SUMMARY
Primary effusion lymphoma (PEL) is caused by Kaposi’s sarcoma-associated herpesvirus (KSHV) and frequently also harbors Epstein-Barr virus (EBV). The expression of KSHV- and EBV-encoded microRNAs (miRNAs) in PELs suggests a role for these miRNAs in latency and lymphomagenesis. Using PAR-CLIP, a technology which allows the direct and transcriptome-wide identification of miRNA targets, we delineate the target sites for all viral and cellular miRNAs expressed in PEL cell lines. The resulting dataset revealed that KSHV miRNAs directly target more than 2000 cellular mRNAs, including many involved in pathways relevant to KSHV pathogenesis. Moreover, 58% of these mRNAs are also targeted by EBV miRNAs, via distinct binding sites. In addition to a known viral analog of cellular miR-155, we show that KSHV encodes a viral miRNA that mimics cellular miR-142-3p function. In summary, this study identifies an extensive list of KSHV miRNA targets, which are likely to influence viral replication and pathogenesis.
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a human γ-herpesvirus that establishes latency in B lymphocytes and causes primary effusion B cell lymphoma (PEL) (Carbone et al., 2001; Cesarman et al., 1995). PEL-derived cell lines serve as an important model system for KSHV latency and transformation. While most PELs are co-infected with the γ-herpesvirus Epstein-Barr virus (EBV) (Cesarman et al., 1995) and EBV infection has been linked to a number of B-cell lymphomas, a role for EBV in the lymphomagenesis of PELs has not been demonstrated and EBV is never found in PEL in the absence of KSHV.
miRNAs are ~22 nucleotide (nt) regulatory RNAs expressed by animals, plants and some viruses, particularly herpesviruses (Gottwein and Cullen, 2008). With few exceptions, miRNA biogenesis proceeds from pri-miRNA transcripts through a pre-miRNA stem-loop intermediate to an imperfect ~22 nt RNA duplex. One strand of this duplex can be incorporated as a mature miRNA in RNA-induced silencing complexes (RISCs). The non-incorporated strand of the duplex is referred to as the star strand and is degraded. Within RISC, miRNAs are bound by Argonaute (Ago) proteins and induce the repression of mRNAs bearing sequences with partial complementarity to the miRNA. This effect is most commonly mediated through base pairing between the “seed region” of the miRNA, spanning nucleotides 2–7, and sites located in the 3′UTRs of target mRNAs (Bartel, 2009; Hafner et al., 2010).
During latency, KSHV expresses 12 pre-miRNAs, which are processed to mature miRNAs called miR-K1 to miR-K12 (Gottwein and Cullen, 2008). The sequences of the KSHV miRNAs are largely conserved between different KSHV isolates and between PEL cell lines (Marshall et al., 2007). However, the seed sequences of herpesviral miRNAs are generally not conserved between herpesviruses that infect evolutionary distant hosts. Consequently, the computational identification of viral miRNA targets is challenging and relatively few targets of KSHV miRNAs have been identified.
Of likely relevance to KSHV-induced lymphomagenesis is the finding that KSHV miR-K11 is a functional analog of cellular miR-155, a consequence of the identical seed region of these miRNAs (Gottwein et al., 2007; Skalsky et al., 2007). While stable antagonism of miR-K11 did not reduce the growth or survival of PEL cells under standard culture conditions (E.G. and B.R.C., unpublished), a role for miR-K11 in KSHV-induced lymphomagenesis in humans seems plausible in analogy to the reported role of cellular miR-155 and a viral analog encoded by the chicken herpesvirus Marek’s disease virus (MDV) in cellular transformation and oncogenesis (Costinean et al., 2006; Linnstaedt et al., 2010; O’Connell et al., 2008; Zhao et al., 2011). A complete mechanistic understanding of the role of miR-155 and its analogs in cancer has remained elusive.
Several cellular targets of KSHV miRNAs have been proposed (Abend et al., 2010; Dolken et al., 2010; Gottwein and Cullen, 2010; Gottwein et al., 2007; Hansen et al., 2010; Lei et al., 2010; Lu et al., 2010a; Lu et al., 2010b; Nachmani et al., 2009; Samols et al., 2007; Skalsky et al., 2007; Ziegelbauer et al., 2009), and biological consequences of target regulation have been explored in some instances. Several KSHV miRNAs have also been proposed to target viral mRNAs, including those encoding RTA (ORF50), primase (ORF56) and MTA (ORF57) (Bellare and Ganem, 2009; Lin and Ganem, 2011; Lu et al., 2010b). Functions that have been attributed to KSHV miRNAs include the regulation of apoptosis (Abend et al., 2010; Ziegelbauer et al., 2009), transcriptional reprogramming (Hansen et al., 2010), regulation of epigenetic genome modification (Lu et al., 2010b), inhibition of cell cycle arrest (Gottwein and Cullen, 2010), changes in cytokine expression (Abend et al., 2010), escape from natural killer cell recognition (Nachmani et al., 2009) and regulation of the entry into lytic KSHV replication (Bellare and Ganem, 2009; Lei et al., 2010; Lu et al., 2010a; Lu et al., 2010b; Ziegelbauer et al., 2009).
The EBV genome contains two miRNA clusters. EBV-positive PEL cell lines express mature miRNAs from all 22 pre-miRNAs of the BART cluster, while BHRF1 miRNAs are not expressed (Cai et al., 2006). The function of the BART miRNAs in PEL has not been addressed and only a few BART miRNA targets have been proposed (Choy et al., 2008; Dolken et al., 2010; Lo et al., 2007; Marquitz et al., 2011; Nachmani et al., 2009).
In addition to the work described above, the identification of KSHV and EBV miRNA targets by Ago-2 immunoprecipitation followed by the quantification of associated transcripts by microarray analysis (RIP-CHIP) has been reported (Dolken et al., 2010). In this work, 114 and 44 mRNAs were identified as candidate targets of the KSHV and EBV miRNAs, respectively, based on increased association with Ago2 in cell lines expressing these miRNAs compared to cell lines lacking KSHV and EBV miRNA expression. While RIP-CHIP can serve as a measure for the association of transcripts with Ago2, miRNA binding site prediction from RIP-CHIP data has to consider the entire transcript and this technique therefore does not allow the direct identification of either the targeting miRNA(s) or specific miRNA binding sites. This is particularly limiting, given that miRNA-mediated repression is often cooperative. This study also identified far fewer mRNA targets than would be expected from the observation that individual miRNAs can target hundreds of mRNAs (Bartel, 2009).
Here, we report the direct and transcriptome-wide identification of miRNA binding sites in EBV-negative and EBV-positive PEL cell lines using PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation). To facilitate accurate target site identification, we first established the identity and sequence of all significantly expressed miRNAs by Illumina sequencing. PAR-CLIP analysis of PEL cell lines led to the identification of hundreds of cellular binding sites for KSHV and EBV miRNAs. This analysis also revealed that KSHV miR-K10a partially mimics the activity of cellular miR-142-3p.
RESULTS
Small RNAs expressed in PEL cell lines
We analyzed miRNA expression in the commonly used PEL cell lines BC-1, BCBL-1 and BC-3 by deep sequencing. BC-1 cells are latently infected with both KSHV and EBV and express EBV BART miRNAs in addition to KSHV miRNAs (Cai et al., 2006). Since the majority of clinical cases of PEL are positive for both KSHV and EBV (Cesarman et al., 1995), the co-expression of KSHV miRNAs and EBV BART miRNAs is potentially relevant to the pathogenesis of PEL. In contrast, BCBL-1 and BC-3 cells are latently infected only with KSHV. While deep sequencing of the KSHV miRNAs in the BCBL-1 and BC-3 cell lines has been reported recently (Lin et al., 2010; Umbach and Cullen, 2010), the small RNA composition of BC-1 cells has so far only been analyzed by low-throughput sequencing (Cai et al., 2005).
