Precise ciliary organization is vital in human health, as disruptions can lead to diseases called ciliopathies. Here, Jackson and colleagues investigate how proteins are selectively transported to and organized in the primary cilium. They identify the ARL3 GTPase effector UNC119 as a myristoyl-binding protein that binds the myristoylated ciliopathy protein NPHP3. The work uncovers a selective, membrane targeting GTPase cycle that delivers myristoylated proteins to the ciliary membrane.
Keywords: ARL3, NPHP, UNC119, myristoylation
Abstract
The membrane of the primary cilium is a highly specialized compartment that organizes proteins to achieve spatially ordered signaling. Disrupting ciliary organization leads to diseases called ciliopathies, with phenotypes ranging from retinal degeneration and cystic kidneys to neural tube defects. How proteins are selectively transported to and organized in the primary cilium remains unclear. Using a proteomic approach, we identified the ARL3 effector UNC119 as a binding partner of the myristoylated ciliopathy protein nephrocystin-3 (NPHP3). We mapped UNC119 binding to the N-terminal 200 residues of NPHP3 and found the interaction requires myristoylation. Creating directed mutants predicted from a structural model of the UNC119–myristate complex, we identified highly conserved phenylalanines within a hydrophobic β sandwich to be essential for myristate binding. Furthermore, we found that binding of ARL3-GTP serves to release myristoylated cargo from UNC119. Finally, we showed that ARL3, UNC119b (but not UNC119a), and the ARL3 GAP Retinitis Pigmentosa 2 (RP2) are required for NPHP3 ciliary targeting and that targeting requires UNC119b myristoyl-binding activity. Our results uncover a selective, membrane targeting GTPase cycle that delivers myristoylated proteins to the ciliary membrane and suggest that other myristoylated proteins may be similarly targeted to specialized membrane domains.
Ciliopathies are rare, autosomal recessive disorders with phenotypes including cystic kidneys, retinal degeneration, neural tube defects, polydactyly, and obesity (for review, see Hildebrandt et al. 2011). The renal ciliopathy nephronophthisis (NPHP) and associated retinal–renal disorder Senior-Loken syndrome (SLS) are caused by mutation of any of 13 currently known disease genes (Wolf and Hildebrandt 2011). The proteins encoded by these genes localize to the microtubule-based primary cilium or the basal body/centrosome at the base of the cilium, leading to the hypothesis that defects in ciliary function underlie the pathology of NPHP/SLS.
The NPHP3 gene is mutated in ∼1% of cases of NPHP (Olbrich et al. 2003). It encodes a protein with a conserved position 2 glycine, which we predict here to be N-terminally myristoylated, and has also been shown to localize to a distinct region of the primary cilium (Shiba et al. 2010). Loss-of-function mutation of Nphp3 results in embryonic lethality, and a hypomorphic mutation of Nphp3 is the causative mutation in the pcy mouse, a classic cystic kidney disease model (Olbrich et al. 2003). Directed protein–protein interaction studies have found that NPHP3 associates with other NPHP proteins, notably NPHP1 (Olbrich et al. 2003) and Inversin/NPHP2 (Bergmann et al. 2008). NPHP3 has been suggested to play a role in both canonical and noncanonical Wnt signaling, although the molecular details of these functions are not understood (Bergmann et al. 2008; Zhou et al. 2010).
The highly conserved small GTPase Arf-like 3 (ARL3) was identified in several screens to identify ciliary genes using different methods (Avidor-Reiss et al. 2004; Efimenko et al. 2005; Pazour et al. 2005). ARL3 localizes to the primary cilium (Zhou et al. 2006), and knockout mice show renal and retinal involvement, a striking resemblance to Senior-Loken syndrome (Schrick et al. 2006). While the guanine nucleotide exchange factor (GEF) for ARL3 is unknown, the protein encoded by the retinitis pigmentosa disease gene Retinitis Pigmentosa 2 (RP2) has been shown to function as an ARL3-specific GTPase-activating protein (GAP) (Veltel et al. 2008). Interestingly, RP2 has been shown to localize to the primary cilium (Hurd et al. 2011) and is implicated in trafficking proteins from the Golgi to the ciliary membrane (Evans et al. 2010). ARL3 has three known GTP-specific interacting proteins (effectors): BART, PDE6D, and UNC119 (Linari et al. 1999; Sharer and Kahn 1999; Van Valkenburgh et al. 2001). Interestingly, UNC119, initially identified in Caenorhabditis elegans as an uncoordinated mutant showing broad neural deficiencies (Maduro and Pilgrim 1995), was identified in several ciliary screens as well, suggesting that ARL3 and UNC119 may function together in some aspect of ciliary biology (Avidor-Reiss et al. 2004; Li et al. 2004; Liu et al. 2007). UNC119 shares extensive sequence homology with PDE6D, which binds to and helps localize prenylated proteins in photoreceptors (Zhang et al. 2004, 2007), and a very recent study demonstrates that UNC119 shares many of the same biochemical properties, including binding to and extracting lipid-modified proteins from membranes (Zhang et al. 2011). Vertebrate genomes contain two UNC119 genes, UNC119a and UNC119b, although the functional differences between the two genes have not been explored. The encoded proteins share a highly similar PDE6D-like domain, while the N-terminal domains have diverged considerably. UNC119a mutant mice show retinal degeneration (Ishiba et al. 2007), and a heterozygous truncation of UNC119a has been found in a patient with a cone–rod dystrophy (Kobayashi et al. 2000).
N-terminal myristoylation of proteins is thought to confer reversible membrane binding activity to the modified protein (Resh 1999). How myristoylated proteins are targeted to the appropriate cellular locations remains unclear. Although the existence of a specific myristoyl-binding “receptor” has been proposed, such a protein has not been identified. Here we report that UNC119 is a myristoyl-binding protein that binds to a subset of myristoylated proteins, including NPHP3 and cystin, and that the small GTPase ARL3 regulates myristoyl–cargo binding. By using a structural model of the UNC119–myristoyl–peptide complex to guide mutagenesis, we identified key residues important for myristate binding. Cell-based NPHP3 ciliary localization assays indicate that ARL3, UNC119b and its myristoyl-binding activity (but not UNC119a), and the ARL3 GAP RP2 are required for proper ciliary targeting of NPHP3. We also showed that the myristoyl-binding activity of UNC119 is required in vivo as well. Furthermore, we identified C5orf30 as an UNC119-binding protein that preferentially binds unliganded UNC119. Finally, morpholino (MO)-mediated knockdown experiments in zebrafish are consistent with a role for the pathway in ciliary function. Based on our results, we propose that ARL3–UNC119b–RP2 constitute a GTPase cycle that targets a specific set of myristoylated proteins to the ciliary membrane.
Results
UNC119 binds the N terminus of ciliopathy protein NPHP3 in a myristoyl-dependent manner
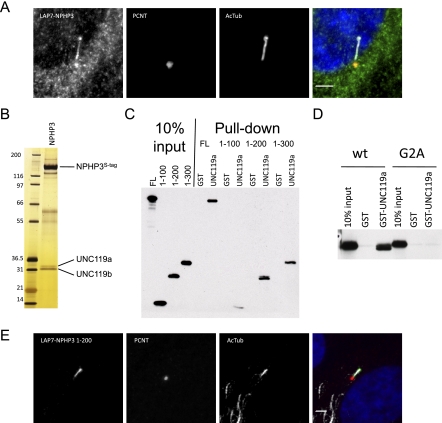
We recently generated a protein–protein interaction network for the NPHP–JBTS–MKS disease spectrum, which included tandem affinity purification of an N-terminally tagged NPHP3 (Sang et al. 2011). Sequence analysis predicts that NPHP3 is N-terminally myristoylated, which would be blocked by our N-terminal fusion. We sought to determine whether a C-terminally tagged, myristoylated NPHP3 would localize differently. Toward this end, we generated a stable retinal pigmented epithelial cell line (RPE) expressing C-terminally LAP-tagged murine NPHP3. In contrast to our N-terminally tagged construct (data not shown), the C-terminally tagged NPHP3 was able to localize to the primary cilium (Fig. 1A), suggesting, as seen for other ciliary proteins, including cystin (Tao et al. 2009), that myristoylation aids in proper NPHP3 targeting. Several attempts to generate a nonmyristoylated, G2A mutant NPHP3 stable cell line suitable for tandem affinity purification were unsuccessful, likely due to poor stability of the mutant protein. An intriguing possibility is that the myristoylation-dependent targeting may be important for accumulating NPHP3 in cilia.
