Abstract
The lengthy course of treatment with currently used anti-mycobacterial drugs and the resulting emergence of drug-resistant strains have intensified the need for alternative therapies against Mycobacterium tuberculosis (Mtb), the etiologic agent of tuberculosis. We show that Mtb and Mycobacterium marinum use Abl and related tyrosine kinases for entry and intracellular survival in macrophages. In mice, the Abl-family tyrosine kinase inhibitor, imatinib (Gleevec®), when administered prophylactically or therapeutically, reduced both the number of granulomatous lesions and bacterial load in infected organs, and was also effective against a rifampicin-resistant strain. Further, when co-administered with current first-line drugs, rifampicin or rifabutin, imatinib acted synergistically. These data implicate host tyrosine kinases in entry and intracellular survival of mycobacteria, and suggest that imatinib may have therapeutic efficacy against Mtb. Because imatinib targets host, it is less likely to engender resistance compared to conventional antibiotics, and may decrease the development of resistance against co-administered drugs.
Keywords: Gleevec®, Drug resistance, tyrosine kinase, XDR TB
Introduction
Mycobacterium tuberculosis, (Mtb) the etiologic agent of tuberculosis (TB), infects one-third of the world’s population, and kills approximately 2 million people per year worldwide (World Health Organization, 2000) with a global case fatality rate of 23% (Bleed et al., 2000). Estimates indicate that more than 90% of all cases of TB and 98% of deaths due to TB occur in developing countries in Southeast Asia, the Western Pacific, and Africa (Raviglione et al., 1995; Snider and La Montagne, 1994). The magnitude and potential impact of this pandemic prompted the World Health Organization (WHO) in 1993 to declare TB a global health emergency. It is estimated that over the next two decades nearly one billion people will become infected, 200 million people will develop disease, and 35 million will die from TB (World Health Organization, 2000).
Although highly effective regimens have been developed for the treatment of TB patients, drugs must be administered for a minimum of six months to cure the disease. Non-adherence with the lengthy course of treatment remains a major problem and has contributed to the emergence of multidrug-resistant and extensively drug-resistant TB (MDR-TB and XDR-TB) strains, which complicates the treatment and control of TB and threatens to exacerbate the epidemic (Dye et al., 2002; Farmer and Kim, 1998). Availability and quality of drugs and altered pharmacokinetics of absorption of some drugs in persons with AIDS has also contributed to the development of drug resistance (Cantwell et al., 1994). Thus, new anti-TB drugs are urgently needed to combat drug resistance, shorten and/or simplify current treatment regimens, provide effective therapy for patients intolerant to current first-line drugs, and provide treatment for patients with latent TB infection.
A key feature of Mtb pathogenesis is the ability of the bacteria to survive and replicate in host phagocytic cells (Russell et al., 2002). Mtb can use as many as eight different cell surface receptors and appears to enter macrophages through conventional phagocytosis (Ernst, 1998). Upon infection, mycobacteria reside within a specialized early phagosomal compartment. Pathogenic mycobacteria prevent fusion with the lysosome, which facilitates evasion of host bactericidal mechanisms, and precludes efficient antigen presentation (Russell et al., 2002). Although there exists a wealth of information on Mtb factors that contribute to entry and intracellular survival within macrophages, information on host factors that contribute to these processes remains more limited.
We have been studying mechanisms by which host tyrosine kinases (TKs), and in particular the Abl-family TKs Abl1 and Abl2, mediate pathogenesis of bacteria and viruses (Lebeis and Kalman, 2009). Abl1 is mutated in human cancers such as Chronic Myelogenous Leukemia (CML), and drugs such as imatinib mesylate (STI-571, Gleevec®), which inhibit Abl1, Abl2 and related TKs are used as therapeutics for CML and other cancers (Druker et al., 2001). Pseudomonas aeruginosa, Shigella flexneri and Chlamydia trachomatis (Burton et al., 2003; Elwell et al., 2008; Pielage et al., 2008) utilize Abl-family TKs during entry, although the precise mechanisms remain unclear. Abl-family TKs also regulate cytoskeletal and trafficking functions in cells, including autophagy (Yogalingam and Pendergast, 2008). In this regard, Shigella flexneri, enteropathogenic Escherichia coli, and orthopoxviruses utilize Abl-family TKs for actin-based motility or release from infected cells, which facilitate spread of the infection (Burton et al., 2005; Reeves et al., 2005; Reeves et al., 2011; Swimm et al., 2004).
The requirement for Abl-family TKs in the pathogenesis of diverse microbes led us to assess their role in Mtb infection. Using cell lines lacking Abl-family TKs and specific inhibitors, we show that Abl-family and related imatinib-sensitive kinases facilitate entry and intracellular survival of Mtb and the related Mycobacterium marinum (Mm). Additionally, imatinib reduces bacterial load and associated pathology in mice infected with Mtb and Mm, including antibiotic-resistant strains. Moreover, imatinib acted in a synergistic manner with the frontline anti-TB drugs rifampicin and rifabutin. Together, our data suggests that modulation of Abl1, Abl2 and related imatinib-sensitive kinases may offer an effective therapeutic strategy for infections caused by mycobacterium species.
Results
Src- and Abl-family TK inhibitors (TKIs) affect intracellular survival of Mtb and Mm in vitro
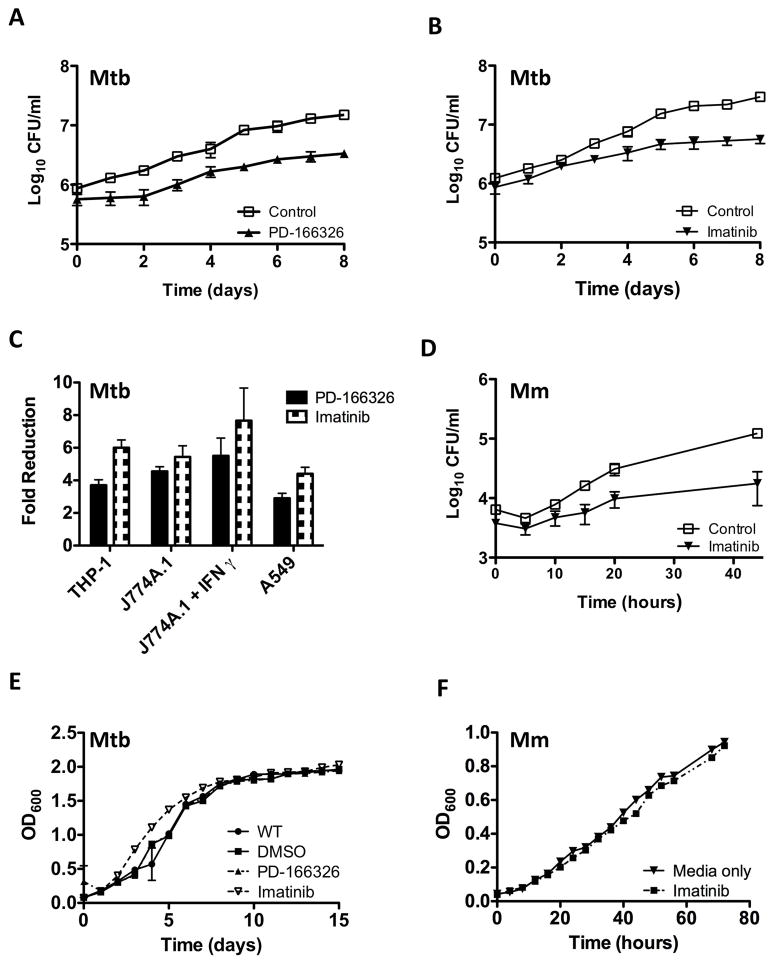
J774A.1 cells, a murine macrophage-like cell line, were infected with Mtb strain H37Rv in media supplemented with PD-166326, an inhibitor of Src- and Abl-family TKs or DMSO, the carrier for PD-166326 (Figure 1A), or imatinib mesylate (imatinib), an inhibitor of Abl- but not Src-family TKs (Figure 1B). PD-166326 or imatinib caused a reduction in CFU/ml by up to 2-fold compared to the carrier at time 0, and up to 7.7 fold at eight days p.i. (Figures 1A and B). Comparable effects of TKIs were evident in several cell lines including A549 cells, a human type II alveolar pneumocyte cell line, and THP-1 cells, a human monocyte cell line (Figure 1C; Supplementary Figures 1A–E). Moreover, the effects of TKIs on J774A.1 cells were independent of stimulation with IFN-γ (Figure 1C and Supp. Figure 1A).
Figure 1. Src- and Abl-family TKIs reduce intracellular survival of Mtb and Mm in vitro.
