Regulation of microtubule polarization is an important aspect of establishing body axes during Drosophila oocyte development. Siomi and colleagues identify the Maelstrom protein (mael) as an essential player in MTOC establishment. Mael forms specific complexes with MTOC components, and its mutation leads to their mislocalization and centrosome segregation defects.
Keywords: maelstrom, microtubule, centrosome, piRNA, axis specification, Drosophila oogenesis
Abstract
The establishment of body axes in multicellular organisms requires accurate control of microtubule polarization. Mutations in Drosophila PIWI-interacting RNA (piRNA) pathway genes often disrupt the axes of the oocyte. This results from the activation of the DNA damage checkpoint factor Checkpoint kinase 2 (Chk2) due to transposon derepression. A piRNA pathway gene, maelstrom (mael), is critical for the establishment of oocyte polarity in the developing egg chamber during Drosophila oogenesis. We show that Mael forms complexes with microtubule-organizing center (MTOC) components, including Centrosomin, Mini spindles, and γTubulin. We also show that Mael colocalizes with αTubulin and γTubulin to centrosomes in dividing cyst cells and follicle cells. MTOC components mislocalize in mael mutant germarium and egg chambers, leading to centrosome migration defects. During oogenesis, the loss of mael affects oocyte determination and induces egg chamber fusion. Finally, we show that the axis specification defects in mael mutants are not suppressed by a mutation in mnk, which encodes a Chk2 homolog. These findings suggest a model in which Mael serves as a platform that nucleates other MTOC components to form a functional MTOC in early oocyte development, which is independent of Chk2 activation and DNA damage signaling.
The cellular processes that elaborate asymmetries along the main body axes are essential to the development of multicellular organisms. In Drosophila, the body axes are established during oogenesis through the differential localization of a large number of specific transcripts, including bicoid (bcd), oskar (osk), and gurken (grk) mRNAs. This localization depends on a polarized microtubule (MT) cytoskeleton and correlates with the formation of an MT-organizing center (MTOC), a small organelle from which most MTs grow (Ephrussi et al. 1991; Chasan and Anderson 1993; Neuman-Silberberg and Schupbach 1993; Theurkauf et al. 1993; Pokrywka and Stephenson 1995; van Eeden and St Johnston 1999; Riechmann and Ephrussi 2001; Januschke et al. 2006; Steinhauer and Kalderon 2006). Thus, the formation of the axis depends on upstream cues that lead to the polarization of MTs.
The Drosophila ovary consists of 16–20 independent strings of egg chambers called ovarioles, which are the functional units of oogenesis. Oogenesis starts with an asymmetric division of a germline stem cell, which occurs at the anterior tip of the germarium located at the anterior tip of an ovariole. This gives rise to a new stem cell and a differentiating daughter cell, the cystoblast (Supplemental Fig. S1). Cystoblasts in turn undergo four mitoses to generate a cluster of 16 germ cells, which are interconnected by cytoplasmic bridges, called ring canals (Spradling 1993). This cluster, named the cyst, is surrounded by a monolayer of somatic follicle cells to form an egg chamber, which then exits the posterior end of the germarium and proceeds through the 14 stages of oogenesis as it moves posteriorly toward the oviduct (Spradling 1993). Two germ cells have four ring canals, and one of these cells is selected to differentiate into the oocyte. This cell remains arrested in meiotic prophase I and moves to the posterior of the cyst in region 3 of the germarium, whereas the other germ cells undergo endoreplication to generate polyploid nurse cells (Spradling 1993). Which cell adopts the oocyte fate may depend on the asymmetric distribution of the fusome, a continuous vesicular organelle linking the 16 cells via the ring canals (Lin et al. 1994; Lin and Spradling 1995). The fusome is partitioned asymmetrically during cystoblast divisions, which requires its stable association with MTs (Grieder et al. 2000). It has been hypothesized that the cell that inherits the most fusome material is the one that will become the oocyte. Shortly after oocyte specification, nurse cell centrosomes migrate toward the oocyte, where they eventually accumulate at the posterior of the nucleus, thus defining the major MTOC of the entire cyst (Mahowald and Strassheim 1970; Grieder et al. 2000). As a cyst moves in region 2b of the germarium, the fusome is replaced by a polarized MT network emanating from a single MTOC that resides at the anterior of the future oocyte. This network extends through the ring canals into the nurse cells (Grieder et al. 2000) and is polarized such that the major routes of transport are directed from the nurse cells toward the oocyte (Theurkauf et al. 1992; Clark et al. 1994, 1997). This asymmetric arrangement of the germ cells generates the first anterior–posterior (A–P) polarity during development. Thus, initial polarization of the oocyte during the oocyte specification phase requires MTs and correlates with the formation of the MTOC in the anterior of the cell (Theurkauf et al. 1993).
In region 3, when oocyte positioning is completed, a reorganization of the oocyte MT network takes place, which shifts the anterior MTOC to the posterior pole (Clark et al. 1997). This polarized MT network is required for asymmetric localization of Grk, a TGF-β homolog (Januschke et al. 2006). The A–P axis of the embryo is then polarized by two signaling events. At stage 6, the Grk signal from the oocyte induces the adjacent follicle cells to adopt a posterior rather than an anterior fate. At stages 7–8, the oocyte MT cytoskeleton undergoes a dramatic reorganization in response to an unknown signal from the overlying posterior follicle cells (Theurkauf et al. 1992; Januschke et al. 2006). The posterior MTOC is disassembled and, accompanying this process, the oocyte nucleus moves away from the posterior pole and localizes at the anterodorsal corner of the oocyte (Januschke et al. 2006). MTs then become repolarized, which directs the transport of mRNAs encoding determinants important for embryonic development; for example, bcd mRNA is transported to the anterior of the oocyte and osk mRNA is transported to the posterior of the oocyte (Brendza et al. 2000; Schnorrer et al. 2000; Arn et al. 2003). At the same time, grk mRNA localizes to the anterodorsal cortex of the oocyte, leading to Grk signaling to the dorsal follicle cells (Neuman-Silberberg and Schupbach 1993, 1996). Thus, the oocyte and the surrounding eggshell are patterned by the reciprocal germline-to-soma signaling cascade. Although the process of early oocyte polarization clearly depends on an intact MT network and correlates with the formation, shift, and disassembly of the MTOC, the mechanisms involved are not well understood.
PIWI-interacting RNAs (piRNAs), a class of small RNAs associated with PIWI proteins of the Argonaute family, silence transposable elements in animal germ cells (Ghildiyal and Zamore 2009; Malone and Hannon 2009; Siomi et al. 2011). In Drosophila ovaries, piRNAs associate with the PIWI subfamily of Argonaute proteins (AGO3, Aubergine [Aub] and Piwi) to form the effector piRISC, which represses retrotransposons and preserves the integrity of the genome (Aravin et al. 2001; Kalmykova et al. 2005; Saito et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Nishida et al. 2007; Li et al. 2009; Malone et al. 2009). A large number of genes have been shown to be involved in piRNA production in Drosophila ovaries. The majority of these piRNA pathway genes are required for assembly of nuage, a perinuclear structure found at the cytoplasmic face of the nuclear envelope in animal germline cells, where multiple components of the piRNA pathway are enriched, enabling piRNA processing to occur (Lim and Kai 2007; Li et al. 2009; Malone et al. 2009). Mutations in these piRNA pathway genes often disrupt axis formation and give rise to disorganized MTs in ovaries, which ultimately lead to the production of spindle-shaped eggs (Gillespie and Berg 1995; Wilson et al. 1996; Clegg et al. 1997; Lasko and Ashburner 1988; Styhler et al. 1998; Tomancak et al. 1998; Cook et al. 2004; Klattenhoff et al. 2007; Pane et al. 2007; Klattenhoff and Theurkauf 2008). Indeed, many piRNA pathway genes, including aub, armitage (armi), maelsrom (mael), spindle-E (spnE), vasa (vas), and zucchini (zuc), had been put in the spindle class of genes before their involvement in piRNA production was revealed (Gillespie and Berg 1995; Wilson et al. 1996; Clegg et al. 1997; Lasko and Ashburner 1988; Styhler et al. 1998; Tomancak et al. 1998; Cook et al. 2004; Pane et al. 2007). Mutations in spindle class genes are highly pleiotropic, resulting in axis patterning defects, which is attributed to failures in MT polarization and mRNA localization (Tearle and Nüsslein-Volhard 1987; Schupbach and Wieschaus 1991; Gonzalez-Reyes et al. 1997). armi, aub, spn-E, and vas mutant oocytes display defects in MT organization, including assembly of the MTOC (Styhler et al. 1998; Cook et al. 2004; Klattenhoff et al. 2007). These findings indicate that in piRNA pathway mutants, the MT network is not polarized, which disrupts the Grk signaling and the initiation of oocyte patterning. Thus, it is possible that piRNA pathway genes are involved in the machinery that impacts MT organization, thereby indicating a link between piRNA production and the polarization of MTs.