Figure 1.
UNC119 proteins bind an N-terminal domain of NPHP3 in a myristoyl-dependent manner. (A) A stably expressed GFP (LAP7) C-terminally tagged full-length mouse NPHP3 in RPE cells was serum-starved, fixed, and immunostained for pericentrin (PCNT) and acetylated tubulin (AcTub). Bar, 3 μm. (B) NPHP3-associated proteins were isolated by tandem affinity purification of LAP-tagged NPHP3 from the LAP7-NPHP3 stable RPE cell line and visualized on silver-stained SDS–polyacrylamide gels. Labeled proteins were identified by mass spectrometry. (C) Full-length (FL) and N-terminal fragments of NPHP3 comprising amino acids 1–100, 1–200, and 1–300 with a C-terminal Myc tag were in vitro translated; incubated with recombinant, purified GST-UNC119a; purified on glutathione beads; resolved by SDS-PAGE; and visualized by immunoblotting for the C-terminal tag. (D) In vitro translated wild-type (wt) or unmyristoylated (G2A) NPHP3 1–200 were incubated with GST-UNC119a protein and assayed for binding as in C. (E) LAP7-NPHP3 1–200 was transfected into RPE cells and processed for imaging as described in A.
To identify the factors that target NPHP3 to cilia, we used tandem affinity purification of our C-terminally tagged NPHP3. Purification of LAP-NPHP3 yielded several strong interacting proteins (Fig. 1B). Excision and tandem mass spectrometry identified the 33-kDa and 31-kDa bands as UNC119a and UNC119b, respectively. We next confirmed the NPHP3:UNC119 interaction by GST pull-down using bacterially expressed GST-UNC119a and myc-tagged NPHP3 in vitro translated in wheat germ extract, a system known to be competent for myristoylation (Fig. 1C; Heuckeroth et al. 1988). Using this assay, we subsequently localized the UNC119-binding domain of NPHP3 to the N-terminal 200 residues (Fig. 1C) and found that when expressed in RPE cells, this domain is sufficient for ciliary localization (Fig. 1E). Attempts to determine the localization of the G2A mutant of the N-terminal NPHP3 fragment were unsuccessful due to poor stability, similar to results obtained with the full-length mutant construct. We next asked whether the interaction required myristoylation. Remarkably, mutation of the myristoylated position 2 glycine of NPHP3 to alanine abrogated binding (Fig. 1D). These data suggested that UNC119 functions as a myristoyl-binding protein.
UNC119 is a myristoyl-binding protein
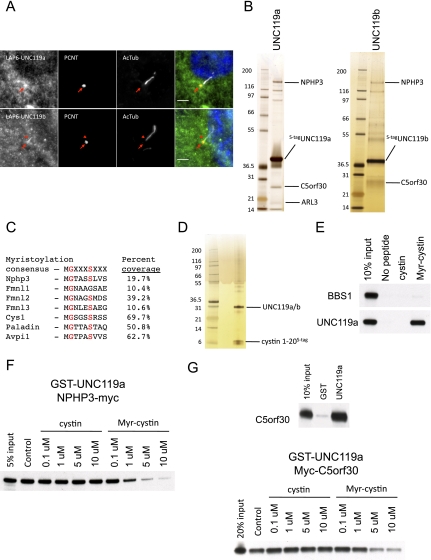
To further investigate whether UNC119 was most centrally a myristoyl-binding protein and to determine other functions of UNC119, we wanted to purify the two UNC119 isoforms and identify their interacting proteins. To identify these proteins, we generated stable RPE cell lines expressing LAP-tagged UNC119a and UNC119b, and also a murine inner medullary-collecting duct cell line (IMCD3) expressing LAP-tagged Unc119a (data not shown). Interestingly, the LAP-UNC119a localized to the centrosome, as determined by colocalization with pericentrin (Fig. 2A). More importantly, the LAP-UNC119b was enriched at the transition zone and extended into the proximal end of the cilium (Fig. 2A). Silver staining of the tandem affinity-purified UNC119a and UNC119b complexes shows an overlapping set of interactors as well as unique interactors (Fig. 2B). Strikingly, one of the most abundant proteins present in each complex is NPHP3, reciprocally validating the NPHP3–UNC119a/b interactions. An affinity purification–mass spectrometry approach was taken for the IMCD3 LAP-Unc119a cell line in an effort to identify less abundant interacting proteins. In addition to Nphp3, the tandem affinity-purified Unc119a complex contained a strong enrichment of proteins (10 in total) either known or predicted to be N-terminally myristoylated (Fig. 2C). These included the known ciliary protein cystin, which is mutated in the mouse cystic kidney model cpk (Hou et al. 2002). Cystin also requires myristoylation for proper ciliary membrane localization (Tao et al. 2009). To validate the interaction with cystin in cells, we generated a stable RPE cell line expressing the first 20 amino acids of cystin tagged on its C terminus, which is sufficient for targeting cystin to the cilium (Supplemental Fig. S1). Tandem affinity purification of this cystin polypeptide yielded virtually stoichiometric amounts of UNC119a and UNC119b (Fig. 2D). To confirm this was a direct interaction in vitro and to characterize the interaction, we synthesized C-terminally biotinylated peptides corresponding to cystin residues 2–20 either unmyristoylated (cystin) or myristoylated (Myr-cystin) on the N terminus to test for direct interaction. As shown in Figure 2E, the myristoylated cystin peptide efficiently coprecipitated in vitro translated UNC119a, whereas the nonmyristoylated peptide failed to interact with UNC119a. We next asked whether the NPHP3:UNC119a interaction could be competed by addition of the Myr-cystin peptide. Addition of increasing amounts of the Myr-cystin peptide, but not the unmyristoylated peptide, to a GST pull-down reaction containing GST-UNC119a and in vitro translated NPHP3 disrupted the NPHP3:UNC119 interaction (Fig. 2F). These data strongly support the idea that UNC119 may function as a highly selective cargo adaptor for myristoylated proteins.
Figure 2.
UNC119 is a myristoyl-binding protein. (A) Stable LAP6-UNC119a and LAP6-UNC119b RPE cells were serum-starved, fixed, and immunostained for pericentrin (PCNT) and acetylated tubulin (AcTub). The arrow indicates the basal body and the arrowhead indicates the cilium. Bar, 3 μm. (B) LAP-tagged UNC119a and UNC119b were tandem affinity-purified from the stable cell lines used in A. The eluates were resolved by SDS-PAGE and visualized by silver staining. Labeled proteins were identified by mass spectrometry. (C) Table showing the top seven proteins predicted to be myristoylated that were identified by tandem affinity purification and mass spectrometry of the stable IMCD3 LAP-Unc119a line and their percent coverage. (D) LAP-tagged cystin amino acids 1–20 were tandem affinity-purified from RPE cells stably expressing LAP5-cystin 1–20. The eluate was resolved by SDS-PAGE and visualized by silver staining. UNC119a and UNC119b were identified by mass spectrometry. (E) Myc-tagged in vitro translated BBS1 or UNC119a were incubated with a C-terminally biotinylated peptide corresponding to cystin residues 2–20, either unmyristoylated (cystin) or myristoylated on its N terminus (Myr-cystin). Complexes were precipitated with streptavidin beads, resolved by SDS-PAGE, and analyzed by immunoblotting with a myc antibody. BBS1 was included as a negative control. (F) GST-UNC119a-binding reactions were performed with C-terminally Myc-tagged full-length NPHP3 as in Figure 1C except that either cystin or Myr-cystin peptide was included at 0.1, 1, 5, or 10 μM. DMSO was added as a control. (G, top panel) GST-UNC119a-binding reactions were performed as in Figure 1C with N-terminally Myc-tagged in vitro translated C5orf30. (Bottom panel) Peptide competition assay with GST-UNC119a and in vitro translated C5orf30 was performed as in the right panel of Figure 1F.