A,B Intracellular survival of Mtb H37Rv (MOI=10) in J774A.1 macrophage-like cells treated with PD-166326 (10 μM), an Src and Abl-family TKI, or the carrier DMSO (0.1%; control) (A) or imatinib (10μM), an Abl-family TKI (B).
C Effects of PD-166326 or imatinib (each at 10 μM) on Mtb H37Rv in THP-1 cells, J774A.1 cells left untreated or treated with IFNγ, or A549 cells. Ordinate represents maximal fold reduction at 8d p.i. with respect to DMSO controls.
D Intracellular survival of Mm 1218R (MOI=1) in J774A.1 cells treated with imatinib (10 μM).
E,F Growth curves of Mtb H37Rv (E) or Mm 1218R (F) in 7H9 broth containing PD-166326 (10 μM), imatinib (10 μM), or DMSO (0.1%). OD600 was measured at times indicated. Data points represent the mean of three separate experiments, and error bars are +/− SEM.
To determine whether TKIs affected other Mycobacterium species, we assessed their effects on intracellular survival of Mycobacterium marinum (Mm) and Mycobacterium smegmatis (Ms) in J774A.1 cells. As with Mtb, PD-166326 or imatinib decreased CFU/ml by up to 9-fold at both early and late time points for both Mm and Ms (Figure 1D and data not shown). TKIs had no effect on the growth of Mtb, Mm, or Ms in 7H9 broth supplemented with PD-166326 or imatinib at concentrations as high as 10 μM (e.g. Figures 1E,F and data not shown). These data suggest inhibitors of imatinib-sensitive host TKs reduce CFUs of several Mycobacterium species in cells at both early and late times after inoculation.
Abl and other imatinib-sensitive TKs mediate entry and intracellular survival of mycobacteria in macrophages
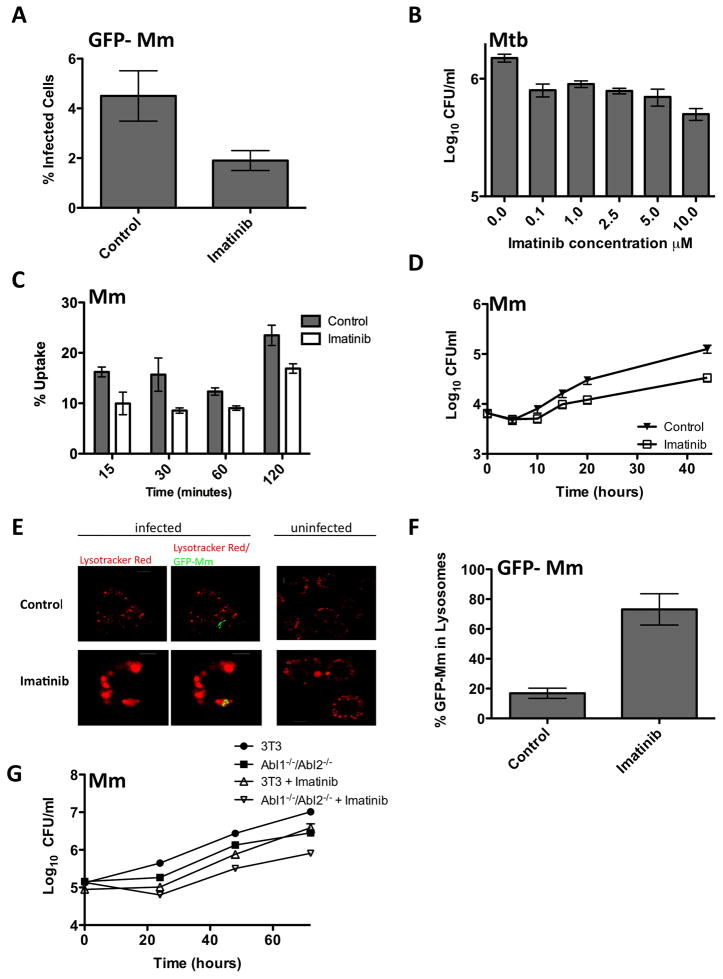
We next determined whether imatinib-sensitive TKs mediate entry or intracellular survival of mycobacteria within the macrophage, or both. To distinguish these possibilities, J774A.1 cells were inoculated with Mm constitutively expressing GFP (GFP-Mm) and treated with imatinib for 2 hours, and then with amikacin for two more hours to kill extracellular bacteria. Imatinib caused a 2.9-fold reduction of the number of GFP-positive cells (Figure 2A). In accordance with these data, imatinib produced 2-fold reduction in CFU/ml when added to THP-1 cells for only 2 hrs at the time of inoculation with Mtb (Figure 2B). A similar fold reduction was evident when imatinib was added to J774A.1 cells for only 15 minutes at the time of inoculation with Mm (Figure 2C). These data suggest that imatinib-sensitive kinases mediate entry of mycobacteria.
Figure 2. Abl-family TKs and other imatinib-sensitive kinases mediate intracellular survival of Mtb and Mm.
A J774A.1 macrophages were infected with GFP-Mm for 2 hours in the presence or absence of imatinib (10 μM), and treated for 2 additional hours with media supplemented with amikacin to kill extracellular bacteria. The number of infected cells, measured as those containing GFP-positive bacilli, was quantified by fluorescence microscopy.
B THP-1 cells were infected with Mtb and treated with imatinib at concentrations ranging from 0.1 to 10 μM for 2 h. Drug was removed, and cells were incubated for an additional 2h with amikacin to kill extracellular bacteria, and then harvested 20 hours later to determine CFU/ml.
C J774A.1 macrophages were treated with imatinib (10 μM) and incubated with Mm at 4°C before returning the cells to 37°C to synchronize entry. At various times thereafter, drug was removed, and the cells treated with amikacin for 2hrs. Cells were then lysed and CFUs determined.
D J774A.1 macrophages were infected with Mm 1218R (MOI=1) for 2h and then incubated with amikacin for an additional 2h. After washing, media with or without 10 μM imatinib was added, and CFU/ml determined at various time points thereafter.
E,F J774A.1 cells were infected and treated as in D, stained 24 h p.i. with lysotracker red, and visualized live. The percent of GFP-Mm colocalizing with the lysotracker red labeled compartment was quantified (250 bacteria from 3 experiments). Scale bar represents 5 μM.
G Fibroblasts derived from Abl1−/−/Abl2−/− mice or from age-matched wild-type mice were infected with Mm 1218R (MOI=60) for 4h. Cells were treated with amikacin for 2h, and CFU/ml determined at various time points thereafter. Imatinib or carrier was present throughout. Data represent the mean +/− SEM from 3 experiments.
To determine whether imatinib had additional effects on intracellular survival independent of entry, J774A.1 cells were infected in media only and drug was added 4 hours post inoculation for the duration of the experiment (Figure 2D). Using this protocol, imatinib reduced CFU/ml by 3.4-fold. To determine whether this reduction in CFU resulted from increased phagolysosomal fusion, macrophages were infected with GFP-Mm, treated with amikacin to kill extracellular bacteria, and then treated with imatinib for an additional 24h. Live macrophages were then stained with the lysosomal marker lysotracker red. In accordance with previous reports (Ertmer et al., 2007), imatinib caused an increase in the area of the lysotracker-labeled compartment, even in uninfected cells (Figure 2E). As shown in Figures 2E,F, 5.8 fold-more GFP-Mm was evident in lysotracker-labeled compartments upon treatment with imatinib, compared to untreated cells. Collectively, these data indicate that an imatinib-sensitive kinase(s) regulate both entry and phagolysosomal trafficking of mycobacteria.
Abl-family TKs and other imatinib-sensitive kinases mediate intracellular survival of Mm
To determine if Abl1 and Abl2 have a direct role in entry or intracellular survival of mycobacteria, we assessed CFU/ml in a fibroblast cell line derived from wild-type mice (3T3) or from mice lacking both c-Abl1 and c-Abl2 (Abl1−/−/Abl2−/−). CFU of Mm decreased by up to 3.6-fold at 72h p.i. in Abl1−/−/Abl2−/− cells compared to 3T3 cells, suggesting that Abl1 and Abl2 are required for intracellular survival of Mm (Figure 2G). We were unable to evaluate Mtb in Abl1−/−/Abl2−/− cells because neither 3T3 cells nor Abl1−/−/Abl2−/− cells were susceptible to infection with this bacterium, even at high MOI (~60). Moreover, no differences between 3T3 cells and Abl1−/−/Abl2−/− cells at the initial time points were evident with Mm infections. These data raise the possibility that entry mechanisms in fibroblasts differed from macrophages, and thus were not evaluated further.
