The cellular heat-shock response involves multiple changes in gene regulation. Using single-base-pair mapping of nucleosomal and subnucleosomal particles in chromatin extracts, Teves and Henikoff explore alterations in the chromatin landscape upon heat shock. They find loss of RNA Pol II and reduced nucleosome turnover in the bodies of genes, providing insight into the interplay between nucleosome dynamics and transcriptional changes.
Keywords: heat shock, epigenome profiling, chromatin
Abstract
Heat shock rapidly induces expression of a subset of genes while globally repressing transcription, making it an attractive system to study alterations in the chromatin landscape that accompany changes in gene regulation. We characterized these changes in Drosophila cells by profiling classical low-salt-soluble chromatin, RNA polymerase II (Pol II), and nucleosome turnover dynamics at single-base-pair resolution. With heat shock, low-salt-soluble chromatin and stalled Pol II levels were found to decrease within gene bodies, but no overall changes were detected at transcriptional start sites. Strikingly, nucleosome turnover decreased genome-wide within gene bodies upon heat shock in a pattern similar to that observed with inhibition of Pol II elongation, especially at genes involved in the heat-shock response. Relatively high levels of nucleosome turnover were also observed throughout the bodies of genes with paused Pol II. These observations suggest that down-regulation of transcription during heat shock involves reduced nucleosome mobility and that this process has evolved to promote heat-shock gene regulation. Our ability to precisely map both nucleosomal and subnucleosomal particles directly from low-salt-soluble chromatin extracts to assay changes in the chromatin landscape provides a simple general strategy for epigenome characterization.
The heat-shock response is a universally conserved reaction to environmental stress that involves rapid transcriptional changes. Inactive monomers of the master heat-shock transcription factor HSF trimerize upon heat shock and translocate to the nucleus, where they bind to the promoters of heat-shock protein (Hsp) genes (Akerfelt et al. 2010). HSF binding triggers the release of paused polymerases already engaged at promoters of Hsp genes, resulting in fast and synchronous activation of HSF targets (O'Brien and Lis 1991; Lee et al. 2008). Gene induction varies according to conditions, but this rapid transcriptional response generates 10-fold to 1000-fold induction of Hsp genes within minutes of temperature elevation (Lindquist 1986). Simultaneously, heat shock results in down-regulation of normal transcription (Jamrich et al. 1977), presumably to prevent accumulation of misfolded translation products (Lindquist 1986). These rapid genome-wide transcriptional responses make the heat-shock system ideal for investigating chromatin alterations that accompany gene regulatory changes.
The basic repeating unit of chromatin is the nucleosome, which consists of an octameric histone protein core that wraps 147 base pairs (bp) of DNA (Luger et al. 1997). Packaging of DNA into nucleosomes can occlude DNA sequences and prevent transcription factor binding, in which case disruption or mobilization of nucleosomes is necessary for gene activation and other processes that require access to regulatory sequences. For example, nucleosomes in the bodies of induced Hsp70 genes are rapidly lost within seconds of heat shock, and this loss depends on HSF, poly(ADP-)ribose polymerase 1 (PARP1), and GAGA factor (GAF) (Petesch and Lis 2008). Furthermore, loss of nucleosomes is required for full activation of Hsp genes (Petesch and Lis 2008). Heat-shock studies in yeast have revealed a functional interplay between multiple nucleosome remodeling complexes in regulating Hsp genes (Shivaswamy and Iyer 2008; Erkina et al. 2010), further implicating nucleosome dynamics in gene induction. Despite the wealth of information on Hsp gene induction, much less is known about the mechanisms for genome-wide down-regulation of transcription during heat shock. Fluorescence analyses of GFP-tagged RNA polymerase II (Pol II) show that Pol II becomes released from DNA upon heat shock, suggesting that direct regulation of Pol II kinetics plays a role in this process (Hieda et al. 2005). In support of this possibility, SINE RNAs that are up-regulated during heat shock (Allen et al. 2004; Mariner et al. 2008) have been shown to disrupt contacts between promoter DNA and Pol II (Yakovchuk et al. 2009). However, some evidence exists for a role of nucleosomes in this process as well. In one mammalian study, histone deacetylases HDAC1 and HDAC2 were shown to mediate global histone deacetylation during heat shock (Fritah et al. 2009), indicating that changes in chromatin “states” correlate with global down-regulation of transcription. Furthermore, repositioning of individual nucleosomes was seen throughout the budding yeast genome in response to heat shock (Shivaswamy et al. 2008). Although these studies revealed effects of gene regulatory changes on chromatin, the mechanistic processes responsible for these changes remain unknown.
To gain insights into the mechanistic role of nucleosomes in gene regulation, we used the heat-shock response as a model system to effect global chromatin changes that occur with transcriptional alterations. Specifically, we asked the following questions: (1) How does the chromatin landscape change with alterations in gene expression? (2) What is the interplay between nucleosome dynamics and changes in transcription? (3) Can we gain insight into the mechanisms that govern global gene repression during heat shock and the possible roles that chromatin plays in this process? To address these questions, we introduced a simple strategy for epigenome characterization based on traditional micrococcal nuclease (MNase) mapping, chromatin salt fractionation, and metabolic labeling of histones to probe the entire low-salt-soluble chromatin landscape and its dynamics at single-base-pair resolution. We show that the low-salt-soluble chromatin fraction is enriched for distinct subnucleosome-sized chromatin particles relative to total MNase-digested chromatin. During heat shock, the landscapes of both nucleosomal and subnucleosomal chromatin components dramatically change at induced genes. In contrast, the genome-wide distribution of subnucleosomal particles at transcription start sites (TSSs) is maintained during heat shock despite global down-regulation of transcription. Furthermore, genome-wide reduction in stalled Pol II occurs concomitantly with a decrease in nucleosome solubility and turnover within bodies of active genes. Using a Pol II elongation inhibitor, we show that the changes in nucleosome turnover that occur during heat shock resemble the turnover changes caused by direct inhibition of transcription elongation. We conclude that heat shock causes reduced Pol II elongation and decreased nucleosome turnover genome-wide.
