Abstract
Rab GTPases localize to distinct sub-cellular compartments and regulate vesicle trafficking in eukaryotic cells. Yeast Rabs Ypt31/32 and Sec4 have 68% homology and bind to common interactors, yet play distinct roles in the transport of exocytic vesicles. The structures of Ypt31/32 have not previously been reported in the uncomplexed state. We describe the crystal structures of GTP and GDP forms of Ypt32 to understand the molecular basis for Rab function. The structure of Ypt32(GTP) reveals that the switch II conformation is distinct from Sec4(GTP) in spite of a highly conserved amino acid sequence. Also, Ypt32(GDP) reveals a remarkable change in conformation of the switch II helix induced by binding to GDI, which has not been described previously.
Keywords: Rab GTPase, Ypt32, Sec4, vesicle trafficking, effectors, X-Ray crystallography
1. Introduction
Rab small GTPases are molecular switches that regulate vesicle trafficking in eukaryotic cells via interactions with effector proteins. Human Rabs comprise about 70 members, while Saccharomyces cerevisiae contains only 11 Rabs [1–5]. Rabs perform a variety of functions including vesicle formation, motility, tethering, fusion, and the various steps are mediated via recruitment of effector proteins [6]. Rab GTPases contain a flexible C-terminal tail that is post-translationally modified at Cys residues to enable attachment to lipid bilayers [3,7].
Rabs oscillate between an active (GTP) and inactive (GDP) states, regulated by GDP dissociation inhibitor (GDI), GTPase activating factors (GAPs), and GDP/GTP exchange factors[8]. Local conformational changes in switch I and switch II, adjacent to the γ-phosphate, distinguish these states. Active Rabs reside in distinct sub-cellular compartments and mediate their biological effects via recruitment of specific effector proteins. Some understanding of the molecular basis for effector recognition has emerged from the crystal structures of Rabs with the Rab-binding domains (RBDs) of effectors [3,9,10]. Generally, the GTP-sensitive switch regions, switch I and II, as well as an invariant tryptophan residue in the interswitch region (between switch I and II) are important determinants of binding to α-helical motifs of RBDs. The exceptions are Early Endosomal Autoantigen 1 [EEA1; [10]] and the Lowe Syndrome protein OCRL1[9], which have non-helical RBDs.
Saccharomyces cerevesiae proteins Ypt31 and Ypt32 are homologs with 87% sequence identities (95% homology) in their globular Ras fold. They regulate vesicle exit from late Golgi compartments [11,12]. In the current model, Ypt31/32 together with phosphatidylinositol 4-phosphate recruit Sec2, which is also an exchange factor for Sec4. The nucleotide exchange action of Sec2 converts Sec4(GDP) to Sec4(GTP), and Sec2 also binds to Sec15, concomitantly releasing Ypt31/32 from the complex, thus facilitating the latter steps of vesicle delivery[13,14]. Both Sec4 and Ypt31/32 also bind to the globular tail domain of the actin-based class V myosin, Myo2 [15–17]. Sec15 is an effector of Sec4 and a component of the multi-protein exocyst, a docking complex that captures vesicles from the Golgi and promotes their fusion[18]. The equivalent interaction in mammalian cells is the Rab11/Sec15, which has been observed in photoreceptor cells of Drosophila melanogaster [19]. Thus, Ypt31/32 and Sec4 regulate consecutive steps in a complex cascade that involves Sec2, myosin V and the exocyst.
Here, we present the crystal structures of Ypt32 in the GTP and GDP bound states. The structure of active Ypt32 is compared with Sec4, which is required for a later step in the trafficking of secretory vesicles [20,21]. Comparisons of their structures reveal significant differences in the conformation of switch II that are influenced by the underlying network of interactions with Rab sub-family specific (RabSF) regions. The structure of Ypt32(GDP) together with the previously determined structure of the Ypt31/GDI complex facilitates a complete description of the pathway leading from an active (membrane-bound) Ypt31/32 conformation to the GDI-bound (cytosolic) structure. Strikingly, the structures reveal that GDI induces a remodeling of the switch II helix prior to membrane extraction.
2. Materials and methods
2.1 Protein expression and purification
The Ypt32 construct of the globular domain in a constitutively active form (Q72L, residues 7–188) was generated as a fusion with maltose binding protein (MBP). The cDNA was cloned into the vector pMBP-parallel 1, which contains an rTEV protease cleavage sequence, at the NcoI (5′end) and SalI (3′end) restriction sites. Expression of the protein, cleavage of MBP, and subsequent purification was performed as described in previous work[22]. However, a second amylose affinity step was not performed, as significant amounts of free Ypt32 attached non-specifically to the resin. As an alternative, brief dialysis in low salt buffer (10mM Tris-Cl, 10mM NaCl, 5mM MgCl2 and 1mM DTT, pH 8) was followed by ion-exchange chromatography using a MonoQ column (GE Healthcare) using a linear gradient to 500mM NaCl. Ypt32 was further purified by size exclusion chromatography (Superdex 200 16/60 column, GE Healthcare) in gel filtration buffer (10mM Tris-Cl, 150mM NaCl, 5mM MgCl2 and 1mM DTT, pH 7.5).
