A perspective on work from the Roshbash laboratory (Abruzzi et al.) discussing the novel insights into how clocks drive daily rhythms in global gene expression.
Keywords: circadian rhythms, Drosophila, transcription
Abstract
In this issue of Genes & Development, Abruzzi et al. (pp. 2374–2386) use chromatin immunoprecipitation (ChIP) tiling array assays (ChIP–chip) to show that physical interactions between circadian (≅24-h) clock machineries and genomes are more widespread than previously thought and provide novel insights into how clocks drive daily rhythms in global gene expression.
A wide variety of life forms exhibit circadian (≅24-h) rhythms in metabolism, physiology, and behavior. These rhythms are governed by cellular “clocks” or pacemakers based on the coordinated expression and interactions of a small set of “clock” genes that appear conserved within a particular kingdom (Young and Kay 2001). A hallmark feature of circadian rhythms is that they persist or free run with periods of ∼24 h in the absence of external temporal cues. Nonetheless, an important adaptive feature of these clocks is that they are entrained (synchronized) to local time by external cues, most notably the daily light/dark cycles. They thus provide organisms with the ability to anticipate environmental transitions, perform activities at biologically relevant times, and undergo appropriate seasonal responses. Malfunctions in the human circadian timing system and/or mutations in clock genes are implicated in many disorders and diseases, including SAD (seasonal affective disorders or “winter” depression), chronic sleep problems, a range of metabolic syndromes, and cancer (for review, see Takahashi et al. 2008).
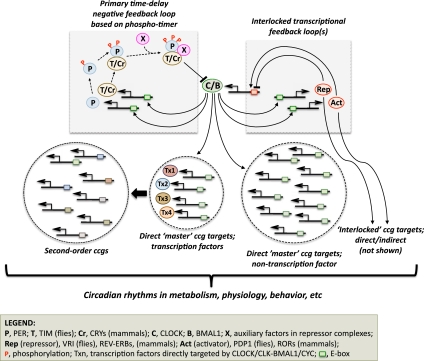
While there is diversity in circadian clock mechanisms, those in eukaryotes can be generally described as biochemical oscillators built on interlocked loops of transcriptional feedback, coupled with phase-specific protein turnover (Fig. 1; for review, see Young and Kay 2001; Gallego and Virshup 2007; Zhang and Kay 2010). At the heart of these circadian transcriptional networks is a “primary” autoinhibitory loop wherein a “master” transcription factor drives expression of one or more repressor proteins. After a time-delay feedback to inhibit the activity of the transcription factor until the repressor is targeted for degradation, another round of transcription ensues (Dunlap 1999). Besides stimulating the production of post-transcriptional repressors, master circadian transcription factors directly regulate the expression of a few “secondary” clock transcription factors (positive and/or negative) that directly feed back into the main circadian circuitry (e.g., regulate expression of the master clock transcription factor), resulting in a small subsystem of intertwined cycling clock transcription factors that appear to stabilize the rhythm-generating mechanism.
Figure 1.
How a master clock transcription factor might drive global rhythms in gene expression, with an emphasis on the Drosophila and mammalian systems. Ovals represent different proteins; some homologs only function as depicted in either Drosophila or mammals. Color-coding of transcription factors and cognate promoter-binding sites (small squares) is meant to signify direct interactions. See the text for more details.
How do these tightly interwoven intracellular networks of self-perpetuating molecular oscillations in the mRNA and protein products of clock genes contribute to the great variety of overt rhythms observed in life forms? It is thought that many of the clock-regulated rhythms in physiology and behavior exhibited by organisms are themselves driven by cyclical gene expression, whereby the transcriptional feedback circuits underlying circadian clock mechanisms also generate, either directly or indirectly, rhythmic expression of numerous downstream targets, collectively termed clock-controlled genes (ccgs) (Fig. 1). For example, ccgs that encode transcription factors could amplify the number of cycling genes within a cell and impart phase and tissue specificity. Also, a ccg that encodes an enzyme functioning in a rate-limiting step could impose circadian regulation on an entire pathway. In addition, the temporal information generated within a clock cell (or more specifically, a cell that contains a clock mechanism) can influence other cells/tissues by driving cyclical expression of an extracellular signaling molecule; like, for example, a secreted neuropeptide.