Results
Low-salt-soluble chromatin is enriched for distinct types of subnucleosomal particles
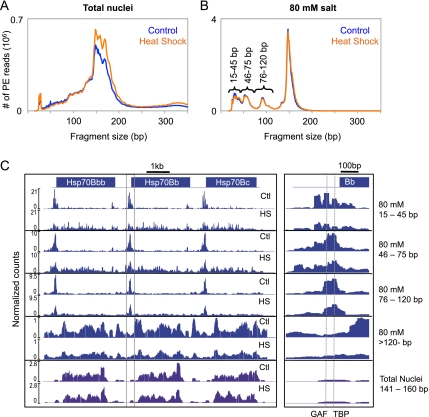
Using Drosophila S2 cells, we digested intact nuclei with MNase to obtain mostly mononucleosomes and extracted the low-salt (80 mM)-soluble fraction, sometimes referred to as classical “active” chromatin (Sanders 1978). We then captured all MNase-protected fragments from both the total MNase-digested nuclei and the 80 mM fraction by using paired-end Solexa sequencing without prior size selection, except for removal of unligated adapters with Ampure magnetic beads (Henikoff et al. 2011). The size distribution of paired-end reads from both total nuclei and the 80 mM fraction reveals that fragments as small as ∼20 bp could be sequenced and mapped to the genome (Fig. 1A,B). Although there is a major peak at 147 bp in the total nuclei fraction representing octameric nucleosomes, peaks of larger fragment sizes are also prominent, likely due to histone H1-containing chromatosomes (Simpson 1978). Interestingly, the size distribution of fragments from the 80 mM salt fraction not only shows a sharp peak for 147-bp nucleosomes, which suggests exclusion of chromatosomes from low-salt-soluble chromatin, but also shows distinct peaks for subnucleosomal components not seen in the distribution of fragments from total nuclei. The peaks suggest distinct types of protection of DNA from MNase by particles that are enriched in 80 mM salt-soluble chromatin.
Figure 1.
Low-salt-soluble chromatin is enriched for distinct subnucleosomal particles. Paired-end read length distribution of MNase-digested total nuclei (A) and the 80 mM salt-extracted fraction (B) before (blue) and after (orange) heat shock. (C) Paired-end reads from the different size classes of the 80 mM salt fraction (blue) and the nucleosome-sized fragments of the total nuclei (purple) were mapped onto the Drosophila melanogaster genome. The mapped reads were converted to normalized counts for each base pair at the Hsp70 gene cluster, and landscapes were displayed with NimbleGen SignalMap. A close-up view of the promoter region of Hsp70Bb that is boxed in C is shown in the right panel, with the dotted lines representing the binding sites for GAF and TBP.
To investigate the distinct size classes in the 80 mM fraction, we parsed the mapped paired-end reads into four length categories based on the peaks in the read size distribution: 15–45, 46–75, 76–120, and >120 bp. For the total nuclei fraction, we used mapped paired-end reads that are 141–200 bp in length to define nucleosomes. We then converted the mapped reads to density counts at single-base-pair resolution and focused on the three tandem copies of the Hsp70 gene at the 87C heat-shock locus (Fig. 1C). In both the total nuclei and the 80 mM fraction, mapped nucleosomes showed a rather complex distribution. With the exception of a few well-positioned nucleosomes, nucleosomes in gene bodies displayed heterogeneous positioning and occupancy profiles. Under normal growth conditions, the subnucleosomal particle DNA fragments (15–45, 46–75, and 76–120 bp) displayed distributions that are very different from those of nucleosomes. At this locus, the particles were found primarily at the 5′ TSSs in regions that are generally depleted of nucleosomes. A closer view of the promoter region revealed several peaks in the 15- to 45-bp class, with two of these peaks corresponding to sites of binding by GAF and TATA-binding protein (TBP) (Fig. 1C, right panel), indicating that we can map distinct small DNA-binding proteins. Subnucleosomal particles also localized to regions with no known gene annotations, suggesting that these particles map to potential regulatory regions in the genome (Supplemental Fig. S1). Furthermore, the square shape of the peaks of these smaller fragments indicates complete protection and tight positioning of the DNA-binding proteins, in contrast to the rounded peaks of the more mobile nucleosomes. This provides evidence for DNA protection from MNase cleavage by small DNA-binding proteins under native conditions.
Changes in the Hsp70 chromatin landscape during heat shock
Previous studies have shown that nucleosomes within the Hsp70 genes are lost within seconds of heat shock (Petesch and Lis 2008), but we wondered how other components of the chromatin landscape change during induction. Therefore, we harvested cells after 15 min at 37°C and followed the chromatin extraction and Solexa library procedures described above. The size distributions of MNase-protected fragments were highly similar between the control and heat-shocked samples for both the total nuclei and the 80 mM salt fraction (Fig. 1A,B). We converted the mapped reads for each size class into density counts and compared heat-shock with control landscapes over the 87C Hsp70 locus (Fig. 1C). Whereas total nuclei showed only slight changes in nucleosome occupancy with heat shock, the 80 mM salt fraction showed a marked decrease. Interestingly, the prominent peaks of shorter fragments at the TSS were diminished with heat-shock induction, whereas occupancy of shorter fragments within gene bodies dramatically increased, which suggests that these shorter fragments derive from the transcriptional machinery itself as it progresses through the Hsp70 gene.