2.2 Crystallization and structure determination
Ypt32 (Q72L) at 10mg/mL was incubated with 5mM GTP (SIGMA) for about an hour on ice. Despite incubation of GTP with the mutant Ypt32(Q72L) at the beginning of crystallization trials, the crystals appeared with GDP, therefore it is likely that Ypt32 had hydrolyzed the γ-phosphate during the several weeks that were required for crystal growth. The crystallization condition was 21–22% PEG4000, 0.1M Tris (pH 8.5) and 0.2M MgCl2. Single crystals of Ypt32(GDP) with maximum dimensions of 0.2 × 0.1 × 0. 05mm were grown by micro-seeding. The crystallization procedure for active Ypt32 was identical, except that guanosine-5′-(βγ-imino)triphosphate (GppNHp), a non-hydrolyzable analog of GTP, was incubated with the protein at the beginning of trials. Crystals were optimized in 0.2M MgCl2, 0.1M HEPES, 15% w/v PEG6000, pH 7. The hanging drop method in Linbro plates (291K) was used for all crystallization experiments. Prior to data collection, crystals were soaked in cryoprotectant (25% xylitol) for approximately 30 seconds and flash cooled at 100K in the cryostream. Data sets were collected at beamline BM14 at the European Synchrotron Research Facility (ESRF, Grenoble), and processed using the XDS package[23].
The crystal structure of Ypt32 in complex with GDP was determined by molecular replacement using MOLREP [24]. A solution with two molecules in the asymmetric unit was found using Ypt31 (PDB code 3cpj, 88% identical) as a search model. The initial electron density map for Ypt32(GppNHp) was obtained by molecular replacement using the refined Ypt32(GDP) structure. Refinement of the models was performed by Refmac [25] using the maximum likelihood method, individual B-factor refinement and TLS refinement, alternating with cycles of manual inspection using Coot [26]. At no point were non-crystallographic symmetry (NCS) restraints applied during refinement. The data collection and refinement statistics are summarized in Table 1.
Table 1.
X-Ray Data Collection and Structure Refinement
| YPT32-GDP | YPT32-GppNHp | |
|---|---|---|
| Wavelength (Å) | 0.9794 | 0.9784 |
| Resolution (Å) | 40–1.7 (1.8–1.7) | 40.0–2.0 (2.15–2.0) |
| Space group | C2 | P212121 |
| Asymmetric unit | 2 molecules | 1 molecule |
| Cell parameters | ||
| a (Å) | 130.0 | 47.09 |
| b (Å) | 45.2 | 49.89 |
| c (Å) | 73.4 | 90.66 |
| β (°) | 110.5 | |
| Rsym (%) | 5.2 (40.1) | 6.9 (44.6) |
| I/σI | 19.5 (3.89) | 23.57 (4.24) |
| Completeness (%) | 99.4 (98.9) | 99.6 (100) |
| Multiplicity | 3.89 (3.9) | 7.1 (7.2) |
| Rwork/Rfree (%) | 16.7/20.3 | 19.4/24.0 |
| R.M.S.D | ||
| Bonds (Å) | 0.011 | 0.010 |
| Angles (°) | 1.33 | 2.01 |
| Ramachandran plot | ||
| Most favoured (%) | 92.2 | 93.4 |
| Disallowed | 0 | 0 |
Values in parentheses are for highest resolution shell.
2.3 Structural alignments by least-squares methods
Pairwise alignments were performed using the secondary structure maching (SSM) algorithm implemented in COOT[26]. The switch regions, which undergo conformational transitions, were excluded from the calculations. The active conformations of Rab GTPases will be referred to as Rab(GTP), although in general, the uncomplexed proteins have been co-crystallized with GppNHp. The refined structures of Ypt32(GDP) were aligned with the previously determined structure of the complex Ypt31/GDI (PDB code 3cpj; [27]). The two molecules in the asymmetric unit of Ypt32(GDP) are identical in conformation, with a root-mean-square (rms) deviation of 0.55Å for 155 common Cα atoms, therefore molecule A was used for structural analyses. Alignment of Ypt32(GDP) with Ypt31 (as part of the GDI complex) resulted in an rms deviation of 0.87Å for 149 common Cα atoms. Similarly, the structure of Ypt32(GTP) was aligned with Sec4(GTP). The two molecules in the asymmetric unit of active Sec4 have an identical conformation with an rms deviation of 0.20Å for all 168 Cα atoms (PDB code 1g17; [28]). Pairwise rms deviations with active Ypt32 were 1.14Å for 163 common Cα atoms in both sets of alignments (molecules A and B of Sec4). Therefore, molecule A of Sec4(GTP) was used as a representative structure for comparisons with Ypt32(GTP). Sec4(GDP) comprises 4 molecules in the asymmetric unit (PDB code 1g16; [28]). Keeping molecule A as the reference, the pairwise rms deviations were relatively low − 0.40Å, 0.57Å and 0.52Å for alignments of molecules B, C and D. However, inspection of their switch II conformations revealed that they fall into two categories, in which molecules A/B are identical with each other, but distinct from C/D. These two sets of conformations and their accompanying crystal contacts have been extensively discussed [28]. Therefore, structural comparisons of GDI-mediated switch II re-arrangements involved analyses of both molecules A and C (section 3.3, below). Finally, pairwise alignments of molecules A and C of Sec4(GDP) with the complex Sec4/GDI resulted in rms deviations of 0.76Å and 0.87Å for 157 Cα atoms, respectively.