In the last 10 years, findings based largely on DNA microarrays have led to numerous insights concerning the extent of circadian gene expression in a range of model organisms (for a recent review see, Doherty and Kay 2010). Applying this technology to animals suggested that 1%–10% of a cell's transcripts undergo daily cycles in expression. The two best-studied animal model systems for circadian rhythms are Drosophila and mice (Zhang and Kay 2010; Hardin 2011). In mammals, the basic helix–loop–helix (bHLH)/PAS (Per–Arnt–Sim) transcription factors CLOCK and BMAL1 form a heterodimer that is at the top of the hierarchy of transcription factors driving cyclical gene expression, whereas a homologous unit operates in Drosophila, termed dCLOCK (CLK) and CYCLE (CYC) (Fig. 1). These transcription factors stimulate gene expression by binding to E-box DNA elements (consensus, CACGTG) in the promoter regions of target genes, including the period (per) genes (dper in Drosophila and mPer1, mPer2, and mPer3 in mammals), key clock factors in mediating feedback repression. The emerging model, based on several lines of evidence discussed below, posited that only a small set of cycling transcripts is directly driven by CLOCK/CLK–BMAL1/CYC and that other cyclical transcription factors, emanating from the interlocked feedback loops and/or downstream ccgs, are largely responsible for most of the rhythmic transcriptomes.
Findings by Abruzzi et al. (2011) in this issue of Genes & Development reveal that, in actuality, an unexpectedly large number of genes are direct targets of CLK–CYC. Similar findings were recently observed with BMAL1 in mammals (Rey et al. 2011), indicating that master clock transcription factors can act in a pervasive yet flexible manner to generate tissue-specific rhythmic patterns of global gene expression. There are many excellent reviews on clock mechanisms, including overviews of relevant DNA microarray studies (Delaunay and Laudet 2002; Etter and Ramaswami 2002; Duffield 2003; Sato et al. 2003; Wijnen et al. 2005; Doherty and Kay 2010). This perspective focuses mostly on the transcriptional aspect of global gene expression, with an emphasis on the Drosophila model system as a foundation for discussing some of the implications by Abruzzi et al. (2011).
The primary circadian negative transcriptional feedback loop: a revolving relationship between master clock transcription factor and phospho-timing repressor
Key components of the primary circadian transcriptional negative feedback loop in Drosophila melanogaster are dPER, TIMELESS (TIM), CLK, and CYC (Fig. 1, top left; Hardin 2011). CLK and CYC heterodimerize and bind E-box-containing DNA elements to stimulate transcription of dper and tim. In turn, the dPER and TIM proteins physically interact in the cytoplasm and translocate to the nucleus, where they participate in repressing CLK–CYC-mediated transcription. For such a system to generate molecular oscillations, it requires a time delay between the activities of the positive (CLK–CYC) and negative (dPER and TIM) elements. Indeed, an elaborate web of post-translational regulatory steps highlighted by multiple protein kinases and phosphatases evokes time-of-day-specific phosphorylation events that control dPER's daily stability, timing of nuclear entry, and duration in the nucleus such that it mainly engages in blocking CLK–CYC activity during the early night to early day (for review, see Bae and Edery 2006).