Distinct genome-wide distributions of low-salt-soluble chromatin particles
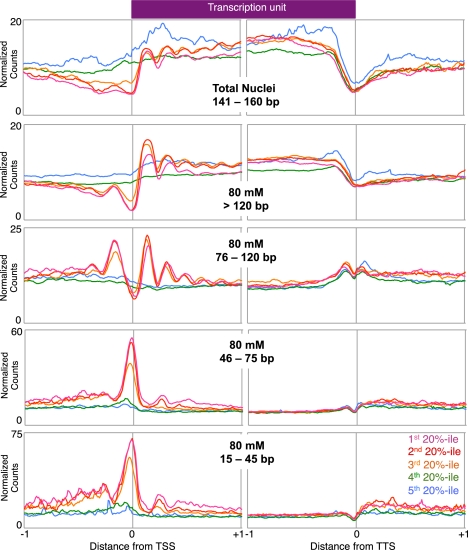
To examine the genome-wide distribution of each size class, we ordered all genes by expression level and averaged the normalized counts for each expression quintile in the 2-kb regions surrounding both the TSS and the transcription termination site (TTS). We also performed the same analysis on the nucleosome-sized fragments from total nuclei to examine global nucleosome occupancy. The genome-wide distribution of nucleosomes from total nuclei agrees with previous studies that show nucleosome depletion at the TSS, positioning of the +1 nucleosome, and a gradual decrease in positioning further downstream for expressed genes (Fig. 2; Mavrich et al. 2008; Weber et al. 2010). In the 80 mM salt-soluble chromatin fraction, both the 76- to 120-bp and >120-bp fragment size classes exhibit similar strongly phased profiles for the three quintiles of expressed genes, in contrast to the flat profiles that characterize the two inactive gene quintiles (Henikoff et al. 2009). Interestingly, the average profile of the 76- to 120-bp fragments resembles the periodicity of phased nucleosomes (>120 bp) but with narrower peaks, consistent with a shorter fragment size, as if derived from partially unwrapped nucleosomes (Li et al. 2005; Weber et al. 2010). Furthermore, the genome-wide profile of the 76- to 120-bp class shows a higher peak at the −1 nucleosome position relative to the >120-bp class, which implies that nucleosomes around active promoters might have a higher tendency to be partially unwrapped than their neighbors.
Figure 2.
Distinct genome-wide distributions of active chromatin particles. Each mapped size class of the 80 mM salt fraction was scaled to normalized counts per 10-bp window. Within each size class, all genes were grouped into quintiles by expression level, and the average normalized counts per 10-bp window were determined for each quintile in the 2-kb region immediately surrounding the TSS and TTS. In the total nuclei fraction, only the 141- to 160-bp fragments were used in order to represent the nucleosomal profile.
We also observed that small fragments (15–45 and 46–75 bp) are enriched at TSSs genome-wide, similar to Hsp70 genes. These subnucleosomal particles are found almost exclusively at expressed genes, which suggests that the proteins that protect TSSs from MNase in low-salt-soluble chromatin are involved in transcription initiation. Using previously determined genomic sites for various DNA-binding proteins, we found that these size classes are enriched for GAF, EZ+PSC, and Zeste sites relative to flanking regions (Supplemental Fig. S2), suggesting that these small fragments result from MNase protection of general transcription factors such as GAF and protein complexes that generally occupy EZ+PSC and Zeste sites.
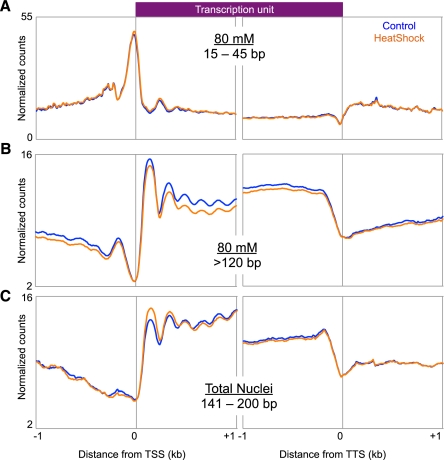
We next asked how heat shock affects the genome-wide distribution of the subnucleosomal particles. To do this, we compared the average gene profiles of the 15- to 45-bp size class in heat-shocked and control cells (Fig. 3A). The plots are superimposable, which indicates that subnucleosomal particles are undisturbed by heat shock. Dividing genes into quintiles based on normal expression level also revealed no differences in the subnucleosomal particle distribution before and after heat shock (Supplemental Fig. S3A). This suggests that heat-shock-mediated down-regulation of expression does not occur at the level of recruitment and initiation, in contrast to heat-shock induction at Hsp70 genes, which is accompanied by major changes in subnucleosomal particle occupancy at the TSS.
Figure 3.
Genome-wide effects of heat shock on distinct chromatin components. Ends analysis of all genes for the 15- to 45-bp size class of the 80 mM fraction (A), all expressed genes for the >120-bp size class of the 80 mM fraction (B), and all expressed genes for the 141- to 200-bp class of the total nuclei fraction (C).
We also examined changes in the nucleosomal component of low-salt-soluble chromatin. We aligned TSSs of normally expressed genes for the nucleosome-sized classes of the 80 mM and total nuclei fractions of heat-shocked and control samples (Fig. 3B,C). We observed a genome-wide reduction in nucleosome solubility in active genes after heat shock, whereas silent genes showed little to no change (Supplemental Fig. S3B). These observations imply that heat shock results in a decrease in nucleosome salt solubility within active gene bodies.