In the majority of piRNA pathway mutations, the earliest phenotype is a persistent DNA damage in germline cells of the germarium, probably due to transposon derepression and mobilization, which can lead to DNA breaks (Klattenhoff and Theurkauf 2008). The axis-specific defects associated with these mutations are suppressed by mutations in mei-41 and mnk, which encode ATR and Checkpoint kinase 2 (Chk2) kinases that function in DNA damage signaling (Klattenhoff et al. 2007). Significantly, the same mutations in mei-41 and mnk also restore MTOC formation during early oogenesis (Klattenhoff et al. 2007). How checkpoint activation causes changes in the cytoskeletal network is currently unknown; however, these findings suggest that the axis specification defects associated with piRNA pathway mutations are a secondary consequence of DNA damage signaling.
The mael gene was originally identified based on its role in Drosophila oocyte axis specification (Clegg et al. 1997). mael mutants exhibit mRNA localization patterns similar to grk mutants in stages 8–10, accumulating bcd mRNA at both poles and osk mRNA in the center of the oocyte (Clegg et al. 1997). Mutations in mael also prevent efficient grk translation in stages 2–6 and disrupt both oocyte nucleus migration to the anterodorsal corner and posterior localization of the MTOC in stage 2–6 oocytes (Clegg et al. 2001). Thus, failure of early Grk signaling in the oocyte has been proposed to be the underlying cause of A–P defects in mael mutants. Although Mael contains a domain with weak similarity to a high-mobility group (HMG) box and a domain called the Mael domain with weak homology with the DnaQ-H 3′-to-5′ exonuclease family (Zhang et al. 2008), its biochemical functions in both axis specification and piRNA production remains to be clarified. Mael localizes to the nucleus as well as to the nuage (Findley et al. 2003; Lim and Kai 2007). The nuage localization of Mael depends on other piRNA genes, including vas, aub, and spn-E (Findley et al. 2003; Lim and Kai 2007).
In the course of biochemically characterizing the role of Mael in Drosophila ovaries, we found that Mael physically interacts with MTOC components. These MTOC components mislocalize in the germarium of mael mutants, leading to centrosome migration defects that are indicative of defects in MT organization. We show that the loss of mael affects oocyte determination and causes egg chamber fusion. The patterning defects associated with mael mutants are not suppressed by mutation of mnk. We propose that, through interaction with MTOC components, Mael contributes to the coordination of MT organization in early oogenesis, which is independent of Chk2 activation and DNA damage signaling and is, therefore, probably independent of its function in piRNA production.
Results
Mael forms complexes with MTOC components
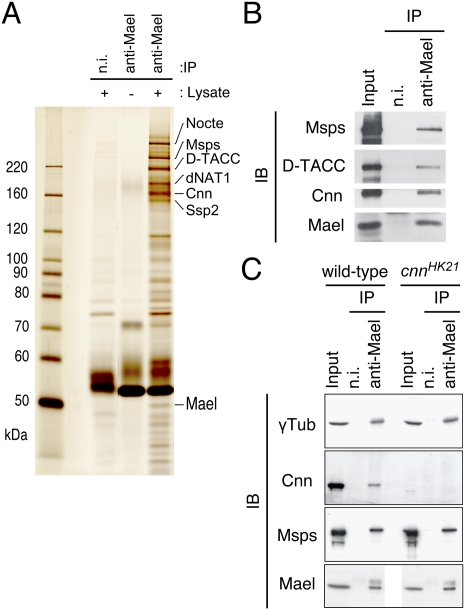
To biochemically characterize its function, we produced a specific monoclonal antibody against Mael. Western blot analysis showed a discrete band in both ovary and cultured Schneider2 (S2) cells but not in mael-depleted S2 cells (Supplemental Fig. S2A), demonstrating the specificity of the antibody. These results also indicated that Mael expression is not germline-specific. Immunostaining revealed that Mael in S2 metaphase cells is cytoplasmic and concentrated at mitotic spindles (Supplemental Fig. S2B,C). Lack of staining in mael-depleted S2 cells and mael mutant ovaries further indicated no off-target binding of the antibody (Supplemental Fig. S2B,D). We performed immunoprecipitation from ovary lysates with the antibody in a buffer containing a strong detergent, Empigen, which will disrupt most protein–protein interactions in vivo, although antigen–antibody associations are maintained (Piñol-Roma et al. 1990). Mael was the only protein precipitated with the antibody under these conditions, further confirming the specificity of the antibody (Supplemental Fig. S2E). In a milder Digitonin-based buffer, Mael was copurified with other protein factors. Mael-associated proteins were resolved by SDS–polyacrylamide gel electrophoresis and visualized by silver staining (Fig. 1A). The eluates were further analyzed by mass spectrometry to identify protein partners that engage with Mael in the ovary. Six proteins were identified as major Mael-interacting proteins: Centrosomin (Cnn), Drosophila transforming acidic coiled-coil protein (D-TACC), Mini spindles (Msps), Short spindle 2 (Ssp2), dNAT1 (CG3845), and No circadian temperature entrainment (Nocte). In animal cells, many of these proteins are associated with centrosomes that act as MTOCs to nucleate MTs with uniform polarity (Kwon and Scholey 2004). Centrosomes are composed of two centrioles that recruit a large number of proteins collectively referred to as pericentriolar material (PCM). Cnn is a core centrosome protein required for normal PCM organization and astral MT assembly (Megraw et al. 2001; Terada et al. 2003). Msps, a member of the conserved MT-associated proteins, localizes to centrosomes and mitotic spindles and is required for the integrity of mitotic spindles (Cullen et al. 1999; Cullen and Ohkura 2001). D-TACC is also a centrosomal protein that interacts with MTs and is found at the spindle poles. Complex formation of D-TACC with Msps has been postulated to stabilize centrosomal MTs (Gergely et al. 2000; Lee et al. 2001). Cnn and the Msps–D-TACC complex are required for efficient MTOC formation. Knockdown of ssp2 produces a short spindle phenotype, suggesting that this gene is also required for the integrity of mitotic spindles (Goshima et al. 2007). The nocte gene encodes a large glutamine-rich protein with unknown function and is involved in maintaining the structure of cilia in circadian clock neurons (Sehadova et al. 2009). dNAT1 is a fly homolog of the vertebrate translational regulator NAT1/DAP5/p97, an eIF4G family member (Yoshikane et al. 2007).
Figure 1.
Mael forms complexes with MTOC components in ovaries. (A) Immunoprecipitation was performed from ovary lysates using an anti-Mael monoclonal antibody. Proteins immunoprecipitated were stained with silver. Mass spectrometry analyses revealed that Nocte, Msps, D-TACC, dNAT1, Cnn, and Ssp2 coimmunoprecipitated with Mael. “n.i.” indicates nonimmune IgG used as a negative control. (B) Western blotting on the Mael immunoprecipitated complex in A using anti-Msps, anti-D-TACC, and anti-Cnn confirmed Mael binding with Msps, D-TACC, and Cnn. Nonimmune IgG (n.i.) was used as a negative control. (C) Immunoprecipitation was performed from wild-type and cnn mutant ovary lysates using an anti-Mael monoclonal antibody. cnnHK21 is a cnn-null allele with a nonsense mutation that truncates the protein at amino acid 106 (Megraw et al. 1999). Mael binds with γTub and Msps in a Cnn-independent manner.
