Formation of G4 structures in the Tel34 probe were prevented by boiling immediately before use and inclusion of LiCl instead of NaCl or KCl in the reaction buffer. RPA showed the same sub-nanomolar affinity for Tel34, a scrambled 34 nt probe, and dT34 (Fig. 1C–E). Tel34 was bound by two POT1 units in a non-cooperative manner whereas no POT1 binding was observed with the non-telomeric DNAs (Fig. 1C,E). Importantly, the affinity of RPA and POT1 for the Tel34 was similar, in the order of 0.5–0.7 nM (Fig. 1E), which is in agreement with previous data on RPA (Kim et al., 1994; Kim et al., 1992; Miyake et al., 2009).
In order to compare RPA to TPP1/POT1, we used the mouse TPP1/POT1a (Fig. S1) and human RPA. This cross-species comparison is justified since human and mouse RPA are virtually identical. Human and mouse RPA70, which contains four of the five OB-folds that mediate the DNA binding of RPA, are identical. Within RPA32, which contributes the fifth OB-fold involved in DNA binding, only 11% of the amino acids are different.
Mouse TPP1/POT1a was purified and the presence of the TPP1/POT1a complex was confirmed by immunoblotting and gel-filtration (Fig. S1C,D). Although the stoichiometry of POT1a and TPP1 could not be discerned from Coomassie-stained gels because of the diffuse pattern of TPP1 (Fig. S1B), antibody super-shift experiments established the presence of TPP1 in the two complexes formed on Tel34 (Fig. 1D, Fig. S1E). These complexes likely contain POT1a as well because TPP1 does not bind DNA on its own. As expected, TPP1/POT1a showed a higher affinity for telomeric DNA than POT1a alone and retained a preference for a 3’ end (Fig. S1F,G).
Despite the improved DNA binding affinity of TPP1/POT1a, side-by-side comparison revealed that RPA and TPP1/POT1a bound Tel34 with a similar apparent affinity (~0.5 nM) (Fig. 1D,E). Together with the much greater abundance of RPA, these data suggest that the intrinsic properties of TPP1/POT1 are unlikely to prevent the binding of RPA to telomeric DNA in vivo.
TIN2 loads TPP1/POT1 onto telomeres
Given these biochemical data, we explored the possibility that TPP1/POT1 might require their interaction with TIN2 to compete with RPA. This hypothesis predicts that deletion of TIN2 elicits the phenotypes associated with loss of POT1a/b. As deletion of TIN2 results in embryonic lethality (Chiang et al., 2004), we generated conditional TIN2 knockout MEFs to determine the outcome of TIN2 loss (Fig. 2). The TIN2 locus was modified by gene targeting, resulting in the insertion of loxP sites before exon 3 and after exon 7 (Fig. 2A). The deletion of exons 3–7 generates a gene that only encodes the first 93 aa of TIN2 from exons 1 and 2. mRNA splicing from exon 2 to exon 8 creates a frame-shift at the splice junction and premature termination of the ORF 12 aa into exon 8. Therefore, the TIN2Δex3–7 allele is expected to encode a severely truncated TIN2 lacking most of the protein, including the TRF1 binding site in exon 6 (Fig. 2A).
Figure 2. Conditional deletion of TIN2.

(A) Schematic showing the Tinf2 genomic locus, the targeting construct, and the alleles generated. Black triangles, LoxP sites with BglI/BglII sites; black boxes, Frt sites; Neo, PGK-Neo gene; DTA, MCI-DTA; black bars, probes for genomic blotting; asterisk, TRF1 interaction motif in exon 6. Right: PCR genotyping of TIN2+/+ and TIN2F/F MEFs after introduction of Cre. (B) Loss of telomeric TIN2 signals from TIN2F/F MEFs treated with pWZL-Cre (92 h). IF for TIN2 (Ab 1447, red) at telomeres detected by FISH (green). (C) Telomeric DNA ChIP for shelterin proteins in TIN2F/F MEFs with or without Cre treatment (92 h). ChIP signals were normalized to the input and the background (PI) was subtracted. Numbers below represent the average decrease in these values after deletion of TIN2 (two experiments). (D) TIN2 deletion diminishes the telomeric IF signals for TRF1 and TRF2. Method as in (B). (E) Effect of TIN2 deletion (H&R-Cre) on soluble and chromatin-bound shelterin proteins. Equal cell equivalents of the whole cell lysate (WC), cytoplasmic proteins (CP), nucleoplasmic proteins (NP), and the chromatin-bound fraction (CB) were analyzed. α-tubulin is cytoplasmic. (F) Immunoblotting for Myc-POT1a and -TPP1 in TIN2F/F MEFs infected with pLPC-puro-Myc-TPP1 or pWZL-Hygro-Myc-POT1a and treated with Cre (92 h) after drug selection. (G) Telomeric ChIP for Myc-POT1a and -TPP1 before and after TIN2 deletion. ChIP assay with TIN2 Ab (1447) and myc Ab as in (F). Input: 20% of the input DNA. (H) Quantification of the ChIP signals in (G) after normalization to input and subtraction of background (PI). (I) Telomeric localization of Myc-TPP1 and -POT1a. IF for myc (red) and telomeric FISH (green) at 92 hr post-Cre.