Treatment of Abl1−/−/Abl2−/− cells with imatinib reduced CFU/ml by an additional 3.5-fold suggesting that cellular targets of imatinib other than Abl-family TKs also mediate intracellular survival of mycobacteria. c-Kit, M-CSF1R, PDGFRα and PDGFRβ are also inhibited by imatinib (Buchdunger et al., 2000; Carroll et al., 1997; Dewar et al., 2005). However we could find no significant difference in the entry or intracellular survival of Mm in PDGFRα−/−, PDGFRβ−/− or PDGFRα−/−β−/− cells or in the presence of neutralizing antibodies to c-Kit or m-CSF-1 compared to control cells (data not shown). Collectively, these data suggest Abl-family TKs together with additional imatinib-sensitive factors regulate intracellular trafficking of Mm.
Imatinib reduces bacterial load in mice infected with Mm
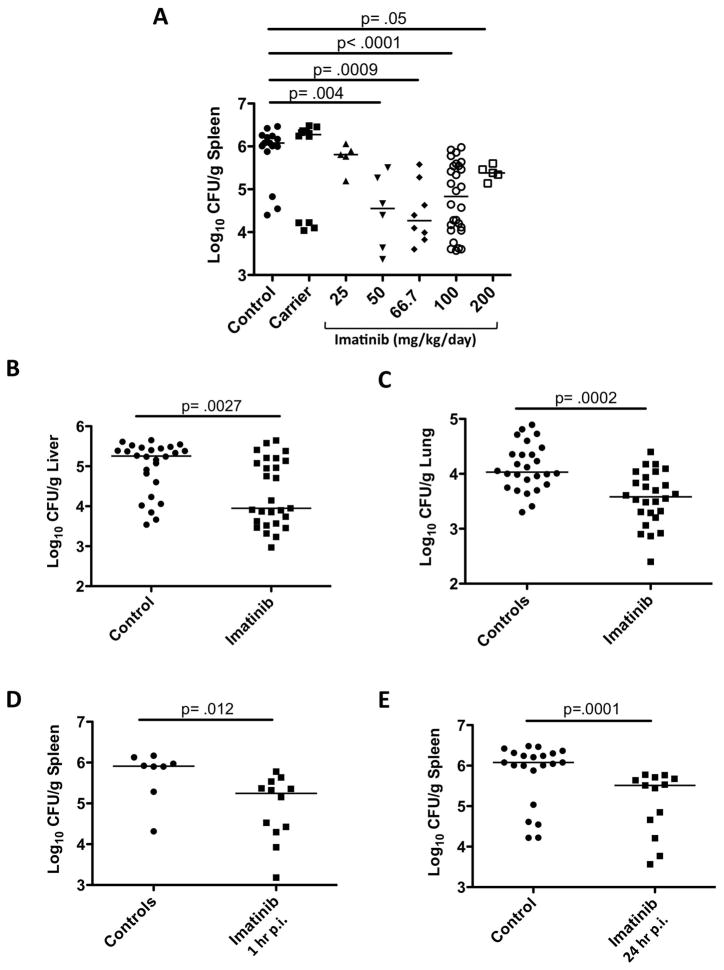
To determine whether Abl-family TKs mediate intracellular survival of mycobacteria in vivo, we assessed the effects of imatinib on mice infected with Mm. In accordance with a previous report, injection of 105 CFU Mm into the tail vein resulted in colonization of spleen, liver, and lung (Robinson et al., 2007). CFU per gram of tissue (CFU/gram) increased to maximal levels by seven days post infection (p.i.), with highest mean levels evident in spleen (6.9×105CFU/gram) compared to liver (9.4×104 CFU/gram), and lung (1.5×104 CFU/gram; Figures 3A–C, controls).
Figure 3. Imatinib reduces bacterial load in mice infected with Mm.
A C57Bl/6 mice were injected in the tail vein with 105 CFU Mm 1218R. Beginning 24h prior to infection, animals were administered water (carrier) or imatinib at concentrations of 25, 50, 66.7, 100 or 200 mg/kg/day. CFU/gram was determined in spleen at d7 p.i.
B,C Mice were infected with Mm as in (A) and pre-treated with carrier or with imatinib (100mg/kg/day) and CFU/g from liver (B) or lungs (C) was determined 7d p.i.
D,E Mice infected with Mm as in A–C except administration of imatinib (100mg/kg/day) commenced 1 h (D) or 24 h (E) p.i. Cumulative data from at least 3 experiments are presented. Each point represents an individual mouse, and the line represents the median CFU; p values determined by a nonparametric Mann-Whitney Rank Sum test.
We next assessed the effects of imatinib, administered at doses ranging from 25 to 200mg/kg/day, or the carrier, water, as a control, beginning 24 h prior to infection and continuing for the duration of the experiment. At concentrations of 50–100mg/kg/day, imatinib reduced the median CFU/gm by 13 to 64 fold (Figure 3A), a statistically significant effect. At lower (25mg/kg/day) or higher (200mg/kg/day) concentrations, imatinib produced a somewhat smaller effect. Statistically significant reductions in bacterial load by imatinib (100mg/kg/day) were also evident in liver and lung (Figures 3b,c). Additionally, administration of imatinib (100 mg/kg/day) reduced bacterial load when delivered beginning either 1h or 24 h p.i. (Figures 3D,E).
Imatinib reduces liver pathology and tail lesions in mice infected with Mm
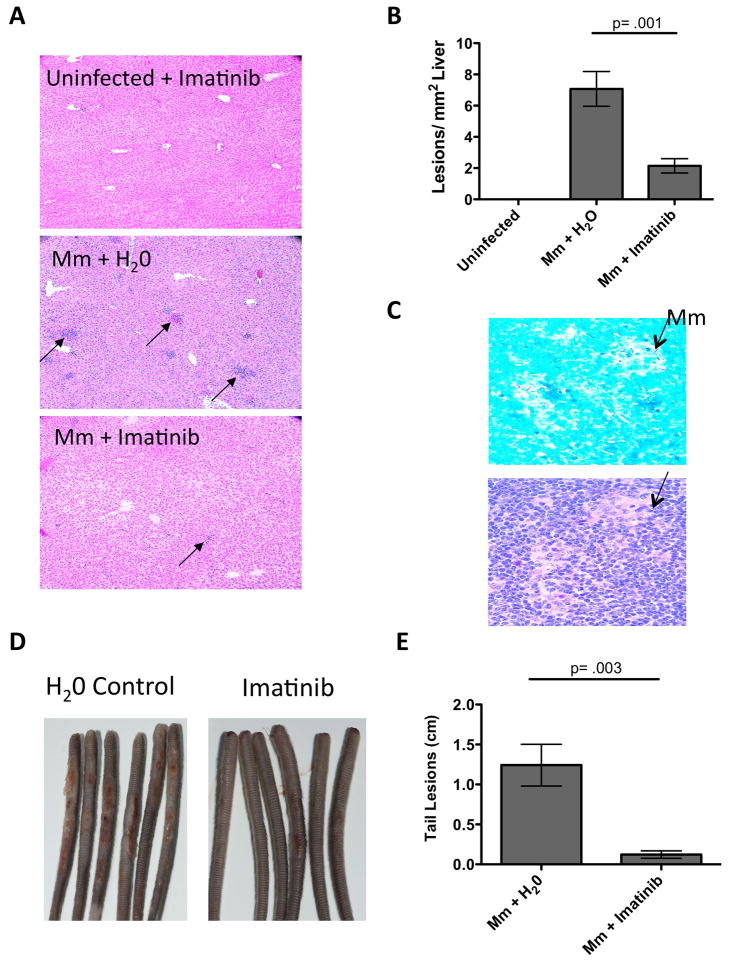
Histological examination of infected liver indicated the presence of granulomatous lesions characterized by monocytic infiltrates seven days p.i. whereas no such lesions were evident in uninfected controls (e.g. compare upper and middle panels in Figure 4A,B). These lesions resembled those seen in livers of mice following i.v. injection with Mtb (Hu et al., 2000). Lesions were also evident in spleen, and were associated with mycobacteria infiltrates, as assessed by acid-fast staining of adjacent sections (e.g. Figure 4C), whereas lesions in liver were not. However, because the load in the spleen was ~10 fold higher than liver, it remains possible that bacteria were more readily observable in adjacent sections. Administration of imatinib (100mg/kg/day) either before or after infection caused a 3.5-fold decrease in the number of lesions per mm2 of liver (e.g. Figures 4A,B). Inoculums of 107 CFU Mm induced lesions on the tail beginning at the site of injection and extending distally. These lesions were reduced in size and extent upon administration of imatinib either prior to or after infection (e.g. Figure 4d,e). Collectively, these data indicate that imatinib reduces not only bacterial load, but also characteristic pathology associated with mycobacterial infection.