C5orf30 is an UNC119-binding protein that preferentially binds unliganded UNC119
In addition to the multiple myristoylated proteins that copurified with UNC119, we found an uncharacterized protein called C5orf30 that lacks a position 2 glycine and would not be predicted to be myristoylated (Fig. 2B). Unlike the ARL3 and UNC119 genes, C5orf30 appears to be only found in vertebrate genomes. When GFP-tagged, C5orf30 localizes to the transition zone and proximal cilium in addition to being found throughout the cytoplasm (Supplemental Fig. S2A), consistent with a possible role in directing ciliary trafficking. In situ hybridization of mouse retinal sections show that C5orf30 is highly expressed in photoreceptors (Supplemental Fig. S2B), suggesting a role for C5orf30 in trafficking of proteins into the photoreceptor. We confirmed that C5orf30 is a direct UNC119-binding protein by GST pull-down assay with in vitro translated C5orf30 and GST-UNC119a (Fig. 2G). We considered several models for how C5orf30 might regulate UNC119 binding to myristoylated proteins. We first determined whether C5orf30 binding to UNC119 was disrupted by the Myr-cystin peptide. Indeed, the UNC119a:C5orf30 interaction was disrupted by addition of the Myr-cystin peptide, but not the nonmyristoylated peptide (Fig. 2G). We wanted to consider the question of whether C5orf30 might reciprocally block myristoylated protein binding. We have yet to produce a form of the C5orf30 protein that would allow a simple competition assay. However, we did compare the relative efficiency of the binding among C5orf30, UNC119, and NPHP3 in our LAP purifications of each protein (Figs. 1B, 2B; data not shown). Consistent with our predictions, NPHP3 bound efficiently to UNC119, but we did not detect C5orf30 by silver staining. Reciprocally, purified C5orf30 binds UNC119, but we could not detect NPHP3. LAP-purified UNC119 bound to both C5orf30 and NPHP3 (Fig. 2A), which we believe to be a mixed population of C5orf30–UNC119 and NPHP3–UNC119 complexes. These data suggest that C5orf30 binds preferentially to unliganded UNC119. An intriguing possibility is that C5orf30 may act as a recycling factor to help extract UNC119 from the membrane after myristoylated cargo release.
Phenylalanine residues lining the hydrophobic β clamshell of UNC119 mediate myristate binding
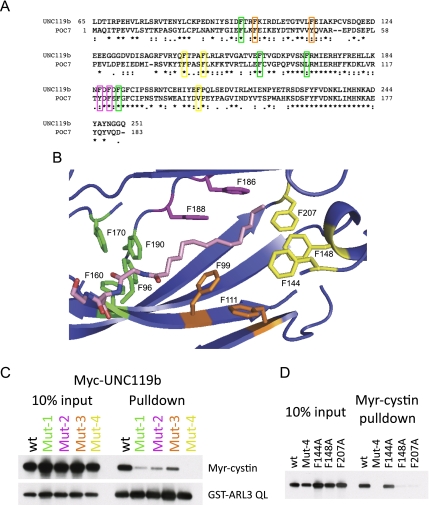
The crystal structure of UNC119a was solved by the Northeast Structural Genomics Consortium (Protein Data Bank [PDB] 3GQQ), revealing a hydrophobic β sandwich fold, similar to other lipid-binding proteins such as PDE6D and RhoGDI. The length of the hydrophobic core is ∼20 Å, which forms a tunnel that would readily accommodate the 14-carbon myristate moiety. Most striking are the 11 highly conserved phenylalanine residues lining the hydrophobic β sandwich (Fig. 3A). We used structural modeling to place a myristate into the hydrophobic core (Fig. 3B), based on positioning with the tunnel domain circumscribed by these phenylalanines. Our model is remarkably similar to, and in full agreement with, the recently published cocrystal structure of UNC119a and a lauroylated peptide (Zhang et al. 2011). Based on our model, we hypothesized that the phenylalanine residues lining the hydrophobic “tunnel” would be important for myristate binding, and that these phenylalanines cluster into four hydrophobic binding sites. To test our hypothesis, we generated four UNC119b mutant constructs, changing two to four phenylalanines to alanines in each of the sites. These constructs were in vitro translated and tested for myristoyl-binding activity using the biotinylated Myr-cystin peptide. As shown in Figure 3C, all four mutants displayed reduced binding to the Myr-cystin peptide, with the Mut-4 mutant (F144A, F148A, and F207A) having the greatest effect. Importantly, each of the phenylalanine mutants were able to bind to ARL3 QL as efficiently as wild-type UNC119b, supporting that the overall architecture of UNC119b is not disrupted by our mutagenesis (Fig. 3C). Next, we mutated each of the phenylalanines in the Mut-4 site individually. We found that while F144 contributes little to Myr-cystin binding, F148 and F207 are critical for binding to the Myr-cystin peptide (Fig. 3D), validating our structural model.
Figure 3.
Mutations in the highly conserved phenyl groove of UNC119 disrupt myristoyl–peptide binding. (A) Sequence alignment of human UNC119b and the Chlamydomonas homolog POC7. Phenylalanine residues mutated in C are indicated and color-coded. (B) Structural model of UNC119a and the myristoylated N-terminal three amino acids of cystin based on the UNC119a crystal structure 3GQQ. The four groups of conserved phenylalanines mutagenized in C are shown and color-coded. Residue numbers correspond to the UNC119b sequence. Portions of the structure were removed to allow visualization of the myristoyl-binding pocket. (C, top immunoblot) Biotinylated Myr-cystin-binding assays were performed as in Figure 2E only using in vitro translated wild-type (wt) or mutant Myc-tagged UNC119b. The bottom immunoblot shows the results of a binding reaction between GST-ARL3 QL and in vitro translated Myc-tagged UNC119b. (D) Biotinylated Myr-cystin-binding assays were performed as in Figure 2E only using in vitro translated wild-type (wt) or mutant Myc-tagged UNC119b.
Ciliary small GTPase ARL3 binds UNC119 and membranes in a GTP-specific fashion in RPE cells
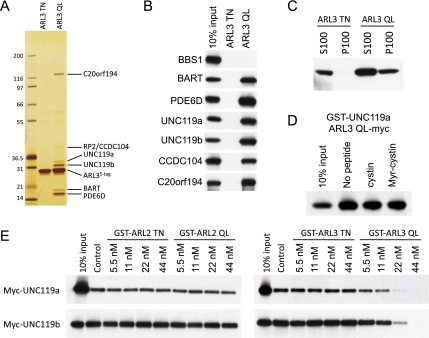
UNC119 is a known GTP-specific effector of ARL2 and ARL3 (Van Valkenburgh et al. 2001). We focused our attention on ARL3 because it localizes to the primary cilium in RPE cells (Zhou et al. 2006), while ARL2 does not (data not shown). We generated stable RPE cell lines expressing LAP-tagged GDP-mimicking ARL3 TN and the GTP-mimicking ARL3 QL mutant proteins. In contrast to other ciliary small GTPases, such as Rab8, both the GDP- and GTP-mimicking mutant forms of ARL3 localized to the primary cilium (Supplemental Fig. S3), suggesting that the ARL3 GTPase cycle may function to release proteins within the cilium, rather than regulating ciliary entry. Tandem affinity purification of LAP-ARL3 TN yielded no strong interacting proteins, while the LAP-ARL3 QL copurified with six additional protein bands (Fig. 4A). Mass spectrometry was used to identify each of the proteins. We found all of the known ARL3 effectors (BART, PDE6D, and UNC119) as well as two novel effectors of unknown function (CCDC104 and C20orf194) and also the ARL3 GAP RP2 (Fig. 4A). The ARL3 effectors were assayed by GST pull-down using either GST-ARL3 TN or GST-ARL3 QL along with the in vitro translated effector (Fig. 4B), and, indeed, each of these candidate effectors bound in a GTP-dependent manner. These novel effectors will be not be discussed further and will be characterized in future work.