The finding that Mael forms a complex in vivo with components of the MTOC suggests that Mael is also a component of the MTOC. Western blot analyses on the immunoprecipitates confirmed that Cnn, Msps, and D-TACC exist in the Mael complex (Fig. 1B; Supplemental Fig. S2F). The immunoprecipitates from mael mutant ovaries showed no bands corresponding to Cnn, Msps, or D-TACC (Supplemental Fig. S2F). These data show that Mael specifically forms complexes with components of the MTOC and suggest its involvement in normal MTOC formation as an MTOC component. To further confirm our findings, we examined whether Mael interacts with other MTOC proteins in ovaries. γTubulin (γTub) is a conserved component of all MTOCs and is required for them to nucleate MT polymerization from heterodimers of αTub and βTub (Wiese and Zheng 1999). We performed Western blot analysis using an anti-γTub antibody on immunopurified Mael complexes and found that Mael indeed interacts with γTub (Fig. 1C). Although γTub was no longer recruited to the centrosome in the absence of Cnn in Drosophila S2 cells (Goshima et al. 2007), γTub associated with Mael in cnn mutant ovaries, indicating that the association between γTub and Mael in the ovary is Cnn-independent (Fig. 1C). This also suggests that γTub–Mael complexes are formed before they become stable components of the MTOC. These results are also in accordance with previous genetic analysis showing that mael is required for proper positioning of the MTOC during oogenesis (Clegg et al. 1997, 2001).
Mael colocalizes with αTub and γTub in the germarium
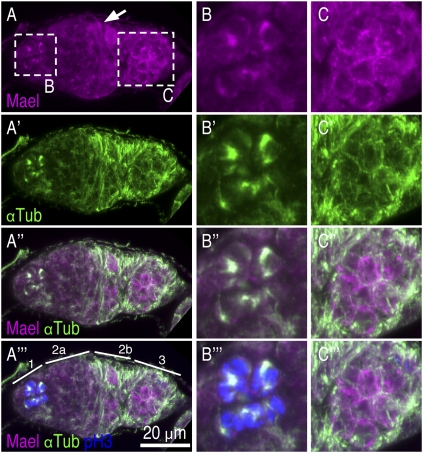
Prior studies showed that in the germarium, Mael was present in all regions with diffuse nuclear and cytoplasmic localization, and its accumulation in the nuage became evident in region 3 (Findley et al. 2003). As a starting point for defining the role of Mael in the MTOC, we re-examined its localization using the antibody we produced and reasoned that Mael could colocalize with components of the MTOC in the ovary. Mael was highly concentrated in dividing cells in the germarium, as judged by the coexpression of the mitotic marker, phospho-histone H3 (Ser 10) (pH3) (Fig. 2; Adams et al. 2001). Lack of staining in mael mutant ovaries indicated that the signals observed depend on the presence of Mael (Supplemental Fig. S2D). In region 1 of the germarium, Mael accumulated in discrete regions of dividing cyst cells and colocalized with αTub, indicating that Mael was concentrated at the MT spindles of early cyst cells (Fig. 2A,B). In region 2, Mael became localized to granules, which did not colocalize with αTub (Fig. 2A). In region 3, Mael became localized to the nuage, which did not colocalize with αTub (Fig. 2A,C). Thus, Mael colocalized with αTub to centrosomes in early dividing cyst cells and then later became accumulated at the nuage in the germarium. Although Mael is known to colocalize with components of the piRNA pathway in the nuage, mass spectrometry analyses of the complex that we isolated revealed no known proteins functioning in the piRNA pathway, at least as major components (Fig. 1A). We also failed to detect PIWI proteins in the complex by Western blot analysis (Supplemental Fig. S3A). Reciprocal immunoprecipitation experiments with antibodies against PIWI proteins further showed that PIWI proteins do not interact with Mael or other MTOC components (Supplemental Fig. S3B–D). Thus, these results suggest that although Mael changes its subcellular localization during early oogenesis, the majority of the protein in ovaries physically interacts with MTOC components (Fig. 1A) but not with components of the piRNA pathway.
Figure 2.
Mael colocalizes with αTub in the germarium. (A–A″′) Ovaries were immunostained with anti-Mael, anti-αTub, and anti-pH3 antibodies. pH3 is a mitotic marker. In region 1 of the germarium, Mael accumulates in discrete regions of dividing cyst cells. The Mael signals overlaps with the γTub signals, suggesting that Mael is localized to the MT spindles in early cyst cells. In region 2, Mael is localized to granules, in which γTub was undetected. In region 3, Mael is accumulated in the nuage, which is also devoid of γTub. A Mael signal in a somatic precursor cell is shown by an arrow. (B–B″′) Magnified views of dividing cyst cells in A–A′″′. (C–C″′) Magnified views of the early egg chamber in A–A″′.
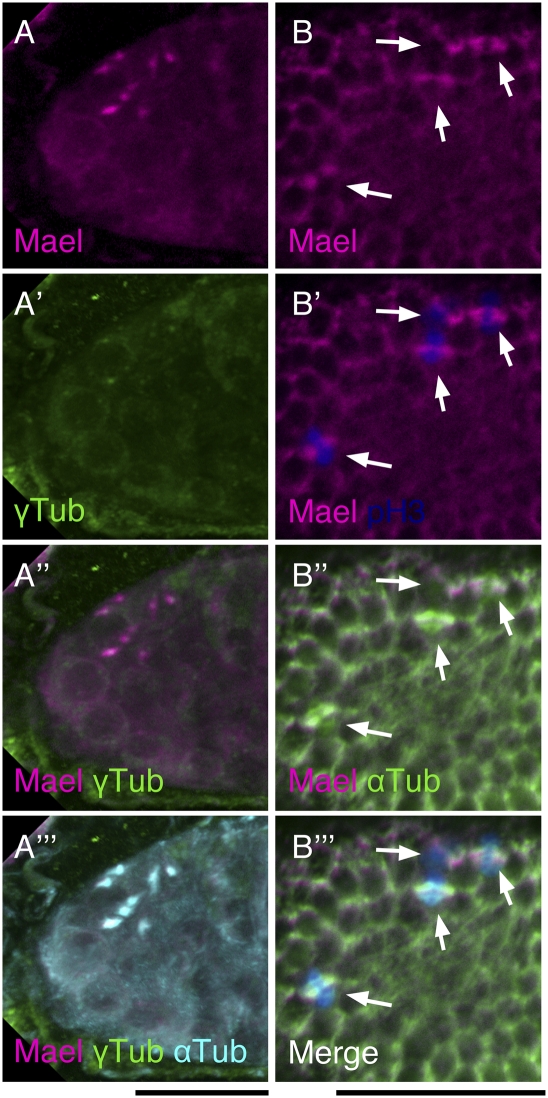
To confirm its centrosomal localization in region 1 of the germarium, we performed double labeling of Mael and γTub in the germarium. γTub is known to be present on the centrosomes in the germarium and on migrating centrioles (Mahowald and Strassheim 1970; Bolívar et al. 2001). Double labeling of Mael and γTub showed their overlap in cyst cells in region 1 (Fig. 3A). Therefore, Mael was concentrated at centrosomes of dividing cyst cells and colocalized with γTub and αTub in region 1 of the germarium. Together, these findings establish that Mael is a component of centrosomes in dividing cyst cells.
Figure 3.
Mael is localized to mitotic spindles in the dividing cyst and follicle cells. (A–A″′) Immunofluorescence was performed using anti-Mael, anti-αTub, and anti-γTub antibodies. Mael is colocalized with αTub and γTub at mitotic spindles of dividing cyst cells. (B–B″′) Immunofluorescence was performed using anti-Mael, anti-αTub, and anti-pH3 antibodies. In dividing follicle cells (pH3-positive cells; indicated by arrows), Mael is colocalized with αTub to mitotic spindles. Bars, 20 μm.
Mael is concentrated in the centrosomal region during follicle cell divisions
Mael was highly expressed in somatic precursor cells (Fig. 2A, arrow; Supplemental Fig. S4). Later, Mael did not accumulate in nuage-like structures in follicle cells but was concentrated at MT spindles of dividing follicle cells in egg chambers, as judged by colocalization with αTub (Fig. 3B). Therefore, Mael colocalizes with these proteins in both dividing cyst cells and dividing follicle cells in the ovary. These findings suggest that Mael is involved in centrosomal function during Drosophila oogenesis.