Fibroblasts from E13.5 TIN2F/F embryos were immortalized with SV40 large T antigen and tested for the expected Cre-mediated recombination by genomic PCR (Fig. 2A). Deletion of TIN2 induced a senescence-like growth arrest, which was negated by exogenous mouse TIN2, indicating that the phenotype resulted from TIN2 loss (Fig. S2A–D). Indirect immunofluorescence (IF) demonstrated the anticipated loss of telomeric TIN2 signals from these cells (Fig. 2B) and the telomeric ChIP suggested that TIN2 levels at telomeres were reduced by ~20-fold (Fig. 2C).
TIN2 was previously shown to promote the association of human TRF1 and TRF2 with telomeres (Ye et al., 2004a; Kim et al., 2004; O'Connor et al., 2006). In agreement, the TRF2 and Rap1 telomeric ChIP values diminished ~3-fold while the TRF1 value was 4-fold lower (Fig. 2C). The IF signals for TRF1 and TRF2 at telomeres were also diminished and although TRF1, TRF2, and Rap1 continued to be chromatin-bound, the overall level of TRF1 detectable in immunoblots was lowered (Fig. 2D,E).
To establish the effect of TIN2 on the telomeric localization of TPP1 and POT1a, Myc-tagged versions were introduced into TIN2F/F MEFs, where they showed the anticipated telomeric localization (Fig. 2F,I). Both Myc-TPP1 and Myc-POT1a lost their telomeric localization after deletion of TIN2, despite unaltered expression (Fig. 2F,I). Furthermore, based on ChIP, the telomeric association of TPP1 and POT1a was reduced to near background levels when TIN2 was absent (Fig. 2G,H). The endogenous TPP1 also disappeared from telomeres when TIN2 was deleted (Fig. 2C). These data argue that TPP1 and POT1a (and most likely POT1b) require TIN2 for their accumulation at telomeres. Similarly, shRNA-mediated knockdown of TIN2 resulted in loss of POT1 from human telomeres (Fig. S2E–G). The TIN2-dependent tethering of TPP1/POT1 to both TRF1 and TRF2, explains why neither TRF1 nor TRF2 KO cells have the phenotypes typical of the POT1a/b DKO (Sfeir et al., 2009; Celli and de Lange, 2005; Lazzerini Denchi and de Lange, 2007).
TIN2 deletion induces the 3’ overhang phenotype typical of POT1b deficiency
A characteristic phenotype associated with the loss of POT1b or TPP1 from telomeres is an increase in the ss TTAGGG repeats. Deletion of TIN2 resulted in the same overhang phenotype observed upon deletion of POT1b (Fig. 3A). The normalized 3’ overhang signal increased by 2–4 fold within two or four days after introduction of Cre. In contrast, the pattern of the total telomeric DNA was not overtly affected by deletion of TIN2.
Figure 3. Excess ss telomeric DNA, endoreduplication, and telomere fusions in TIN2 KO.
(A) In-gel hybridization assay for ss telomeric DNA after TIN2 deletion. Left: TelC signals under the native condition. Right: same gel re-hybridized after in situ denaturation of the DNA. Overhang signals (left) were normalized to the total telomeric signals (right) and compared to TIN2F/F MEFs without Cre. Two independent experiments are shown. (B) FACS analysis for DNA content (propidium iodide) of TIN2F/F and TIN2+/+ MEFs after pWZL-Cre infection (day 8). Both MEFs contained tetraploid cells prior to deletion of TIN2. (C) Diplochromosomes in Cre-treated TIN2F/F MEFs. DNA stained with DAPI (blue) and telomeric FISH (green). (D) Telomere clustering in a polyploid TIN2-deficient cell. Staining for 53BP1 IF (red) and telomeric FISH (green). DNA is stained with DAPI (blue). The enlarged image illustrates clustered telomeres. (E) CO-FISH illustrating examples of telomere fusions and T-SCEs in metaphases of TIN2-deficient cells. Red: leading-end telomeres; green: lagging-end telomeres; blue: DAPI DNA stain. (F) Summary of telomere phenotypes induced by TIN2 deletion determined by CO-FISH as shown in (E). Values are averages of 3–4 independent experiments (1000–2000 telomeres/experiment) and SDs. Sister fusions were scored on long arm telomeres in metaphase spreads with separated chromosome arms.