Stalled Pol II decreases globally during heat shock
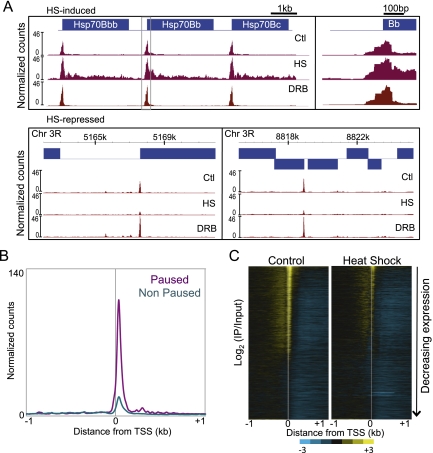
Transcriptional changes during heat shock can be detected cytologically in Drosophila polytene salivary gland chromosomes within minutes (Belyaeva and Zhimulev 1976; Bonner and Pardue 1976; Ashburner and Bonner 1979); however, standard expression profiling is relatively insensitive to rapid transcriptional changes because newly synthesized mRNA is diluted by steady-state mRNA. A 15-min heat shock represents only 1/80th of the 20-h S2 cell cycle, which likely accounts for our inability to detect significant expression changes after heat shock by microarray-based profiling, except for heat-shock genes (data not shown). A more direct genome-wide approach to observing transcriptional levels is to map Pol II bound to DNA. We showed previously that low-salt-soluble chromatin includes ∼50-bp particles that are enriched 30 bp downstream from the TSS in genes that had been found to be regulated at the level of Pol II elongation (paused genes), in contrast to genes matched for expression but lacking paused Pol II (nonpaused genes) (Weber et al. 2010). To confirm that these particles represent paused Pol II itself, we performed native chromatin immunoprecipitation (ChIP) with an antibody against the Pol II C-terminal domain (CTD), using 80 mM salt-extracted chromatin as input (low-salt-soluble Pol II). DNA was isolated and subjected to paired-end sequencing. When cells were grown under normal conditions, low-salt-soluble Pol II was enriched immediately downstream from TSSs of Hsp70 genes, but nowhere else in the gene bodies (Fig. 4A), consistent with previous studies identifying Hsp genes as containing paused Pol II (O'Brien and Lis 1991). We then averaged the normalized counts for all paused and nonpaused genes (Muse et al. 2007) in a region of 2 kb surrounding the TSS and found that Pol II was indeed strongly enriched at promoters of paused genes (Fig. 4B). We also used 5,6-dichlorobenzimidazole (DRB) to induce Pol II stalling followed by ChIP-seq and found a 40% increase in low-salt-soluble Pol II at the 87C Hsp70 locus (Fig. 4A), further confirming that the soluble chromatin fraction is enriched for paused Pol II at Hsp70 promoters.
Figure 4.
Effects of heat shock on low-salt-soluble Pol II. (A) The mapped reads of the Pol II ChIP for control, heat-shocked, and DRB-treated samples were converted to normalized counts for each base pair at the Hsp70 gene cluster, and landscapes were displayed with NimbleGen SignalMap. A close-up view of the boxed promoter region of Hsp70Bb is shown in the right panel. Landscapes for heat-shock-repressed genes are also shown for two regions in chromosome 3R. (B) Average normalized counts for all paused (purple) and nonpaused (cyan) genes in a 2-kb region surrounding the TSS. (C) Heat maps generated by Java TreeView of the log2 of Pol II ChIP to 80 mM input ratio in the 2-kb region surrounding the TSS for control (left) and heat-shocked (right) samples.
To visualize the genome-wide distribution of our low-salt-soluble Pol II ChIP, we arranged the genes by decreasing level of expression and displayed them as a heat map of the 2-kb region surrounding the TSS (Fig. 4C). This showed a striking alignment of low-salt-soluble Pol II just downstream from the TSS that strongly correlated with expression level, suggesting that we captured the generally “stalled” Pol II that includes paused, backtracked, and arrested Pol II species (Levine 2011). The large fraction of genes characterized by stalled Pol II in the soluble chromatin fraction is consistent with previous studies of paused genes (Muse et al. 2007; Core et al. 2008; Gilchrist et al. 2008, 2010; Min et al. 2011) and supports the proposal that this fraction is enriched in paused Pol II (Weber et al. 2010).
To explore how the stalled Pol II changes genome-wide during heat shock, we performed Pol II ChIP and paired-end sequencing as described above using as input the 80 mM salt fraction extracted from cells heat-shocked for 15 min at 37°C. We found that during heat shock, stalled Pol II is present as irregular closely spaced peaks throughout the bodies of Hsp70 genes (Fig. 4A). Most of these stalled Pol II ChIP peaks lined up with the 15- to 45-bp fragment peaks seen in the heat-shocked 80 mM fraction used as input, confirming that a major component of this subnucleosomal chromatin is Pol II itself. This result also implies that Pol II stalls at multiple sites within bodies of the highly induced Hsp70 gene, reminiscent of a recent report in which mapping of nascent transcripts in yeast showed that stalling throughout gene bodies is a common feature of highly expressed yeast genes (Churchman and Weissman 2011).
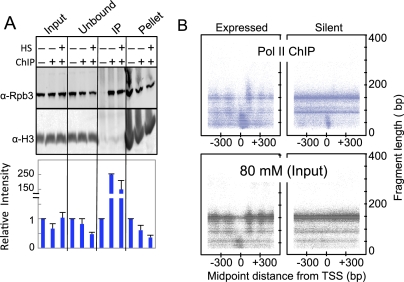
Heat shock causes loss of Pol II from constitutively active puffs in Drosophila salivary glands (Jamrich et al. 1977). When we focused on non-HSF target genes containing prominent stalled Pol II peaks under normal conditions, we found that these peaks were greatly decreased upon heat shock but were maintained during DRB Pol II inhibition (Fig. 4A). Therefore, we wondered whether the stalled Pol II that we mapped genome-wide is released from constitutively active genes in S2 cells. Indeed, a heat map display revealed genome-wide decreases in stalled Pol II during heat shock (Fig. 4C). To confirm that this decrease resulted from Pol II release rather than reduced solubility of Pol II-bound DNA, we performed quantitative Western blot analysis using a Pol II antibody for the input, unbound, ChIP, and pellet fractions of samples before and after heat shock (Fig. 5A). We detected a 50% reduction in insoluble Pol II during heat shock, which indicates that stalled Pol II became released from the DNA. However, we also detected a decrease in the unbound fraction without significant increase in global Pol II levels in the input fraction. Because the starting material in these experiments is nuclei, it is possible that Pol II that is unaccounted for has shuttled into the cytoplasm or degraded upon heat shock. Nevertheless, this result suggests that reduced Pol II affinity is a mechanism for heat-shock-mediated down-regulation of expression.
Figure 5.
Properties of the stalled Pol II. (A) Quantitative Western blot from a representative Pol II ChIP experiment. Pol II was probed using an antibody raised against the Rpb3 subunit. Histone H3 was used as a loading control. For both Pol II and H3, bands were quantified using ImageJ software. The ratio of Rpb3 to H3 for each lane was determined, and the values for each fraction (Input, Unbound, IP, and Pellet) were divided by the value for the ChIP no-antibody control. (B) For all paired-end reads, the distance between the midpoint of the fragment and the TSS was determined and compared with the length of the fragment. All genes were separated into two groups (expressed and silent), and a random sample of 100,000 data points from each was used in the heat maps.