Protein components of the MTOC are mislocalized in mael egg chambers
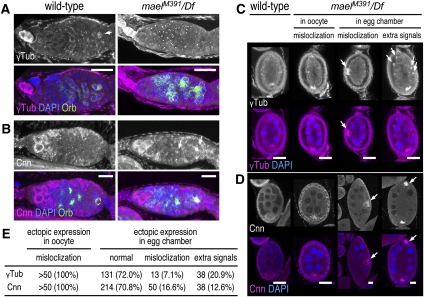
An early event in oocyte differentiation is the establishment of a single MTOC in the cyst. At the end of four cystocyte divisions, the centrosomes are inactivated and the centrioles are transported into the pro-oocyte where a single MTOC in the cyst is established. From this focus in the oocyte, MTs emanate into all of the 15 nurse cells. This polarized MT array is essential for the transport of maternal factors required for oocyte differentiation and development (for reviews, see Cooley and Theurkauf 1994; Knowles and Cooley 1994; Mahajan-Miklos and Cooley 1994; Theurkauf 1994). Centrosomes have been reported to migrate into the pro-oocyte during cyst development in the germarium. Our finding that Mael interacts with centrosome components suggests that centrosome migration may be perturbed in mael mutant egg chambers. To test this, we examined the localization of γTub and Cnn in the mael germarium (Fig. 4). In the wild-type germarium, γTub accumulated at dot-like structures, which were presumably centrosomes in region 1. These started to migrate into pro-oocytes in region 2 and then accumulated in pro-oocytes in region 3. γTub also accumulated at dot-like structures in region 1 of the mael germarium, but it remained at these structures and did not accumulate in pro-oocytes in regions 2–3 (Fig. 4A). In the wild-type germarium, Cnn accumulated in the cytoplasm of germline stem cells in region 1, but its expression in regions 2–3 was barely detected. Cnn was also accumulated in the cytoplasm of germline stem cells in region 1 of the mael germarium, but its expression was detected at significant levels in pro-oocytes in regions 2–3 (Fig. 4B). Thus, both γTub and Cnn were mislocalized and ectopically expressed in the mael germarium.
Figure 4.
Protein components of the MTOC are mislocalized in mael mutant ovaries. (A) γTub localization in the germarial region of wild-type (left panels) and mael mutant (right panels) ovaries. Anti-Orb antibody was used as a marker for developing oocytes. γTub is accumulated in the oocyte in the wild-type germarium (arrow), but was absent from the mael mutant oocyte in the germarium. (B) Cnn localization in the germarial region of wild-type (left panels) and mael mutant ovaries (right panels). Cnn, which is undetectable in the oocyte in the wild-type germarium, was detected in the oocytes in the mael mutant germarium (arrows). (C) γTub localization in wild-type and mael mutant egg chambers. In the wild type, γTub is specifically accumulated at the anterior region in the oocyte. However, in the mael mutant, γTub was often mislocalized to the posterior region of the oocytes. γTub was also ectopically accumulated within the egg chambers (arrows) of mael. (D) Cnn localization in wild-type and mael mutant egg chambers. Cnn, which is accumulated at the posterior region in the wild-type oocyte, was detected as speckles in the mael mutant oocytes. Cnn was ectopically accumulated within the egg chambers (arrows). (E) The percentage of mael egg chambers that show ectopic localization of Cnn and γTub. Bars, 20 μm.
We also observed mislocalization of γTub and Cnn in oocytes in developing mael egg chambers. In the wild type, γTub accumulated in oocytes with a punctuated pattern (Fig. 4C,E). However, γTub was highly concentrated at the posterior region of mael oocytes (Fig. 4C,E). Cnn localized to the posterior region of oocytes in stages 6–8 of mael egg chambers as in the wild type (Fig. 4D,E). However, Cnn was detected with granules in mael egg chambers. In addition, γTub and Cnn were ectopically expressed in ∼39% and 29%, respectively, of all mael egg chambers. Some egg chambers contained two strong signals at both poles, while others contained misplaced oocytes (Fig. 4C–E). We also occasionally observed mael egg chambers that contained abnormal accumulation of γTub in the nurse cells (Fig. 4C). These observations provide a functional involvement of mael in the proper localization and expression of MTOC proteins during Drosophila oogenesis.
Loss of mael function affects the distribution and dynamics of oocyte MTs
The dynamics of MT accumulation and the temporal–spatial changes in the MTOC have been observed in the developing oocyte by staining MTs with antibodies against αTub (Theurkauf et al. 1992). From stage 1 to stage 6, MTs extend into the nurse cells from the MTOC, situated at the posterior of the oocyte (Theurkauf et al. 1992, 1993). Mislocalization and ectopic expression of γTub and Cnn in mael egg chambers prompted us to examine the MT cytoskeleton in mael ovaries. We compared the MT cytoskeleton of the germariums and of stage 1–8 egg chambers in mael and wild-type ovaries to look for phenotypic differences. In the wild-type, an MTOC was detected at the posterior region of the stage 1 oocyte (region 3 in the germarium), whereas an MTOC was not detected in the stage 1 mael mutant oocytes (Fig. 5A,B). The MT cytoskeleton in the mael germarium was clearly disorganized (Fig. 5B). In the wild type, αTub was concentrated in stage 3–6 oocytes, in the location where MTOCs are formed (Theurkauf et al. 1992). MTs extended from an MTOC at the posterior region of stage 3–6 oocytes through the ring canals and into nurse cells (Fig. 5C). However, high concentrations of MTs at the posterior region of stage 3–6 oocytes in mael mutant ovaries were not observed (Fig. 5C). This is not due to reduced expression of αTub in mael mutant ovaries, as judged by Western blot analysis (Supplemental Fig. S5).
Figure 5.
MTOC formation is defective in mael mutant ovaries. MT distribution in wild-type and mael mutant (maelM391/Df) oocytes was visualized with anti-αTub antibody (white). DNA was visualized with DAPI (blue). (A) MTs in the control germarium and stage 1 egg chamber. The MTOC was observed at the posterior region of the oocyte in region 3 of the germarium. A red asterisk indicates the nucleus of the oocyte that is denoted by a dotted line. (B) MTs in the mael mutant germarium and stage 1 egg chamber. The MTOC was not detected when mael function was lost. (C) MTs in stage 3–8 egg chambers in control and mael mutant ovaries. The MTOC was detected at the posterior region of stage 3–6 control oocytes, but not in the mael mutant oocytes. In the wild type, MTs are then rolled up (“diaphragm” pattern) during stage 7 (Supplemental Fig. S1) and project from the oocyte nucleus in an anterior–posterior direction at stage 8 (“open diaphragm” pattern). This reorganization of MTs was not observed in the mael mutant. Bars, 20 μm.
At stage 7, a shift in the polarity of MT accumulation occurs in oocytes. At this time, the oocyte nucleus migrates to the anterior of the oocyte. Nuclear migration is an MT-dependent process that in many systems is dependent on MTOCs associated with the nucleus (Reinsch and Gonczy 1998). After migration of the nucleus to a position that will mark the anterior–dorsal position of the developing oocyte, MTs are rolled up in the shape of a “diaphragm” in the center of the oocyte. In stages 8–9, the “diaphragm” has opened and MT bundles project from the anterior of the oocyte in the A–P direction in a horseshoe-like pattern (Supplemental Fig. S1; Januschke et al. 2006). In the wild type, in stages 7 and 8, an MTOC was normally reorganized and the oocyte nucleus migrated anteriorly (Fig. 5C). However, in mael mutant oocytes, MT accumulation and reorganization were not observed (Fig. 5C). Thus, mael MT cytoskeletons are not organized to form an MTOC even in the germarium and are unable to reorganize and rearrange into an elaborate pattern of subcortical MT bundles that mediate oocyte nucleus migration in stages 6–8 of the oocyte.
To gain further insights into the involvement of Mael in MT organization and dynamics, we examined cell division of Mael-depleted S2 cells. We observed that Mael depletion delayed the progression to anaphase and thereby increased the percentage of metaphase cells (∼4% of the population) (Supplemental Fig. S6). These results, together with the concentration of Mael at mitotic spindles in S2 cells (Supplemental Fig. S2D), suggest that Mael is required for the efficient assembly of metaphase spindles in S2 cells and further implicate Mael in MT organization.