Figure 4.
ARL3 binding, but not ARL2 binding, releases myristoylated cargo from UNC119. (A) Silver-stained gel showing the eluates from tandem affinity purifications of LAP-tagged ARL3 TN and ARL3 QL. Indicated proteins were identified by mass spectrometry. (B) Binding reactions were performed using GST-ARL3 TN or GST-ARL3 QL along with the in vitro translated Myc-tagged proteins indicated on the left side of the panel. The Myc-tagged interacting proteins were detected by immunoblotting with a myc antibody. (C) Stable RPE cell lines expressing LAP-ARL3 TN or LAP-ARL3 QL were fractionated into a soluble (S100) and insoluble (P100) membrane fractions. These fractions were resolved by SDS-PAGE and immunoblotted for the GFP tag. (D) Peptide competition assays were performed as in Figure 2G with ARL3 QL-myc in vitro translated protein and 5 μM Myr-cystin peptide. (E) Preformed complexes of biotinylated Myr-cystin and either in vitro translated Myc-tagged UNC119a or UNC119b captured on streptavidin-sepharose beads were incubated with increasing amounts of either GST-ARL2 TN, GST-ARL2 QL, GST-ARL3 TN, or GST-ARL3 QL. The complexes were washed and analyzed by immunoblotting with a myc antibody.
ARF family and ARF-like GTPases are known to bind membranes GTP-specifically (Gillingham and Munro 2007; Jin et al. 2010). We tested whether ARL3 is able to similarly bind membranes by fractionating the LAP-ARL3 TN and LAP-ARL3 QL stable RPE cell lines into soluble (S100) and insoluble (P100) membrane fractions and immunoblotting for LAP-ARL3. As shown in Figure 4C, ARL3 QL pelleted in the membrane fraction, while ARL3 TN did not, indicating that ARL3 indeed binds membranes GTP-specifically.
ARL3 QL releases myristoylated peptides from UNC119
We next considered whether the ARL3 interaction would regulate UNC119 interactions with myristoylated proteins. First, we tested whether the cystin N-terminal peptide would disrupt the known interaction between UNC119 and the GTP-bound small GTPase ARL3. As shown in Figure 4D, addition of the cystin peptide to a binding reaction containing GST-UNC119a and in vitro translated ARL3 QL had no effect. This result confirms the specificity of the competitive binding seen between NPHP3 and the myristoylated cystin peptide. Moreover, like many ARF/ARL family proteins, ARL3 contains a position 2 glycine and would itself be predicted to be myristoylated. Myristoylation of these factors would help direct the ARF/ARL GTPase to the membrane. Here, it appears that ARL3 binds UNC119 only via its GTP effector function and not via a myristoylation signal.
We next tested whether ARL3 QL would release myristoylated cargo from UNC119. We preformed complexes of in vitro translated UNC119a or UNC119b and the biotinylated Myr-cystin peptide on streptavidin-sepharose beads and then added recombinant GST-ARL3. Remarkably, addition of increasing amounts of GST-ARL3 QL, but not the addition of GST-ARL3 TN, leads to a dose-dependent release of UNC119 from the immobilized Myr-cystin peptide (Fig. 4E). Interestingly, we found that despite binding to UNC119 similarly to ARL3 QL, ARL2 QL was unable to release UNC119 from the Myr-cystin peptide (Fig. 4E). These data suggest that ARL3 QL facilitates release of myristoylated cargo from UNC119 into the appropriate membrane domain. Given that PDE6D shares sequence similarity with UNC119 and also binds to ARL2/ARL3 and lipid-modified proteins, it is likely that ARL2 or ARL3 will similarly regulate PDE6D cargo binding.
The ARL3 pathway is required for NPHP3 ciliary localization
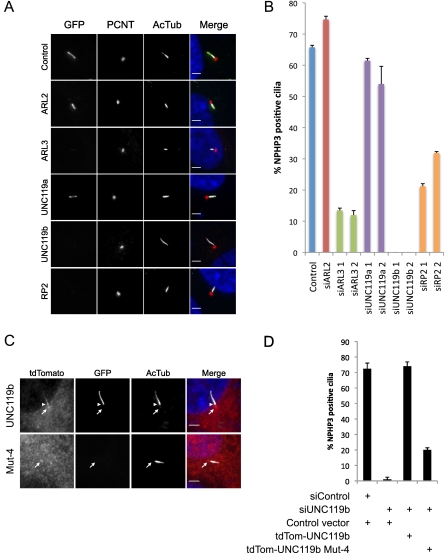
Having established a biochemical relationship between ARL3, UNC119, and myristoylated ciliary proteins (NPHP3 and cystin), we sought to test the model that ARL3/UNC119 are responsible for targeting myristoylated ciliary proteins to the primary cilium. To test this idea, we generated a stable RPE cell line expressing LAP-tagged NPHP3 1–200. We chose this N-terminal fragment of NPHP3 over the full length because it exhibited notably more efficient and consistent ciliary localization, making this variant more suitable for a ciliary targeting assay. We transfected these cells with either control scrambled siRNA or siRNAs targeting ARL2, ARL3, UNC119a, UNC119b, or RP2 (Fig. 5A,B). We used two distinct siRNAs for ARL3, UNC119a, UNC119b, and RP2 to minimize the chance of off-target effects. All siRNAs depleted the endogenous transcripts to <15% of control levels (Supplemental Fig. S4). As expected, depletion of ARL2 had no effect. Strikingly, knockdown of ARL3, UNC119b, and RP2 each dramatically reduced the percentage of cilia with localized NPHP3 1–200. We observed very low amounts of our LAP-NPHP3 1–200 protein in knockdown cilia when imaged at longer exposures (<10-fold wild-type levels), likely reflecting a combination of the level of overexpression of our reporter construct and possibly incomplete knockdown of the targeted protein. Interestingly, depletion of UNC119a had no effect even though it is able to bind NPHP3. We suspect that this is due to the difference in localization between UNC119a and UNC119b in RPE cells, but may report more broadly on functional differences between UNC119a and UNC119b (see the Discussion). Depletion of ARL3 and UNC119b in a stable cell line expressing GFP-tagged cystin 1–20 yielded similar results (data not shown). Further, knockdown of ARL3, UNC119a, UNC119b, or RP2 showed no effect on the trafficking of ciliary G-protein-coupled receptors (Supplemental Fig. S5), indicating that delivery of nonmyristoylated proteins to the cilium is not disrupted by the depletion of ARL3 pathway components.
Figure 5.
ARL3, UNC119b, and RP2 are required for ciliary localization of NPHP3. (A) RPE cells stably expressing LAP7-NPHP3 1–200 transfected with siRNAs against the indicated genes and immunostained with pericentrin (PCNT) and acetylated tubulin (AcTub) antibodies. Bar, 3 μm. (B) Quantification of the percentage of cilia containing LAP7-NPHP3 1–200 after transfection with the indicated siRNAs. Experiments were performed in duplicate and are presented as average plus standard deviation. (C) RPE cells stably expressing LAP7-NPHP3 1–200 were transfected with tdTomato-tagged wild-type or Mut-4 UNC119b, then transfected with siRNAs targeting the UNC119b 3′ UTR. The cells were then immunostained for pericentrin and acetylated tubulin and imaged. The arrow indicates the basal body, and the arrowhead indicates the cilium. Bar, 3 μm. (D) Quantification of the percentage of cilia containing LAP7-NPHP3 1–200 after transfection with the indicated plasmids and siRNAs. Experiments were performed in duplicate and are presented as average plus standard deviation.