Small RNAs between ~18 and 25 nts were used for cDNA library construction and sequenced on the Illumina GAII platform. The genomic origin of aligned small RNA reads is presented in Figure 1A–C and Table S1. As previously reported (Umbach and Cullen, 2010), we observed that the majority (i.e. ~83%) of miRNA reads in BC-3 cells were of KSHV origin. In contrast, the percentages of KSHV miRNA reads obtained from BCBL-1 and BC-1 cell lines were much lower (~30% and ~27% of miRNA reads, respectively). We were able to validate higher expression of a subset of KSHV miRNAs as well as lower expression of the cellular miRNA miR-16 in BC-3 cells using primer extension analysis (Figure 1D and Figure 3A).
Figure 1. Small RNA sequencing.
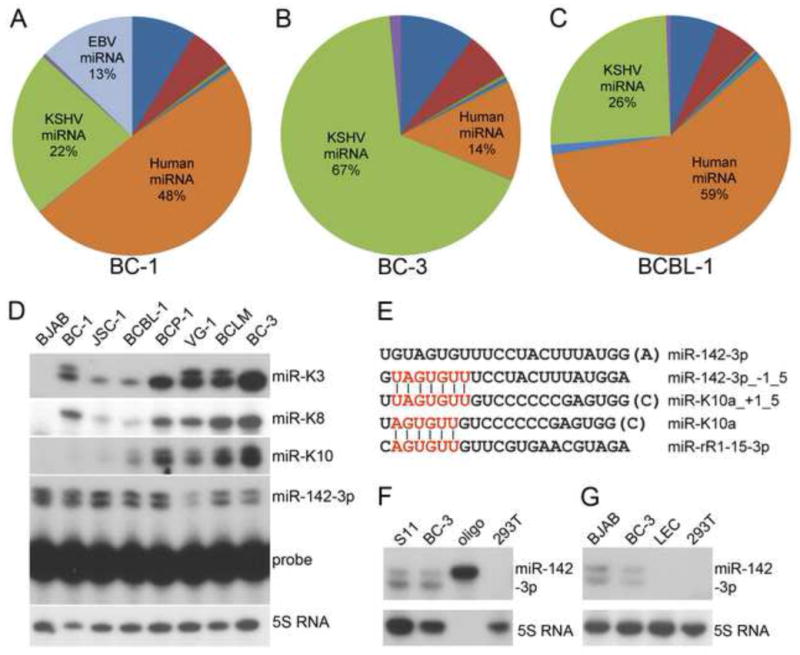
(A–C) Genomic location of aligned small RNA reads. For a detailed breakdown, see Table S1. (D) Primer extension analysis of miRNA expression in an extended set of PEL cell lines. miR-K10a was easily detected in BC-1 and JSC-1 cells upon longer exposure (not shown). (E) Alignment of miR-K10a and miR-142-3p sequences. −/+1_5 indicates a 5′ terminal offset from the miRBase sequence by 1 nucleotide. The seed sequence of miR-K10a is also identical to that of rhesus lymphocryptovirus miR-rR1-15-3p (Umbach et al., 2010). (F) Primer extension analysis of miR-142-3p in BC-3 cells and the murine B cells line S11. 293T was used as a negative control (G) primer extension analysis of miR-142-3p expression (top panel) in BJAB, BC-3 and lymphatic endothelial cells (LEC). 5S RNA served as loading control. See also Tables S1–4.
Figure 3. Characteristics of BC-1 and BC-3 PAR-CLIP libraries.
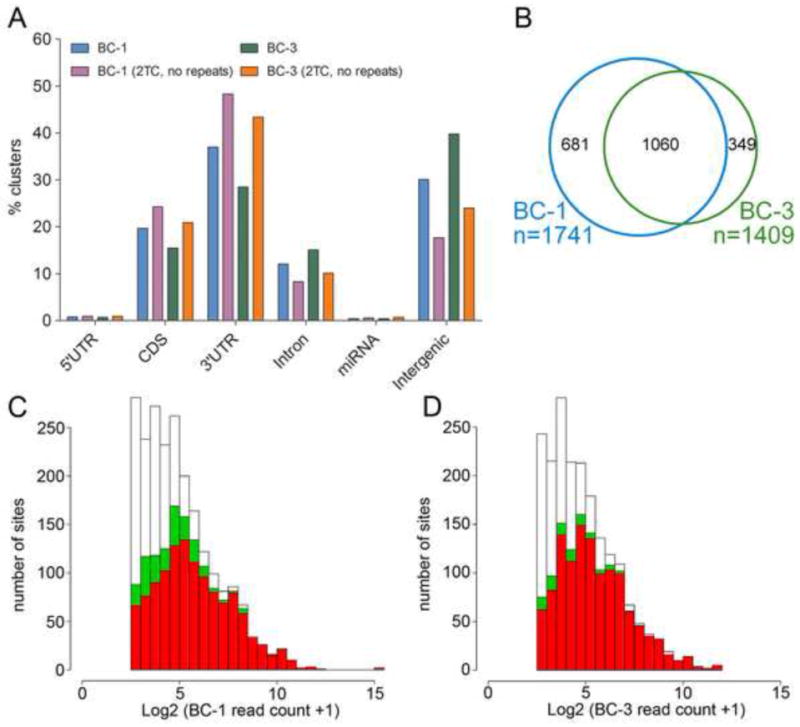
(A) Location of PAR-CLIP clusters within the human genome. “2TC, no repeats”: only clusters not mapping to repeats and with T to C conversions at ≥ 2 locations were analyzed. (B) Venn diagram showing the overlap of mRNAs with 3′UTR sites for KSHV miRNAs between BC-1 and BC-3 libraries. Only KSHV miRNAs expressed in both cell lines were considered. (C) Read depth for KSHV miRNA targets sites found in BC-1 (white), read depth for the subset of these clusters that were also recovered in BC-3 with ≥ 1 (green) or ≥2 (red) locations with T to C conversions. (D) as in (C), except that sites from BC-3 are in white and the sites that are also recovered in BC-1 are in green and red. See also Tables S5, 6.
The relative abundance of KSHV miRNA reads differed slightly between this manuscript and the data reported previously for BC-3 (Umbach and Cullen, 2010) and more substantially from those reported for BCBL-1 (Lin et al., 2010). Absolute read numbers were therefore only cautiously interpreted as indicators of the expression level for each miRNA. Nevertheless, this dataset identifies the precise sequence of all significantly expressed viral and cellular miRNAs in these cell lines. In addition, several other findings obtained from this dataset are pointed out below.
KSHV miRNAs
Significantly expressed miRNAs of KSHV origin are summarized in Table 1. KSHV miRNAs were recovered at substantially different frequencies in the same cell line, and frequencies of individual miRNAs also differed between cell lines. Primer extension analysis of miR-K3, miR-K8-3p and miR-K10 expression in an extended set of PEL cell lines confirmed that the expression of these miRNAs differs substantially between cell lines (Figure 1D). We and others detected KSHV miRNA sequences derived from both arms of most of the 12 KSHV pre-miRNAs (Table 1) (Lin et al., 2010; Umbach and Cullen, 2010). Our dataset also confirmed the absence of miR-K9 in BC-3 cells (Umbach and Cullen, 2010).
Table 1.
KSHV miRNA expression profile in PEL cell lines. −/+1_5 indicates a 5′ terminal offset from the miRBase sequence by 1 nucleotide. Nucleotides in brackets were not present in all reads considered.