Loss of mael function affects the number of germ cells per egg chamber
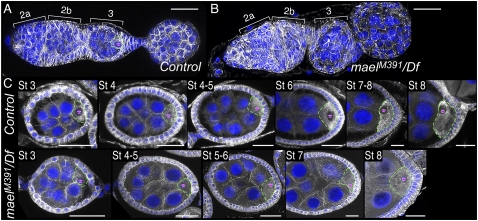
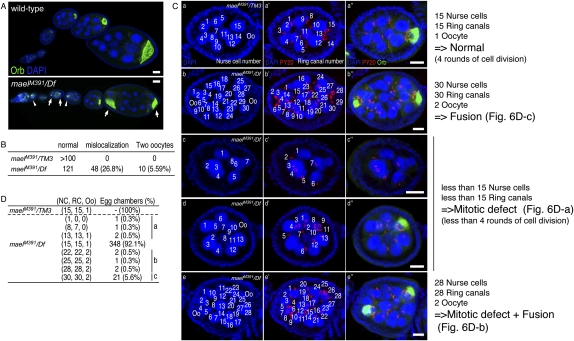
Mael mutant ovaries have a defective MT cytoskeleton with no clear MT accumulation or MTOC throughout the early stages of the oocyte. In addition, a significant number of mael egg chambers show ectopic expression of γTub and Cnn. Thus, we re-examined the number and position of the oocytes in mael mutant egg chambers. We observed that ∼27% of egg chambers in mael mutant ovaries have misplaced oocytes (stained with anti-Orb antibody) and ∼5.5% of mael egg chambers have two oocyte-like nuclei with >15 nurse cells (Fig. 6A,B). In the egg chambers with supernumerary germ cells, an MTOC was not detected in either of the two oocyte-like cells (Supplemental Fig. S7). The posterior localization of the oocyte is thought to be directed by cell sorting via DE-cadherin (DE-cad) (Godt and Tepass 1998; González-Reyes and St Johnston 1998). In mael mutant ovary egg chambers, DE-cad did not appear to be enriched on the oocyte cortex or apically concentrated in follicular epithelia compared with the wild type (Supplemental Fig. S8), indicating that this abnormal localization of DE-cad may be responsible for the misplaced oocytes in mael egg chambers.
Figure 6.
Defective egg chamber development in the mael mutant. (A) Wild-type (top) and mael mutant (bottom) oocytes were visualized by immunohistochemical analyses using an anti-Orb antibody (green). DNA was stained with DAPI (blue). In the wild type, the oocyte is localized at the posterior region of the egg chamber. In contrast, in the mael mutant, egg chambers possessing a mislocalized oocyte (arrowheads) or two oocytes (arrows) were observed. (B) The percentage of abnormal egg chambers in the mael mutant. (C) The number of nurse cells and ring canals in the mael mutant egg chambers. Ring canals (red) and oocytes (green) were visualized using PY20 and anti-Orb antibody, respectively. DNA was stained with DAPI (blue). (D) The population of abnormal egg chambers in the mael mutants. NC, RC, and Oo indicate the number of nurse cells, ring canals, and oocytes, respectively. (Groups a–c) See the text. Bars, 20 μm.
We observed two types of egg chamber in mael mutants with abnormal numbers of germ cells. Egg chambers with >16 germ cells could be generated by either the fusion of two egg chambers or mitotic abnormality of the cystoblast. A normal developing egg chamber contains 16 germ cells with an oocyte and 15 ring canals in the egg chamber (Fig. 6C). If an egg chamber was fused with the anterior egg chamber due to defective follicle cell development, the fused egg chamber should have 32-cell cysts with 30 ring canals, which is twice the number of the wild type. It is also possible that an egg chamber with 32-cell cysts arises from an extra mitotic division, as reported for the tribbles gene mutation (Mata et al. 2000). In this case, however, the egg chamber should have 31 ring canals (Supplemental Fig. S9; Huynh 2006). To investigate a possible mechanism in mael mutant ovaries that leads to the production of egg chambers with >16 germ cells, we examined the number of ring canals in mael egg chambers. About 5.6% of egg chambers showed 30 nurse cells with 30 ring canals and two oocyte-like nuclei (30 + 2) (Fig. 6C [panel b], D [panel c]). Therefore, these mael mutant egg chambers were fused to each other. Approximately 1.1% of mael egg chambers showed <15 nurse cells and 15 ring canals with or without an oocyte, indicating that these abnormal egg chambers with reduced cell numbers are generated by the arrest of cystoblast mitotic divisions (Fig. 6C [panels c,d], D [panel a]). About 1.3% of egg chambers showed a compound phenotype that contains <30 nurse cells and two oocyte-like nuclei with two distinct populations of ring canals (Fig. 6C [panel e], D [panel b]). The compound phenotype was generated by fusion of egg chambers and defective cystocyte division. These phenotypes are similar to those observed in γTub mutant ovaries (Tavosanis and Gonzalez 2003). We thus conclude that mael is required for both germ and somatic follicle cell development, possibly by coordinating components of the MTOC, such as γTub, and thereby regulating proper MT organization.
Mutation in mael triggers Chk2-independent defects in MT polarity
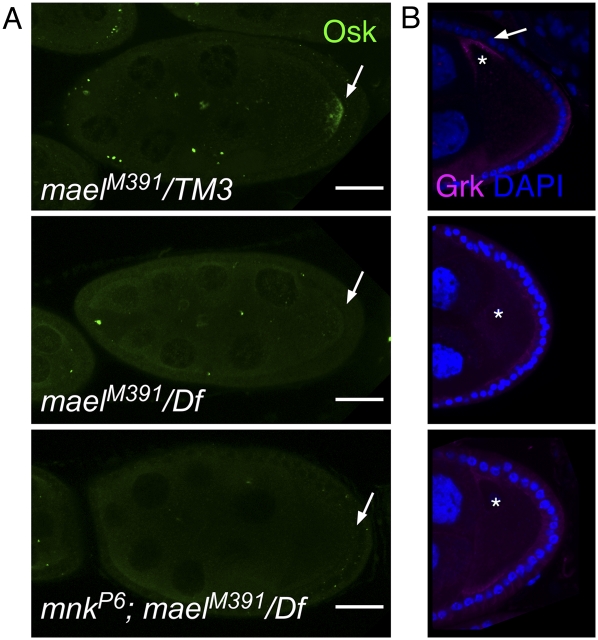
Most Drosophila piRNA pathway mutations lead to germline DNA damage and disrupt axis specification through activation of Chk2, which functions in DNA damage signaling (Cook et al. 2004; Chen et al. 2007; Pane et al. 2007). The axis specification defects of the oocyte that are linked to any of several piRNA pathway mutations are genetically suppressed by a loss of function mutation in mnk, which encodes a Chk2 homolog required for DNA damage signaling (Hari et al. 1995; Brodsky et al. 2004). Defects in MT organization in the mael oocyte may be secondary to DNA damage due to defects in the piRNA pathway. To test this, we generated double mutants with mnk and analyzed the localization of axis determinants Osk and Grk. Loss of mael function disrupted the posterior localization of Osk and the dorsal anterior localization of Grk in the oocyte, and the localization of both proteins was not restored in mnk;mael double mutants (Fig. 7). Thus, the axis specification defect of mael oocytes does not appear to be triggered by germline-specific DNA breaks and the activation of damage signaling through Chk2.
Figure 7.
mnk does not suppress the mael phenotype. (A) Mutations in mnk do not suppress the Osk localization defect in mael mutant ovaries. (Top) In the control oocyte (stage 9), Osk is localized at the posterior region of the oocyte (arrow). (Middle) In mael egg chambers, the localization of Osk was dispersed throughout the oocyte. (Bottom) In mnk;mael double mutant egg chambers, the mislocalization of Osk was not rescued. (B) In the control, Grk is localized at the dorsal anterior cortex (arrow) near the oocyte nucleus (asterisk). (Bottom) In mnk;mael double mutant egg chambers, the localizations of Osk and Grk are also dispersed throughout the oocyte. (Middle) In mael egg chambers, Grk was not accumulated. (Bottom) In mnk;mael double mutant egg chambers, the mislocalization of Grk was not rescued. DAPI (blue) marks the cell nuclei. Bars, 20 μm.