Myristoyl-binding activity of UNC119b is required to localize NPHP3 to the primary cilium
Having established that the N-terminal fragment of NPHP3 requires UNC119b for proper ciliary localization, we wondered whether this localization would require UNC119b myristoyl-binding activity. To test this hypothesis, we performed a series of siRNA/rescue experiments in our stable LAP-NPHP3 1–200 cell line using tdTomato-tagged UNC119b constructs and a siRNA targeting the UNC119b 3′ untranslated region (UTR) (Fig. 5C,D). As expected, the wild-type tdTomato-UNC119b construct was able to completely rescue localization of NPHP3 to the cilium in UNC119b-depleted cells. Strikingly, the myristoyl-binding mutant tdTomato-UNC119b Mut-4 failed to rescue to the same extent, indicating that NPHP3 ciliary targeting indeed requires the myristoyl-binding activity of UNC119b. Interestingly, the myristoyl-binding mutant UNC119b was found only at the basal body/transition zone (Fig. 5C, open arrowhead), whereas the wild-type version was present in the proximal cilium (Fig. 5C, closed arrowhead) as well as the basal body/transition zone. An intriguing possibility is that when lacking its myristoylated protein cargo, UNC119b may only traffic to the basal body, and that cargo binding may trigger ciliary entry of UNC119b.
Myristoyl-binding activity of UNC119b is required in C. elegans
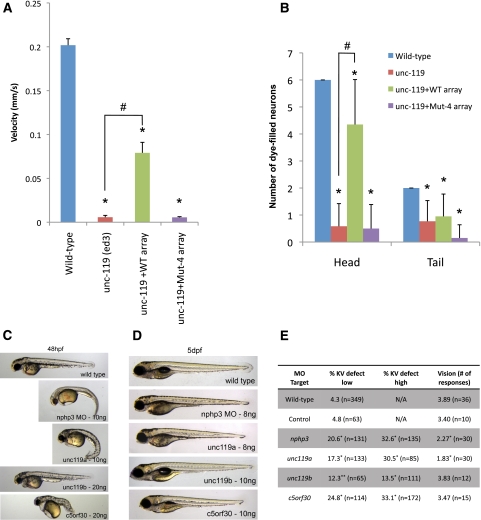
Unc-119 mutant worms exhibit defects in locomotion (Maduro and Pilgrim 1995), and sensory neurons in these mutants fail to fill with lipophilic dye (see below). We asked whether human UNC119b with mutations in the myristoyl-binding site (Mut-4) would be able to rescue C. elegans unc-119 phenotypes. Wild-type human UNC119b, but not Mut-4, was able to rescue both the locomotor and head sensory neuron dye-filling defects of unc-119 mutant animals, while Mut-4 appeared to have a dominant-negative activity in dye-filling sensory neurons in the tail (Fig. 6A,B). These data suggest that UNC119 family proteins require myristoyl-binding activity in vivo.
Figure 6.
Myristoyl-binding activity is required to rescue C. elegans unc-119 (ed) mutants, and knockdown of nphp3, unc119a, and c5orf30 in zebrafish results in cilia-related phenotypes. (A) Locomotor activity of wild-type, unc-119 (ed3) mutant, and unc-119 (ed3) mutant worms rescued with either wild-type or Mut-4 human UNC119b. An asterisk (*) and hash mark (#) denote values that are significantly different (at P < 0.05) from wild-type and unc-119(ed3) values, respectively. (B) The ability of head and tail sensory neurons of animals of the indicated genotypes to fill with DiI dye was analyzed and graphed. An asterisk (*) and hash mark (#) denote values that are significantly different (at P < 0.05) from wild-type and unc-119(ed3) values, respectively. (C) Representative images of MO-injected zebrafish embryos at 48 h post-fertilization (hpf). (D) Representative images of low-dose MO-injected zebrafish at 5 d post-fertilization (dpf) used in the vision assay. Note that a lower amount of MO was necessary, since embryos with a curved body cannot be used in the vision assay. (E) Table reporting results from KV analysis and vision assay. (*) P < 0.01; (**) P < 0.05, Fisher's exact test.
Ciliary function in zebrafish requires nphp3, unc119a, and c5orf30
Zebrafish have become a useful model organism used to study basic ciliary function. MO-mediated knockdown of genes involved in ciliary function typically result in curved body, defects in the size of the ciliated embryonic structure known as the Kupffer's vesicle (KV), and reduced visual function (Yen et al. 2006; Pretorius et al. 2010; Baye et al. 2011). Zebrafish embryos injected at the one- to four-cell stage with gene-specific MOs were assessed for overall morphology, KV size, and visual function. Previous work has noted ciliary roles for nphp3 and unc119a (most similar to human UNC119b) (Manning et al. 2004; Zhou et al. 2010). We found that knockdown of zebrafish nphp3, unc119a, and c5orf30 each resulted in a MO dose-dependent curved body axis and KV defects (Fig. 6C,E). Moreover, we found that nphp3 and unc119a knockdown results in reduced visual function (Fig. 6D,E). Consistent with the results from our cell-based NPHP3 targeting assay and the differences in localization seen between human UNC119a and UNC119b, knockdown of zebrafish unc119b (most similar to human UNC119a) yielded only a mild KV defect but no other obvious cilia-related phenotypes. Finally, in zebrafish, we identified three genes with high homology with ARL3: arl3 (95%), the syntenic gene to human ARL3, and two arl3-like factors, arl3l1 (75%) and arl3l2 (77%), that are not found in humans or mice. Knockdown of the syntenic arl3 using two independent MOs resulted in no obvious cilia-related defects, suggesting possible subfunctionalization of arl3 family members within the zebrafish (data not shown). Taken together, these data demonstrate that nphp3, unc119a, and c5orf30 have a strong role in ciliary function in the zebrafish.
Discussion
We describe here data showing that the highly conserved UNC119 proteins recognize the myristoylated N terminus of a subset of proteins, including the ciliopathy protein NPHP3 and cystin. Mutagenesis based on our structural model of the UNC119–myristate complex allowed us to identify four groups of conserved phenylalanine residues that are important for myristoyl–peptide binding, with two key phenylalanines being critical for binding. We then turned our attention to the ciliary small GTPase ARL3. By using a proteomic approach, we identified a comprehensive list of GTP-specific effector proteins, including the known ARL3 effectors BART, PDE6D, and UNC119, as well as two proteins of unknown function (CCDC104 and C20orf194). By fractionating our cell lines expressing GDP- and GTP-mimicking forms of ARL3, we found that, similar to other ARF and ARF-like family members, ARL3 bound to membranes GTP-specifically. Next, we found that ARL3 binding, but not ARL2 binding, released a myristoylated peptide from UNC119. Based on knockdown experiments using our cell-based assay monitoring ciliary localization of the myristoylated N-terminal UNC119-binding fragment of NPHP3, we suggest that the ARL3 GTPase pathway (ARL3-GTP, UNC119b, and GAP RP2) is required to target myristoylated ciliary proteins to the ciliary membrane.
Model of myristoylated ciliary protein trafficking
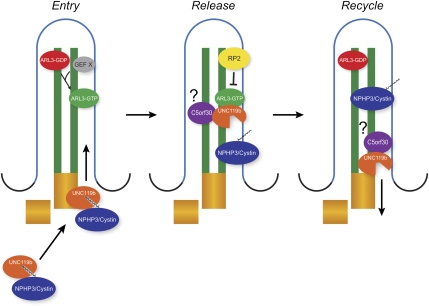
Based on the data presented, we propose a model (Fig. 7) in which UNC119b first binds a subset of myristoylated cargo proteins in the cytoplasm. Because myristoylation occurs cotranslationally (Wilcox et al. 1987), this binding may happen shortly after the initiation of translation to prevent mistargeted myristate–membrane interactions (see below). The UNC119b–myristoylated cargo complex would then be targeted to the cilium by a currently unknown mechanism. Also working at an early step in the process, the ciliary small GTPase ARL3 would be activated by a currently unknown GEF. This activation would potentially occur within the cilium because both the GTP- and GDP-locked forms are ciliary, although the site of this activity is not truly clear. Once in the cilium, membrane-bound ARL3-GTP binds to UNC119b, releasing the myristoylated cargo into the cilium/ciliary membrane. The ARL3 GAP RP2, which is known to reside within the ciliary membrane (Hurd et al. 2011), activates the ARL3 GTPase activity, thus releasing UNC119b (Veltel et al. 2008). Interestingly, we obtained multiple peptides identifying RP2 as present in our purification of NPHP3 (data not shown). It is tempting to speculate that the cargo itself may lend specificity to target membrane deposition by containing sequences that bind the GAP. Based on our competitive binding experiments with C5orf30 and the Myr-cystin peptide, we propose that C5orf30 binds preferentially to unliganded UNC119. Based on this activity, and our observation that C5orf30 can reside in the primary cilium (Supplemental Fig. S3A), C5orf30 may aid in either the release of cargo or recycling of UNC119b.