A large proportion of Pol II ChIP-mapped reads were nucleosomal in size (Supplemental Fig. S4). We sought to determine whether these nucleosomes represent specific associations with stalled Pol II or nonspecific background in the ChIP. For both the control Pol II ChIP and the corresponding input 80 mM fraction, we measured the distance between the midpoint of each fragment and the TSS and plotted this distance versus the length of each fragment in a 1-kb region surrounding the TSS for expressed and silent genes separately (Fig. 5B). In the 80 mM salt fraction (the input for Pol II ChIP), the profiles of all size classes became evident in the expressed genes, including the phased ∼150-bp nucleosomes, the phased ∼90-bp fragments from internal cleavage of nucleosomes, and the <60-bp fragments predominating at the TSS (Fig. 5B, bottom left). Inactive genes of the 80 mM fraction showed no phasing of nucleosomes and a reduced level of subnucleosomal particles at TSSs (Fig. 5B, bottom right). In comparison, the Pol II ChIP midpoint-versus-length plot for expressed genes showed an enrichment of small fragments especially downstream from the TSS (Fig. 5B, top left). Interestingly, we also observed larger (∼200- to 250-bp) fragments centered near the +1 nucleosome, which would represent fragments spanning both stalled Pol II and the +1 nucleosome in the Pol II ChIP but not detectable in the input material (Fig. 5B, left panels). In inactive genes, the ChIP midpoint-versus-length plot showed an enrichment of stalled Pol II at the TSS of some genes, whereas the remaining fragments displayed similar profiles as with the 80 mM input (Fig. 5B, top right). Taken together, these observations suggest a physical interaction between stalled Pol II and the +1 nucleosome.
Inhibition of Pol II elongation decreases nucleosome turnover within gene bodies
Transcription levels are correlated with levels of nucleosome turnover within gene bodies in Drosophila (Deal et al. 2010), but a causal relationship has not been established. If transcriptional elongation is responsible for nucleosome turnover in gene bodies, then reduced elongation during heat shock will lead to a decrease in the level of turnover. To test this prediction, we applied the CATCH-IT (covalent attachment of tags to capture histones and identify turnover) metabolic labeling procedure (Deal et al. 2010), which measures the kinetics of nucleosome turnover, in control and heat-shocked cells. Cells were fed with Azidohomoalanine (Aha)-containing medium either at room temperature (control) or for 15 min at 37°C. Aha-containing proteins within isolated nuclei were then conjugated with an alkyne-adapted biotin linker through a copper-catalyzed cycloaddition reaction. Following MNase digestion, chromatin was extracted in 350 mM NaCl, and Aha-biotin-containing nucleosomes were affinity-purified using streptavidin-coated magnetic beads with stringent washing to remove H2A/H2B dimers. DNA from immunoprecipitated tetramers and from the 350 mM input fraction was isolated and prepared for paired-end sequencing as described above. At the Hsp70 gene cluster, we observed that nucleosomes underwent a dramatic increase in turnover after 15 min of heat shock (Fig. 6A), suggesting that a large decrease in nucleosome occupancy within seconds of heat shock shown in a previous study (Petesch and Lis 2008) involves increased turnover. To determine the genome-wide changes in nucleosome turnover caused by heat shock, we subtracted the control from heat-shock values and displayed the changes in a heat map with genes ordered by decreasing expression (Fig. 7A, left) or by changes in stalled Pol II due to heat shock (Supplemental Fig. S5A). These analyses revealed a decrease in nucleosome turnover for most expressed genes, while some previously silent Hsp genes showed increases in turnover, as well as a correlation between changes in stalled Pol II and nucleosome turnover. We then performed CATCH-IT on DRB-treated cells to ask whether turnover changes result from changes in transcriptional elongation. Indeed, inhibition of elongation with DRB led to a conspicuous reduction in nucleosome turnover (Fig. 7A, right), with a remarkable resemblance between the heat maps for heat-shocked and DRB-treated cells. Furthermore, when the changes in turnover were displayed as a heat map with genes ordered by changes in stalled Pol II due to DRB treatment, we observed that the greatest turnover changes occur within gene bodies even though the major changes in stalled Pol II occur immediately downstream from the TSS (Supplemental Fig. S5B). This suggests that much of the turnover within gene bodies is caused by the presumably insoluble elongating Pol II. The similar effects of heat shock and DRB inhibition of Pol II on turnover strongly suggest that down-regulation of expression during heat shock occurs after recruitment of the transcriptional machinery.
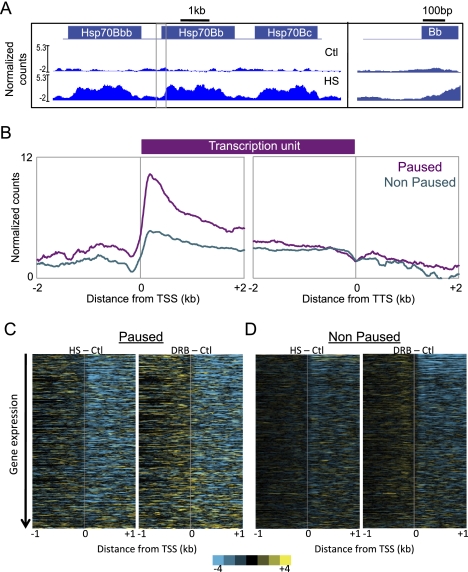
Figure 6.
Nucleosomes within bodies of paused genes have high turnover relative to nonpaused genes. CATCH-IT was performed before and after heat shock, and before and after DRB treatment. Input-normalized control samples were subtracted from heat-shocked samples (HS-Ctl) and DRB-treated samples (DRB-Ctl) to obtain changes in nucleosome turnover caused by heat-shock and DRB treatment, respectively. (A) The Hsp70 gene locus was visualized as described as in Figure 1C. (B) Average normalized counts for all paused (purple) and nonpaused (cyan) genes in a 2-kb region surrounding the TSS and TTS of the input-normalized CATCH-IT experiment under normal growth conditions. (C) Heat maps generated by Java TreeView show changes in nucleosome turnover resulting from heat-shock and DRB treatments in paused and nonpaused genes ordered by decreasing expression under normal conditions.