Discussion
In this study, we showed that Mael is an MTOC component and that dynamic organization of MTs does not occur in developing mael oocytes, which correlates with mislocalization of other MTOC components. We also observed that loss of mael affects the number and position of the oocytes in egg chambers and induces fusion of egg chambers. These results indicate that Mael specifically regulates MTOC formation, and thereby plays a key role in coordinating dynamic MT organization during Drosophila oogenesis.
Initial polarization of the oocyte during the oocyte specification phase in the germarium requires replacement of the fusome by a polarized MT network, which correlates with the formation of the MTOC (Lin et al. 1994; Lin and Spradling 1995; Grieder et al. 2000). Mael is concentrated in the centrosomal region and is colocalized with αTub and γTub during cyst cell divisions. γTub does not migrate to a developing oocyte in mael germariums, suggesting that Mael is required for the migration of centrioles from the cytoplasm of cysts to pro-oocytes in the germarium. Currently, the detailed mechanism by which Mael functions in MT organization is not clear. The simplest hypothesis is that Mael might serve as a platform that nucleates other MTOC components to form a functional MTOC. A previous report has shown that weaker mutant alleles of γTub affect the number of nurse cells and oocytes within the egg chamber (Tavosanis and Gonzalez 2003). These γTub mutant defects are very similar to those found in mael mutants in this study. γTub is involved in the nucleation of MTs and is present in the centrosomes and MTOCs in many different systems (Jeng and Stearns 1999; Moritz and Agard 2001). It was hypothesized that reduced activity of γTub could activate the oocyte determination program in one of the nurse cells by ectopically presenting MTOC material (Tavosanis and Gonzalez 2003). Our findings that γTub does not accumulate at centrosomes in the mael germarium and is ectopically expressed in the mael egg chamber suggest that Mael regulates localization of γTub at centrosomes through its complex formation and is thereby involved in properly organizing or positioning the MTOC.
PIWI proteins function in transposon silencing via association with piRNAs and maintain genome integrity during germline development. Recent studies have suggested that PIWI proteins in sea urchin (Seawi) and Xenopus (Xiwi) can interact with the MTs of the meiotic spindle (Rodriguez et al. 2005; Lau et al. 2009), while fly ovarioles with mutations in any of several piRNA pathway genes, including spn-E, aub, and armi, have disorganized MTs (Chen et al. 2007; Klattenhoff et al. 2007; Pane et al. 2007). This raises the possibility for either a functional role of PIWI proteins in the machinery that impacts on MT organization (in addition to transposon silencing), a role of the MT cytoskeleton in piRNA generation, or both. Our findings further corroborate a link between components of the piRNA pathway and proper MT organization. Although we found that Mael forms a complex with MTOC components, we were unable to identify components of the piRNA pathway in this complex. This is in contrast to observations of the mouse Mael homolog, which functions in the piRNA pathway (similar to fly Mael) and interacts with mouse PIWI proteins in the testes (Costa et al. 2006). Mouse Mael in the testes is almost exclusively cytoplasmic with accumulation at nuage (Soper et al. 2008). In contrast, fly Mael is located in both the nucleus and the cytoplasm in the ovary and is known to shuttle between them. Thus, one possibility is that in fly ovaries, there may exist nuclear Mael complexes involved in both piRNA generation and transposon silencing, which are distinct from the cytoplasmic complex containing MTOC components that we identified in this study.
Female flies with mutations in several genes in the piRNA pathway often lay eggs with axis patterning defects because of MT cytoskeletal changes that result in the mislocalization of bic, grk, and osk mRNAs within the egg chamber (Chen et al. 2007; Klattenhoff et al. 2007; Pane et al. 2007). These defects have been linked to the Chk2 DNA damage checkpoint that may be activated by increased retrotransposon transcript levels in mutants defective in piRNA biogenesis. However, because a mutation in mnk does not suppress the mislocalization of Osk and Grk in the mael oocyte, the axis specification defect of mael oocytes does not appear to be triggered by the activation of germline-specific DNA breaks and damage signaling through Chk2. In addition, a mutation in the mei-W68 locus, which encodes the Drosophila Spo11 homolog and induces meiotic double-strand breaks in chromosomes (McKim and Hayashi-Hagihara 1998), cannot suppress the axis specification defect of mael oocytes (Findley et al. 2003). Therefore, these results suggest that the axis specification defects of mael oocytes are not a secondary consequence of DNA damage signaling. However, it has been shown that in mael mutant ovaries, Vas is post-translationally modified (Findley et al. 2003), which is another hallmark of checkpoint activation (Ghabrial and Schupbach 1999). These results together imply that, acting not only through Chk2, the functions of Mael in MT organization are in parallel with its function in piRNA generation and transposon silencing. There are mutants—including zuc and spn-E, which are piRNA pathway genes—whose axis defects cannot be rescued by mnk mutations (Pane et al. 2007). Vas also appears modified in these mutants, although the relationship with activated checkpoint-modified Vas is unclear (Pane et al. 2007).
Given that Mael is a new component of the MTOC in the Drosophila ovary, identification of a domain within Mael that is responsible for binding to other MTOC components could aid in understanding how Mael nucleates and regulates MTOC formation. Because Mael contains an evolutionarily highly conserved domain of unknown function, termed the Mael domain (Zhang et al. 2008), determination of its crystal structure should prove valuable in elucidating mechanisms of both MTOC formation and piRNA generation processes.
Materials and methods
Drosophila strains
For ovary staining and molecular work, y w or maelM391/TM3 was used as a control. The mutant alleles and allelic combinations used in this work were maelM391/Df(3L)BSC554 (described as maelM391/Df) and cnnHK21. maelM391/TM3 Sb (Clegg et al. 1997; Findley et al. 2003) and mnkP6/CyO were kind gifts from T. Kai (National University of Singapore), and w1118; Df(3L)BSC554/TM6C, Sb1 and cn1 cnnHK21 bw1/CyO, l(2)DTS5131 were obtained from the Bloomington Drosophila Stock Center at Indiana University.
Production of an anti-Mael monoclonal antibody
An anti-Mael monoclonal antibody was raised specifically against the N terminus of the protein. A 200-amino-acid N-terminal fragment of Mael, fused with glutathione S-transferase (GST), was used as the antigen to immunize mice. The anti-Mael monoclonal antibody was produced essentially as described previously (Ishizuka et al. 2002) and was purified from culture supernatant of hybridoma cells under standard procedures using Thiophilic-Superflow resin (BD Biosciences).
Immunoprecipitation
Ovaries were dissected manually from adult flies in cold phosphate-buffered saline (PBS) and homogenized in 300 μL of Digitonin-based binding buffer containing 30 mM HEPES-KOH (pH7.3), 150 mM potassium acetate (KOAc), 4 mM magnesium acetate (MgOAc), 1 mM dithiothreitol (DTT), 50 ng/mL Digitonin, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 0.5% aprotinin. In Supplemental Figure S2E, immunoprecipitation was performed using Empigen-based binding buffer containing 1% Empigen BB (Sigma), 1 mM EDTA, 1 mM DTT, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 0.5% aprotinin in PBS. After centrifugation at 13,500 rpm for 1 min at 4°C, the supernatant was collected into a new microcentrifuge tube and kept on ice. The pellet was rehomogenized in 300 μL of binding buffer and recentrifuged as described above. The second aliquot of supernatant was combined with the first. These steps were repeated three times. Collected supernatant was used as the sample solution for immunoprecipitation. The protein concentration of the sample solution was adjusted each time to 5 mg/mL with binding buffer, and incubated with 5 μg of purified anti-Mael monoclonal antibody immobilized on Gamma-Bind beads (Amersham Bioscience) for 2 h at 4°C with rotation. After incubation, the bead fractions were extensively washed at least five times using binding buffer, and the protein complexes were eluted from the beads with sample buffer containing SDS by heating for 10 min at 70°C and then loaded onto SDS-PAGE gels. After electrophoresis, proteins were visualized by silver staining using Silver-Quest (Invitrogen) or processed for Western blot analyses. Protein bands detected by silver staining were excised for mass spectrometric analysis.