Figure 7.
Model of ARL3–UNC119b–RP2-dependent ciliary targeting of NPHP3/cystin. UNC119b and its ciliary myristoylated cargo are first trafficked to the cilium by a currently unknown mechanism. In the cilium, ARL3 is activated by an unknown GEF. Membrane-bound ARL3-GTP then binds to the UNC119b/myristoylated cargo complex and releases the cargo into the cilium. Next, the ciliary GAP RP2 activates ARL3 GTPase activity, which releases UNC119b from ARL3. UNC119b then exits the cilium for another round of trafficking.
As we were preparing our paper, Zhang et al. (2011) published a study reporting that UNC119 binds the myristoylated Gα protein Transducin and showed biochemically that UNC119 facilitates its extraction from rod outer segment membranes. While we considered a possible GDI-like “extraction” mechanism for UNC119, our results showing that NPHP3 fails to localize to the primary cilium when UNC119b is depleted, rather than accumulating in the ciliary membrane, suggest a membrane targeting function. It will be interesting to determine what factors dictate UNC119's role in membrane extraction versus delivery.
UNC119a versus UNC119b
The UNC119a and UNC119b proteins share a highly similar PDE6D-like domain, comprising more than three-quarters of each protein, which we showed here binds to the myristate moiety of a subset of N-terminally myristoylated proteins. Despite this high degree of similarity, we found that UNC119b, but not UNC119a, is required for proper ciliary NPHP3 localization. In contrast, the N-terminal ∼50 residues of the two orthologs have diverged considerably. Interestingly, we found that when expressed in RPE cells, UNC119a localized to the centrosome, while UNC119b localized to the transition zone and proximal cilium. The ciliary targeting sequence thus may lie within the divergent N terminus of UNC119b, although experiments to test this hypothesis will be necessary. We also noted differences in the banding pattern of our silver-stained gels comparing the purified UNC119a and UNC119b complexes. The divergent N-terminal domains may additionally contribute to the selectivity of myristoyl–peptide binding, accounting for the differences observed between the two purified complexes. Furthermore, in zebrafish, knockdown of unc119a lead to three cilia-related phenotypes (curved body, KV defect, and vision impairment), while unc119b depletion displayed only a mild KV defect, supporting differential roles for unc119a and unc119b in vivo. Additional studies addressing the binding partners, specificity, and mouse knockout phenotype of UNC119b would be important follow-on studies.
UNC119 specificity and functional orthologs
Approximately 0.5% of the human genome is predicted to be myristoylated (Maurer-Stroh et al. 2004), and >50 proteins in the flagellar and basal body (FABB) proteome, a list of candidate ciliary proteins (Li et al. 2004), possess position 2 glycines (our observation). A subset of these myristoylated proteins contains a cysteine at position 3, predicted to also be palmitoylated, including RP2. Based on our structural model, the β sandwich fold is unlikely to accommodate a second long hydrophobic lipid chain, and thus doubly acylated proteins are unlikely to be candidates for the ARL3–UNC119 targeting pathway. Notably, all of the predicted myristoylated proteins identified in our UNC119 purifications lacked a position 3 cysteine. Our purifications of UNC119a and UNC119b suggest that they recognize only a subset of the total myristoylated protein population, although future purifications from different cell types or tissues may expand the current repertoire. It is intriguing to consider whether there are functional equivalents of UNC119 for other myristoylated proteins.
UNC119 as a chaperone
In addition to acting as a ciliary targeting factor for a subset of myristoylated proteins, UNC119 may also act as a chaperone, burying the myristate until the cargo reaches its appropriate target membrane. Interestingly, other myristoylated proteins, such as HIV gag and c-abl, are able to bury their myristate moieties within an internal protein domain (Hantschel et al. 2003; Tang et al. 2004). In the “myristate switch” mechanism, the HIV gag myristate is exposed only when the protein reaches its destination at the apical membrane (Tang et al. 2004). It may be that proteins unable to bury their myristate will require UNC119 or UNC119-like proteins for proper membrane localization. Here, especially as these proteins are conserved to Chlamydomonas, the function of an independent myristate-binding protein would appear to be an ancient device for membrane targeting.
Strong evolutionary conservation of the ARL3 pathway
The ARL3 and UNC119 genes are conserved from humans to the green algae Chlamydomonas (called ARL3 and POC7, respectively). The Chlamydomonas genome also contains at least two RP2-like proteins (Stephan et al. 2007). While the ciliary cargos identified here (NPHP3 and cystin) are not obviously conserved, myristoylated proteins appear to be enriched in biochemical purifications of Chlamydomonas flagellar proteins (Pazour et al. 2005). This remarkable degree of conservation suggests an ancient origin of the myristoylation-based membrane targeting mechanism described here.
New candidates for renal and retinal degeneration (NPHP/SLS) genes
NPHP and SLS are autosomal recessive genetic disorders characterized by cystic kidneys and cystic kidneys plus retinal degeneration, respectively. Currently, there are 13 genes known to cause NPHP or SLS when mutated, with mutation of several of these genes leading to both diseases. These known disease alleles account for only ∼30% of diagnosed cases of NPHP, suggesting that many more disease genes remain to be identified. Based on the data presented here, we propose that UNC119b and C5orf30 are strong candidates for NPHP or SLS disease genes. Additionally, patients with retinitis pigmentosa or the pediatric retinal disease Leber's congenital amaurosis would be candidates, having lesions in these genes. Future studies will be necessary to confirm these two genes as bona fide ciliopathy genes.
Materials and methods
Plasmids
Gateway entry clones for human ARL3, BART, BBS1, CCDC104, PDE6D, and UNC119a were obtained from Invitrogen. C5orf30, cystin 1–20, C20orf194, and UNC119b were synthesized by DNA 2.0. C-terminally LAP-tagged NPHP3 was generated by removing the stop codon from the previously described murine NPHP3 vector. NPHP3 truncations were made by PCR using the murine NPHP3 as a template. The G2A NPHP3 constructs were generated by PCR-based mutagenesis. ARL3 TN and ARL3 QL were made by PCR-based mutagenesis of T31 to N and Q71 to L, respectively. Mutagenesis of UNC119b was performed by GeneWiz. Multisite Gateway entry clones were generated for UNC119b and UNC119b Mut-4 using the pDONR221-5,2 vector, and were generated for tdTomato using the pDONR221-1,5r vector; LR plus was used to place tdTomato and UNC119b constructs into pDEST47 following the manufacturer's recommendations (Invitrogen). The above entry clones were put into one or more of the following destination vectors by LR recombination: pG-LAP5, pG-LAP6, pG-LAP7, pCS2-DEST+6xMyc, pCS2-DEST+C-term 3xMyc, and pGEX6p1-DEST. The pCS2-DEST+C-terminal 3xmyc vector was generated by inserting sequences coding for three in-frame myc epitopes into the XhoI and XbaI sites of pCS2+DEST.
Cell line generation
RPE and IMCD3 stable cell lines were generated as previously described (Sang et al. 2011). Briefly, a parental RPE or IMCD3 cell line containing a stably integrated FRT cassette was transfected with pOG44 encoding FLP recombinase (Invitrogen) and the appropriate LAP vector. Cells were then selected with the appropriate antibiotic.