Figure 4. Imatinib reduces liver pathology and tail lesions in mice infected with Mm.
A H&E-stained sections of livers of mice left uninfected and treated with 100mg/kg/day imatinib, or infected and treated with H20 or with 100mg/kg/day imatinib 1d prior to infection. Arrows depict individual lesions. Magnification x100.
B Quantitation of number of lesions present in liver sections per mm2. Data were derived from 27 liver sections from mice in 3 experiments. Data are presented as mean +/− SEM. p values were determined by Mann-Whitney rank sum test.
C Acid fast (upper) or H&E (lower) staining of adjacent sections of spleen taken from infected and untreated mice from Figure 3A. Arrows indicate Mm. Magnification x400.
D Images of tails from mice infected i.v. with 107 CFU/ml Mm 1218R and treated with carrier (H2O) or imatinib (100mg/kg/day) for seven days beginning 24 h p.i.. Each mouse received one injection. Images are representative of 3 independent experiments.
E Effects of imatinib (100mg/kg/day) on the extent of lesions on tail. Size of lesions on a particular tail were measured and summed, and the values depicted in cm. Data are represented as the mean cumulative lesion size/tail from 15 tails from each experimental group. +/− SD; p values were determined by Mann-Whitney rank sum test.
Effects of imatinib on rifampicin-resistant Mm
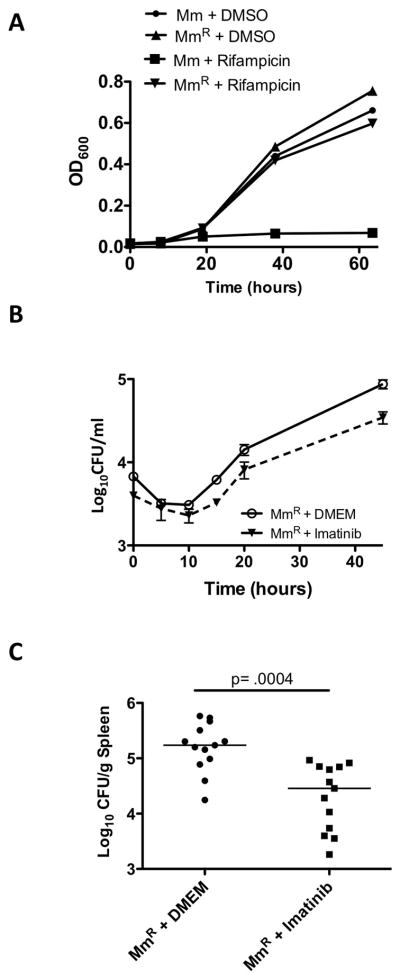
To determine whether imatinib was effective against antibiotic-resistant mycobacteria, we generated a spontaneous rifampicin-resistant Mm mutant (MmR), which contained an H to Y substitution at amino acid 134 in rpoB, which corresponds to the H526Y substitution in Mtb rpoB, the cause of high-level resistance to rifamipicin in clinical isolates (Zhang, 2000). MmR did not display a growth defect in the presence or absence of 1μg/ml rifampicin in 7H9 media compared to wild-type Mm (Figure 5A). In J774A.1 cells, imatinib caused a similar fold reduction in growth of MmR and the parental strain (Figure 5B). In mice, administration of imatinib reduced the load of MmR in the spleen to a similar extent as that seen with Mm (Figure 5C). These data suggest that imatinib is effective against both antibiotic-susceptible and antibiotic-resistant strains of Mm.
Figure 5. Imatinib reduces bacterial load in macrophages and mice infected with rifampicin-resistant Mm.
A Growth curves of Mm wild-type strain (Mm) or a rifampicin-resistant Mm strain (MmR) in 7H9 media left untreated or treated with 1 μg/ml rifampicin. Cell density was measured by OD600. Data in A and B are presented as mean +/− SEM.
B Intracellular survival of MmR (MOI=1) in J774A.1 macrophages in the presence or absence of imatinib (10 μM).
C Effects of imatinib (100mg/kg/day) on mice infected with 105 CFU Mm strain MmR. Cumulative data from 3 independent experiments are presented. The line represents the median CFU; p values were calculated by a nonparametric Mann-Whitney Rank Sum test.
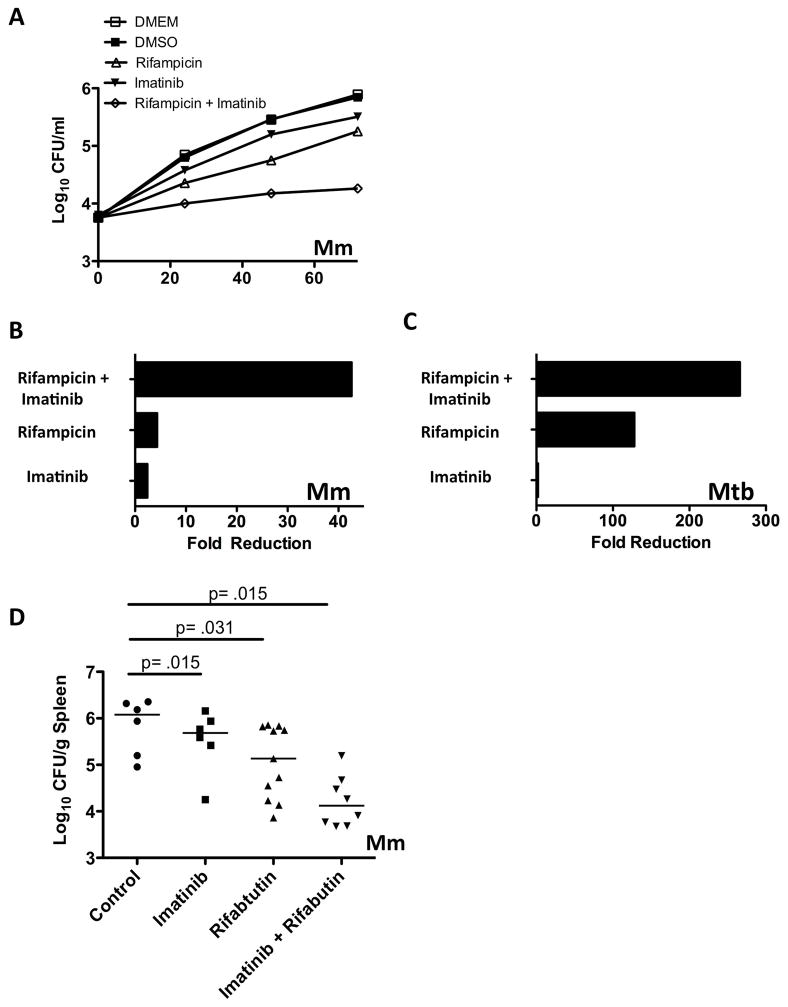
Imatinib and antibiotics synergistically reduce bacterial load
We next determined the effects of imatinib in conjunction with first-line anti-TB drug drugs. J774A.1 cells were infected with Mm and treated with imatinib, or rifampicin, or with a combination of both. For these experiments, the rifampicin concentration chosen only partially inhibited bacterial growth. As shown in Figures 6A and B, the sum of the maximum fold reduction with the combination exceeded the sum of fold effects with either drug alone, indicating a synergistic effect. Similar effects were observed with Mtb, although the maximum fold effect of rifampicin at the concentration used (0.125 μg/ml) was larger than that for Mm (Figure 6C). Together, these data suggest that in combination, imatinib and rifampicin produced a synergistic reduction in survival of mycobacteria in macrophages.
Figure 6. Imatinib and antibiotics act in synergy to reduce mycobacteria survival.
A Intracellular survival of Mm (MOI=1) in J774A.1 macrophages left untreated or treated with imatinib (10 μM), or rifampicin (0.5 μg/ml), or both drugs together. CFU/ml were determined at times indicated.
B Maximal fold reduction in intracellular survival of Mm in J774A.1 cells with various treatments compared to controls at d3 p.i.
C Maximal fold reduction in intracellular survival of Mtb H37Rv in THP-1 cells (MOI=10) with imatinib (10 μM), rifampicin (0.125 μg/ml), or both drugs together.
D Mice were administered imatinib (100mg/kg/day), or rifabutin (2.5mg/kg/day i.p.) or both drugs together. CFU in the spleen were determined at day seven p.i. Line represents the median. (n=3; data from a representative experiment are presented). The line represents the median CFU; p values were calculated by a nonparametric Mann-Whitney Rank Sum test.