Western blot analysis
Whole embryo, larval, and pupal bodies were collected and homogenized in lysis buffer containing 30 mM HEPES-KOH (pH 7.3), 150 mM KOAc, 4 mM MgOAc, 1 mM DTT, 50 ng/mL digitonin, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 0.5% aprotinin. After centrifugation at 13,500 rpm for 10 min at 4°C, the supernatants were used as samples. Ovary lysates were prepared as described above. Western blot analysis was performed as described previously (Miyoshi et al. 2005). Culture supernatant of hybridoma cells producing anti-Mael monoclonal antibody was used without dilution as a primary antibody. Anti-Cnn was a kind gift from T.C. Kaufman (Indiana University) and used at 1:2000. Anti-D-TACC was a kind gift from J.W. Raff (University of Oxford) and used at 1:10,000. Anti-Msps was a kind gift from H. Ohkura (University of Edinburgh) and used at 1:10,000 dilution. Anti-γTub (GTU88; Sigma, #T6557) was used at 1:2000. Anti-Ago1, anti-Ago2, anti-Ago3, anti-Piwi, and anti-Aub antibodies have been described previously (Miyoshi et al. 2005; Gunawardane et al. 2007; Saito et al. 2006; Nishida et al. 2007). Anti-αTub was obtained from the Developmental Studies Hybridoma Bank and used at 1:1000.
Immunohistochemistry
Ovaries were dissected manually from adult flies in cold PBS. For MT detection, dissected ovaries were fixed immediately in cold methanol for 10 min at −20°C, rehydrated in PBS for 15 min, permeabilized with 0.5% NP-40 in PBS containing 0.1% Tween 20 and 2% bovine serum albumin (BSA) for 30 min, and then incubated overnight at 4°C with anti-αTub antibodies. For detection of αTub, anti-αTub antibody (rat YL1/2; Serotec) and FITC-conjugated anti-αTub (Sigma, #F2168) were used at a 1:150 dilution in Figures 2–4 and at a 1:50 dilution in Figure 6. Anti-Cnn and anti-γTub (Sigma, #T3559) antibodies were used at 1:500 and 1:100 dilutions, respectively. Anti-Orb, anti-Grk, and anti-DE-cad antibodies were obtained from the Developmental Studies Hybridoma Bank and used at 1:400, 1:40, and 1:20 dilutions, respectively. Anti-Osk and anti-Vas were a kind gift from A. Nakamura (RIKEN, CDB) and used at a 1:3000 and 1:1000 dilutions, respectively. PY20 (BD Transduction Laboratories) and anti-pH3 (Upstate Biotechnologies) antibodies were used at 1:100 and 1:1000 dilutions, respectively. Alexa Fluor488-conjugated anti-mouse IgG, Alexa Fluor488-conjugated anti-mouse IgG2a, Alexa Fluor546-conjugated anti-mouse IgG2b, Alexa Fluor633-conjugated anti-mouse IgG, Alexa Fluor546-conjugated anti-rabbit IgG, Alexa-Fluor488 conjugated anti-guinea pig IgG, and Alexa Fluor488-conjugated anti-rat IgG (Molecular Probes) were used as secondary antibodies. Ovaries were mounted in VectaShield with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Vector Laboratories). All images were collected using a Zeiss LSM510 laser-scanning microscope. Image processing and annotation were performed using Adobe Photoshop (Adobe) and ImageJ (NIH) software.
Cell culture and RNAi
mael cDNA was amplified by RT–PCR from Drosophila ovarian total RNA using the following primers: 5′-GGGGGATCCATGGCTCCTAAGAAGCATAGT-3′ and 5′-GGGCTCGAGTTATTTTTTAAGTTTCCCATC-3′. PCR products were digested with BamHI and XhoI, and then subcloned into pBlueScript SK. The entire inserted sequence was amplified by PCR using the resultant plasmid as a template with T7 (5′-TAATACGACTCACTATAGGG-3′) and T3 (5′-ATTAACCCTCACTAAAGGGAA-3′) promoter primers. cnn, cdc27 cDNAs were amplified by RT–PCR from Drosophila ovarian total RNA using the following specific primers: for cnn, 5′-TAATACGACTCACTATAGGGCAGATGGCGG-3′ and 5′-TAATACGACTCACTATAGGGCTTCTCC TCC-3′; for cdc27, 5′-TAATACGACTCACTATAGGGGCCGAGGACA-3′ and 5′-TAATACGACTCACTATAGGGCTTCATTGCA-3′. It should be noted that the 5′ end of each primer contained the T7 RNA polymerase promoter sequence (5′-TAATACGACTCACTATAG-3′). PCR products were purified using the QiaQuick gel extraction kit (Qiagen) according to the manufacturer's instructions. Purified PCR products were used to produce dsRNAs using Megascript T7 and T3 transcription kits (Ambion). The resultant RNAs were purified according to the manufacturer's instruction, heated for 5 min at 95°C, and then left to cool to room temperature. S2 cells were grown at 26°C in Schneider's Insect medium (Sigma) supplemented with 10% fetal calf serum (Gibco), 50 μg/mL streptomycin, and 50 μg/mL penicillin. For RNAi in S2 cells, ∼5 × 106 cells were suspended with 5 μg of dsRNA and incubated for 4 d at 26°C.
Mitotic index
To determine the mitotic index, S2 cells were processed for immunofluorescence microscopy as described above. The cells were labeled with anti-pH3, as a marker of mitotic cells and with DAPI to visualize DNA (Goshima et al. 2007). The percentage of mitotic cells was determined using the image processing software ImageJ.
Live-cell imaging
Aequorea coerulescens GFP (AcGFP) cDNA was amplified by PCR from pAcGFP1 vector (Clontech) using the primers 5′-TTTTTCCATGGTGAGCAAGGGCGCCG-3′ and 5′-GATACATTCACGTGCTAGAATTCCTGCCTTGTACAGC-3′. αTubulin84B (αTub) cDNA was amplified by RT–PCR from Drosophila ovarian total RNA using the primers 5′-GCTGTACAAGGCAGGAATTCTAGCACGTGAATGTATC-3′ and 5′-TTTTTGCTAGCTTAGTACTCCTCAGCGCCCT-3′. A cDNA encoding an AcGFP-αTub fusion protein was then amplified by an “overlap PCR” method using AcGFP and αTub PCR products with primers 5′-TTTTTCCATGGTGAGCAAGGGCGCCG-3′ and 5′-TTTTTGCTAGCTTAGTACTCCTCAGCGCCCT-3′. The PCR product was digested with NcoI and NheI, and then subcloned into a vector downstream from the metallothionein promoter. The AcGFP-αTub-expressing plasmid vector was transfected into S2 cells using Cellfectin (Invitrogen) according to the manufacturer's instructions. The AcGFP-αTub fusion protein was induced by adding CuSO4 to the culture medium (final concentration ∼0.5 mM). For time-lapse video fluorescence microscopy imaging, AcGFP-αTub trnsfected S2 cells were plated in poly-L-lysine-coated glass-bottom culture dishes (Matsunami) on day 4 after RNAi treatment. All mages were collected in 35- to 55-sec intervals using a Zeiss LSM710 laser-scanning microscope. Image processing and annotation were performed using ZEN software (Carl Zeiss).
Acknowledgments
We are grateful to G. Goshima, T. Kai, T. Kaufman, H. Kose, A. Nakamura, H. Ohkura, and J.W. Raff for providing antibodies, fly stocks, protocols, and advice. Special thanks to I. Sagawa for mass spectrometry. We also thank members of the Siomi laboratory for discussions and helpful comments on the manuscript. This work was supported by MEXT (Ministry of Education, Culture, Sports, Science, and Technology of Japan) grants to H.S., M.C.S., M.K.N., and K.S. M.C.S. is supported by CREST (Core Research for Evolutional Science and Technology) from JST (Japan Science and Technology Agency).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.174110.111.