PER proteins in flies and mammals are thought to inhibit CLOCK/CLK–BMAL1/CYC activity in an indirect manner by acting as scaffolds to facilitate the timely assembly/delivery of other cofactors such as kinases and chromatin remodeling factors (collectively depicted as X in Fig. 1) that more directly inhibit, in a catalytic and/or stoichiometric manner, the activity of these master clock transcription factors (Yu et al. 2006; Kim et al. 2007; Duong et al. 2011). Results in Drosophila using chromatin immunoprecipitation (ChIP) show that dPER participates in a concerted two-step inhibitory mechanism (Menet et al. 2009). The first wave of inhibition involves an on-DNA repression phase whereby dPER interacts with chromatin-bound CLK–CYC, followed by an off-DNA phase where dPER associates in a 1:1 complex with CLK–CYC. What underlies the transition from on-DNA to off-DNA is not yet clear. Although there is remarkable commonality in the primary circadian transcriptional negative feedback loops operating in Drosophila and mammals (Gallego and Virshup 2007), there are some differences. For example, there is no firm role for a TIM homolog in mammalian clocks. However, CLOCK–BMAL1 also drives E-box-dependent cyclical expression of cryptochrome 1 (cry1) and cry2, whose protein products participate with mPERs in transcriptional repression.
Recent findings in Drosophila suggest that much of the intrinsic circadian timing mechanism in animal clocks is encoded in the daily phospho-kinetics of PER (Chiu et al. 2011). Similar findings were also shown for FREQUENCY (FRQ) (Querfurth et al. 2011), the key repressor in the Neurospora clock, an early system that played a pivotal role in our understanding of the molecular underpinnings for circadian rhythms (Baker et al. 2011). Thus, “phospho-timing repressors” such as PER proteins have a dual role in clock mechanisms by acting as the primary biochemical timer setting the pace of the clock and relaying this temporal information into cyclical gene expression by phase-specific interactions with a master clock transcription factor.
Numerous studies support the designation of CLOCK/CLK–BMAL1/CYC as master regulators in circadian rhythm generation. Mutations that inactivate either Clk or cyc function abolish molecular and behavioral circadian rhythms in flies (Allada et al. 2001). Ectopic expression of Clk in nonclock cells can produce ectopic clocks (Zhao et al. 2003), and manipulating the transcriptional potency of CLK–CYC modulates clock speed (Kadener et al. 2008). It is thought that the levels of CLK are the limiting clock protein in the Drosophila oscillator, at least in the primary negative feedback loop (Bae et al. 2000). In mice, inactivation of Bmal1 is the only single-gene mutation that abolishes overt behavioral rhythms (Bunger et al. 2000). With regard to CLOCK in mammals, the situation is a bit more complex, as the highly related homolog NPAS2 might be a functional replacement for CLOCK, at least in some tissues (DeBruyne et al. 2007).
Clock mechanisms gain more transcriptional feedback loops and phase control of gene expression
Studies in a wide variety of model organisms indicate that primary circadian transcriptional negative feedback loops are interlocked with one or more “secondary” transcriptional feedback loops (Zhang and Kay 2010), an observation first made with Clk expression in Drosophila more than 10 years ago (Glossop et al. 1999). Clk mRNA levels undergo daily rhythms that are anti-phase to those of dper and tim (Bae et al. 2000). Alternating phases of repression by VRILLE (VRI) and activation by PAR domain protein 1ε (PDP1ε), both basic leucine zipper (bZip) transcription factors, drives daily rhythms in Clk expression; in turn, via E-box elements, CLK directly stimulates the rhythmic expression of vri and Pdp1ε, generating an interlocked circadian transcriptional feedback loop (Fig. 1, top right; Cyran et al. 2003; Glossop et al. 2003). Similar to Clk in Drosophila, cyclical expression of Bmal1 is generated by alternating phases of repression by REV-ERBs and activation by RORs, members of a subfamily of orphan nuclear receptors whose expression is also rhythmic and directly stimulated by CLOCK–BMAL1 (Fig. 1, top right; for review, see Zhang and Kay 2010). Moreover, Bmal1 and mPer1–3 transcript rhythms are anti-phase to each other—analogous to Clk and dper/tim in flies—and are controlled primarily by different cis-acting DNA elements: Baml1 is controlled by the REV/ROR response element (RRE; “evening”-responsive element), and mPer1–3 are controlled by E-box-containing DNA elements (“morning”-responsive elements). Additional interlocked transcriptional loops have been observed in the clock mechanisms of flies and mammals (Zhang and Kay 2010; Hardin 2011), including a midday D-box-responsive element in mammals (Ueda et al. 2005).