Table 1. Significantly expressed KSHV miRNAs. Columns identify the miRNA name, which arm of the precursor the miRNA sequence is derived from, miRNA sequence, read counts and the %-age of total or KSHV miRNA reads these reads represented
| BC1 | BC3 | BCBL1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | Arm | Sequence | Count | % total | % KSHV | Count | % total | % KSHV | Count | % total | % KSHV |
| miR-K1-star | 3p | GCAGCACCTGTTTCCTGCAACC | 27 | 0.00% | 0.01% | 449 | 0.01% | 0.01% | 72 | 0.00% | 0.01% |
| miR-K1 | 5p | ATTACAGGAAACTGGGTGTAAG(CTG) | 12151 | 0.97% | 3.57% | 64825 | 1.64% | 1.96% | 57761 | 1.32% | 4.26% |
| miR-K2-star | 3p | GATCTTCCAGGGCTAGAGCTG | 250 | 0.02% | 0.07% | 659 | 0.02% | 0.02% | 309 | 0.01% | 0.02% |
| miR-K2 | 5p | AACTGTAGTCCGGGTCGAT(CTGA) | 5862 | 0.47% | 1.72% | 47772 | 1.21% | 1.45% | 37418 | 0.85% | 2.76% |
| miR-K3-star | 3p | TCGCGGTCACAGAATGTGACA | 31 | 0.00% | 0.01% | 201 | 0.01% | 0.01% | 201 | 0.00% | 0.01% |
| miR-K3 | 5p | TCACATTCTGAGGACGGCAGCGA(CG) | 4034 | 0.32% | 1.19% | 5546 | 0.14% | 0.17% | 5546 | 0.13% | 0.41% |
| miR-K3_+1_5 | 5p | ATCACATTCTGAGGACGGCAGCGA | 1163 | 0.09% | 0.34% | NA | NA | NA | NA | NA | NA |
| miR-K4-3p | 3p | TAGAATACTGAGGCCTAGCTG(A) | 40506 | 3.22% | 11.91% | 1093017 | 27.62% | 33.09% | 309943 | 7.06% | 22.84% |
| miR-K4-5p | 5p | AGCTAAACCGCAGTACTCTAGG | 340 | 0.03% | 0.10% | 13072 | 0.33% | 0.40% | 14667 | 0.33% | 1.08% |
| miR-K5 | 3p | TAGGATGCCTGGAACTTGCCGG(T) | 34494 | 2.74% | 10.15% | 18391 | 0.46% | 0.56% | 11647 | 0.27% | 0.86% |
| miR-K6-3p | 3p | TGATGGTTTTCGGGCTGTTGAG(C) | 133588 | 10.63% | 39.29% | 668532 | 16.90% | 20.24% | 584933 | 13.33% | 43.10% |
| miR-K6-5p | 5p | CCAGCAGCACCTAATCCATCGG | 723 | 0.06% | 0.21% | 22725 | 0.57% | 0.69% | 795 | 0.02% | 0.06% |
| miR-K7 | 3p | TGATCCCATGTTGCTGGCGC(TCA) | 10806 | 0.86% | 3.18% | 194585 | 4.92% | 5.89% | 128153 | 2.92% | 9.44% |
| miR-K7-star | 5p | AGCGCCACCGGACGGGGATTTATG | 218 | 0.02% | 0.06% | 6260 | 0.16% | 0.19% | 859 | 0.02% | 0.06% |
| miR-K8 | 3p | CTAGGCGCGACTGAGAGAGC(AC) | 38752 | 3.08% | 11.40% | 204612 | 5.17% | 6.19% | 24851 | 0.57% | 1.83% |
| miR-K8-star | 5p | ACTCCCTCACTAACGCCCCGCT | 230 | 0.02% | 0.07% | 5312 | 0.13% | 0.16% | 336 | 0.01% | 0.02% |
| miR-K9 | 3p | CTGGGTATACGCAGCTGCGT(AA) | 286 | 0.02% | 0.08% | NA | NA | NA | 242 | 0.01% | 0.02% |
| miR-K9-star | 5p | ACCCAGCTGCGTAAACCCCG(CT) | 305 | 0.02% | 0.09% | NA | NA | NA | 793 | 0.02% | 0.06% |
| miR-K10a | 3p | TAGTGTTGTCCCCCCGAGTGG(C) | 283 | 0.02% | 0.08% | 11156 | 0.28% | 0.34% | 7908 | 0.18% | 0.58% |
| miR-K10b | 3p | TGGTGTTGTCCCCCCGAGTGG(C) | 30 | 0.00% | 0.01% | 697 | 0.02% | 0.02% | 467 | 0.01% | 0.03% |
| miR-K10a_+1_5 | 3p | TTAGTGTTGTCCCCCCGAGTGG(C) | 92 | 0.01% | 0.03% | 6461 | 0.16% | 0.20% | 4539 | 0.10% | 0.33% |
| miR-K10b_+1_5 | 3p | TTGGTGTTGTCCCCCCGAGTGG(C) | 28 | 0.00% | 0.01% | 713 | 0.02% | 0.02% | 398 | 0.01% | 0.03% |
| miR-K10-star | 5p | GGCTTGGGGCGATACCACCACT | NA | NA | NA | 341 | 0.01% | 0.01% | 1175 | 0.03% | 0.09% |
| miR-K11 | 3p | TTAATGCTTAGCCTGTGTCCG(AT) | 4124 | 0.33% | 1.21% | 101848 | 2.57% | 3.08% | 56219 | 1.28% | 4.14% |
| miR-K11-star | 5p | GGTCACAGCTTAAACATTTCTAGG | 18 | 0.00% | 0.01% | 546 | 0.01% | 0.02% | 1106 | 0.03% | 0.08% |
| miR-K12-star | 3p | TGGGGGAGGGTGCCCTGGTTG(A) | 233 | 0.02% | 0.07% | 79473 | 2.01% | 2.41% | 17691 | 0.40% | 1.30% |
| miR-K12 | 5p | AACCAGGCCACCATTCCTCTCCG | 3 | 0.00% | 0.00% | 823 | 0.02% | 0.02% | 776 | 0.02% | 0.06% |
As reported (Umbach and Cullen, 2010), the 5′ termini of the mature KSHV miRNAs were mostly uniform, with some notable exceptions (Table 1). Specifically, we confirmed the presence of four distinct seed sequences of miR-K10-3p, as reported previously for BC-3 (Umbach and Cullen, 2010), in all three cell lines. Seed variation of miR-K10-3p is due to differential processing and adenosine (A) to inosine (I) editing within the seed of both 5′-processing variants of miR-K10 (Pfeffer et al., 2005; Umbach and Cullen, 2010). By primer extension analysis, we confirmed that miR-K10 has two different 5′ ends in a panel of seven PEL cell lines (Figure 1D). Interestingly, BC-1 libraries, but not BCBL-1 and BC-3 libraries, also contained a novel 5′ seed variant of miR-K3, in addition to the variant shared with BC-3 and BCBL-1 (Table 1). Using primer extension analysis, we detected expression of miR-K3_+1_5 in BC-1 cells and, surprisingly, also in two other PEL cell lines (Figure 1D). It remains to be addressed whether differences in miR-K3 processing between PEL cell lines reflect genomic sequence polymorphisms. Significant A to I editing of viral miRNAs other than miR-K10 were not observed (not shown) and the presence of previously reported low abundance small RNAs was not analyzed due to the limited sequencing depth of the dataset (Lin et al., 2010; Umbach and Cullen, 2010).
We and others have previously shown that miR-K11 shares a seed region with miR-155, resulting in regulation of an overlapping set of target mRNAs by these two miRNAs (Gottwein et al., 2007; Skalsky et al., 2007). Interestingly, miR-K10a_+1_5 has a 7mer seed region identical to that of cellular miRNA miR-142-3p_−1_5, whose sequence differed by one 5′-terminal nucleotide from that reported in miR-base (v16) (Figure 1E) (Griffiths-Jones et al., 2006; Wu et al., 2009). Using primer extension analysis, we confirmed the expression of miR-142-3p and miR-142-3p_−1_5 in a panel of PEL cell lines (Figure 1D). While pri-miR-142-3p has been reported to undergo A to I editing within the miRNA stem loop (Yang et al., 2006), we did not detect editing of mature miR-142-3p in our libraries (data not shown), despite high numbers of miR-142-3p reads. The detection of identical 5′ variants of miR-142-3p in the murine B cell line S11 (Figure 1F) indicated that the seeds of both miR-142-3p 5′ variants are evolutionary conserved and therefore likely important for miR-142-3p function.