Loss of TIN2 induces polyploidization through endoreduplication
A prominent phenotype of POT1a/b or TPP1 deficiency is the formation of polyploid cells formed through endoreduplication (Hockemeyer et al., 2006; Davoli et al., 2010; Kibe et al., 2010). Similarly, TIN2-deficient cells showed an increase in ploidy, resulting in FACS profiles showing discrete peaks at 8-, 16-, and 32N DNA content (Fig. 3B). Consistent with endoreduplication, TIN2 deficient cells had diplochromosomes (Fig. 3C) and showed telomere clustering (Fig. 3D). Both phenomena are observed in the POT1a/b DKO cells and are consistent with the persistent association of sister chromatids through multiple rounds of DNA replication.
After replication, leading- and lagging-end telomeres fuse in TIN2-deficient cells
POT1a/b DKO cells lack the prominent G1-type telomere fusions typical of TRF2-deficient cells. The telomere fusions in POT1a/b DKO cells most often arise after DNA replication as evidenced by chromatid-type fusions. Importantly, these fusions can involve both products of DNA replication: telomeres duplicated by leading-strand DNA synthesis (leading-end telomeres) and those duplicated by lagging-strand DNA synthesis (lagging-end telomeres). As a consequence, POT1a/b DKO cells display fused sister chromatids which are exceedingly rare in TRF2-deficient cells (Hockemeyer et al., 2006). Sister telomere fusions also occur in TPP1 knockout cells, presumably because of the loss of POT1a/b (Kibe et al., 2010; Tejera et al., 2010).
Metaphase chromosomes from TIN2-deficient cells were examined using Chromosome-Orientation FISH (CO-FISH) to identify leading- and lagging-end telomeres (Fig. 3E,F). TIN2 deletion induced a significant level of telomere fusions. A substantial fraction of the fusions were generated after DNA replication as they involved the fusion of duplicated chromatids (Fig. 3E,F). Among these, sister telomere fusions were prominent indicating that TIN2 loss resulted in deprotection of both leading- and lagging-end telomeres. Leading-to-lagging end fusions were also noted among the chromatid-type fusions between two different chromosomes (Fig. 3E).
TIN2 deletion also resulted in chromosome-type fusions, which could either indicate a fusion in G1 or result from duplication of the chromatid-type fusions formed in the preceding G2. As shown below, the chromosome-type fusions in the TIN2 KO cells are most likely the result of diminished loading of TRF2.
There was no prominent loss of the telomeric signals after TIN2 deletion, the telomeres did not show the fragile site phenotype associated with TRF1 loss, and Telomeric DNA containing Double Minute Chromosomes (TDMs) were not induced (Fig. 3E). TIN2 deficiency led to a modest increase in the rate of telomere sister chromatid exchanges (T-SCEs) (~5% compared to 0.5% in the control) but the statistical significance of this phenotype is marginal (p=0.06, Student’s t-test; Fig. 3E,F).
Activation of ATR and ATM at telomeres lacking TIN2
As expected, TIN2 deletion resulted in the activation of a DNA damage response, evident from the accumulation of 53BP1 at telomeres, the proliferative arrest, and phosphorylation of Chk1 and Chk2 (Fig. 4A,B; Fig. S2). Exogenous TIN2 repressed the accumulation of 53BP1 at telomeres of TIN2 KO cells, whereas expression of Myc-TPP1 or Myc-POT1a had no effect (Fig. S3A,B). Compound TIN2/ATR and TIN2/ATM double knockout cells indicated that the DNA damage response involved both the ATM and the ATR kinases (Fig. 4; Fig. S3C–E). This contrasts with the specific induction of either ATM or ATR signaling upon individual deletion of other shelterin components. The ATR response was evident from the phosphorylation of Chk1, which was absent when TIN2 and ATR were co-deleted from TIN2F/FATRF/F cells (Fig. 4B). Furthermore, deletion of TIN2 resulted in significantly fewer TIFs per nucleus when ATR was absent (Fig. 4A,C). The DNA damage response elicited by TIN2 loss also involved the ATM kinase, as shown by phosphorylation of Chk2, which was diminished when ATM was absent, and a reduced TIF response in ATM-deficient cells (Fig. 4B,C). Consistent with signaling involving both ATM and ATR, the inhibition of both kinases with caffeine lowered the frequency of TIFs more than the absence of either kinase alone (Fig. S3F). In contrast, the absence of DNA-PKcs did not affect the DNA damage response at telomeres lacking TIN2 (Fig. S3G).