2.4 Mutagenesis of Ypt32 and Sec4
In order to produce constructs pRS415 YPT32 and pRS425 YPT32, the 1.4 kb fragment of BamHI-PstI from YEp352 YPT32 [29] was inserted into BamHI-PstI sites of pRS415 or pRS425, respectively. For Sec4, An XbaI-MfeI fragment was subcloned from YEp24 SEC4 [29] into pRS416 at XbaI and EcoRI sites. The XbaI-SphI fragment from pRS416 SEC4 was inserted into pRS415 at XbaI and SphI sites to generate pRS415 SEC4. To generate pRS425 SEC4, the NotI-SalI fragment from pRS415 SEC4 was ligated into the NotI-SalI sites of pRS425. To make the ypt32-E110T mutant, the YPT32 gene was subjected to site-directed mutagenesis using the following primers: (5′-CGA AAA TTG CAA CCA CTG GCT TAC Cac ACT GAG AGA GAA CGC AG-3′) and (5′-CTG CGT TCT CTC TCA GTg tGG TAA GCC AGT GGT TGC AAT TTT CG-3′). For the sec4-T117E variant, the SEC4 gene was mutagenized using the following primers: (5′-CTA ATA TCA AGC AAT GGT TTA AAg agG TTA ATG AGC ATG CGA ACG-3′) and (5′-CGT TCG CAT GCT CAT TAA Cct cTT TAA ACC ATT GCT TGA TAT TAG-3′). The mutated nucleotides are in lower case.
PDB accession codes
The structures of Ypt32(GDP) and Ypt32(GppNHp) were deposited in the PDB data bank with the accession codes 3rwo and 3rwm, respectively.
3. Results and Discussion
3.1 Ypt32 active and inactive structures
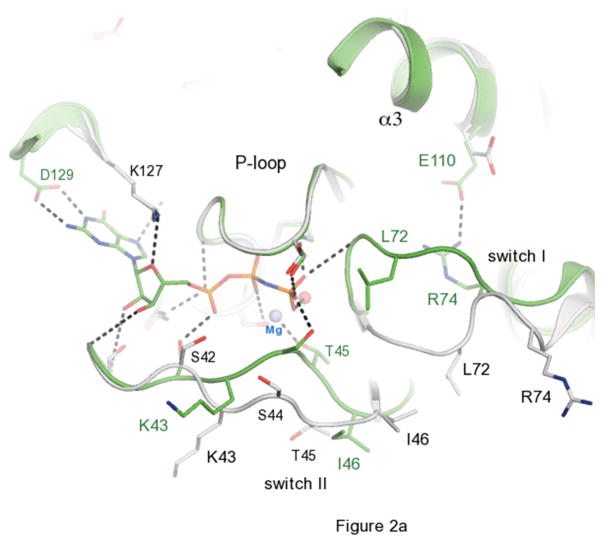
The overall fold of Ypt32 belongs to the small Ras-related GTPase superfamily that consists of a central 6-stranded mixed β-sheet containing five parallel and one anti parallel β-strands flanked by five α-helices (Fig 1). A superposition of the active and inactive structures reveals extensive conformational displacements in both switch I and switch II regions. In the GppNHp bound active form, the polypeptide backbone from 42–46 shifts toward the phosphate arm, mediated in part by the conserved Thr45 side chain, which interacts with the γ-phosphate via the bridging Mg2+ ion. The conformational change in switch II is even more dramatic. It moves up to 6Å closer towards the nucleotide so that the backbone NH of Gly71 points toward the γ-phosphate (dotted line, Fig 2a).
Figure 1.