The expression of miR-142-3p is restricted to hematopoietic cells and overexpression of miR-142 in murine hematopoietic progenitor cells enhances T-cell differentiation in vitro (Chen et al., 2004). Targets and specific functions of miR-142-3p, however, remain largely unexplored. The expression pattern of miR-142-3p suggests that miR-K10a_+1_5 may be redundant with miR-142-3p_−1_5 in B cells. Because KSHV also causes Kaposi’s sarcoma, a tumor driven by KSHV infected endothelial cells, we tested whether miR-142-3p is expressed in primary human lymphatic endothelial cells (LECs). Expression of miR-142-3p in LECs was not detectable by either primer extension analysis (Figure 1G) or Illumina sequencing (not shown), suggesting that miR-K10a_+1_5 is not redundant with miR-142-3p expression in endothelial cells, a cell type relevant to the biology and pathogenesis of KSHV. Using the miRNA sequencing data described above and miRBase (v16), we identified several other KSHV miRNAs with potential cellular analogs (Table S2).
EBV and cellular miRNA expression profile
Consistent with previous reports (Cai et al., 2006; Grundhoff et al., 2006; Pfeffer et al., 2004), we detected mature miRNAs derived from all known BART pre-miRNAs but no mature BHRF1 miRNAs in BC-1 cells (Table S3). While the EBV BART-miRNAs together accounted for only 13% of the miRNA reads in BC-1, reads for some individual EBV miRNAs were abundant with BART-16-5p, miR-BART-22 and miR-BART-6-3p together accounting for ~56% of the EBV miRNA reads. Candidates for cellular analogs of the EBV BART miRNAs have been listed previously (Chen et al., 2010). All three PEL cell lines showed high expression of several cellular miRNAs, despite the overall lower expression of cellular miRNAs in BC-3 cells. Cellular miRNA seed sequences that were among the top 15 most abundant cellular seed sequences in all three libraries are listed in Table S4.
PAR-CLIP
We selected BC-1 and BC-3 cells for the identification of miRNA binding sites using PAR-CLIP, a technology which allows the transcriptome-wide identification of miRNA target sites, when applied to Argonaute (Ago) proteins (Hafner et al., 2010). Briefly, PEL cells were incubated with the photoactivatable ribonucleoside 4-thiouridine (4SU), which is incorporated into newly synthesized RNAs. Cells were UV-irradiated at 365 nm, which efficiently crosslinks 4SU to associated RNA binding proteins (Hafner et al., 2010). Cells were then lysed and immunoprecipitated for endogenous Ago2. Other Ago proteins were not included, because indistinguishable results have been obtained previously for the analysis of different Ago proteins (Hafner et al., 2010). Immunoprecipitates were digested with RNaseT1, protected RNAs were radiolabeled and Ago2-RNA complexes purified by SDS-PAGE. Associated RNA fragments were recovered and processed for Illumina sequencing. Due to altered base-pairing properties of formerly crosslinked 4SU bases, resulting libraries contain thymidine (T) to cytidine (C) conversions at crosslinking sites, thereby distinguishing signal from noise (Hafner et al., 2010). Crosslinking is not observed in regions that undergo perfect base-pairing to the miRNA seed, and T to C conversions are typically observed 5′ of the miRNA seed match (Hafner et al., 2010).
Reads obtained for each library were analyzed using mapping and clustering strategies described in Figure 2 and Supplemental Experimental Procedures. We obtained 12,887,172 reads for BC-1 cells and 14,029,755 reads for BC-3 cells, ~40% (BC-1) and ~26% (BC-3) of which mapped to the human or viral genomes at a single location. 75% (BC-1) and 49% (BC-3) of aligned reads had T to C conversions. We used the PARalyzer toolkit to identify Ago2 binding sites (Corcoran et al., 2011). Reads that overlapped by at least 1nt were grouped together. Next, clusters were generated based on the distributions of T to C transitions (signal) and non-converted Ts (noise) as well as read depth. This approach results in clusters that are smaller and consequently allows for higher resolution of closely spaced binding sites compared to the approach reported previously (Hafner et al., 2010). The breakdown of the resulting clusters is presented in Figure 3A and Table S5. Clusters were most frequently located in 3′UTR regions (37% for BC-1 and 29% for BC-3 cells) and intergenic regions (30% in BC-1 and 39% in BC-3 cells). A smaller percentage of clusters mapped to coding regions and introns. When we required T to C conversion events at more than one location within clusters and excluded clusters mapping to repeats, the percentage of clusters in 3′UTRs and in coding regions increased (Figure 3A). In contrast, clusters in intergenic and intronic regions of the genome became less frequent. Together, these observations confirm that miRNA binding sites are preferentially located in 3′UTRs and indicate that at least a subset of intronic and intergenic clusters likely represent transient interactions that are captured by PAR-CLIP.
Figure 2. Computational analysis of PAR-CLIP data.
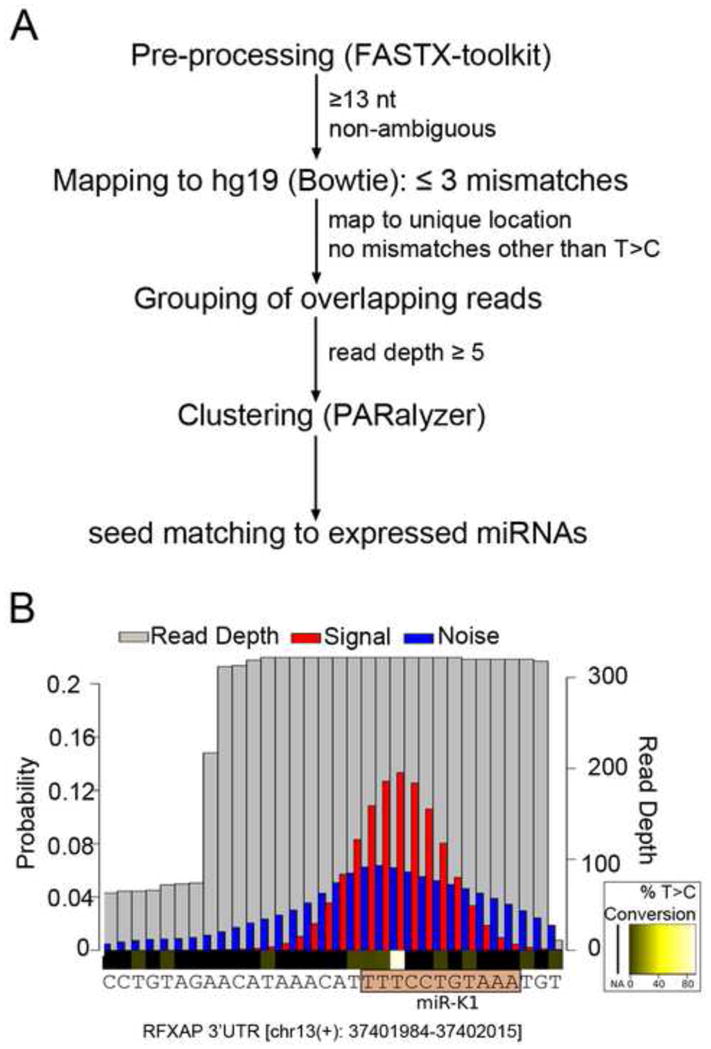
(A) Flowchart outlining the computational pipeline used for PAR-CLIP analysis. (B) Example of PARalyzer cluster identification. A BC-1 derived cluster mapping to the 3′UTR of RFXAP with a seed match to miR-K1 is shown. Shown are read depth (grey) and the distributions of T to C conversions (red, signal) and non-converted Ts (blue, noise). The seed match to miR-K1 is highlighted. This seed match is validated further below.
Finally, we examined clusters for the presence of canonical seed matches to all miRNAs that are significantly expressed in the matched small RNA libraries described above. The minimal seed matches considered were “7mer1A” and “7mer2-8” matches (Bartel, 2009). Clusters with seed matches to expressed miRNAs were considered candidate target sites. Of the clusters mapping to human 3′UTRs, 69% (BC-1) and 70% (BC-3) had seed matches to expressed miRNAs (Tables S6). Similarly, ~65% (BC-1 and BC-3) of clusters that mapped to human coding regions had seed matches to expressed miRNAs. Of note, assignment of the targeting miRNA is sometimes not entirely unambiguous, when clusters contain matches to more than one miRNA or for miRNAs with seed homology. Our unrelated work using WT and miRNA-mutant EBV allowed us to estimate the rate of incorrect assignment to be ≤ 15% (Skalsky et al., unpublished, and Supplemental Discussion).