Figure 4. TIN2 loss induces ATM and ATR signaling.
(A) TIFs detected by IF-FISH in MEFs of the indicated genotypes. MEFs were fixed at 92 hr after H&R Cre and processed as in Fig. 3D. (B) Immunoblots of phospho-Chk1, total Chk1, and Chk2 in MEFs of the indicated genotypes at 48 and 92 hr after H&R Cre. UV treated (30 min after 25 J/m2) TIN2F/F MEFs serve as a control. (C) Quantification of the effect of ATM and ATR deletion on TIFs induced by TIN2 deletion. TIN2F/F (top), TIN2F/FATRF/F (middle), and TIN2F/FATM−/− MEFs (bottom) were scored for 53BP1 TIFs per nucleus (n>100) after H&R-Cre (92 h). Averages of 3 independent experiments and SDs. P values from one-way analysis of variance (ANOVA) and Bonferroni’s Multiple Comparison Test.
RPA at telomeres and RPA-coated chromatin bridges after TIN2 loss
TIN2 loss also recapitulates the phenotype of POT1a/b deletion with regard to the association of RPA with telomeres. Accumulation of RPA at telomeres was previously observed 4 hours after removal of POT1a from POT1b-deficient cells (Gong and de Lange, 2010). Approximately 15% of the TIN2-deficient cells showed RPA foci at telomeres whereas RPA was not observed at telomeres in TIN2-proficient cells (Fig. 5A). The presence of RPA at telomeres was noteworthy because the cells were not in S phase as surmised from the generally low level of RPA staining at non-telomeric loci. After POT1a deprivation, RPA also becomes detectable at telomeres in non-replicating cells although to a lesser extent than in S phase (Gong and de Lange, 2010). Approximately 40% of the POT1a-deprived G1 cells showed 5 or more RPA foci that were inferred to be at dysfunctional telomeres based on their co-localization with 53BP1 (Gong and de Lange, 2010). Thus, the level of RPA accumulation at non-replicating telomeres is lower after TIN2 loss compared to POT1a removal. This difference may be due to timing differences in the two methods used because Cre-mediated deletion of TIN2 required several days before analysis whereas POT1a loss was studied with a rapidly acting degron fusion. Nonetheless, this data establish that RPA can associate with telomeres after TIN2 removal.
Figure 5. RPA at telomeres after TIN2 deletion.
(A) Co-localization of RPA with telomeres in TIN2F/F MEFs infected with Cre (92 h). RPA32 IF (red) and telomeric DNA FISH (green). Arrowheads: RPA signals at telomeres. Below: quantification of cells with 5 or more telomeric RPA signals as averages from 3 experiments (n≥100 nuclei) and standard errors. Asterisks: micronuclei. The cells used also expressed Myc-RPA32 but the myc-tag was not used for IF. (B) Model for RPA exclusion through TIN2-tethered TPP1/POT1. POT1 is shown to have a greater on-rate (arrow) due to its TPP1 connection to the TIN2/TRF1/TRF2 complex on the duplex telomeric DNA. Although only the most terminal shelterin complex is depicted, POT1 in shelterin distal from the telomere terminus may well be physically close to ss DNA due to higher order structure of the telomeric DNA. Tethered POT1 may also prevent RPA binding to the ss telomeric DNA in the D loop when telomeres are in the t-loop configuration (not shown).
We noticed that RPA often stained the chromatin bridges connecting individual nuclei (Fig. S4). Chromatin bridges occur in immortalized MEFs experiencing telomere fusions and might represent stretched chromatin from fused chromosomes that persist through cytokinesis. Alternatively, they could represent unresolved recombination events, but T-SCEs are infrequent in the TIN2 KO setting. The presence of RPA indicates that at least part of this DNA is single-stranded. Most of the RPA bridges also showed telomeric FISH signals consistent with their being derived from dysfunctional telomeres (Fig. S4). We also noted very prominent RPA signals on the micronuclei that are often formed in MEFs experiencing telomere dysfunction (see asterisk in Fig. 5A), suggestive of extensive DNA processing in these compartments.
Collectively, the data on the TIN2 deficient cells reveal a key role for TIN2 in facilitating telomere protection by POT1a/b (Fig. 5B).