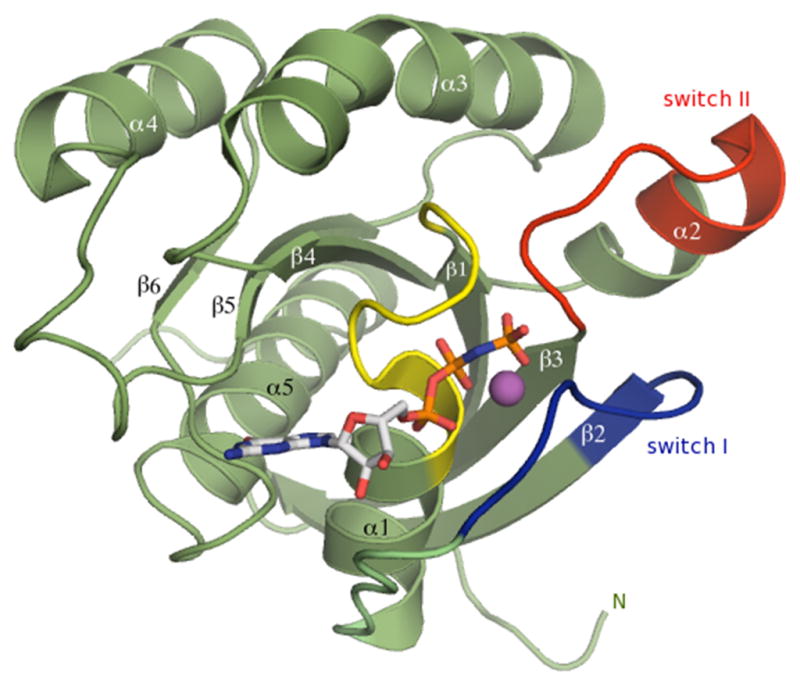
Structure of Ypt32 small GTPase in the GTP-bound conformation. The nucleotide is shown as a stick model, the P-loop is yellow, switch I is blue, and switch II is red. The Mg2+ ion is a purple sphere.
Figure 2.
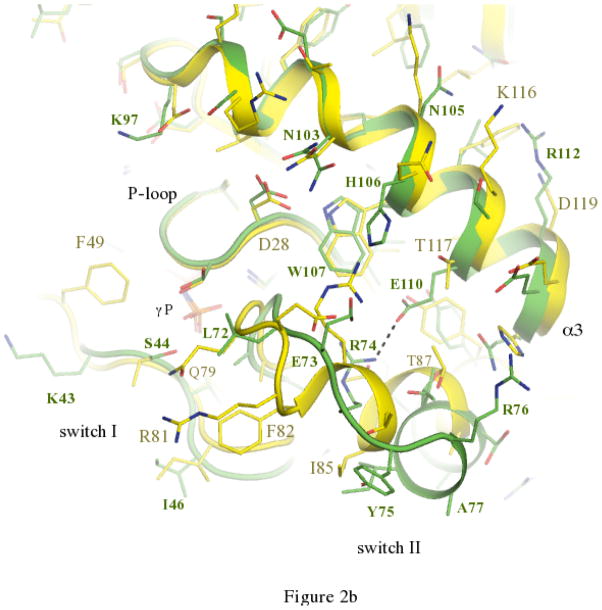
The activation cycle of Ypt32. (a) Comparisons of active and inactive conformations of Ypt32. Side chains of key residues in switch regions are shown as ball and stick – all other segments are represented as cartoon. Ypt32(GTP) is green, and Ypt32(GDP) is grey. The hydrogen bond between γ-phosphate and the backbone NH of Gly71 is shown. (b) Structural comparisons of active Ypt32 and Sec4 at the switch regions. Ypt32 is green, and Sec4 is yellow. The black dashes show the salt bridge between Arg74 and Glu110 in Ypt32. The side chain of Thr79 (Ypt32) is modeled in two conformational states. Similarly, Asn103 (Ypt32) displays side chain flexibility.
3.2 Comparisons of active Ypt32 and Sec4
The trafficking of vesicles to the plasma membrane is a stepwise process involving Rabs, myosin and the multi-protein exocyst. Sec2 plays a pivotal role in vesicle delivery, acting both as an effector of Ypt32 and a guanine nucleotide exchange factor (GEF) for Sec4[30]. Upon activation, Sec4 (a Rab8/Rab10 homologue) regulates vesicle tethering in yeast via interactions with its effector Sec15[31], which itself is a component of the exocyst. Myo2 is essential for yeast viability, and it is an effector for both Ypt31/32 and Sec4. Thus, Ypt31/32 and Sec4 recruit the same proteins (Sec2, Myo2) to secretory vesicles at different stages during their stepwise delivery to plasma membranes. The previously determined structures of Sec4 [28] afford an opportunity to compare its active conformation with Ypt32 (Fig 2b; see Materials and Methods). The sequence of their switch I regions are highly conserved, and their switch II regions are identical apart from two conservative substitutions. Excluding the prenylated tails, Ypt32 and Sec4 share 48% identities (68% homology) in their globular domains (Fig 3). Comparisons of switch I from Ypt32 (42–49) and Sec4 (48–56) reveal an identical conformation, but slight variations in composition result in divergent structural features displayed for effector recruitment. In particular, Lys43 (Ypt32) and Ile50 (Sec4) reside at an equivalent position in Sec4.
Figure 3.