The presence of multiple interlocked transcriptional feedback loops might endow circadian oscillatory mechanisms with enhanced robustness and/or expand the repertoire of high-amplitude cycling transcription factors available to drive downstream rhythms in global gene expression (e.g., Liu et al. 2008). At a minimum, the inclusion of multiple cis-regulatory regions responsive to different positive and negative transcription factors expanded the known modules available for generating rhythmic transcripts. Furthermore, the differential combinations of these modules offered an attractive explanation for how cyclical gene expression with varying phases could be attained (e.g., Ukai-Tadenuma et al. 2008).
Some lessons (mis)learned from DNA microarrays and global circadian gene expression patterns
DNA microarrays have been instrumental in trying to connect central clock mechanisms to output rhythms in physiology and behavior by identifying—on a global scale—transcripts that undergo daily changes in abundance in a manner that is dependent on the presence of a functional clock. This topic has been the subject of several excellent reviews (e.g., Delaunay and Laudet 2002; Duffield 2003; Sato et al. 2003; Wijnen et al. 2005; Doherty and Kay 2010; Zhang and Kay 2010), and only a very brief summary will be highlighted here, with an emphasis on findings in Drosophila and mammals that favored the suggestion that only a small subset of cycling transcripts are direct targets of CLOCK/CLK–BMAL1/CYC.
An early realization from the multiple studies performed with Drosophila and mice was that, besides canonical clock factors, there was little overlap in the subset of circadian transcripts identified between independent studies sampling similar, if not identical, material. This also revealed that, in general, the highest-amplitude circadian cyclers are clock transcripts and that many of the downstream ccgs have low amplitudes wherein their classification as bona fide circadian is more prone to differences in algorithms and/or slight variations in experimental conditions (e.g., Wijnen et al. 2005; Keegan et al. 2007). Thus, it appears that the transcriptional oscillatory potential generated within the core clock mechanism is somewhat dissipated for the majority of ccgs. Moreover, although about the same fraction of transcripts manifest circadian rhythms in abundance in different tissues (1%–10%), there is little overlap and they cluster around different phases (e.g., Storch et al. 2002). This variety of phases and transcripts in different tissues suggested that in animals, very few ccgs are direct targets of CLOCK/CLK–BMAL1/CYC. Indeed, microarray studies tended to support this notion, as there appeared to be some, but not extensive, enrichment of E-box elements in the 5′ promoter regions of cycling genes (Doherty and Kay 2010). Moreover, studies in Drosophila based on identifying genes whose expression increased with induction of CLK in the presence of the translation inhibitor cycloheximide to favor the identification of primary targets suggested that, of the ccgs identified in prior microarray studies, only a few are likely to be directly stimulated by CLK–CYC, with many being core clock factors (McDonald and Rosbash 2001; Kadener et al. 2007).
Together, the findings suggest a model whereby CLOCK/CLK–BMAL1/CYC is perched at the top of the circadian transcriptional hierarchy in animals because it is the key nodal point that both drives and responds to the main phospho-timing repressor complexes (i.e., PER-containing complexes). Moreover, its status at the top is further solidified because its main task is to produce a small cascade of high-amplitude cycling clock transcription factors (e.g., VRI and PDP1 in Drosophila, and Rev-erbs and RORs in mammals) that both provide feedback to help maintain the rhythmic engine and serve as a conduit for downstream cyclical gene expression. As such, much of the global rhythmic gene expression would be directly imparted by the “secondary” cycling clock transcription factors and/or downstream ccgs encoding transcription factors (Fig. 1, bottom far left and bottom far right), an idea supported by findings that as a functional class, transcription factors are overly represented as ccgs in microarray studies from a wide variety of organisms (Doherty and Kay 2010). Such a model would largely free up CLOCK/CLK-BMAL1/CYC from directly handling tissue-specific chores.