TIN2 contributes to TRF2-mediated repression of ATM signaling
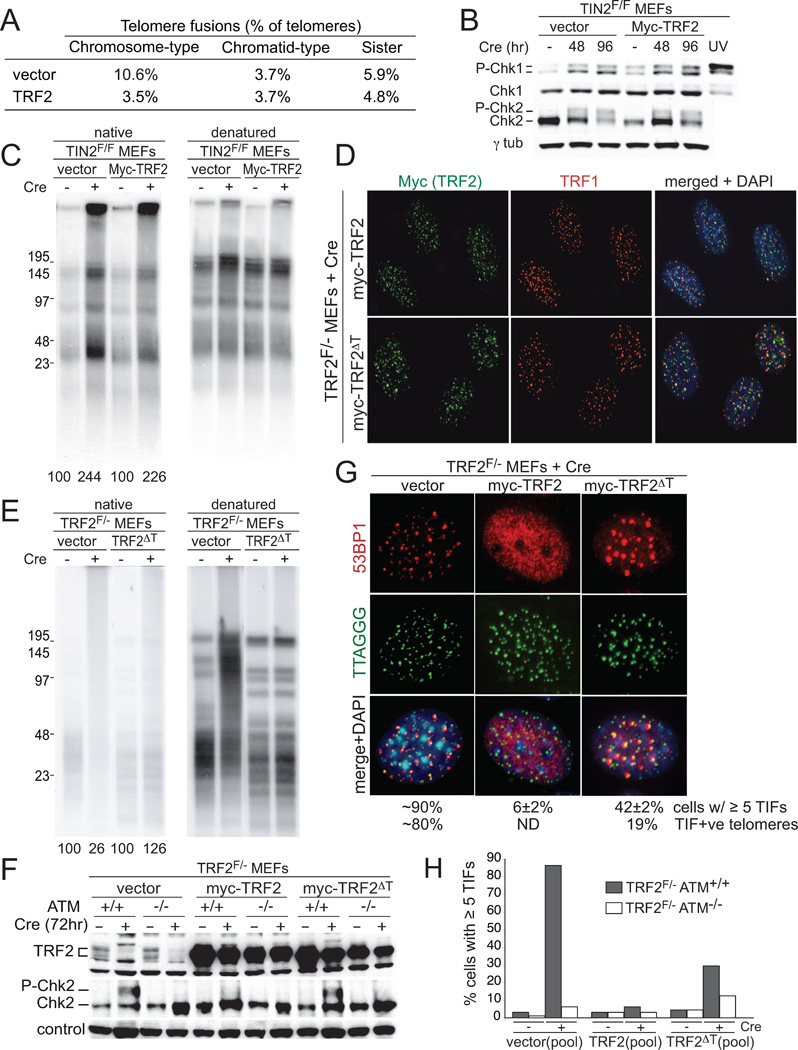
The activation of the ATM kinase pathway and formation of chromosome-type fusions are suggestive of diminished TRF2 function in the TIN2 KO cells. To determine whether these phenotypes resulted from reduced association of TRF2 with telomeres or were due to the absence of TIN2 from telomere-bound TRF2, two approaches were taken. In the first approach, Myc-tagged TRF2 was overexpressed in TIN2F/F MEFs to determine whether an increased level of TRF2 expression could ameliorate these phenotypes (Fig. 6A–C). In the second approach, we complemented conditional TRF2 KO cells with an allele of mouse TRF2 lacking aa 350–365, which is the predicted TIN2 interaction motif (Chen et al., 2008) (Fig. 6D–H). This deletion mutant, TRF2ΔT, binds to telomeres and recruits Rap1 but lacks the ability to interact with TIN2 based on a far-western assay (Fig. 6D, Fig. S5A,B). Consistent with previous telomeric ChIP of TIN2 and TRF1 in TRF2 KO cells (Hockemeyer et al., 2007), cells expressing TRF2ΔT show diminished accumulation of TIN2 at telomeres while TRF1 appears largely unaffected (Fig. S5B).
Figure 6. TRF2 requires TIN2 for repression of ATM but not NHEJ.