To address the overlap in candidate targets between the two libraries, we compared human candidate 3′UTR targets of KSHV miRNAs expressed in both cell lines. We restricted this analysis to 3′UTRs, because recent publications suggest that miRNA binding sites located in 3′UTRs result in greater target inhibition than those located in coding regions (Gu et al., 2009; Hafner et al., 2010). We identified 1741 candidate target mRNAs of the KSHV miRNAs in BC-1 cells and 1409 candidate target mRNAs in BC-3 cells, with 60% of the BC-1 sites and ~75% of the BC-3 sites detected in both libraries (Figure 3B). To address whether the overlap between the two libraries may be limited primarily by library depth rather than biological differences, we plotted read numbers for clusters with KSHV miRNA seed matches from BC-1 and superimposed clusters that were also recovered from BC-3 cells (Figure 3C). It is evident that sites detected in one library were typically also recovered in the other library if they had higher read numbers, while overlap was more limited for sites with lower read depth. The same was true for targets found in BC-3 (Figure 3D). Therefore, limited library depth may at least partially account for the incomplete overlap between KSHV miRNA target sites recovered from both cell lines.
Many published miRNA targets are confirmed by PAR-CLIP
To confirm that the PAR-CLIP libraries correctly identified miRNA targets, we took advantage of our previous finding that miR-K11 is a functional analog of cellular miR-155 (Gottwein et al., 2007; Skalsky et al., 2007). To date, 153 3′UTR targets of miR-155 have been validated minimally using 3′UTR indicator assays (Table S7). Of these, 121 were expressed in BC-1 and/or BC-3 cells based on published (Fan et al., 2005) and/or newly generated microarray data (Tables S7 and S8). 47 of these 121 mRNAs were identified as targets for miR-K11 in at least one of the PAR-CLIP libraries. In cases where these targets were also predicted as targets of miR-155 by Targetscan (Grimson et al., 2007), PAR-CLIP identified the conserved site(s) of interaction. In addition, PAR-CLIP identified >300 mRNAs as candidate targets of miR-K11 that have not previously been reported to be targets of miR-155 or miR-K11. Despite the large number of miR-K11 targets recovered, ~70 previously proposed targets of miR-155 with expression in BC-1 and BC-3 were not confirmed as targets of miR-K11 by PAR-CLIP. These mRNAs may 1) be targets of miR-155 in other settings, but not targets of miR-K11 in PEL; 2) may have been missed in our analysis, for example due to limitations in library depth; or 3) may not be authentic miR-155/miR-K11 targets. Nevertheless, the recovery of ~40% of the reported miR-155 targets strongly suggests that these libraries correctly identify many valid miRNA targets.
Our dataset also confirmed 12 out of 29 validated KSHV miRNA targets with expression in our dataset (>40%). Confirmed interactions include those with BACH1, FOS, CDKN1A (p21), TNFRSF12A (TWEAKR), RAD21 and RBL2 mRNAs, whose regulation had previously been validated at the level of protein expression (Abend et al., 2010; Gottwein and Cullen, 2010; Gottwein et al., 2007; Li et al., 2009; Lu et al., 2010b). A list summarizing the recovery of previously published targets of the KSHV and EBV miRNAs is presented in Table S9. This table also includes a comparison with the dataset published by Dölken et al. (Dolken et al., 2010), which is further discussed in the Supplemental Discussion.
PAR-CLIP targets of KSHV miRNAs are downregulated at the mRNA level
miRNA-mediated regulation often leads to a drop in mRNA expression (Lim et al., 2005). We therefore investigated whether PAR-CLIP targets were downregulated in microarray data from KSHV-negative B-cell pools individually expressing physiological levels of miR-K1, miR-K4-3p and miR-K11 ( Gottwein et al., 2007 and Figure 4A). In addition to the reported comprehensive validation of the array data for miR-K11 (Gottwein et al., 2007), we confirmed that changes in mRNA expression corresponded to changes in protein levels for two candidate targets of the KSHV miRNAs: Tumor protein D52 (TPD52) was identified as a candidate target for miR-K4-3p and miR-K11 by PAR-CLIP, but its regulation in the microarray data was statistically significant only for miR-K4-3p. As anticipated, TPD52 was downregulated at the protein level by miR-K4-3p (Figure 4B). TPD52 expression was also lower in cells expressing miR-K11, suggesting that PAR-CLIP correctly identified TPD52 as a target of miR-K11. The catalytic subunit of protein phosphatase 2A (PPP2CA) was identified as a target for miR-K1 by both PAR-CLIP and microarray analysis and was also regulated by miR-K1 at the protein level (Figure 4B).
Figure 4. PAR-CLIP targets of KSHV miRNAs are downregulated at the mRNA level.
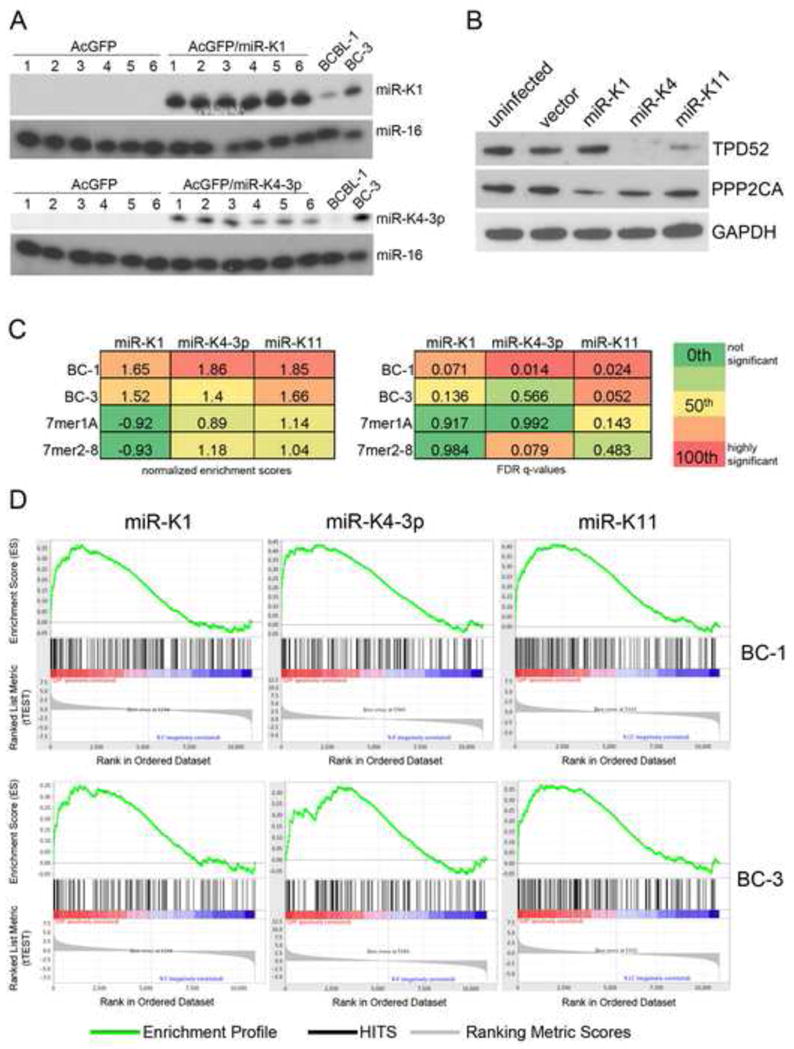
(A) Primer extension analysis of BJAB cell pools stably expressing KSHV miR-K1 (upper) or miR-K4-3p (lower). miR-16 served as a loading control. (B) Western Blot analysis of TPD52 and PPP2CA expression in untransduced BJAB cells and transductants stably expressing the control vector or the indicated miRNA. GAPDH expression served as loading control. (C) Normalized enrichment scores (NES) and FDR q-values for the enrichment of PAR-CLIP targets of the indicated miRNA from BC-1 or BC-3 libraries in the corresponding microarray data. Also shown are data for the enrichment of either 7mer1A or 7mer2-8 seed matches in the array data. (D) Gene set enrichment plots showing the enrichment of PAR-targets of the indicated miRNA from BC-1 or BC-3 libraries in the corresponding rank-ordered gene lists. See also Table S10.