Sequence alignment of yeast Rabs. Ypt11 has been excluded from the list, since it contains large insertions relative to other yeast Rabs. All of the sequences are from Saccharomyces cerevisiae. Secondary structure elements are above the sequence. Background colouring indicates the P-loop (green), switch I (blue) and switch II (pink), using the structure of Ypt32 as the reference. Rab sub-family specific regions (RabSF) are marked below the sequences.
In contrast to switch I, the conformations of switch II are highly divergent despite a conserved sequence (Fig 2b). Globally, the switch II region of Ypt32 moves away from α3 relative to the same region in Sec4 such that the equivalent Cα atoms of Arg76 (Ypt32) and Arg83 (Sec4) lie 7.8Å apart. A key determinant of the conformational difference is the presence of a salt bridge in Ypt32 between Glu110 in α3 and Arg74 in switch II (2.7Å). This interaction requires spatial segregation of the loop 69–75, which precedes an α-helix in switch II (Fig 2b; Supplementary Fig 1). The helix α3 is divergent in Rab GTPases and forms part of the Rab subfamily (RabSF) motif 3 [32]. The equivalent Glu110 locus is Thr117 in Sec4, which is packed against Thr87 and Ty90 in the α-helical segment of switch II. It is interesting to note that Glu110 is conserved in all yeast Rabs except for Sec4, which has a threonine residue (Fig 3; red circle).
Rab GTPases Ypt31/32 are essential for yeast cell viability, as evidenced by the inability of dual gene knockouts (ypt31Δ+ypt32Δ) to grow on solid media (Supplementary Fig 2). Similarly, a single gene knockout of Sec4 (sec4Δ) is also unable to support growth. Exploiting viability as an assay for overall function, the mutation Glu110→Thr in Ypt32 (Ypt32ET) and Thr117→Glu in Sec4 (Sec4TE) were generated. Yeast strains harboring the single-site mutations are able to grow, therefore the mutations have no effects on cell viability. Furthermore, the mutant Ypt32ET is unable to support the growth of sec4Δ, and vice versa. Thus, the structural determinants of Rab function extend beyond a single site, i.e., the single locus in RabSF3 is not the unique determinant of Rab identity (Ypt31/32 vs. Sec4). Whether the mutations had sub-lethal effects on vesicle trafficking, such as the strength of interactions with effectors (Myo2, Sec15) could not be determined from the viability assay. Lethal phenotypes or functional complementation may require additional (multi-site) mutagenesis, such as Lys43 (Ypt32) vs. Ile50 (Sec4) in switch I (Fig 4). Interestingly, the L7 region of RabSF3 (Glu120-Leu128), immediately adjacent to Thr117 (Sec4), was previously found to be an important determinant of Ypt1 and Sec4 functionality[33,34]. However, it is worthy noting that Sec4 grafts of switch I and RabSF3 onto Ypt1 were also unable to support growth in sec4Δ yeast [34], although a complete graft of switch II + RabSF3 was able to complement the sec4Δ knockout [33]. Further insight into the secretory pathway will require the crystallization of binary Rab/effector complexes - Ypt32/Myo2, Sec4/Myo2, Sec4/Sec15 - to understand structure and function.
Figure 4.
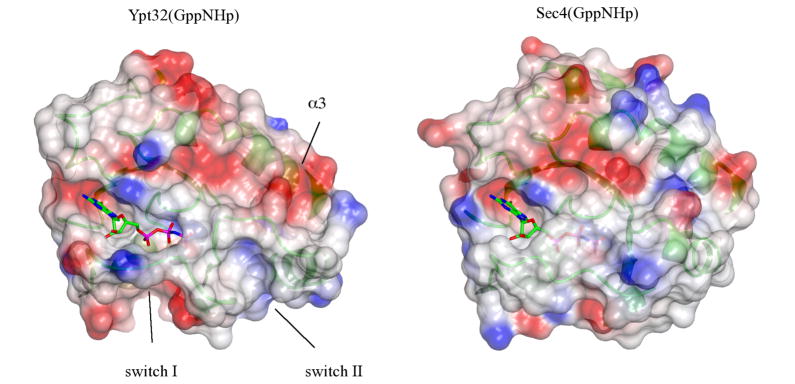
Surface electrostatics rendering of active Ypt32 and Sec4. Grey is neutral, blue is positively charged, and red is negative, and the nucleotide is a ball-and-stick model. The surface is slightly transparent to allow visualization of the underlying secondary structures (green ribbons). Switch I shows a positive interface in Ypt32 relative to a hydrophobic Sec4, due to the presence of Lys43 vs. Ile50.