(A) Overexpression of TRF2 reduces chromosome-type fusions in TIN2 KO cells. Telomere fusions were scored as in Figs. 3E and F. (B) Immunoblots for Chk1 and Chk2 phosphorylation. Cells as in (A) and processing as in Fig. 4B. (C) No effect of TRF2 overexpression on telomeric overhang in TIN2 KO cells. Cells as in (A) and processing and quantification as in Fig. 3A. (D) Telomeric localization of the TRF2ΔT mutant. TRF2F/−MEFs were infected with the indicated TRF2 alleles, selected, cloned, and treated with Cre. TRF2 was detected with the Myc Ab in combination with IF for TRF1. (E) Repression of telomere fusions by TRF2ΔT. Cells as in D were analyzed at 120 hr post-Cre as in Fig. 3A. (F) Chk2 phosphorylation in TRF2ΔT cells. TRF2F/−ATM+/+ and TRF2F/−ATM−/− cells were infected with the indicated retroviruses, selected, and treated with Cre as indicated. Immunblotting for Chk2 and TRF2 at 72 hr post-Cre. Loading control: non-specific band in the Chk2 blot. (G) TIFs in TRF2ΔT cells. The indicated clonal lines (as in (D)) were treated with Cre (96 h) and analyzed for TIFs as in Fig. 4A. Note fewer but larger 53BP1 TIFs in TRF2ΔT cells. (H) Quantification of the TIF response in the indicated cells at 96 hr after Cre. Pools of TRF2F/− ATM+/+ and TRF2F/−ATM−/− infected as indicated were used for TIF assays as in Fig. 4A.
As expected, the telomeric overhang phenotype associated with TIN2 deletion was unaltered upon overexpression of TRF2 and no increase in the telomeric overhang occurred in a clonal line expressing TRF2ΔT instead of wild type TRF2 (Fig. 6C,E). Furthermore, Chk1 phosphorylation was induced after TIN2 loss, despite the overexpression of TRF2 (Fig. 6B). These results are consistent with the requirement for TIN2 in the tethering of TPP1/POT1a/b and argue against a role for TRF2 in the recruitment of POT1a/b as had been proposed for human POT1 (Yang et al., 2005).
TIN2-deficient MEFs with overexpressed TRF2 showed a strong reduction in chromosome-type telomere fusions as determined in metaphase spreads (Fig. 6A). In addition, the TRF2ΔT allele repressed the occurrence of telomere fusion products in gel-electrophoretic analysis of telomeric restriction fragments of TRF2F/− cells treated with Cre (Fig. 6E). Thus, the ability of TRF2 to repress NHEJ of telomeres does not require its interaction with TIN2.
In terms of the contribution of TIN2 to the repression of ATM signaling by TRF2, however, an intermediate effect was observed. The phosphorylation of Chk2 in TIN2 KO cells overexpressing wild type TRF2 and in TRF2 KO cells containing the TRF2ΔT allele was only slightly reduced (Fig. 6B,F). Furthermore, TRF2ΔT did not fully repress TIF formation in TRF2 KO cells (Fig. 6G,H). Thus, the interaction of TRF2 with TIN2 appears to improve its ability to repress ATM kinase signaling but has no effect on its protection of telomeres from NHEJ.
DISCUSSION
The role of TIN2 within shelterin
This characterization of the telomere defects in TIN2-deficient cells completes the catalogue of telomere dysfunction phenotypes associated with loss of individual shelterin components (Table 1). Each shelterin subunit is unique with regard to the spectrum of aberrant events at chromosome ends elicited upon its loss. This phenotypic diversity bears witness to the functional compartmentalization within shelterin.
Table 1.
Phenotypes associated with the deletion of individual shelterin components
| Telomere fusions |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DDR |
Endoredu- plication |
Chromosome- type |
Chromatid-type Leading Lagging |
3’ over- hang |
Fragile telomeres |
T-SCEs (in Ku null) |
||||
| Gene | viable | ATR | ATM | |||||||
| POT1a | − | + | − | + | + | + | + | − | − | − |
| POT1b | + | − | − | − | − | − | − | + | − | − |
| POT1a/b | − | ++ | − | ++ | + | + | + | + | − | + |
| TPP1 | − | ++ | − | ++ | + | + | + | + | − | n.t. |
| TIN2 | − | ++ | + | ++ | + | + | + | + | − | ? |
| TRF1 | − | +a | − | + | − | − | − | − | + | n.t. |
| TRF2 | − | − | ++ | + | ++ | +b | − | − | − | + |
| Rap1 | + | − | − | − | − | − | − | − | − | + |
ATR signaling requires progression through S phase
Leading-end chromatid fusions occur only in absence of MRN or ATM.
Importantly, TIN2 deletion mimics the loss of TPP1 or POT1a/b in inducing the same combination of phenotypes: the activation of ATR signaling, endoreduplication, post-replicative telomere fusion involving both leading- and lagging-end telomeres, and the induction of excessive single-stranded TTAGGG repeat DNA (Table 1). It therefore appears evident that TIN2, like TPP1, is required to allow the POT1 proteins to fulfill their functions.