Microscopy
Cells were plated on coverglass and induced to ciliate by withdrawing serum. Twenty-four hours later, the cells were cold-treated for 15 min on ice to depolymerize nonciliary microtubules, fixed 10 min in 4% formaldehyde in PBS, and washed three times with PBS. Coverslips were blocked in immunofluorescence buffer (PBS plus 30 mg/mL BSA, 0.1% NP-40) plus 5% normal donkey serum (Jackson ImmunoResearch). Cilia and basal bodies/centrosomes were visualized by immunostaining with 6-11b-1 anti-acetylated tubulin (1:2000; Sigma) and rabbit anti-pericentrin (1:2000; Abcam) antibodies in immunofluorescence buffer, respectively. Cy3 anti-rabbit and Cy5 anti-mouse secondary antibodies (Jackson ImmunoResearch) were added at 1:2000 in immunofluorescence buffer for 30 min after five washes with immunofluorescence buffer. Hoechst 33342 (AnaSpec, Inc.) was added for 5 min, and the coverslips were washed five times with immunofluorescence buffer and mounted in Fluoromount G. Images were acquired on an Everest deconvolution workstation (Intelligent Imaging Innovations) equipped with a Zeiss AxioImager.Z1 microscope, a CoolSnapHQ cooled CCD camera (Roper Scientific), and a 40× PlanApochromat NA 1.3 objective or 63× PlanApochromat NA 1.4 objective.
Tandem affinity purification
Tandem affinity purifications were performed essentially as described (Mukhopadhyay et al. 2010). Briefly, cells stably expressing a LAP-tagged construct were lysed in 50 mM HEPES (pH 7.4), 300 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.5 mM DTT, 0.3% NP-40, 0.2 mM PMSF, and 1× Roche Complete protease inhibitors. Lysates were cleared at 27,000g, and then again at 100,000g. Complexes were immunoprecipitated with rabbit anti-GFP antibodies and eluted with either TEV (LAP6) or PreScission (LAP5 and LAP7) protease. Complexes were again captured by using S protein agarose (Novagen), and finally eluted in 1× SDS sample buffer.
Recombinant proteins
Recombinant GST, GST-ARL3 TN, GST-ARL3 QL, and GST-UNC119a were expressed in Rosetta 2 cells (EMD Biosciences), induced with 0.2 mM IPTG at overnight 18°C, and purified using glutathione-sepharose according to the manufacturer's recommendations (GE Healthcare Life Sciences). In vitro translated proteins were generated by using the TNT HighYield Wheat Germ Protein expression system following the manufacturer's recommendations (Promega).
GST pull-down assays
For each pull-down reaction, 2.5 μg of GST or GST fusion protein was added along with 5–10 μL of in vitro translated protein and 5 μL of glutathione-sepharose in binding buffer (300 μL 25 mM HEPES at pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1% Triton X-100) for 1 h at 4°C. The beads were washed five times with the same buffer and eluted with SDS sample buffer. The eluates were resolved by SDS-PAGE and analyzed by immunoblotting with anti-myc-HRP antibodies (GeneTex). For competition assays, binding reactions were performed as described above, only 0.1 μM, 1 μM, 5 μM, or 10 μM peptide was included.
Peptide-binding assays
Peptides were synthesized and purified by reverse-phase chromatography by Elim Biopharmaceuticals, corresponding to the human cystin protein. More than 95% of the Myr-cystin peptide contains the myristoyl moiety. Peptide-binding reactions were performed by adding 450 fmol of C-terminally biotinylated peptide with 5–10 μL of Wheat Germ Extract IVT (Promega) protein and 5 μL of streptavidin-sepharose (GE Healthcare Life Sciences) in 300 μL of binding buffer for 1 h at room temperature. The beads were washed five times with binding buffer and eluted with SDS sample buffer. For the peptide release assays, 450 fmol of biotinylated Myr-cystin peptide was incubated with 5–10 μL of in vitro translated protein and 5 μL of streptavidin-sepharose in 300 μL of binding buffer for 1 h at room temperature. After two washes with binding buffer, GST-ARL2 or GST-ARL3 was added to 5.5 nM, 11 nM, 22 nM, or 44 nM, with a final volume of 300 μL. After a 1-h incubation, the complexes were washed and analyzed as described above.
Structural modeling
To build our structural model, we used the structure of truncated UNC119 (PDB code 3GQQ) as a reference. Docking of the myristoylated peptide was performed in two steps. First, the myristoyl group was placed into the cavity surrounded by the phenyl grove of UNC119, and energy was minimized using the interactive modeling program Benchware3D (Tripos). The choice of orientation of the myristoyl polar tail was made so that space is available to connect the peptide. The peptide was then built out of the myristate ester and extended on the protein surface to complement the surface charge and shape of UNC119. The peptide was then truncated back to the five N-terminal residues to yield the model presented here. The illustration of the model was made using MacPyMOL.
Cellular fractionation
Stable cell lines expressing LAP-tagged ARL3 TN or ARL3 QL were fractionated by first lysing the cells in hypotonic buffer and douncing. Nuclei were pelleted at 1400g and discarded. The supernatant was then separated into a soluble fraction (S100) and an insoluble pellet (P100) by centrifugation at 100,000g. Equal amounts of each were then separated by SDS-PAGE and analyzed by immunoblotting with an anti-GFP antibody (Abcam).
NPHP3 trafficking assay and rescue experiments
The LAP7-NPHP3 1–200 stable cell line was plated on coverslips overnight and transfected with siRNA with Lipofectamine RNAi Max (Invitrogen) following the manufacturer's recommendations. After 48 h, the cells were serum-starved with 0.2% FBS-containing medium. Twenty-four hours later, the cells were processed for immunofluorescence as described above. Images from each sample were collected and analyzed using identical settings. Percent ciliary NPHP3 was determined by first counting the number of NPHP3-positive cilia (viewing only the Cy3 channel for pericentrin and GFP channel for NPHP3), and then counting the total number of cilia as determined by Cy5 acetylated tubulin immunostaining. Rescue experiments were performed by first transfecting cells with the rescue plasmid using the Amaxa Nucleofection Kit V and plating them on coverglass. The following day, the cells were transfected with siRNAs as described above. After 24 h, the cells were serum-starved for 24 h to induce ciliation, processed for imaging, and analyzed as described above. The following siRNAs used were purchased from Dharmacon: ON-TARGETplus nontargeting control, ON-TARGETplus SMARTpool targeting ARL2, ON-TARGETplus UNC119a oligos J-009734-07 and J-009734-08, ON-TARGETplus ARL3 oligos J-011813-06 and J-011813-07, ON-TARGETplus UNC119b oligos J-035010-15 and J-035010-16, and ON-TARGETplus RP2 oligos J-012350-08 and J-012350-09. UNC119b oligo J-035010-15 targets the 3′ UTR and was used in the rescue assay.
C. elegans studies
Strains
The wild-type strain was C. elegans Bristol, strain N2. Additional strains were DP38 unc-119(ed3) (Caenorhabditis Genetics Center) and CX3553 lin-15(n765ts); kyIs104 [str-1p::gfp+lin-15(+)] (Troemel et al. 1997).
Molecular biology
Ce unc-119p::Hs unc119b cDNA::Ce unc-119 3′ UTR constructs were generated by sequential PCR fusions (Hobert 2002). The fusions contain 2.56 kb and 1.27 kb of sequences upstream of and downstream from, respectively, the Ce unc-119 coding region (Maduro et al. 2000). Primer sequences are available on request. PCR fusion products were sequenced to confirm the absence of PCR-induced mutations. Transgenic strains were generated by injecting plasmids at 100 ng/μL together with unc-122::dsRed as the coinjection marker.
Dye-filling assay
Animals were dye-filled with DiI as described previously (Perkins et al. 1986; Herman and Hedgecock 1990). Individual animals were placed on agarose slides and examined under 400× magnification on a Zeiss Axioplan microscope.
Tracking of locomotion
Well-fed 1-d-old adult animals were placed on a freshly seeded plate and their movements were recorded for 15 min as described (Biron et al. 2008; Beverly et al. 2011). Velocities of individual animal movements were extracted using custom-written scripts in MatLab (The MathWorks, Inc.).