We next used gene set enrichment analysis (GSEA) (Subramanian et al., 2005) to analyze the distribution of PAR-CLIP 3′UTR targets of miR-K1, miR-K4-3p and miR-K11 in the microarray-derived rank ordered gene lists (Figure 4C and Figure 4D). Importantly, PAR-CLIP targets were significantly enriched among downregulated genes. In contrast, enrichments of candidate targets predicted by seed matching alone were not statistically significant when the entire gene list was considered (Figure 4C and 4D). Thus, PAR-CLIP clearly outperformed seed matching in the prediction of functional miRNA targets from microarray data, despite the fact that PAR-CLIP targets were derived from unrelated B-cell lines. PAR-CLIP derived targets that contributed to these enrichments are listed in Tables S10.
PAR-CLIP successfully identifies KSHV miRNA targets
The data described above strongly suggests that PAR-CLIP identified functional targets of the KSHV miRNAs. We next asked whether PAR-CLIP-identified 3′UTR target interactions could be validated using 3′UTR indicator assays. We focused this analysis mostly on 3′UTRs of mRNAs encoding proteins with functions of potential relevance to KSHV biology and pathogenesis (Table S11). Thirty-six combinations of 3′UTR indicators and KSHV miRNAs with PAR-CLIP interactions were tested for regulation. Nine of these interactions were independently also predicted by microarray analysis of the BJAB cell pools expressing miR-K1, miR-K4-3p, or miR-K11 described above (i.e. for miR-K1: NMI, RAD21, BCL11A, RFXAP; for miR-K4-3p: TPD52, GRB2, MCC, YWHAB; for miR-K11: WEE1). Targets were representative of the full range of seed match types observed in the libraries (Tables S6 and S11).
Resulting data are summarized as a heatmap in Figure 5A and presented as individual bargraphs in Figure S1. Expression controls for the KSHV miRNAs are also presented in Figure S1. Of the 36 miRNA-3′UTR pairs, 28 showed significant downregulation of 3′UTR indicator activity, i.e. > 77% of PAR-CLIP predicted target sites could be validated as regulatory interactions using this assay. Validated interactions included several targets with minimal seed base pairing (NMI, CSNK1A1), several targets that were detected by PAR-CLIP in only one library, targets with ambiguous seed match assignment (e.g. ZFYVE9: miR-K9) and also targets with only one location of T to C conversion events in the relevant cluster (BCL11A, SOS1, WEE1), suggesting that these criteria do not necessary limit successful target validation. Eight 3′UTR-miRNA combinations did not respond as expected (green boxes in Figure 5A, Figure S1). Potential reasons for the observed lack of regulation are discussed in the Supplementary Discussion.
Figure 5. Validation of PAR-CLIP targets by 3′UTR indicator assays.
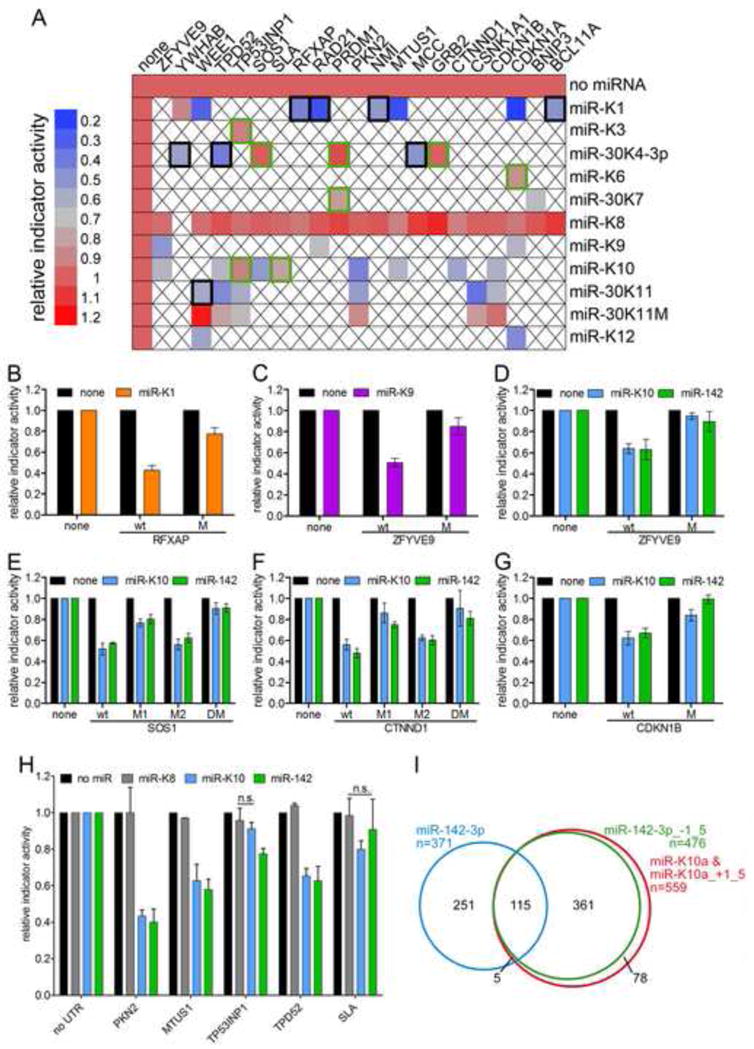
(A) Heatmap summarizing 3′UTR indicator data obtained with combinations of 3′UTR indicators (columns) and miRNA expression vectors (rows) as indicated. Expression of miR-K1 (YWHAB), miR-K8 (all other 3′UTRs) or seed mutant miR-K11 served as negative controls. Other than controls, 37 interactions were tested, all predicted by PAR-CLIP, with the exception of CDKN1B/miR-K11. Black boxes mark mRNAs that were downregulated in microarray data. Regulation was statistically significant (p<0.05, n≥3), except where indicated by green boxes. Empty boxes: not tested. (B) The miR-K1 seed match in the 3′UTR of RFXAP was inactivated by mutation (CTGTAA to CAGTTA). (C) The seed match to miR-K9-3p in the 3′UTR of ZFYVE9 was inactivated by mutation (TACCCA to TTCCGA) (D–G) Seed matches to miR-K10/miR-142-3p in the ZFYVE9 (D), SOS1 (E), CTNND1 (F) and CDKN1B (G) 3′UTRs were inactivated (ACACTA to AGACAA) alone (M, M1, M2) or in combination (DM). (H) Additional miR-K10 candidate targets were tested for regulation by miR-142-3p. Expression of miR-K8 served as negative control. All error bars represent s.d. (n≥3). See also Figure S1. (I) Overlap of mRNAs with 3′UTR clusters assigned to seed variants of miR-K10 and miR-142-3p.
We next sought to determine if targets of miR-K10a are shared by miR-142-3p, as predicted from the identical seed sequences of miR-K10_+1_5 and miR-142-3p_−1_5. We tested nine 3′UTR indicator vectors with PAR-CLIP-predicted 3′UTR targets for miR-K10a_+1_5p and miR-142-3p_−1_5 for regulation by miR-142 (Figure 5D–H). For eight of nine 3′UTRs tested, expression of miR-K10 and miR-142 resulted in similar levels of downregulation of indicator activity, suggesting that these two miRNAs do indeed share targets, as predicted by their seed homology and PAR-CLIP analysis. The 3′UTR indicator vector for SLA did not respond significantly to either miR-K10 or miR-142-3p expression (Figure 5A, H). To test whether PAR-CLIP indeed correctly identified miRNA binding sites, we introduced 2nt seed mutations into nine PAR-CLIP-predicted binding sites. To further strengthen the point that miR-K10a and miR-142-3p indeed act through the same binding sites, 7 of these were predicted to be targeted by miR-K10a/miR-142-3p. In all cases where miRNA expression resulted in significant downregulation of indicator expression, seed mutation rescued indicator expression (Figure 5B–G), further demonstrating that PAR-CLIP indeed correctly identified the site(s) of interaction. In addition, these experiments also demonstrate that KSHV encodes at least two functional mimics of cellular miRNAs expressed in hematopoietic cells, i.e. miR-142-3p_−1_5 and miR-155.