3.3 GDI binding to Ypt31/32
Ypt31/32 regulate the post-Golgi secretory pathway and share 87% sequence identities (95% homology) within their globular regions [12]. The structure of Ypt31 has been determined in complex with GDI (PDB code 3cpj) [27]. Upon superposition of Ypt31/GDI complex with Ypt32(GDP), the rms deviation of 155 common Cα atoms is 0.95Å using the SSM algorithm in COOT[26]. Thus, the structure of Ypt32(GDP) enables visualization of the complete pathway for GDI-mediated extraction of Ypt31/32 from membranes to the cytosolic fraction. In our structure of uncomplexed Ypt32(GDP), switch I is well ordered due to non-crystallographic symmetry contacts in the asymmetric unit (not shown). Although switch I is likely flexible, our interpretation is that a pre-existing solution conformation of switch I has been captured during crystallization of Ypt32(GDP).
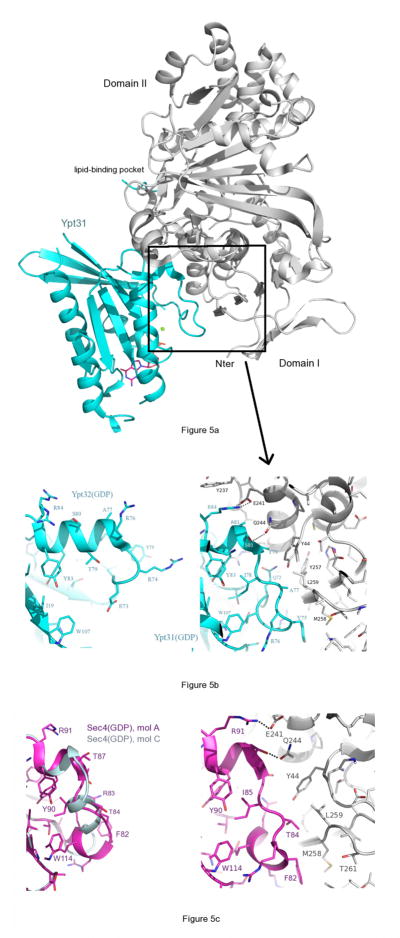
In contrast to switch I, which is disordered in the GDI-bound state, switch II is ordered and undergoes a dramatic conformational re-arrangement upon binding to Domain I of GDI (Fig 5). One turn of the C-terminal end of the switch II helix unwinds and the loop is pushed closer toward α3 by GDI. The Rab-binding platform (RBP) in Domain I of GDI (rectangle, Fig 5a) binds to the newly formed N-terminus of α2 such that the side chain of Gln244 (αI of GDI) forms a hydrogen bond to the backbone NH of Ser80 (Ypt32; Fig 5b). Interactions between the RBP and conserved residues in Ypt31 have been discussed in detail previously [27]. The important observation here is that GDI binding to Ypt31/32(GDP) is not possible without a significant remodeling of the switch II helix. This remodeling of switch II has not been described, since the uncomplexed structures of Ypt31/32 were not previously known. The two molecules in the asymmetric unit of Ypt32(GDP) are identical, with an rms deviation of 0.55Å for 170 common Cα atoms (see Materials and Methods). Although the switch regions are involved in crystal contacts, the conformations of switch II are indistinguishable in the two molecules of Ypt32(GDP), and no NCS restraints were applied during refinement. The only significant difference is a relative rotation of the Arg74 side chain. Overall, the segment Gly71-Ile78 is remodeled via favourable interactions that include hydrophilic and hydrophobic contacts against the backbone and side chains of switch II. It is interesting to note that the switch II helix is already one turn shorter in Sec4(GDP) and Ypt1(GDP). The equilibrium dissociation constants (Kd) for the interactions between GDI and full-length Rabs are 18.9μM for Ypt31/GDI, 1.5μM for Ypt32/GDI and 0.33μM for Sec4/GDI [27]. One reasonable explanation for these differences is the increased thermodynamic penalty associated with remodeling of switch II in Ypt31/32, which would contribute to lower affinities (higher Kd) in Ypt31/32 relative to Sec4.
Figure 5.
GDI-mediated remodeling of the switch II helix of Ypt32(GDP). GDI is coloured gray in all figures. (a) Structure of Ypt31/GDI generated from PDB file 3cpj. The Rab-binding platform (RBP) is indicated by the rectangle. The lipid-binding pocket is behind GDI in this view. (b) Left panel, uncomplexed Ypt32(GDP); right panel, the complex Ypt31(GDP)/GDI, which corresponds to the region indicated by the rectangle. The two panels are shown with the same view and scale, thus emphasizing the loss of a helical turn in switch II upon binding of GDI. Arg84 (left panel) is observed in two conformations, and becomes ordered upon forming a salt bridge with Glu241 (GDI). A hydrogen bond is also shown between Q244 and the backbone NH of Ser80 (Ypt32). (c) Uncomplexed Sec4(GDP) is shown on the left panel (magenta, molecule A; light blue, molecule C), and as the complex Sec4/GDI (right panel, PDB code 3cph). The side chains of molecule C are not shown, for clarity. Structural alignments were performed as described in section 2.3 of Materials and Methods.