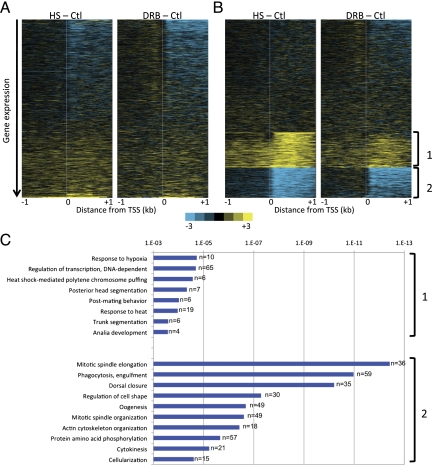
Figure 7.
Genome-wide changes in nucleosome turnover during heat shock. (A) Heat maps generated by Java TreeView show changes in nucleosome turnover resulting from heat-shock and DRB treatments, where all genes are ordered by decreasing expression under normal conditions. (B) Data from A for HS-Ctl were subjected to unsupervised k-means clustering with k = 3 (Cluster version 3), and the DRB-Ctl heat map was arranged according to the HS-Ctl clustering. (C) GO term analysis of groups 1 and 2 from the k-means clustering of the CATCH-IT data.
To further explore the relationship between transcriptional elongation and nucleosome turnover, we analyzed the CATCH-IT profiles of paused and nonpaused genes under normal conditions (Fig. 6B). Notably, paused genes showed consistently higher nucleosome turnover within gene bodies compared with nonpaused genes. We then examined the effects of heat shock and DRB on nucleosome turnover for these two gene sets. Both treatments led to substantial decreases in nucleosome turnover within gene bodies of both paused and nonpaused genes of matched expression (Fig. 6C), which argues that Pol II elongation drives nucleosome turnover regardless of how Pol II is regulated.
Finally, we asked about the molecular functions of the subset of genes that showed higher turnover during heat-shock treatment. To identify genes undergoing nucleosome turnover changes during heat-shock treatment, we applied unsupervised k-means clustering with k = 3 to the heat-shock minus control data and applied this clustering to the DRB minus control data (Fig. 7B). Heat map analysis showed a surprisingly similar pattern in nucleosome turnover changes in both treatments, suggesting that transcriptional inhibition has consistent effects on nucleosome dynamics regardless of the cause. Genes with increased turnover downstream from the TSS during both heat-shock and DRB treatments were enriched for gene ontology (GO) terms for transcription regulation, and heat-shock and hypoxia responses (Fig. 7C, group 1). The largest class of genes that showed a significant increase in nucleosome turnover is involved in DNA-dependent regulation of transcription (P = 6 × 10−4, n = 65), which suggests that nucleosome turnover at these genes is an inherent feature of the heat-shock response. In contrast, the genes that showed reduced nucleosome turnover are predominantly those with developmental and other functions that are expected to be perturbed during heat shock (Fig. 7C, group 2). Discovering concordant nucleosome turnover changes at genes in the appropriate functional categories for the heat-shock response itself suggests that regulation of turnover is an evolved response to environmental perturbation.
Discussion
We showed here that heat-shock induction involves genome-wide changes in chromatin solubility, stalled Pol II occupancy, and nucleosome turnover in bodies of expressed genes. Using new methods for epigenome profiling and analysis, we mapped both nucleosomes and subnucleosomal particles from the soluble chromatin fraction at single-base-pair resolution from control and heat-shocked cells. In addition, we determined the stalled Pol II profiles derived from this fraction and showed that loss of Pol II at active genes during heat shock correlates with reduced nucleosome turnover, thus providing evidence for a causal relationship between Pol II transit and nucleosome turnover in gene bodies. A similar pattern of reduced nucleosome turnover was also observed following drug inhibition of Pol II elongation, which implies that Pol II sometimes evicts nucleosomes that it encounters during elongation. Loss of stalled Pol II, and subsequent reduction in genic nucleosome turnover, might reflect a general mechanism for repression of diverse genes during heat shock, while maintenance of transcription factor binding at the promoters would facilitate efficient recovery (Zanton and Pugh 2006).
Several studies that have profiled nucleosomes genome-wide used MNase digestion followed by size selection for nucleosome-protected fragments (Mavrich et al. 2008; Li and Arnosti 2010; Kent et al. 2011; Xi et al. 2011). However, by using salt fractionation of native chromatin to selectively extract the soluble fraction, omitting size selection, and sequencing the paired ends of fragments of all sizes, we were able to investigate structural properties and dynamics for both nucleosomal and subnucleosomal components of the epigenome. For example, the footprint of subnucleosomal particles can be inferred from the square peaks of short fragments at the promoters of Hsp70 genes, implying that the bound particles confer complete protection of DNA by being precisely positioned over their binding sites. In contrast, the spontaneous unwrapping and rewrapping of nucleosomes allow for MNase encroachment into nucleosomal DNA (Li et al. 2005), thereby giving nucleosomal peaks rounded shapes that signify average protection. Furthermore, the precise mapping of subnucleosomal particles has allowed us to use the low-salt fraction as the starting material for native Pol II ChIP experiments, which indicates that the stalled Pol II can be specifically extracted from the soluble chromatin fraction. By sequencing all MNase-protected fragments from the ChIP and analyzing their midpoint-to-length relationships, we uncovered direct evidence in support of a proposed interaction between the stalled Pol II and the highly dynamic TSS nucleosome (Gilchrist et al. 2010; Weber et al. 2010). Thus, salt fractionation combined with a modified library preparation protocol and a simple computational analysis provides a powerful tool for investigating the structure, profile, and dynamics of the transcriptionally active epigenome.