We next assessed the effects of imatinib and antibiotics on Mm infection in vivo. Rifampicin has been shown to induce levels of cytochrome P450s that act directly on imatinib and reduce its serum concentration in human plasma (Bolton et al., 2004). To circumvent this problem, we instead used rifabutin, which is a much less potent inducer of cytochrome P450s (Reinach et al., 1999). As shown in Figure 6D, imatinib and rifabutin together reduced the median CFU/ml to a level that exceeded the sum of either drug alone. Similar effects were seen in the liver (data not shown). Together, these data suggest that imatinib acts in a synergistic manner with first-line antibiotics when delivered in combination in vivo.
Imatinib reduces bacterial load in mice infected with Mtb
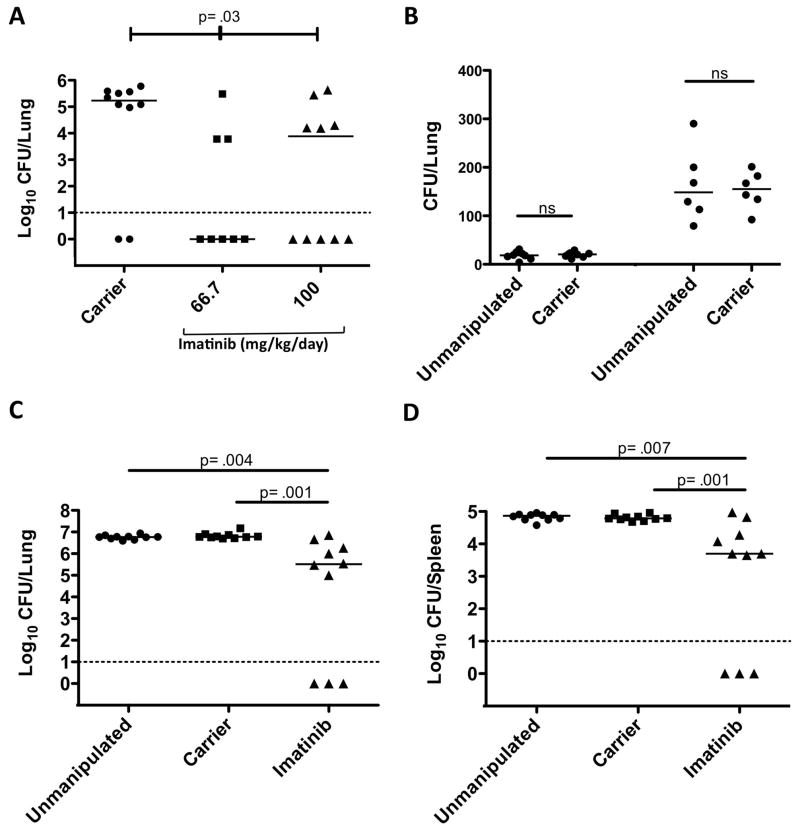
We next determined whether imatinib affected intracellular growth and survival of Mtb in vivo. We assessed the effects of imatinib, administered at doses of either 66 or 100mg/kg/day, or the carrier water as a control, beginning 24 h prior to infection and continuing for the duration of the experiment. Mice were infected with a low dose (~25–50 CFU) of aerosolized Mtb (Erdman strain) and lungs were harvested to assess the bacterial burden and disease pathology 4 weeks post infection. Significance was assessed by a Kruskal-Wallis nonparametric rank sum test, wherein the data from the groups are pooled and ranked according to CFU; the statistic determines whether the observed distribution of rankings could arise by chance. As shown in Figure 7A, imatinib reduced the median CFU by 21 to 57 fold, a statistically significant effect (p=0.035), though no statistically significant difference was evident between the groups treated with different doses of drug (p=0.45). Taken together, lung CFUs were below the level of detection of plating (~10 CFUs; dotted line in Figure 7A) in 56% of animals treated with imatinib, with 78% below 105 CFUs, compared to 20% below the detection level and 80% greater than 105 CFUs in the carrier-treated group. Histopathological examination of the lungs from a subset of mice, including those with CFUs below the level of detection, showed lymphocytic infiltrates (data not shown).
Figure 7. Imatinib reduces bacterial load in mice infected with Mtb.
A C57Bl/6 mice were infected with 50–100 CFU of aerosolized Mtb Erdman. Beginning 24h prior to infection, animals were administered water (carrier) or imatinib at concentrations of 66.7 mg/kg/day or 100 mg/kg/day. CFU was determined in right superior lobe of the lung at 28 days p.i. Solid lines represent the median CFU; dotted line represents the limit of detection (10 CFU). p values were determined by a nonparametric Kruskal-Wallis test.
B 24h prior to infection C57Bl/6 mice were administered carrier pumps. Un-manipulated or carrier treated mice were infected with a low dose (2.5×105 CFU; left) or high dose (1×107 CF; right) of aerosolized Mtb Erdman and CFU determined in the whole lung at 24h p.i. The solid line represents the median CFU.
C–D C57Bl/6 mice were infected with 2.5×105 CFU of aerosolized Mtb Erdman. Beginning 24h prior to infection, animals were left untreated, or, administered carrier (water) or imatinib at a concentration of 66.7 mg/kg/day. CFU was determined by plating homogenates of the whole lung (C) or spleen (D) at 28 days p.i. The solid line represents the median CFU; dotted line represents the limit of detection (10 CFU). p values were determined by a nonparametric Mann-Whitney test.
The observation that 20% of the carrier-treated mice displayed CFUs below the level of detection was unusual and led us to consider the possibility that the osmotic pump interfered with the deposition of bacteria to the lungs. To test this possibility, un-manipulated (no pump and no drug) or carrier-treated mice were exposed to aerosolized Mtb at a concentration of 2.5×105/ml CFU or a higher concentration of 1×107/ml CFU. As shown in Figures 7B, deposition of Mtb at 24 hrs p.i. was equivalent in the lungs of un-manipulated mice compared to those with carrier pumps for both concentrations of bacteria. Together, these data suggest that the pumps were without effect, but the reasons for the undetectable CFU in carrier-treated animals in Figure 7A remain unclear.
To determine whether the undetectable CFU in carrier-treated animals was an experimental anomaly, and to confirm that there existed a statistically significant difference in CFUs between the control and imatinib-treated groups, we carried out a second 28d experiment in which we counted CFUs from the entire lung rather than a single lobe, so as to avoid underestimating the number of bacteria. We also included un-manipulated animals as controls. As shown in Figure 7C, with a measured inoculum of approximately 50 CFU, there was no statistically significant difference between un-manipulated mice (median CFU, 5.8×106) and carrier treated mice (median CFU, 6×106). Moreover, neither group had outliers below the level of detection (10 CFU). Importantly, the median CFU was reduced by 185-fold in the imatinib-treated group compared with the carrier-controls, a statistically significant difference (p=0.001), with 30% of the animals displaying CFUs below the level of detection. Examination of CFUs from spleen followed a similar pattern as the lungs, with the median CFU reduced by 12-fold in imatinib-treated animals compared to the carrier-controls (Figure 7D). Moreover, animals with CFUs below detectable levels in the lung likewise had undetectable levels in the spleen. Collectively, these data suggest that imatinib is effective against Mtb in acutely infected animals.
Discussion
Using specific TKIs and cell lines derived from knockout animals, we provide evidence for a role of host TKs, including Abl1, Abl2, and additional imatinib-sensitive kinases, in the entry and intracellular survival of Mtb and related pathogen Mm in macrophages. Pathogenic mycobacteria likely use multiple kinases, in a redundant fashion, perhaps as a means to infect cell types that express different classes of kinases. TKs, including members of the Abl family, are used by diverse bacterial and viral pathogens (reviewed in (Lebeis and Kalman, 2009)). Notably, redundant usage of TKs has been reported for several of these (Lebeis and Kalman, 2009). A recent study showed that siRNA against Abl1 transcript appeared to facilitate survival of Mtb strain H37Rv in J774A.1 macrophages (Jayaswal et al., 2010). However, experiments presented here using specific inhibitors as well as cells derived from knockout animals suggest that multiple kinases, including both Abl1 and Abl2, mediate intracellular survival within macrophages. The apparent disparity may be explained by redundant kinase usage, which our experiments suggest. A recent report indicated that non-specific serine/threonine kinase inhibitors affect MDR-Mtb growth in a primary macrophage cell line, though it remains to be determined whether these compounds affect additional targets in the bacteria or the host or both (Kuijl et al., 2007).