Direct interactions between master clock transcription factors and genomes are unexpectedly widespread and complex
To better understand the relationship between circadian clocks and global gene expression, Abruzzi et al. (2011) use ChIP tiling array assays (ChIP–chip) to interrogate the daily interactions of CLK, CYC, PER, and RNA polymerase II (Pol II) with the fly genome. Surprisingly, they identify ∼1500 regions in the genome that CLK binds to in adult heads, which, reassuringly, are enriched for canonical and degenerate E-boxes. At a minimum, ∼60% of these binding sites were shown to undergo daily fluctuations in their association with CLK, of which 500 could be unambiguously ascribed to a single gene. Peak timing of CLK binding largely occurs between Zeitgeber time 14 (ZT14) and ZT16 (where ZT0 is lights on and ZT12 is lights off in a 12-h light:12-h dark cycle), consistent with earlier work probing dper and tim promoters (Menet et al. 2009). Further analysis showed that most, if not all, genes wherein CLK rhythmically binds also interact with CYC in a coincident manner, followed by a 4- to 6-h phase delay in dPER binding, similar in regulation to canonical clock genes directly targeted by CLK–CYC (Menet et al. 2009). Analogous findings were recently shown for a ChIP–chip analysis of BMAL1 binding in mouse livers (Rey et al. 2011). There are ∼2000 binding sites for BMAL1 in the liver, and some 60% show rhythmic binding with a phase distributed around ZT4–ZT8, consistent with prior work measuring cycles in the mRNA levels of clock genes directly targeted by BMAL1. As might be expected based on prior microarray studies, in both Drosophila and mice, clock genes were among the strongest bound and rhythmic. This suggests that the relatively high-amplitude cycling observed in DNA microarrays for cycling clock transcripts compared with other ccgs is at least partly due to enhanced binding by CLOCK/CLK–BMAL1/CYC. Moreover, Abruzzi et al. (2011) show a strong correlation between robust mRNA cycling and daily rhythms in Pol II binding, a surrogate marker for active transcription. The molecular underpinnings that define the relationship between CLK–CYC binding and Pol II activity will be of interest to determine. It should be noted that promoter-specific differences in responses to PER-mediated repression could also modulate amplitudes in gene expression rhythms.
Abruzzi et al. (2011) also show some unexpected specificity in alternative transcription start sites, leading to rhythmic expression of some isoforms but not others. By comparing CLK ChIP–chip results from head extracts between wild-type flies and an eyeless strain, they identified genes that are likely targeted by CLK in a tissue-specific manner. More refined analysis on isolated circadian clock neurons is anticipated (Nagoshi et al. 2010). For example, some of the clock-relevant kinases, which were thought to be constitutively expressed based on analyzing whole heads, undergo robust expression rhythms in specific pacemaker neurons (Kula-Eversole et al. 2010).
As with DNA microarrays, the most overly represented class of cycling CLK targets are transcription factors—∼10% of the total. Likewise, in the case of BMAL1 DNA binding in mouse livers, transcriptional regulators were the most enriched functional cluster (Rey et al. 2011). Clearly, the findings that CLK and BMAL1 have many more direct targets than previously thought do not negate the possibility that many cycling genes are driven by a multitude of downstream rhythmic transcription factors. Indeed, the results suggest that master circadian transcription factors are workhorses, directly engaged in driving high-amplitude rhythms in clock transcriptional regulators—generally lower-amplitude rhythms of a large set of downstream transcription factors—in addition to controlling the rhythmic fates of many hundreds of individual genes with a wide variety of functions (Fig. 1). How all this bodes for tissue-specific expression of global gene expression is not clear, but these findings almost certainly demand that master clock transcription factors are quite amenable to being guided by tissue-specific factors. Also, CLK–CYC likely directly dominates only the spectrum of rhythmic transcripts peaking in the early night. Are other phases dominated by a few transcription factors and cis-acting elements or many?