In addition to the phenotypes typical of POT1a/b loss, TIN2-deficient cells show ATM kinase signaling and a modest level of chromosome-type telomere fusions, which are phenotypes ascribed to TRF2 loss (Table 1). Our data argue that the chromosome-type telomere fusions are due to the diminished loading of TRF2 on telomeres lacking TIN2, rather than a genuine contribution of TIN2 to this function of TRF2. In contrast, we find that TRF2 relies on its association with TIN2 for maximal repression of the ATM kinase pathway. None of the other shelterin components, including the TRF2 binding partner Rap1, appear to contribute to the repression of ATM signaling by TRF2.
In contrast to this contribution of TIN2 to the function of TRF2, TIN2 deletion does not show the fragile telomere phenotype associated with the telomeric replication problems resulting from loss of TRF1 (Martinez et al., 2010; Sfeir et al., 2010). Thus, the lowered amount of TRF1 on telomeres lacking TIN2 (Fig. 2) is likely sufficient for the efficient replication of the telomeric repeat array. Previous work using either a TRF1 dominant negative allele or tankyrase to remove TRF1 also showed that TRF1 levels can be reduced substantially without the induction of deleterious telomeric phenotypes (Karlseder et al., 2002; van Steensel and de Lange, 1997; Smith and de Lange, 2000).
In sum, the TIN2-tether is required for all the protective functions of the TPP1/POT1a/b complexes and the binding of TIN2 to TRF2 contributes to the ability of TRF2 to repress ATM signaling. In contrast, although TRF1 is stabilized on telomeres by TIN2, the telomere-bound TRF1 operates independently of its association with TIN2.
RPA exclusion and repression of ATR signaling by TPP1/POT1
Zou and colleagues raised the possibility that TPP1/POT1 are assisted by hnRNPA1, which, unlike TPP1/POT1, has the ability to remove RPA from single-stranded telomeric DNA in vitro (Flynn et al., 2011). The subsequent displacement of hnRNPA1 by POT1 is proposed to require recruitment of hnRNPA1 to TERRA RNA, which provides optimal binding sites for hnRNPA1 but not for POT1 (Nandakumar et al., 2010). It was further proposed that the step-wise removal of RPA by hnRNPA1 and hnRNPA1 by TERRA is orchestrated by variation in TERRA levels during S phase such that the replicated telomeres end up bound by POT1, rather than RPA or hnRNPA1. However, this model does not explain why POT1, once loaded at the end of S phase, is not displaced by RPA. POT1 also does not show the predicted increase on telomeres at the end of S phase and in G2 (Verdun et al., 2005). Finally, the hnRNPA1/TERRA model for RPA competition predicts that the dependence on POT1 should be greater in G2 and G1 than in most of S phase when RPA is repressed by hnRNPA1. Yet, removal of POT1 from telomeres initiates an RPA-dependent ATR signaling within a few hours regardless of whether the cells are in S, G2, or G1 phase (Gong and de Lange, 2010). Thus, while hnRNPs may well play a role in the protection of telomeres, it is likely that other mechanisms are required to exclude RPA from the single-stranded telomeric DNA.
We propose that TPP1/POT1 acts as an effective competitor for RPA when bound to TIN2 (Fig. 5B). The crux of the TIN2-tether model for POT1 function is that through its association with TIN2, the TPP1/POT1 heterodimer has two binding sites at chromosome ends, one on the single-stranded DNA and one on a nearby TIN2/TRF1/TRF2 complex. This dual binding will increase the on-rate of POT1 binding to the ss TTAGGG repeats and thereby greatly enhance the stability of the TPP1/POT1 complex on the ss DNA.
We envisage that TPP1/POT1 heterodimers are associated with TIN2 bound to the TRF1/TRF2 units anywhere on the duplex telomeric repeat array. This view is in agreement with ChIP data and the ability of POT1 to accumulate on telomeres independent of its ss DNA binding activity (Loayza and de Lange, 2003). As TIN2/TRF1/TRF2 are more abundant than TPP1/POT1 (Takai et al., 2010), there is ample TIN2 for the binding of all TPP1/POT1. Most of the tethered POT1 is not likely to be positioned adjacent to the ss telomeric DNA. However, POT1 at a distal site may well be in close proximity to, and interact with, the ss telomeric DNA owing to a higher order structure of the telomeric chromatin. When telomeres are in the t-loop configuration, the ss DNA in the D loop is also likely to require engagement of POT1 to prevent ATR signaling. In this setting, POT1 tethered to the shelterin on the duplex DNA formed by strand-invasion of the 3’ overhang will be positioned to engage the ss DNA in the D loop. Further in vitro tests will provide insight into the validity of the TIN2-tethering model for RPA exclusion by TPP1/POT1.