Our data suggest that Abl or related TKs affect both entry of mycobacteria into cells and intracellular survival. While a role for Abl or related TKs in entry remains unexplored, several observations suggest a role for these TKs in vessicular trafficking. Imatinib causes accumulation of acidified vesicles that contain lamp-1 and LC3, which are markers for lysosomes and autophagosomes, respectively (Ertmer et al., 2007; Yogalingam and Pendergast, 2008), and accumulation of Mm in the lysosome (Figure 2). Notably, stimulation of autophagy with rapamycin or starvation reduces intracellular survival of mycobacteria in macrophages (Gutierrez et al., 2004). Together, these data suggest that imatinib-sensitive kinases mediate inhibition of phagolysosomal fusion by mycobacteria.
Our data suggest that imatinib similarly affects Mtb, M. smegmatis and Mm in diverse cell types. Thus, imatinib-sensitive TKs may serve as a “node” and mediate entry by multiple receptors or alter receptor mediated trafficking of a phagosome utilized by several mycobacteria species. Mm is the closest genetic relative to the Mtb complex based on 16S rRNA sequencing and 85% identity to orthologous regions of the Mtb genome (Stinear et al., 2008; Tonjum et al., 1998). Mm causes both systemic and granulomatous disease in cold-blooded animals as well as subcutaneous granulomas in humans due to its preference for lower temperatures (Clark and Shepard, 1963). Notably, Mm exhibits a five-fold faster generation time compared to Mtb and requires only BSL2 containment, thereby expediting data acquisition and optimization of drug dose and delivery methodologies. The observation that M. smegmatis is similarly affected by imatinib suggests that the effects of the drug are independent of the RD1 locus, which is associated with virulence. This observation also raises the possibility that imatinib might likewise be effective against other pathogenic mycobacteria, including M. avium, M. intracellulare, M. ulcerans and M. leprae.
Some differences exist between Mtb and Mm. Mm escapes the phagosome, and our data and that of others indicate that Mm forms actin “tails” after 48h that facilitate infection of adjacent cells (Stamm et al., 2003). Recent reports suggest that Mtb also escapes the phagosome (van der Wel et al., 2007), although no actin motility has been observed. Notably, the effects of imatinib on infectivity of Mm in vitro are evident prior to 48h, and no reduction in actin tails was apparent when imatinib was applied at 48h, nor in cells lacking Abl-family TKs (data not shown). Thus, differences in actin motility between Mtb and Mm in vitro appear independent of host TKs.
Imatinib may have significant utility in treating infections caused by mycobacteria. Imatinib reduced CFUs in organs of mice infected with Mtb (Figure 7) or Mm (Figure 3). The effect of imatinib on bacterial load was dose dependent with optimal concentrations of 66.7–100mg/kg/day, the approximate dose used to treat CML in humans (Goldman and Druker, 2001). Clearance of Mtb to levels below the limit of detection was evident in some animals (Figures 7A,C, and D). Clearance was not observed in Mm experiments, perhaps owing to the shorter time course, higher inoculum (105 CFU for Mm versus ~50 CFU for Mtb), or route of inoculation. For reasons that are unclear, considerable variance was evident in CFUs for both Mtb and Mm infections with imatinib administration, even within a single experiment, although median CFUs were consistently lower (Figures 3 and 7). Other disease indicators in animals infected with Mm, including lymphocytic or monocytic aggregates in the liver and superficial lesions on the tail, were reduced upon treatment with imatinib (Figure 4).
Based on in vitro studies, reductions in CFU may reflect a reduction in the number of bacteria entering macrophages or their intracellular survival or both. We also show that imatinib and the first-line antibiotics rifampicin, when provided together, can act in a synergistic fashion to reduce survival of Mtb and Mm in macrophages (Figure 6A,B, and data not shown); imatinib and rifabutin act similarly in vivo. It remains possible that in vivo, treatment with imatinib results in an increased number of extracellular bacteria, which may be exposed to higher concentrations of antibiotics. Alternatively, localization of intracellular bacteria in a lysosomal compartment upon treatment with imatinib may render them more susceptible to antibiotics.
Besides Abl-family TKs, imatinib inhibits receptor TKs responsible for mast cell development stem cell differentiation (c-Kit), macrophage differentiation and function (m-CSF1R), and cell growth (PDGFR) (Buchdunger et al., 2000; Carroll et al., 1997; Dewar et al., 2005). Thus, we cannot rule out the possibility that additional immunomodulatory effects contribute to the observed phenotype. In this regard, imatinib appeared more efficacious at lower concentrations (Figure 3A), suggesting that the drug may exert concentration-dependent effects on factors important for control of the infection.
A primary concern when considering drugs directed at host targets remains their toxicity toward the host. Thus, host molecules that are essential for survival or required to mount an effective immune response may prove difficult to target. Imatinib has exhibited remarkably low toxicity in patients, though some recent reports indicate side effects on the heart and the immune system with long-term exposure (Kerkela et al., 2006; Mumprecht et al., 2006). Adjusting the dosing regimen or limiting the period of exposure may mitigate such effects. Imatinib may prove most effective in patients when the bacteria are actively growing (“active TB”), or with reactivated TB, which may warrant acute, rather than sustained, exposure to the drug. We have not pursued in vivo studies with dual Abl- and Src-family TKIs such as Sprycel (BMS-354825) or PD-166326 because these drugs cause significant immunotoxicity (EMEA, 2008), which has proven insurmountable in an orthopoxvirus infection model (Reeves et al., 2011).
Here we provide evidence that host TKs including Abl1, Abl2, and other imatinib-sensitive kinases regulate the pathogenesis of Mtb and Mm in vitro and in vivo. Data from macrophages and mice indicate that infections caused by Mm and Mtb infections are similarly sensitive to imatinib. Similar imatinib-sensitive kinases may govern trafficking of Mm and Mtb in macrophages. However, in vivo the mechanism by which imatinib reduces Mtb CFUs may be more complex than with Mm. In the Mm model, effects of imatinib are evident within 3–7 days, and reductions in CFU likely reflect entry or intracellular trafficking of bacterium or innate immune effects. Such effects may also be important in Mtb infections, but at 28 days, adaptive responses may also contribute. We are currently investigating how imatinib impacts the innate and adaptive responses in animals infected with Mycobacteria sp. Nevertheless, because of its relatively rapid growth rate and limited biocontainment requirements, Mm may be useful for initial phases of drug discovery for acute mycobacteria infections in vitro and in vivo.
Additionally, our data suggests that imatinib may prove useful against both antibiotic-susceptible and antibiotic-resistant strains of Mtb including MDR-TB and XDR-TB (e.g. Figure 5). Because imatinib targets host TKs and not bacterial factors, it is less likely to engender resistance compared to conventional antibiotics. To evade the block and develop resistance, Mtb would have to significantly alter its pathogenic strategy. Moreover, when delivered in conjunction with current first-line anti-TB drugs, decreasing the total bacterial burden below the spontaneous mutation frequency with imatinib may serve to reduce the probability of developing antibiotic-resistant disease thereby increasing the clinical lifespan of those drugs.
Methods
Cell culture
Fibroblast cell lines derived from wild-type mice or from Abl1−/− Abl2−/− mice (55), and the mouse macrophage cell line J774A.1 were maintained in Dulbecco’s modified Eagle Medium (DMEM). Human type II alveolar pneumocyte cell (A549) were grown in Eagle’s minimal essential medium, and human monocytic-like cells (THP-1) in RPMI1640. Media was supplemented with 10% FBS and 2mM L-glutamine. For some experiments, THP-1 cells differentiated by adding 10ng/ml Phorbol 12-myristate 13-acetate (PMA) (Sigma), and J774A.1 were activated with 100 U/ml INF-γ (BD Biosciences Pharmingen, San Diego, CA).
Bacterial strains, growth conditions, and mutants
Mtb strain H37Rv (TMC102) and Mm strain 1218R (ATCC 927), a fish outbreak isolate, were grown in Middlebrook 7H9 broth (7H9) (BBL Microbiology Systems, Cockeysville, MD) supplemented with ADC (Difco Laboratories, Detroit, MI) and 0.05% Tween 80 (Mtb) (Sigma, St. Louis, MI) or 0.025% Tween 80 (Mm). Middlebrook 7H11 agar was used for Mtb, and Middlebrook 7H10 agar for Mm. Both were supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Difco Laboratories, Sparks, MD). Mtb stocks were grown at 37°C in 5% CO2 with shaking at 70 rpm for seven days to an optical density of 0.6 (OD600), as measured by a Novaspec II spectrophotometer (Amersham Pharmacia Biotech, Uppsala, Sweden). Cells were pelleted, resuspended in fresh 7H9 broth, aliquoted, and stored at −70°C. After freezing, an aliquot was thawed for viable count assays on 7H11 agar. Mm stocks were grown at 30°C for 3 days to an OD600 of 0.8,, and the cells pelleted, re-suspended in fresh 7H9 broth, aliquoted, and stored at −80°C. After freezing, an aliquot was thawed to perform viable count assays on 7H10 agar supplemented with OADC. To generate rifampicin-resistant Mm mutant, strain 1218R was grown on 7H10 agar plates with 1 μg rifampicin/ml (Sigma) for seven days. Colonies were restreaked to confirm resistance, and mutants assessed in growth curves. The mutation was identified by amplifying and then sequencing the rifampicin-resistance-determining region of rpoB using primers 5′GACGACATCGACCACTTC3′ and 3′TAGTCCACCTCTGACGAG5′.