Because so many CLK targets show low-amplitude (1.5-fold to twofold) binding rhythms yet are well expressed, Abruzzi et al. (2011) suggest that for many genes, master clock transcription factors do not operate as a daily “on–off” switch, but rather, in addition to other ongoing transcription programs, provide a “boost” at the right time of day. Nonetheless, the production of cyclical mRNA by gene expression rhythms requires that the transcript is not long-lived. Clearly, the dynamics governing mRNA rhythms (amplitude and phase) can also be influenced by post-transcriptional mechanisms, such as regulated degradation. In the end, however, we imagine that for oscillations in mRNA abundance to have functional meaning, they must be represented at the protein level. An important challenge is to determine how many of the daily rhythms in transcript information manifest themselves as oscillations with physiological consequences at the level of protein abundance. It is possible that interactions between multiple low-amplitude cycling proteins within a common pathway, especially some that function in a rate-limiting capacity, could amplify the signal. The future promises additional exciting and unanticipated answers to how cellular clocks that drive global gene expression lead to circadian rhythms in a wide variety of physiological and behavioral rhythms.
Acknowledgments
I am supported by NIH grants NS42088 and NS34958. Many of the key studies that contributed to the understanding of clock mechanisms and global rhythmic gene expression could not be cited here due to space limitations.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.180984.111.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M 2011. Drosophila CLOCK target gene characterization: Implications for circadian tissue-specific gene expression. Genes Dev (this issue). doi: 10.1101/gad.178079.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M 2001. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24: 1091–1119 [DOI] [PubMed] [Google Scholar]
- Bae K, Edery I 2006. Regulating a circadian clock's period, phase and amplitude by phosphorylation: Insights from Drosophila. J Biochem 140: 609–617 [DOI] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I 2000. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK–CYC transcription factor and the PER–TIM complex. J Neurosci 20: 1746–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC 2011. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. doi: 10.1111./j.1574-6976.2011.00288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341 [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM 2007. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci 10: 543–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay F, Laudet V 2002. Circadian clock and microarrays: Mammalian genome gets rhythm. Trends Genet 18: 595–597 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Kay SA 2010. Circadian control of global gene expression patterns. Annu Rev Genet 44: 419–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield GE 2003. DNA microarray analyses of circadian timing: The genomic basis of biological time. J Neuroendocrinol 15: 991–1002 [DOI] [PubMed] [Google Scholar]
- Dunlap JC 1999. Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ 2011. A molecular mechanism for circadian clock negative feedback. Science 332: 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter PD, Ramaswami M 2002. The ups and downs of daily life: Profiling circadian gene expression in Drosophila. Bioessays 24: 494–498 [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148 [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE 1999. Interlocked feedback loops within the Drosophila circadian oscillator. Science 286: 766–768 [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE 2003. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37: 249–261 [DOI] [PubMed] [Google Scholar]
- Hardin PE 2011. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet 74: 141–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21: 1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M 2008. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol 6: e119 doi: 10.1371/journal.pbio.0060119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan KP, Pradhan S, Wang JP, Allada R 2007. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol 3: e208 doi: 10.1371/journal.pcbi.0030208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27: 5014–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci 107: 13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023 doi: 10.1371/journal.pgen.1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107: 567–578 [DOI] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M 2009. Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev 24: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M 2010. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13: 60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner AC, Gin E, Malzahn E, Hofer T, Brunner M 2011. Circadian conformational change of the neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell 43: 713–722 [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595 doi: 10/1371/journal.pbio.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Kay SA, Hogenesch JB 2003. DNA arrays: Applications and implications for circadian biology. J Biol Rhythms 18: 96–105 [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83 [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL 2008. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet 9: 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37: 187–192 [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Kasukawa T, Ueda HR 2008. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol 10: 1154–1163 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Young MW 2005. Molecular and statistical tools for circadian transcript profiling. Methods Enzymol 393: 341–365 [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA 2001. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA 2010. Clocks not winding down: Unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776 [DOI] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R 2003. Drosophila clock can generate ectopic circadian clocks. Cell 113: 755–766 [DOI] [PubMed] [Google Scholar]