The structure of uncomplexed Sec4(GDP) revealed the presence of two distinct conformations for the four molecules in the asymmetric unit[28]. The conformation that molecules A and B adopt is identical to the conformation of Sec4 in its complex with GDI (Fig 5c), and resembles the switch II conformation of Ran(GDP)[28]. Therefore, despite crystal contacts by the switch regions of Sec4(GDP), there was strong circumstantial evidence that GDI binds to a pre-existing Sec4(GDP) conformation in solution. Small conformational changes are nevertheless induced upon binding – an example is the repositioning of Phe82, which is the equivalent of Tyr75 in Ypt32 (Fig 5c). However, the Sec4 rearrangement involves a movement of 2.6Å in the Cα position of Phe82, therefore the magnitude of the conformational changes are smaller when compared with Ypt31/32. Thus, Sec4 binding to GDI can be considered more of a ‘rigid dock’ at switch II relative to Ypt32.
In summary, Ypt31/32 affinity to GDI is an order of magnitude weaker than Sec4/GDI [27]. Remodeling Ypt31/32 by GDI to induce the conserved switch II conformation found in all complexes likely contributes to the weaker affinities. It is important to note that affinities are also a function of the interactions between GDI and the prenylated tails, which vary in detail among the Rab/GDI complexes [27].
Supplementary Material
Supplementary Figure 1 Electron density at the GTP-dependent salt bridge of Ypt32. Density is contoured at 1σ, and dashes represent the bond between Arg74 and Glu110. Several water molecules (red spheres) are observed in this region. The flexibility of Thr79 is visible with the presence of the two conformers, and the quality of the model is apparent from the clear backbone electron density for switch II.
Supplementary Figure 2 Theypt32-E110T and sec4-T117E mutants show normal growth. (A) Similar to wild-type SEC4, the sec4-T117E mutant cannot suppress the lethality of ypt31Δ+ypt32Δ (Left panel). Plasmids transformed into a ypt31Δ+ypt32Δ strain (LWY10906), pRS415 [CEN, LEU2] (mock), pRS415 YPT32, pRS415 ypt32-E110T, pRS415 SEC4, pRS415 sec4-T117E, pRS425 [2m, LEU2] (mock), pRS425 YPT32, pRS425 ypt32-E110T, pRS425 SEC4, or pRS425 sec4-T117E. Transformed colonies were re-streaked onto SC-Leu+5FOA to counter select against pRS316 [URA3] YPT31 (middle panel; pNS220, Jedd et al., 1997 JCB) and on SC-Leu-Ura to show that the cells received a [LEU2] plasmid (right panel). (B) Colonies (pRS415 YPT32 or pRS415 ypt32-E110T transformed) from SC–Leu+5FOA (A) were cultured in liquid and serial dilutions spotted on YEPD plates, and incubated at 18, 24, 30 or 37°C for 2 days. (C)ypt32-E110T cannot suppress the lethality of sec4ypt31Δ+ypt32Δ. (Left panel) Indicated plasmids were transformed into a sec4D strain (LWY10832). Transformed colonies were re-streaked onto SC-Leu+5FOA to counter select against pRS416 [URA3] SEC4 (middle panel) and on SC-Leu-Ura to show that the cells received a [LEU2] plasmid (right panel). The dark patches seen at 3 o’clock of the SC-Leu+5FOA plate, are due to scratches in the agar, not yeast growth. (D) Colonies (pRS415 SEC4 or pRS415 sec4-T117E transformed) from SC–Leu+5FOA (B) were cultured in liquid and serial dilutions spotted on YEPD plates, and incubated at 18, 24, 30 or 37°C for 2 days.
Highlights.
Despite having a common effector - Myo2 - active Ypt32 and Sec4 have divergent 3-D structures
switch II helix of Ypt32(GDP) is remodeled by GDI during membrane extraction
Acknowledgments
We would like to Dr. Hassan Belrhali of BM14 at the ESRF for his help in data collection. This work was supported by Science Foundation Ireland [grant number 03/IN.3/B371 to ARK], an Irish Research Council Science, Engineering and Technology (IRCSET) fellowship to AS and R37 GM62261 from the National Institutes of Health to LSW.