In vitro Mtb infections
THP-1 cells were seeded at 106 cells per well in a 24-well microculture plate and differentiated with PMA. After 48 h, the cells were washed once, and fresh RPMI media added. A549 cells or J774A.1 were seeded at 3×105 cells per well in 24-well plates and incubated for 48h prior to infection. For infections, frozen Mtb stocks were thawed and centrifuged, the supernatant removed, and the pellet resuspended in media without antibiotics. Bacteria were centrifuged again, resuspended in fresh medium, washed twice with 20ml of medium, transferred to a 50ml Oakridge tube containing ~30 3-mm glass beads, and vortexed for 15s to disperse clumps and obtain a single cell suspension.
Mammalian cells infected in triplicate with Mtb (MOI=10) for 2h in RPMI without FBS, after which extracellular Mtb were killed by adding 200 μg/ml amikacin (Sigma) and incubating at 37°C with 5% CO2 for an additional 2h. Monolayers were washed five times with RPMI, and the cultures incubated at 37°C with 5% CO2. At 24 hour intervals, supernatants were removed and the monolayers lysed in 0.1% Triton-X-100 (Sigma) for 15 min. Three sets of serial 10-fold dilutions of the lysates from each well were prepared in 0.05% Tween 80 (Sigma) and portions plated on 7H11 agar. Colonies were counted after 3 to 4 weeks incubation at 37°C in 5% CO2.
In vitro Mm infections
J774A.1, Abl1−/−/Abl2−/− cells, or 3T3 cells were plated at 105 cells/ml in 24 well plates, and allowed to adhere overnight. A frozen vial of 1218R Mm was thawed, the sample centrifuged, and the pellet re-suspended in DMEM with 10% FBS and used to inoculate fresh media at 105 CFU/ml. One ml of the Mm-containing DMEM/FBS was added to each well (MOI=1) and the plates were incubated at 30°C with 5% CO2 for 2h, rinsed twice with fresh PBS, and incubated with 200 μg/ml amikacin for an additional 2h to kill remaining extracellular Mm. Cells were then washed twice with PBS and the media replaced with DMEM/FBS. At 4h post inoculation (time 0) and 5, 10, 15, 20, 25, or 48h thereafter, the supernatant was removed and the monolayers lysed in 0.1% Triton-X-100 (Sigma) for 10 min. Three sets of serial 10-fold dilutions of the lysates from each well were plated on 7H10 agar, and colonies counted after seven days incubation at 30°C. Data presented are from at least three independent experiments.
Determination of Mm entry
The percentage of infected J774A.1 cells after four hours was measured using a Mm strain constitutively expressing plasmid encoded GFP under control of the secA promoter. Infected cells distinguished by live imaging with a scientific-grade cooled charge-coupled device (Cool-Snap HQ with ORCA-ER chip) on a multi-wavelength, wide-field, three-dimensional microscopy system (Intelligent Imaging Innovations, Denver, CO), based on a Zeiss 200M inverted microscope using a 63x lens with a numerical aperture of 1.4 (Carl Zeiss, Thornwood, NY). To measure CFUs with imatinib present, J774A.1 cells were plated at 105 per well for one day, and then chilled to 4°C for 20 minutes prior to addition of bacteria and drug. To synchronize entry, Mm (MOI= 1) was added at 4°C, and the bacteria and cells were centrifuged at 700g for 10 minutes at 4°C. The culture dish was then returned to 37°C for 15, 30, 60, or 120 minutes. At each time point, cells were washed twice with PBS and incubated for an additional 2h with media containing 200ug/ml amikacin. Monolayers were washed twice with PBS and lysed with 0.1% Triton-X 100 and CFU/ml determined as above. Percent uptake was measured by dividing the number of internalized bacteria by the CFU/ml of the inoculum.
Measurement of Mm in lysosomes
To determine intracellular localization, J774A.1 cells were plated in 8 well Lab-Tek chambered coverglass plates at 105 cells/well (Nunc, Nalge Nunc International), infected with GFP-Mm (MOI 1) for 2h, and then treated with 200ug/ml Amikacin for 2 additional hours. Cells were washed, and media alone or supplemented with imatinib was added, and the cells were incubated at 30°C. After 24 hours, 50nM of lysotracker red (Invitrogen) was added, and cells were imaged as described above. The GFP fluorescence that colocalized with lysotracker red was quantified.
In vivo Mtb infections
Age matched un-manipulated mice, carrier controls containing water pumps, or mice containing pumps with imatinib were infected with Mtb Erdman (Trudeau Institute, Saranac, NY) of Mtb using a nose-only aerosolization system (In-Tox Products, Albuquerque, NM). Mice were exposed for 20 min to a nebulized suspension of bacteria at a density 2.5×105/ml CFU or 1×107/ml CFU that was optimized to deliver a dose of approximately 25 or 200 CFU to the lungs. Mice were sacrificed at 1d or 28d post infection. The whole lung or right superior lobe of the lung was homogenized in PBS containing 0.05% Tween-80 and serial dilutions were plated onto 7H11 agar. The plates were incubated at 37°C and colonies counted after three weeks.
In vivo Mm infections
Six-week old male C57Bl/6 mice were injected in the tail vein with active growing cultures at ~105 CFU/mouse. The number of bacteria injected for each experiment was determined by retrospective plating. The average dose was 2.5×105 CFU/mouse. For experiments to determine effects of drugs on tail lesions, mice were infected at 107 CFU/ml. Seven days after infection, lung, liver, and spleen were harvested, weighed and homogenized (Fisher Scientific, Tissuemiser) in one ml PBS. Each homogenate was diluted and spread on 7H10 agar. Colonies were scored after seven days of incubation at 30°C. Total weight of the organ and colonies/ml of the homogenized organ were used to determine CFU/gram. For histology studies, the liver was removed, fixed in 10% formalin, and embedded in paraffin. Sections were cut and stained with hematoxylin and eosin (H&E) or Ziehl-Neelsen (acid-fast bacillus), and imaged on a Nikon Eclipse 80i microscope.
Delivery of drugs in vivo
For experiments with imatinib, the mesylate salt was dissolved in water and loaded into Alzet pumps (Braintree Scientific, 1007D) capable of dispensing a continuous flow of drug at 25, 50, 100 or 200 mg/kg/day. Pumps were inserted subcutaneously into anesthetized 6-week old male C57Bl/6 mice (Jackson Laboratories). At these doses, we observed no weight loss or other adverse effects in uninfected animals. Moreover, such doses have been used to treat cancer in mice, and are equivalent to those used in humans, with adjustments for differences in pharmacodynamics and pharmacokinetics (Wolff and Ilaria, 2001). For pre-treatment experiments, water- or imatinib-containing pumps were inserted 24 h prior to intravenous injection of Mtb Erdman, Mm 1218R or rifampicin-resistant Mm 1218R (MmR), and delivery was maintained for the duration of the experiment. For same day and post-treatment experiments, mice were infected with Mm for either 1h or 24h, respectively, before inserting the pump. For both pre- and post-treatment experiments, the mice were euthanized on d7 for Mm or d28 for Mtb, and CFU measured as described above. For synergy experiments mice were injected intraperitoneally (i.p.) with 2.5 mg/kg rifabutin (sigma) solubilized in 100% DMSO once a day for seven days.
Statistical analysis
Statistical analysis was done using non-parametric tests including the Mann-Whitney Rank Sum test, the Wilcoxan Rank Sum test, or the Kruskal-Wallis test. In all of these tests, the data are pooled and the values ranked. The statistic calculates the probability that the observed ranking occurred by chance. Values less than or equal to 0.05 were considered statistically significant.
Supplementary Material
A,B Growth curves in the presence of PD-166326 (10μM) or carrier (DMSO, 0.1%) were conducted in J774A.1 cells treated with IFNγ (A) or A549 cells (B) infected with Mtb H37Rv (MOI=10).
C–E Growth curves were carried out in the presence of imatinib (10 μM) or carrier (H20) in J774A.1 cells treated with IFNγ (C), in THP-1 cells (D), or in A549 cells (E). Cells were infected with Mtb H37Rv (MOI=10). CFU/ml were determined at designated time points. Data are representative of three individual experiments, and are presented as mea+/− SEM.