While more experiments will be required to elucidate functional differences between miR-K10a and miR-142-3p, it is relevant to point out that, because the first nucleotide of miR-K10a is a uridine, 7mer1A matches to miR-K10a are also 7mer2-8 matches to miR-K10a_+1_5/miR-142-3p_−1_5. Consequently, only ~15% of the PAR-CLIP-predicted targets for both miR-K10a 5′ seed variants are predicted to be unique targets of miR-K10a (Figure 5I). We therefore anticipate that, together, the two seed variants of miR-K10a effectively mimic miR-142-3p_−1_5. In contrast, less than 30% of the PAR-CLIP predicted targets for both miR-142-3p variants are common to both seed variants of this miRNA (Figure 5I).
Properties of the KSHV miRNA targets
To predict which cellular pathways are likely to be regulated by KSHV miRNAs, we tested whether PAR-CLIP targets of the KSHV miRNAs are enriched in specific cellular pathways with “Gene Ontology (GO) FAT” terms using DAVID (Huang da et al., 2009). A small subset of pathways with significant enrichment of KSHV miRNA candidate targets is listed in Figure 6A. The complete list of pathways with significant enrichments can be found in Table S12A. Based on the large number of KSHV miRNA targets and the fact that we find significant enrichments in multiple cellular pathways, it is evident that the KSHV miRNAs together serve many different functions. KSHV miRNA targets were especially enriched for transcriptional activators and repressors, proteins with roles in signal transduction and protein and vesicular trafficking.
Figure 6. Properties of KSHV miRNA targets.
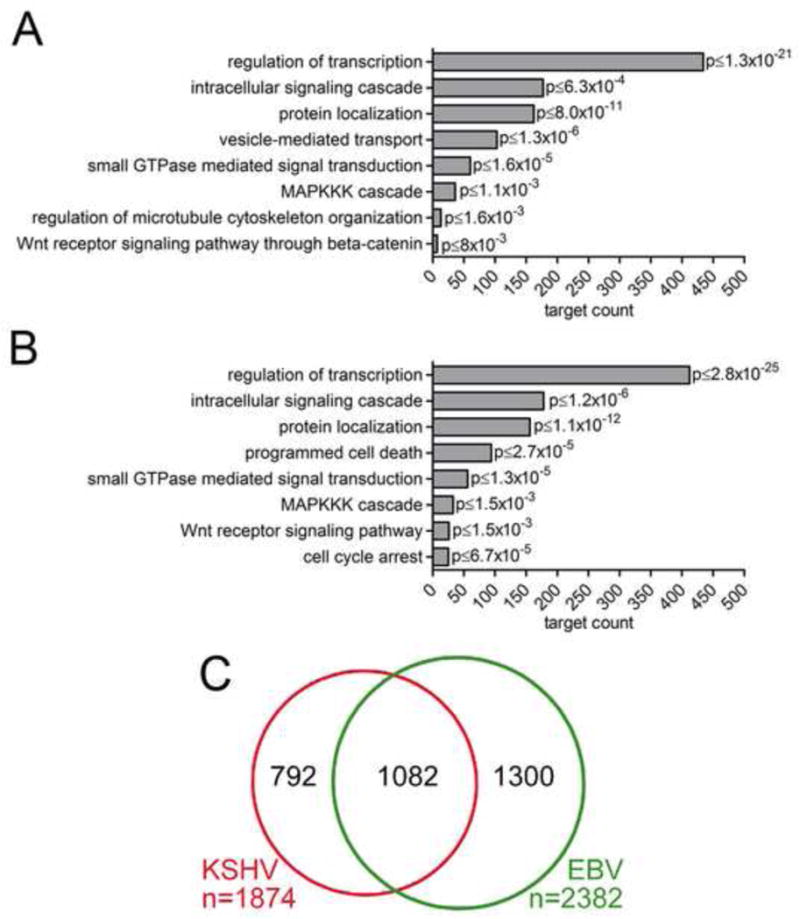
(A) Selected pathways with significant enrichments of KSHV miRNA targets. p values shown were calculated against the background of the human genome, but remained significant (p<0.05) when calculated against mRNAs expressed in both PEL cell lines or transcripts with PAR-CLIP clusters. (B) Selected pathways with significant enrichments of EBV miRNA targets. p values were calculated against human genome, all pathways shown remained significant when calculated against expressed mRNAs or transcripts with PAR-CLIP clusters in BC-1 cells. (C) Overlap of KSHV and EBV 3′UTR target mRNAs with T to C conversions at minimally 2 locations identified in BC-1. See also Tables S8, 12.
We next conducted a similar analysis for 3′UTR targets of the EBV BART miRNAs. A small subset of overrepresented terms is shown in Figure 6B and the full set of terms with enrichments and associated target mRNAs is presented in Table S12B. Interestingly, many of the pathways with enrichments overlapped with those targeted by the KSHV miRNAs, suggesting functional similarities between the KSHV and EBV miRNAs. This finding prompted us to investigate the overlap between cellular targets of the KSHV miRNAs and the EBV miRNAs. 58% of the 3′UTR targets of KSHV miRNAs also had clusters with seed matches to EBV BART miRNAs. This distribution is very unlikely to be due to chance (p<10−8). This was estimated by random sampling of the same number of targets from all target mRNAs recovered in the BC-1 PAR-CLIP library (Supplemental Experimental Procedures). Consequently, it appears that the KSHV and EBV miRNAs have independently evolved to share many targets and therefore may have overlapping functions, despite the distinct seed sequences of the KSHV and EBV miRNAs. However, validation of these interactions and functional experiments will be required to confirm that this overlap in PAR-CLIP targets indeed translates into significant functional similarities. Similar functions of the KSHV and EBV miRNAs are perhaps expected, given that KSHV and EBV share tropism for B cells in vivo and have similar requirements for latent infection.
The large majority of the targets of either KSHV or EBV miRNAs were cellular mRNAs. However, several clusters with seed matches to either viral and/or cellular miRNAs were detected in the KSHV and EBV genomes (Tables S6 and data not shown). Because nearly all cells used for PAR-CLIP do not express the KSHV transcripts characteristic of lytic replication, these clusters were mostly located in mRNAs with known expression during KSHV latency (Table S13). Clusters detected in both cell lines included sites for miR-K10/miR-142-3p in the 3′UTRs of vIL-6 and v-IRF3 as well as two clusters each for miR-K10/miR-142-3p and let7 located within the mRNAs encoding the viral latent proteins LANA, v-cyclin and v-FLIP. The functional significance of these sites remains unknown. We did not detect evidence of any of the reported interactions with lytic mRNAs (Bellare and Ganem, 2009; Lin and Ganem, 2011), but such interactions might have been missed due to the very low abundance of these mRNAs.
DISCUSSION
During latency, KSHV expresses few proteins but multiple viral miRNAs. While a substantial amount of work has addressed the role of the latent proteins, the functions of KSHV-encoded miRNAs remain largely unknown. Here we report the comprehensive identification of miRNA target sites of all significantly expressed miRNAs in EBV-negative and EBV-positive PEL cell lines. The resulting dataset confirmed many interactions with known functional relevance and identified KSHV miRNA binding sites in >2000 candidate targets of the KSHV miRNAs. Based on the recovery of published targets, we estimate that our dataset identifies minimally 40% of canonical 3′UTR targets of the KSHV miRNAs in B cells. We believe this estimate to be conservative, because many published interactions are supported by only minimal evidence and may therefore prove to be false. Moreover, not every possible interaction is expected to occur in every cellular context. Assuming that at least a subset of the binding sites found in coding regions are functional and that KSHV miRNAs may also engage in some interactions that are not dependent on perfect seed base-pairing, the actual number of cellular binding sites for KSHV miRNAs will certainly reach several thousand. Consequently, it appears that the KSHV miRNAs have evolved a highly complex regulatory network with the human transcriptome. Despite this, it also remains possible that only a relatively small subset of these interactions provide a strong selective advantage to KSHV, while many other interactions could be largely inconsequential to KSHV biology. Such particularly important targets could include those with multiple KSHV miRNA binding sites, for example CDKN1A (p21) or Wee1.