Abbreviations
- Myo2p
Myosin 2p
- RBD
Rab-binding domain
- RabSF
Rab sub-family specific regions
Footnotes
Conflict of interest
The authors have no financial or commercial conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–15. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MT, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic. 2009;10:1377–89. doi: 10.1111/j.1600-0854.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–41. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 5.Pereira-Leal JB. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic. 2008;9:27–38. doi: 10.1111/j.1600-0854.2007.00667.x. [DOI] [PubMed] [Google Scholar]
- 6.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 7.Leung KF, Baron R, Seabra MC. Thematic review series: lipid posttranslational modifications. geranylgeranylation of Rab GTPases. J Lipid Res. 2006;47:467–75. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cellular and molecular life sciences: CMLS. 2005;62:1657–70. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. The EMBO journal. 2011;30:1659–70. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra A, Eathiraj S, Corvera S, Lambright DG. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10866–71. doi: 10.1073/pnas.1000843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benli M, Doring F, Robinson DG, Yang X, Gallwitz D. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 1996;15:6460–75. [PMC free article] [PubMed] [Google Scholar]
- 12.Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–80. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Developmental cell. 2010;18:828–40. doi: 10.1016/j.devcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medkova M, France YE, Coleman J, Novick P. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol Biol Cell. 2006;17:2757–69. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casavola EC, Catucci A, Bielli P, Di Pentima A, Porcu G, Pennestri M, Cicero DO, Ragnini-Wilson A. Ypt32p and Mlc1p bind within the vesicle binding region of the class V myosin Myo2p globular tail domain. Molecular microbiology. 2008;67:1051–66. doi: 10.1111/j.1365-2958.2008.06106.x. [DOI] [PubMed] [Google Scholar]
- 16.Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N. Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol Biol Cell. 2008;19:4177–87. doi: 10.1091/mbc.E08-02-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Developmental cell. 2011;20:47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brocker C, Engelbrecht-Vandre S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Current biology: CB. 2010;20:R943–52. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nature structural & molecular biology. 2005;12:879–85. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 20.Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–38. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 21.Stroupe C, Brunger AT. Crystal structures of a Rab protein in its inactive and active conformations. J Mol Biol. 2000;304:585–98. doi: 10.1006/jmbi.2000.4236. [DOI] [PubMed] [Google Scholar]
- 22.Jagoe WN, Jackson SR, Lindsay AJ, McCaffrey MW, Khan AR. Purification, crystallization and preliminary X-ray diffraction studies of Rab11 in complex with Rab11-FIP2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:692–4. doi: 10.1107/S1744309106023074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta crystallographica. Section D, Biological crystallography. 2010;66:22–5. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. Section D, Biological crystallography. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O. A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism. J Biol Chem. 2008;283:18377–84. doi: 10.1074/jbc.M709718200. [DOI] [PubMed] [Google Scholar]
- 28.Stroupe C, Brunger AT. Crystal structures of a Rab protein in its inactive and active conformations. Journal of molecular biology. 2000;304:585–98. doi: 10.1006/jmbi.2000.4236. [DOI] [PubMed] [Google Scholar]
- 29.Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–80. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. Journal of molecular biology. 2000;301:1077–87. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 33.Dunn B, Stearns T, Botstein D. Specificity domains distinguish the Ras-related GTPases Ypt1 and Sec4. Nature. 1993;362:563–565. doi: 10.1038/362563a0. [DOI] [PubMed] [Google Scholar]
- 34.Brennwald P, Novick P. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature. 1993;362:560–563. doi: 10.1038/362560a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Electron density at the GTP-dependent salt bridge of Ypt32. Density is contoured at 1σ, and dashes represent the bond between Arg74 and Glu110. Several water molecules (red spheres) are observed in this region. The flexibility of Thr79 is visible with the presence of the two conformers, and the quality of the model is apparent from the clear backbone electron density for switch II.
Supplementary Figure 2 Theypt32-E110T and sec4-T117E mutants show normal growth. (A) Similar to wild-type SEC4, the sec4-T117E mutant cannot suppress the lethality of ypt31Δ+ypt32Δ (Left panel). Plasmids transformed into a ypt31Δ+ypt32Δ strain (LWY10906), pRS415 [CEN, LEU2] (mock), pRS415 YPT32, pRS415 ypt32-E110T, pRS415 SEC4, pRS415 sec4-T117E, pRS425 [2m, LEU2] (mock), pRS425 YPT32, pRS425 ypt32-E110T, pRS425 SEC4, or pRS425 sec4-T117E. Transformed colonies were re-streaked onto SC-Leu+5FOA to counter select against pRS316 [URA3] YPT31 (middle panel; pNS220, Jedd et al., 1997 JCB) and on SC-Leu-Ura to show that the cells received a [LEU2] plasmid (right panel). (B) Colonies (pRS415 YPT32 or pRS415 ypt32-E110T transformed) from SC–Leu+5FOA (A) were cultured in liquid and serial dilutions spotted on YEPD plates, and incubated at 18, 24, 30 or 37°C for 2 days. (C)ypt32-E110T cannot suppress the lethality of sec4ypt31Δ+ypt32Δ. (Left panel) Indicated plasmids were transformed into a sec4D strain (LWY10832). Transformed colonies were re-streaked onto SC-Leu+5FOA to counter select against pRS416 [URA3] SEC4 (middle panel) and on SC-Leu-Ura to show that the cells received a [LEU2] plasmid (right panel). The dark patches seen at 3 o’clock of the SC-Leu+5FOA plate, are due to scratches in the agar, not yeast growth. (D) Colonies (pRS415 SEC4 or pRS415 sec4-T117E transformed) from SC–Leu+5FOA (B) were cultured in liquid and serial dilutions spotted on YEPD plates, and incubated at 18, 24, 30 or 37°C for 2 days.