Statistical analyses
Statistical analyses were performed using the SPSS 19 statistical analysis package.
Zebrafish studies
The following start site antisense MOs were designed and purchased from GeneTools: unc119a, AGCTCTGACTGTTCATCTCTCGGAA; unc119b, GCTGGTACAAGAATAGCTCATTATC; C5orf30, ACGCTCCACTGGCATCCATTTCCAT; and nphp3, TCACCAGAGAGGATGCCGTACCCAT. The MOs were air pressure-injected into one- to four-cell embryos at the amounts indicated in Figure 6.
Analysis of KV size
The size of the KV was assessed on live embryos at the six- to 12-somite stage of development as previously described (Yen et al. 2006). Briefly, embryos with a KV smaller than the width of the notochord were scored as abnormal.
Vision startle response assay
The light-induced vision startle response behavioral assay was performed as previously described (Pretorius et al. 2010). In brief, 5-d-post-fertilization (dpf) embryos were exposed to a 1-sec block in bright light intensity as a visual stimulus. A positive visual response was recorded if the embryo made an abrupt change in swimming behavior immediately upon light change. Five trials were performed, spaced 30 sec apart, followed by the mechanical stimulus of probing embryos with a blunt needle. Only embryos that responded to the mechanical stimulus and had a straight body were included in the vision analysis.
Acknowledgments
We thank Qui Phung, Susan Sa, John Beck, and Svetha Swaminathan for technical support; Matthew Beverly for assistance with worm-tracking analysis; the Caenorhabditis Genetics Center for strains; and members of the Jackson laboratory for helpful discussions. This work is supported in part by the Foundation Fighting Blindness (V.C.S.), National Institutes of Health grants (R01CA112369 to D.C.S., R01EY110298 and R01EY017168 to V.C.S., R37 GM56223 to P.S., and DC010090 to A.O.M.). V.C.S. is an Investigator of the Howard Hughes Medical Institute. K.J.W., S.M., L.S., M.K., W.W., and P.K.J. are employees of Genentech, Inc.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.173443.111.
References
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS 2004. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell 117: 527–539 [DOI] [PubMed] [Google Scholar]
- Baye LM, Patrinostro X, Swaminathan S, Beck JS, Zhang Y, Stone EM, Sheffield VC, Slusarski DC 2011. The N-terminal region of centrosomal protein 290 (CEP290) restores vision in a zebrafish model of human blindness. Hum Mol Genet 20: 1467–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. 2008. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal–hepatic–pancreatic dysplasia. Am J Hum Genet 82: 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly M, Anbil S, Sengupta P 2011. Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans. J Neurosci 31: 11718–11727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron D, Wasserman S, Thomas JH, Samuel AD, Sengupta P 2008. An olfactory neuron responds stochastically to temperature and modulates Caenorhabditis elegans thermotactic behavior. Proc Natl Acad Sci 105: 11002–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P 2005. Analysis of xbx genes in C. elegans. Development 132: 1923–1934 [DOI] [PubMed] [Google Scholar]
- Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME 2010. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Genet 19: 1358–1367 [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S 2007. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol 23: 579–611 [DOI] [PubMed] [Google Scholar]
- Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G 2003. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell 112: 845–857 [DOI] [PubMed] [Google Scholar]
- Herman RK, Hedgecock EM 1990. Limitation of the size of the vulval primordium of Caenorhabditis elegans by lin-15 expression in surrounding hypodermis. Nature 348: 169–171 [DOI] [PubMed] [Google Scholar]
- Heuckeroth RO, Towler DA, Adams SP, Glaser L, Gordon JI 1988. 11-(Ethylthio)undecanoic acid. A myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J Biol Chem 263: 2127–2133 [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N 2011. Ciliopathies. N Engl J Med 364: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730 [DOI] [PubMed] [Google Scholar]
- Hou X, Mrug M, Yoder BK, Lefkowitz EJ, Kremmidiotis G, D'Eustachio P, Beier DR, Guay-Woodford LM 2002. Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease. J Clin Invest 109: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Margolis BL 2011. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin β2. J Cell Sci 124: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiba Y, Higashide T, Mori N, Kobayashi A, Kubota S, McLaren MJ, Satoh H, Wong F, Inana G 2007. Targeted inactivation of synaptic HRG4 (UNC119) causes dysfunction in the distal photoreceptor and slow retinal degeneration, revealing a new function. Exp Eye Res 84: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G 2000. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol Vis Sci 41: 3268–3277 [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552 [DOI] [PubMed] [Google Scholar]
- Linari M, Hanzal-Bayer M, Becker J 1999. The δ subunit of rod specific cyclic GMP phosphodiesterase, PDEδ, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett 458: 55–59 [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN Jr, Rux JJ, Speicher DW, Pierce EA 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Gordon M, Jacobs R, Pilgrim DB 2000. The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J Neurogenet 13: 191–212 [DOI] [PubMed] [Google Scholar]
- Manning AG, Crawford BD, Waskiewicz AJ, Pilgrim DB 2004. unc-119 homolog required for normal development of the zebrafish nervous system. Genesis 40: 223–230 [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL, Wildpaner M, Hayashi N, Eisenhaber F 2004. MYRbase: Analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol 5: R21 doi: 10.1186/gb-2004-5-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24: 2180–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf MT, Sasmaz G, Trauer U, Reinhardt R, et al. 2003. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet 34: 455–459 [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB 2005. Proteomic analysis of a eukaryotic cilium. J Cell Biol 170: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117: 456–487 [DOI] [PubMed] [Google Scholar]
- Pretorius PR, Baye LM, Nishimura DY, Searby CC, Bugge K, Yang B, Mullins RF, Stone EM, Sheffield VC, Slusarski DC 2010. Identification and functional analysis of the vision-specific BBS3 (ARL6) long isoform. PLoS Genet 6: e1000884 doi: 10.1371/journal.pgen.1000884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD 1999. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. 2011. Mapping the NPHP–JBTS–MKS protein network reveals ciliopathy disease genes and pathways. Cell 145: 513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS 2006. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol 168: 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharer JD, Kahn RA 1999. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J Biol Chem 274: 27553–27561 [DOI] [PubMed] [Google Scholar]
- Shiba D, Manning DK, Koga H, Beier DR, Yokoyama T 2010. Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton (Hoboken) 67: 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG 2007. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic 8: 1323–1330 [DOI] [PubMed] [Google Scholar]
- Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci 101: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao B, Bu S, Yang Z, Siroky B, Kappes JC, Kispert A, Guay-Woodford LM 2009. Cystin localizes to primary cilia via membrane microdomains and a targeting motif. J Am Soc Nephrol 20: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI 1997. Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell 91: 161–169 [DOI] [PubMed] [Google Scholar]
- Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA 2001. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: Characterizing ARL1-binding proteins. J Biol Chem 276: 22826–22837 [DOI] [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A 2008. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol 15: 373–380 [DOI] [PubMed] [Google Scholar]
- Wilcox C, Hu JS, Olson EN 1987. Acylation of proteins with myristic acid occurs cotranslationally. Science 238: 1275–1278 [DOI] [PubMed] [Google Scholar]
- Wolf MT, Hildebrandt F 2011. Nephronophthisis. Pediatr Nephrol 26: 181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HJ, Tayeh MK, Mullins RF, Stone EM, Sheffield VC, Slusarski DC 2006. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum Mol Genet 15: 667–677 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W 2004. Photoreceptor cGMP phosphodiesterase δ subunit (PDEδ) functions as a prenyl-binding protein. J Biol Chem 279: 407–413 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W 2007. Deletion of PrBP/δ impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci 104: 8857–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, et al. 2011. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci 14: 874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA 2006. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 17: 2476–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Dai J, Attanasio M, Hildebrandt F 2010. Nephrocystin-3 is required for ciliary function in zebrafish embryos. Am J Physiol Renal Physiol 299: F55–F62 doi: 10.1152/ajprenal.00043.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]