Using this tool, we also found that active genes that lose stalled Pol II during heat shock show no overall changes in nucleosomal occupancy, comparable with a yeast study showing that heat shock does not affect global nucleosome occupancy (Shivaswamy et al. 2008). However, we also observed a genome-wide decrease in the low-salt-soluble nucleosomal fraction. How would a decrease in transcription affect nucleosome solubility? Previously, we showed that low-salt solubility largely corresponds to nucleosomes containing the conserved histone variant H2A.Z (Weber et al. 2010), which in Arabidopsis is responsive to changes in ambient temperature (Kumar and Wigge 2010). Therefore, it is possible that transcriptional repression results in decreased H2A.Z occupancy within bodies of expressed genes, and, as a result, nucleosome solubility decreases.
As we found to be the case for nucleosome occupancy, heat shock appears to have no effect on the average occupancies of subnucleosomal particles, in contrast to its major effect at the TSSs of heat-shock genes. Such maintenance of factor binding at the promoter region of transcribed genes suggests that down-regulation during heat shock occurs 3′ of the TSS. Evidence in support of downstream regulation during heat shock comes from the observation that SINE RNAs induced by heat shock disrupt elongating Pol II contacts with promoter DNA but do not disrupt transcription factor recruitment or binding in vitro (Yakovchuk et al. 2009). In addition, most of the factors involved in the preinitiation complex remain bound to promoters after heat shock in budding yeast (Zanton and Pugh 2006), further supporting the notion that repression occurs after recruitment.
The fact that low-salt extraction specifically enriches for stalled Pol II suggests inherent differences between stalled and actively elongating polymerases. Actively elongating Pol II is highly insoluble under standard extraction methods (Courvalin et al. 1976; Kimura et al. 1999), and nucleosomes of some transcribed genes remain insoluble even after high-salt treatments that solubilize most of the nucleosomes (Henikoff et al. 2009), which implies an association between these nucleosomes and the large, hydrophobic complexes involved in transcription. This further suggests that stalled Pol II disengages from these large complexes or from transcription factories (Eskiw et al. 2008; Papantonis et al. 2010), making it more soluble during extraction. The large fraction of genes with Pol II signals that we observed suggests that low-salt extraction in general captures stalled Pol II. Such widespread Pol II stalling implies a discontinuous elongation process, as has been shown in vitro (Herbert et al. 2006) and in vivo in Saccharomyces cerevisiae (Churchman and Weissman 2011). Furthermore, our study showed that heat shock causes partial release of stalled Pol II from DNA just downstream from TSSs, which is an attractive mechanism for transcriptional repression. Pol II release after heat shock has been previously observed in Drosophila salivary gland chromosomes (Jamrich et al. 1977). A similar Pol II release process has been proposed in mammals, where heat shock results in increased release of bound Pol II from DNA (Hieda et al. 2005).
We also showed that heat shock results in increased nucleosome turnover at heat-shock-induced genes and decreased turnover elsewhere over gene bodies. These observations imply a causal relationship between transcriptional repression and nucleosome turnover that was previously inferred from correlations between expression levels and H3.3 (Mito et al. 2005) or turnover rates (Deal et al. 2010) over gene bodies. Furthermore, the similarity between heat-shock- and DRB-induced nucleosome turnover patterns suggests that this causal relationship is an inherent general feature of eukaryotic transcription, although it is unknown whether these effects on transcriptional elongation involve the insoluble RNA polymerase fraction, which cannot be probed by ChIP. In vitro studies have revealed that a nucleosome can survive transcription by a single transcribing Pol II but becomes evicted when bursts of multiple Pol IIs pass through (Kulaeva et al. 2009, 2010; Jin et al. 2010). This in vitro observation, taken together with our direct measurements of changes in nucleosome turnover in vivo, suggests that both the level of transcription and the rate of Pol II bursting are major determinants of nucleosome eviction in a large portion of the soluble chromatin.
It seems counterintuitive that paused genes would show higher turnover within bodies than nonpaused genes (Fig. 4), insofar as pausing has been proposed to be a mechanism for preventing Pol II bursting (Levine 2011). However, this and other proposals have only considered events around the TSS, whereas our study is unique in observing features of paused genes throughout gene bodies. One proposal for the immediate disruption of downstream nucleosomes at the 87A Hsp70 locus during heat shock (Petesch and Lis 2008) is that the sudden acceleration of Pol II transit elicits a wave of positive supercoiling ahead that causes nucleosomes to unwrap before the topoisomerase swivel can relieve the strain (Zlatanova and Victor 2009). The fact that ordinary genes show high nucleosome occupancy when they are active (Cui et al. 2010) suggests that transcriptional elongation has evolved such that eviction of nucleosomes upon sudden acceleration of RNA polymerases is minimized. By this rationale, nucleosome eviction would also be minimized by favoring a more uniform rate of polymerase transit, thus reducing damaging acceleration events. Therefore, we speculate that the starts and stops that must occur when RNA Pol II pauses during its transit result in enhanced nucleosome eviction, followed by replacement, which is what we profiled in our CATCH-IT experiments. Paused genes would simply undergo more frequent damaging acceleration events than nonpaused genes and so show a higher rate of nucleosome turnover. In this way, events that occur generally within gene bodies and events around the TSSs of paused genes can be seen as resulting from common properties of the Pol II transcriptional apparatus.
Materials and methods
Cell culture and low-salt extraction
Drosophila S2 cells were grown to log phase in HYQ-SFX insect medium (Invitrogen) supplemented with 18 mM L-Glu and harvested as previously described (Henikoff et al. 2009). For heat-shocked samples, cells were incubated for 15 min at 37°C and immediately harvested. DRB treatment was performed as described previously using 125 μM final concentration in the medium for 10 min immediately before the heat-shock experiment began (Petesch and Lis 2008). The following cell treatments were used: control (normal conditions), heat shock for 15 min, DRB treatment for 10 min, and DRB for 10 min followed by 15 min of heat shock. Nuclear isolation, MNase digestion, and low-salt extraction in 80 mM buffer (70 mM NaCl, 10 mM Tris at pH 7.4, 2 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, 0.5 mM PMSF) were performed as described previously (Henikoff et al. 2009).