This report identifies targets of viral miRNAs at sufficient depth to allow meaningful pathway analysis, which revealed a number of pathways that are likely to be regulated by these miRNAs. These included transcriptional regulation, signal transduction, vesicular trafficking, and the regulation of cell cycle and apoptosis. Interestingly, we found that 58% of genes targeted by KSHV miRNAs were also bound by EBV miRNAs. Because KSHV and EBV have a similar biology, this observation may help to identify targets of key importance to these viruses. It is important to point out that PAR-CLIP merely captures interactions, including those that may be transient or may not result in functionally relevant levels of regulation (see above and Supplemental Discussion). Thus, this manuscript is intended to provide a resource for further validation and functional investigation. Clearly, loss of function experiments in the context of the virus and gain of function experiments with ectopically expressed miRNAs will be needed to validate the inhibitory effect of the KSHV miRNAs on individual targets and on the pathways listed in this manuscript.
Finally, our data revealed that KSHV encodes miRNAs that mimic the function of two hematopoietic microRNAs, miR-155 and miR-142-3p. Because miR-142-3p is likely expressed in lymphoid targets of KSHV, miR-K10a may have evolved to overexpress one of the miR-142-3p seeds in B cells and thus enforce the repression of a subset of miR-142-3p targets. Alternatively miR-K10a may mimic a subset of miR-142-3p functions in cell types lacking endogenous miR-142 expression, including endothelial cells. The observation that miR-142-3p/miR-K10 interact with a number of latent transcripts also raises the possibility that KSHV uses this abundant cellular miRNA to fine tune latent gene expression in lymphoid cells.
EXPERIMENTAL PROCEDURES
Small RNA and PAR-CLIP library construction and bioinformatics analysis
Small RNA and PAR-CLIP libraries were constructed as described (Hafner et al., 2010; Umbach and Cullen, 2010). Resulting reads were pre-processed using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit) and aligned to the human, KSHV and/or EBV genomes using Bowtie (Langmead et al., 2009). PAR-CLIP libraries were prepared as described (Hafner et al., 2010). For PAR-CLIP, ~1.5 × 109 BC-3 cells and ~3 × 109 BC-1 cells were labeled with 100μM 4-Thiouridine (4SU, Sigma) for ~18 hours. At the time of harvest, cells were at or below 106 cells/ml. PAR-CLIP data were analyzed using PARalyzer, with slightly different mapping parameters (Corcoran et al., 2011). Briefly, we employed a low stringency mapping strategy, allowing up to three mismatches per read to maximize the number of unique sites. Reads of at least 13nts were kept if they mapped to a unique location after discounting T to C mismatches. Consecutive nucleotides with higher likelihood of T to C conversion than non-conversion and a read depth of at least 5 are considered “interaction sites” and were extended to clusters, which include all adjacent positions with a read depth of at least 5. Clusters were searched for miRNA seed matches to all miRNAs with significant expression in the matched small RNA sequencing samples. Seed matches were designated according to standard nomenclature (Bartel, 2009): for e.g. “7mer1A” seed matches, nts 2–7 of the miRNA are perfectly base paired with the target and the match is followed by an A in the 3′UTR. For e.g. “7mer2-8” seed matches, nts 2–8 of the miRNA are perfectly base paired. We designated different seed types for up to 12mer seed matches. Seed matches that were more extensive were also designated as “12mer” seed matches. Next generation sequencing data were submitted to GEO (GSE32113) and the complete, analyzed small RNA and PAR-CLIP datasets can be downloaded from http://bugs.mimnet.northwestern.edu/labs/gottweinlab/index.html.
Plasmids
Plasmids were either previously reported or based on the previously published vectors pLCE, for miRNA expression, and pLSG/pLSR for 3′UTR indicator expression (Gottwein and Cullen, 2010; Gottwein et al., 2007; Zhang et al., 2009). Detailed cloning procedures can be found in Supplemental Experimental Procedures.
Cell lines, microarrays, and cell based assays
All cell lines were cultured as previously described (Gottwein and Cullen, 2010; Gottwein et al., 2007). BC-1 and BC-3 cells were from ATCC, other PEL cell lines were obtained from Dirk Dittmer (UNC), with permission from the original investigators. Lentivirally transduced BJAB cell pools expressing miR-K1, miR-K4-3p or miR-K11 were described previously or generated as described using published constructs (Gottwein et al., 2007). Microarray analysis of BJAB cell pools was performed on Human Operon v3.0.2 arrays as described for miR-K11 (Gottwein et al., 2007). Microarray analysis of BC-1 and BC-3 cell lines was done on Affymetrix Gene 1.0 ST arrays. Indicator assays, primer extension analyses and Western Blotting were carried out as described previously (Gottwein and Cullen, 2010; Gottwein et al., 2007) with modifications detailed in the Supplemental Experimental Procedures. Primary antibodies specific for TPD52 (1:500, antibody H-45, sc-67063) and GAPDH (1:5000, antibody 0411, sc-47724) were from Santa Cruz, primary antibody for PPP2CA (1:1000, antibody 52F8, catalog number 2259) was from Cell Signaling Technologies. Microarray and Illumina sequencing data were submitted to GEO (accession number GSE32113).
Supplementary Material
HIGHLIGHTS.
miRNA target sites of all miRNAs expressed in KSHV infected B cells were identified
KSHV-encoded miRNAs target more than 2000 cellular mRNA transcripts
The KSHV miRNA miR-K10a mimics cellular miR-142-3p function
Acknowledgments
The authors would like to thank Dr. Dirk Dittmer for cell lines, John Whitesides in the Duke Center for AIDS Research BSL3 Flow Cytometry Core Facility for flow cytometry and Eleonora Forte for comments on the manuscript. Microarrays were performed in the Duke Microarray Facility. Sequencing was performed by the Duke Genome Sequencing & Analysis Core Facility. This research was supported by NIH K99-CA-137860-01A1/R00-CA-137860-02 (to E.G.) and by NIH R01-AI067968 to B.R.C. R.L.S. was supported by NIH T32-CA009111. M.H. is supported by a fellowship of the Charles Revson, Jr. Foundation. T.T. is an HHMI investigator, and work in his laboratory was supported by NIH grants GM073047, MH08442, NIH Challenge Grant RC1CA145442, and the Starr Foundation.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Discussion, one figure and thirteen tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi’s sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. 2010;84:12139–12151. doi: 10.1128/JVI.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Capello D, Gaidano G. Genetic pathways and histogenetic models of AIDS-related lymphomas. Eur J Cancer. 2001;37:1270–1275. doi: 10.1016/s0959-8049(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: Definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Fan W, Bubman D, Chadburn A, Harrington WJ, Jr, Cesarman E, Knowles DM. Distinct subsets of primary effusion lymphoma can be identified based on their cellular gene expression profile and viral association. J Virol. 2005;79:1244–1251. doi: 10.1128/JVI.79.2.1244-1251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol. 2010;84:5229–5237. doi: 10.1128/JVI.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010;24:195–205. doi: 10.1101/gad.553410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12:193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J Virol. 2009;83:12751–12758. doi: 10.1128/JVI.01325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin HR, Ganem D. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc Natl Acad Sci U S A. 2011;108:5148–5153. doi: 10.1073/pnas.1102033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Li Z, Chu CY, Feng J, Sun R, Rana TM. MicroRNAs encoded by Kaposi’s sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010a;11:784–790. doi: 10.1038/embor.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 2010b;84:2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V, Parks T, Bagni R, Wang CD, Samols MA, Hu J, Wyvil KM, Aleman K, Little RF, Yarchoan R, et al. Conservation of virally encoded microRNAs in Kaposi sarcoma--associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis. 2007;195:645–659. doi: 10.1086/511434. [DOI] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR. In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol. 2010;84:695–703. doi: 10.1128/JVI.02013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Strelow LI, Wong SW, Cullen BR. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology. 2010;405:592–599. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V. Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek’s Disease Lymphomas. PLoS Pathog. 2011;7:e1001305. doi: 10.1371/journal.ppat.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