References
- Adams RR, Maiato H, Earnshaw WC, Carmena M 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol 153: 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Arn EA, Cha BJ, Theurkauf WE, Macdonald PM 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell 4: 41–51 [DOI] [PubMed] [Google Scholar]
- Bolívar J, Huynh JR, López-Schier H, González C, St Johnston D, González-Reyes A 2001. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development 128: 1889–1897 [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24: 1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasan R, Anderson K 1993. Maternal control of dorso–ventral polarity and pattern in the embryo. In The development of Drosophila melanogaster (ed. Bate M. and Martínez-Arias A.), pp. 387–424 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Chen Y, Pane A, Schüpbach T 2007. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I, Giniger E, Ruohola-Baker H, Jan L, Jan Y 1994. Transient posterior localisation of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr Biol 4: 289–300 [DOI] [PubMed] [Google Scholar]
- Clark I, Jan LY, Jan YN 1997. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124: 461–470 [DOI] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H 1997. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: Posterior localization of grk mRNA. Development 124: 4661–4671 [DOI] [PubMed] [Google Scholar]
- Clegg NJ, Findley SD, Mahowald AP, Ruohola-Baker H 2001. Maelstrom is required to position the MTOC in stage 2–6 Drosophila oocytes. Dev Genes Evol 211: 44–48 [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Cooley L, Theurkauf WE 1994. Cytoskeletal functions during Drosophila oogenesis. Science 266: 590–596 [DOI] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ 2006. Mouse MAELSTROM: The link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet 15: 2324–2334 [DOI] [PubMed] [Google Scholar]
- Cullen CF, Ohkura H 2001. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol 3: 637–642 [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deák P, Glover DM, Ohkura H 1999. Mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol 146: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50 [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871 [DOI] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW 2000. D-TACC: A novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J 19: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A, Schupbach T 1999. Activation of a meiotic checkpointregulates translation of Gurken during Drosophila oogenesis. Nat Cell Biol 1: 354–357 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA 1995. homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev 9: 2495–2508 [DOI] [PubMed] [Google Scholar]
- Godt D, Tepass U 1998. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature 395: 387–391 [DOI] [PubMed] [Google Scholar]
- González-Reyes A, St Johnston D 1998. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development 125: 3635–3644 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St Johnston D 1997. Oocyte determination and the origin of polarity in Drosophila: The role of the spindle genes. Development 124: 4927–4937 [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder N, De Cuevas M, Spradling A 2000. The fusome organizes the microtubules network during oocyte differentiation in Drosophila. Development 127: 4253–4264 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821 [DOI] [PubMed] [Google Scholar]
- Huynh JR 2006. Fusome as a cell–cell communication channel of Drosophila ovarian cyst. In Cell–cell channels (ed. Baluska F, et al. ), pp. 217–235 Landes Bioscience-Springer Verlag, New York [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A 2006. The centrosome–nucleus complex and microtubule organization in the Drosophila oocyte. Development 133: 129–139 [DOI] [PubMed] [Google Scholar]
- Jeng R, Stearns T 1999. γ-Tubulin complexes: Size does matter. Trends Cell Biol 9: 339–342 [DOI] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA 2005. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55 [DOI] [PubMed] [Google Scholar]
- Knowles BA, Cooley L 1994. The specialized cytoskeleton of the Drosophila egg chamber. Trends Genet 10: 235–241 [DOI] [PubMed] [Google Scholar]
- Kwon M, Scholey JM 2004. Spindle mechanics and dynamics during mitosis in Drosophila. Trends Cell Biol 14: 194–205 [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M 1988. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335: 611–617 [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD 2009. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J 28: 2945–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol 3: 643–649 [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwic MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling A 1995. Fusome asymmetry and oocyte determination in Drosophila. Dev Genet 16: 6–12 [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling A 1994. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120: 947–956 [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L 1994. Intercellular cytoplasm transport during Drosophila oogenesis. Dev Biol 165: 336–351 [DOI] [PubMed] [Google Scholar]
- Mahowald AP, Strassheim JM 1970. Intercellular migration of centrioles in the germarium of Drosophila melanogaster. An electron microscopic study. J Cell Biol 45: 306–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rørth P 2000. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101: 511–522 [DOI] [PubMed] [Google Scholar]
- McKim KS, Hayashi-Hagihara A 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: Evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev 12: 2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC 1999. The Centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126: 2829–2839 [DOI] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC 2001. Zygotic development without functional mitotic centrosomes. Curr Biol 11: 116–120 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Agard DA 2001. γ-Tubulin complexes and microtubule nucleation. Curr Opin Struct Biol 11: 174–181 [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T 1993. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF α-like protein. Cell 75: 165–174 [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T 1996. The Drosophila TGFα-like protein Gurken: Expression and cellular localization during Drosophila oogenesis. Mech Dev 59: 105–113 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Choi YD, Dreyfuss G 1990. Immunological methods for the purification and characterization of hnRNP particles. Methods Enzymol 181: 317–325 [DOI] [PubMed] [Google Scholar]
- Pokrywka N, Stephenson E 1995. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol 167: 363–370 [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P 1998. Mechanisms of nuclear positioning. J Cell Sci 111: 2283–2295 [DOI] [PubMed] [Google Scholar]
- Riechmann V, Ephrussi A 2001. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev 11: 374–383 [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, DI Carlo M, Suprenant KA, Bonder EM 2005. Seawi—a sea urchin piwi/argonaute family member is a component of MT–RNP complexes. RNA 11: 646–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, Bohmann K, Nusslein-Volhard C 2000. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol 2: 185–190 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R 2009. Temperature entrainment of Drosophila's circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron 64: 251–266 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic C, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A 1993. Developmental genetics of oogenesis. In The development of Drosophila melanogaster (ed. Bate M and Martinez-Arias A), pp. 1–70 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Steinhauer J, Kalderon D 2006. Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn 235: 1455–1468 [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578 [DOI] [PubMed] [Google Scholar]
- Tavosanis G, Gonzalez C 2003. γ-Tubulin function during female germ-cell development and oogenesis in Drosophila. Proc Natl Acad Sci 100: 10263–10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tearle R, Nüsslein-Volhard C 1987. Tübingen mutants stocklist. Drosoph Inf Serv 66: 209–226 [Google Scholar]
- Terada Y, Uetake Y, Kuriyama R 2003. Interaction of aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 162: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE 1994. Microtubules and cytoplasm organization during Drosophila oogenesis. Dev Biol 165: 352–360 [DOI] [PubMed] [Google Scholar]
- Theurkauf W, Smiley S, Wong M, Alberts B 1992. Reorganization of the cytoskeleton during Drosophila oogenesis: Implications for axis specification and intercellular transport. Development 115: 923–936 [DOI] [PubMed] [Google Scholar]
- Theurkauf W, Alberts M, Jan Y, Jongens T 1993. A central role for microtubules in the differentiation of Drosophila oocytes. Development 118: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A 1998. Oocyte polarity depends on regulation of gurken by Vasa. Development 125: 1723–1732 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- van Eeden F, St Johnston D 1999. The polarisation of the anterior–posterior and dorsal–ventral axes during Drosophila oogenesis. Curr Opin Genet Dev 9: 396–404 [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y 1999. γ-Tubulin complexes and their interaction with microtubule-organizing centers. Curr Opin Struct Biol 9: 250–259 [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM 1996. aubergine enhances oskar translation in the Drosophila ovary. Development 122: 1631–1639 [DOI] [PubMed] [Google Scholar]
- Yoshikane N, Nakamura N, Ueda R, Ueno N, Yamanaka S, Nakamura M 2007. Drosophila NAT1, a homolog of the vertebrate translational regulator NAT1/DAP5/p97, is required for embryonic germband extension and metamorphosis. Dev Growth Differ 49: 623–634 [DOI] [PubMed] [Google Scholar]
- Zhang D, Xiong H, Shan J, Xia X, Trudeau VL 2008. Functional insight into Maelstrom in the germline piRNA pathway: A unique domain homologous to the DnaQ-H 3′–5′ exonuclease, its lineage-specific expansion/loss and evolutionarily active site switch. Biol Direct 3: 48 doi: 10.1186/1745-6150-3-48 [DOI] [PMC free article] [PubMed] [Google Scholar]