Highlights.
Abl-related tyrosine kinases regulate mycobacterial entry and intracellular survival
Abl-family kinase inhibitor imatinib reduces Mtb levels in mice
Imatinib is effective against drug-resistant mycobacteria
Imatinib works in synergy with front-line anti-TB drugs
Acknowledgments
We thank members of the Kalman laboratory and L. Uebelhoer, V. Faundez, and M. Sherman for helpful discussions, I. Williams and C. Parkos for pathology expertise, K. Easley for statistical advice, and J. Posey for GFP-Mm strain. This work was supported by NCICA016672 (to W.B), and by R01A107246201, R01AI056067–01 and a grant from the Bio-Merieux Foundation (all to D.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bleed D, Dye C, Raviglione MC. Dynamics and control of the global tuberculosis epidemic. Curr Opin Pulm Med. 2000;6:174–179. doi: 10.1097/00063198-200005000-00002. [DOI] [PubMed] [Google Scholar]
- Bolton AE, Peng B, Hubert M, Krebs-Brown A, Capdeville R, Keller U, Seiberling M. Effect of rifampicin on the pharmacokinetics of imatinib mesylate (Gleevec, STI571) in healthy subjects. Cancer Chemother Pharmacol. 2004;53:102–106. doi: 10.1007/s00280-003-0722-9. [DOI] [PubMed] [Google Scholar]
- Buchdunger E, Cioffi CL, Law N, Stover D, Ohno-Jones S, Druker BJ, Lydon NB. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J Pharmacol Exp Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- Burton EA, Oliver TN, Pendergast AM. Abl kinases regulate actin comet tail elongation via an N-WASP-dependent pathway. Mol Cell Biol. 2005;25:8834–8843. doi: 10.1128/MCB.25.20.8834-8843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton EA, Plattner R, Pendergast AM. Abl tyrosine kinases are required for infection by Shigella flexneri. Embo J. 2003;22:5471–5479. doi: 10.1093/emboj/cdg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell MF, Snider DE, Jr, Cauthen GM, Onorato IM. Epidemiology of tuberculosis in the United States, 1985 through 1992. Jama. 1994;272:535–539. [PubMed] [Google Scholar]
- Carroll M, Ohno-Jones S, Tamura S, Buchdunger E, Zimmermann J, Lydon NB, Gilliland DG, Druker BJ. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood. 1997;90:4947–4952. [PubMed] [Google Scholar]
- Clark HF, Shepard CC. Effect of Environmental Temperatures on Infection with Mycobacterium Marinum (Balnei) of Mice and a Number of Poikilothermic Species. J Bacteriol. 1963;86:1057–1069. doi: 10.1128/jb.86.5.1057-1069.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar AL, Cambareri AC, Zannettino AC, Miller BL, Doherty KV, Hughes TP, Lyons AB. Macrophage colony-stimulating factor receptor c-fms is a novel target of imatinib. Blood. 2005;105:3127–3132. doi: 10.1182/blood-2004-10-3967. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Capdeville R, Ford JM, Baccarani M, Goldman JM. Chronic myelogenous leukemia. Hematology (Am Soc Hematol Educ Program) 2001:87–112. doi: 10.1182/asheducation-2001.1.87. [DOI] [PubMed] [Google Scholar]
- Dye C, Espinal MA, Watt CJ, Mbiaga C, Williams BG. Worldwide incidence of multidrug-resistant tuberculosis. J Infect Dis. 2002;185:1197–1202. doi: 10.1086/339818. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMEA EMA. European Public Assessment Report. London: European Medicines Agency; 2008. Sprycel, INN-dasatinib; pp. 1–46. [Google Scholar]
- Ernst JD. Macrophage receptors for Mycobacterium tuberculosis. Infect Immun. 1998;66:1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, Elsasser HP, Schatzl HM. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- Farmer P, Kim JY. Community based approaches to the control of multidrug resistant tuberculosis: introducing “DOTS-plus”. Bmj. 1998;317:671–674. doi: 10.1136/bmj.317.7159.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JM, Druker BJ. Chronic myeloid leukemia: current treatment options. Blood. 2001;98:2039–2042. doi: 10.1182/blood.v98.7.2039. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hu C, Mayadas-Norton T, Tanaka K, Chan J, Salgame P. Mycobacterium tuberculosis infection in complement receptor 3-deficient mice. J Immunol. 2000;165:2596–2602. doi: 10.4049/jimmunol.165.5.2596. [DOI] [PubMed] [Google Scholar]
- Jayaswal S, Kamal MA, Dua R, Gupta S, Majumdar T, Das G, Kumar D, Rao KV. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA screen. PLoS Pathog. 2010;6:e1000839. doi: 10.1371/journal.ppat.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkela R, Grazette L, Yacobi R, Iliescu C, Patten R, Beahm C, Walters B, Shevtsov S, Pesant S, Clubb FJ, et al. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12:908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJ, Geluk A, et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- Lebeis SL, Kalman D. Aligning antimicrobial drug discovery with complex and redundant host-pathogen interactions. Cell Host Microbe. 2009;5:114–122. doi: 10.1016/j.chom.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Mumprecht S, Matter M, Pavelic V, Ochsenbein AF. Imatinib mesylate selectively impairs expansion of memory cytotoxic T cells without affecting the control of primary viral infections. Blood. 2006;108:3406–3413. doi: 10.1182/blood-2006-04-018705. [DOI] [PubMed] [Google Scholar]
- Pielage JF, Powell KR, Kalman D, Engel JN. RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog. 2008;4:e1000031. doi: 10.1371/journal.ppat.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviglione MC, Snider DE, Jr, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, et al. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med. 2005;11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- Reeves PM, Smith SK, Olson VA, Thorne SH, Bornmann W, Damon IK, Kalman D. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and SRC family tyrosine kinases. J Virol. 2011;85:21–31. doi: 10.1128/JVI.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinach B, de Sousa G, Dostert P, Ings R, Gugenheim J, Rahmani R. Comparative effects of rifabutin and rifampicin on cytochromes P450 and UDP-glucuronosyl-transferases expression in fresh and cryopreserved human hepatocytes. Chem Biol Interact. 1999;121:37–48. doi: 10.1016/s0009-2797(99)00089-7. [DOI] [PubMed] [Google Scholar]
- Robinson N, Wolke M, Ernestus K, Plum G. A mycobacterial gene involved in synthesis of an outer cell envelope lipid is a key factor in prevention of phagosome maturation. Infect Immun. 2007;75:581–591. doi: 10.1128/IAI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. J Cell Biol. 2002;158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider DE, Jr, La Montagne JR. The neglected global tuberculosis problem: a report of the 1992 World Congress on Tuberculosis. J Infect Dis. 1994;169:1189–1196. doi: 10.1093/infdis/169.6.1189. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swimm A, Bommarius B, Li Y, Cheng D, Reeves P, Sherman M, Veach D, Bornmann W, Kalman D. Enteropathogenic Escherichia coli use redundant tyrosine kinases to form actin pedestals. Mol Biol Cell. 2004;15:3520–3529. doi: 10.1091/mbc.E04-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Welty DB, Jantzen E, Small PL. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J Clin Microbiol. 1998;36:918–925. doi: 10.1128/jcm.36.4.918-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Wolff NC, Ilaria RL., Jr Establishment of a murine model for therapy-treated chronic myelogenous leukemia using the tyrosine kinase inhibitor STI571. Blood. 2001;98:2808–2816. doi: 10.1182/blood.v98.9.2808. [DOI] [PubMed] [Google Scholar]
- World Health Organization; World Health Organization. Fact Sheet. Geneva, Switzerland: 2000. Tuberculosis. [Google Scholar]
- Yogalingam G, Pendergast AM. Abl kinases regulate autophagy by promoting the trafficking and function of lysosomal components. J Biol Chem. 2008;283:35941–35953. doi: 10.1074/jbc.M804543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YTA. Molecular Genetics of Mycobacteria. Washington, DC: ASM Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A,B Growth curves in the presence of PD-166326 (10μM) or carrier (DMSO, 0.1%) were conducted in J774A.1 cells treated with IFNγ (A) or A549 cells (B) infected with Mtb H37Rv (MOI=10).
C–E Growth curves were carried out in the presence of imatinib (10 μM) or carrier (H20) in J774A.1 cells treated with IFNγ (C), in THP-1 cells (D), or in A549 cells (E). Cells were infected with Mtb H37Rv (MOI=10). CFU/ml were determined at designated time points. Data are representative of three individual experiments, and are presented as mea+/− SEM.