EXPERIMENTAL PROCEDURES
Analysis of the abundance and biochemical features of RPA and TPP1/POT1a
Flag-human POT1, His6-mouse POT1a, His6-mouse POT1b, and Flag-mouse TPP1 were purified from baculovirus-infected cells as described (Palm et al., 2009; Takai et al., 2010). Recombinant human RPA was isolated as described (Binz et al., 2006) with minor modifications. Probe labeling reactions and gel-shift assays were performed as described (Palm et al., 2009) with minor modification. For additional information, see the Supplemental Materials.
Tinf2 gene targeting, mouse strains, and generation of MEFs
The targeting construct was constructed from a bacterial artificial chromosome (BAC (Children’s Hospital Oakland Research Institute)) using the recombineering method. Targeted ES cell clones from Albino C57BL/6J were screened by genomic blotting of BglI-digested DNA and injected into C57BL/6J blastcysts. Mice carrying the TIN2 Flox allele were generated using the FLPe deleter strain (The Jackson Laboratory). Additional information is available in the Supplemental Materials.
Analysis of shelterin components and telomere (dys-) function
Detailed experimental procedures are available in the Supplemental Material. Immunoblotting for shelterin and cell fractionation was performed as described previously (Takai et al., 2010). Telomeric DNA FISH was combined with IF as described previously (Lazzerini Denchi and de Lange, 2007). For RPA, the in situ cell fractionation protocol of Mirzoeva and Petrini was used (Mirzoeva and Petrini, 2001). ChIP was performed as described (Loayza and de Lange, 2003) with minor modification. Telomeric overhang signals and telomeric restriction fragment patterns were analyzed by in-gel analysis as previously described (Celli and de Lange, 2005). The procedures for telomeric FISH and CO-FISH on metaphase spreads were as described (Sfeir et al., 2009).
HIGHLIGHTS.
Biochemical data do not explain how TPP1/POT1 excludes RPA from ss telomeric DNA
Deletion of TIN2 shows that TPP1/POT1 depends on TIN2 for telomere binding
Without the TIN2 tether, TPP1/POT1 fails to block RPA binding and ATR activation
In addition, TIN2 contributes to TRF2-dependent repression of ATM signaling
Supplementary Material
ACKNOWLEDGEMENTS
We thank Devon White for expert mouse husbandry and the RU Transgenics facility for help with generating mouse strains. We thank Marc Wold for the RPA expression plasmid and Yihu Xie and Nikola Pavletich for the gel-filtration analysis of TPP1/POT1a. Members of the de Lange laboratory are thanked for advice and comments. This work was supported by a postdoctoral fellowship from the ACS to DF, a postdoctoral fellowship from the Japan Society for the promotion of Science and the Toyobo Biotechnology Foundation to TK, and grants (AG016642 and GM49046) from the NIH to TdL. TdL is an ACS Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Saintome C, Ourliac-Garnier I, Riou JF, Londono-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA. Human replication protein A binds singlestranded DNA in two distinct complexes. Mol Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA, Mastrangelo IA. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol Cell Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Chiang YJ, Kim SH, Tessarollo L, Campisi J, Hodes RJ. Telomereassociated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol Cell Biol. 2004;24:6631–6634. doi: 10.1128/MCB.24.15.6631-6634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011 doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39:665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Gong Y, de Lange T. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell. 2010;40:377–387. doi: 10.1016/j.molcel.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. Embo J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian POT1 requires interaction with TPP1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- Kenny MK, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T. Telomere Protection by TPP1 Is Mediated by POT1a and POT1b. Mol Cell Biol. 2010;30:1059–1066. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- Kim C, Snyder RO, Wold MS. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Kim SM, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Lavrik OI, Kolpashchikov DM, Weisshart K, Nasheuer HP, Khodyreva SN, Favre A. RPA subunit arrangement near the 3'-end of the primer is modulated by the length of the template strand and cooperative protein interactions. Nucleic Acids Res. 1999;27:4235–4240. doi: 10.1093/nar/27.21.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004a;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004b;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: A nonamer 5'-TAGGGTTAG-3' minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Namiki Y, Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci U S A. 2006;103:580–585. doi: 10.1073/pnas.0510223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroussi E, Lavi S. Replication protein A is the major single-stranded DNA binding protein detected in mammalian cell extracts by gel retardation assays and UV cross-linking of long and short single-stranded DNA molecules. J Biol Chem. 1993;268:7147–7154. [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsangaris E, Adams SL, Yoon G, Chitayat D, Lansdorp P, Dokal I, Dror Y. Ataxia and pancytopenia caused by a mutation in TINF2. Hum Genet. 2008;124:507–513. doi: 10.1007/s00439-008-0576-7. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wold MS, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, Van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004a;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004b;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008;36:e14. doi: 10.1093/nar/gkm1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.