Pol II native ChIP
A 10% aliquot of the 80 mM salt fraction was saved, and the remainder was immunoprecipitated overnight with Pol II antibody 8WG16 (Abcam, ab817) at 4°C with constant agitation. Samples were next mixed with 50 μL of 80 mM buffer-prewashed Protein G Dynabeads (Invitrogen, 100.04D) for 2 h at 4°C. The bound Dynabeads were washed twice with 80 mM buffer without Triton X-100 and resuspended in 200 μL of the same buffer. For DNA extraction, 1/50 vol of 5 M NaCl and 0.5 M EDTA were added to both the resuspended beads and saved input material, and total RNA was digested with 0.5 μg of RNase (Roche, 10928100) for 10 min at 37°C. SDS was added to a final concentration of 0.5%, and protein was digested with 20 μg of Proteinase K (Invitrogen 25530-049) for 10 min at 70°C. Samples were extracted twice with phenol/chloroform followed by ethanol precipitation of DNA using 20 μg of glycogen (Roche, 14267332) as carrier. The precipitated DNA was resuspended in 20 μL of 0.1× TE (pH 8) and quantified using the Picogreen quantification kit for modified Solexa paired-end library preparation.
CATCH-IT
CATCH-IT was performed essentially as described (Deal et al. 2010) with the following modifications: Immediately following Aha addition, samples were either incubated for 15 min at room temperature (control) or heat-shocked at 37°C. For experiments with DRB treatment, DRB was added to the cells at 125 μM final concentration during the last 10 min of methionine depletion, with the same final concentration maintained during the 15-min incubation with Aha. Nuclei extraction, biotin coupling, MNase digestion, and streptavidin immunoprecipitation were performed as described previously. The input and streptavidin ChIP DNA were prepared for paired-end Solexa sequencing using the modified library preparation.
Sequencing and data analysis
Solexa sequencing libraries were constructed as described (Henikoff et al. 2011). Cluster generation and 25 rounds of paired-end sequencing were performed by the FHCRC Genomics Shared Resource using the Illumina Hi-Seq 2000. Base calling, data processing, and analysis were performed as described (Henikoff et al. 2011). Gene tracks from the sequencing data were visualized using SignalMap (NimbleGen, Inc.). Ends analysis, heat maps, and k-means clustering were performed as described (Henikoff et al. 2009). Midpoint-versus-length analysis was performed as described (Henikoff et al. 2011). GO analysis was performed on each k-means cluster using the GeneCodis 2.0 program (Carmona-Saez et al. 2007; Nogales-Cadenas et al. 2009) with a hypergeometric test and false discovery rate calculation to correct the P-values for multiple testing. Data have been deposited with Gene Expression Omnibus (GSE30755).
Acknowledgments
We thank A. Marty (FHCRC Genomics Shared Resource) for Illumina sequencing; C. Weber, R. Deal, and P. Talbert for critical comments; and J. Henikoff for help on sequencing data analysis. This material is based on work supported by the National Science Foundation Graduate Research Fellowship (under grant number DGE-0718124 awarded to S.T.), NIH grants U54 CA143862 and R01 ES020116, and the Howard Hughes Medical Institute (to S.H.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177675.111.
References
- Akerfelt M, Morimoto RI, Sistonen L 2010. Heat shock factors: Integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11: 816–821 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Bonner JJ 1979. The induction of gene activity in drosophilia by heat shock. Cell 17: 241–254 [DOI] [PubMed] [Google Scholar]
- Belyaeva ES, Zhimulev IF 1976. RNA synthesis in the Drosophila melanogaster puffs. Cell Differ 4: 415–427 [DOI] [PubMed] [Google Scholar]
- Bonner JJ, Pardue ML 1976. The effect of heat shock on RNA synthesis in Drosophila tissues. Cell 8: 43–50 [DOI] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A 2007. GENECODIS: A Web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 8: R3 doi: 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin JC, Bouton MM, Baulieu EE, Nuret P, Chambon P 1976. Effect of estradiol on rat uterus DNA-dependent RNA polymerases. Studies on solubilized enzymes. J Biol Chem 251: 4843–4849 [PubMed] [Google Scholar]
- Cui P, Zhang L, Lin Q, Ding F, Xin C, Fang X, Hu S, Yu J 2010. A novel mechanism of epigenetic regulation: Nucleosome-space occupancy. Biochem Biophys Res Commun 391: 884–889 [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina TY, Zou Y, Freeling S, Vorobyev VI, Erkine AM 2010. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res 38: 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw CH, Rapp A, Carter DR, Cook PR 2008. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J Cell Sci 121: 1999–2007 [DOI] [PubMed] [Google Scholar]
- Fritah S, Col E, Boyault C, Govin J, Sadoul K, Chiocca S, Christians E, Khochbin S, Jolly C, Vourc'h C 2009. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol Biol Cell 20: 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky J, Krassovsky K, MacAlpine DM, Henikoff S 2011. Epigenetic characterization at single base-pair resolution. Proc Natl Acad Sci doi: 10.1073/pnas.1110731108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM 2006. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell 125: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieda M, Winstanley H, Maini P, Iborra FJ, Cook PR 2005. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res 13: 135–144 [DOI] [PubMed] [Google Scholar]
- Jamrich M, Greenleaf AL, Bautz EK 1977. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci 74: 2079–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD 2010. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol 17: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K 2011. Chromatin particle spectrum analysis: A method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res 39: e26 doi: 10.1093/nar/gkq1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Tao Y, Roeder RG, Cook PR 1999. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: Little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol 19: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM 2009. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol 16: 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Hsieh FK, Studitsky VM 2010. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci 107: 11325–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA 2010. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LM, Arnosti DN 2010. Fine mapping of chromatin structure in Drosophila melanogaster embryos using micrococcal nuclease. Fly (Austin) 4: 213–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12: 46–53 [DOI] [PubMed] [Google Scholar]
- Lindquist S 1986. The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008. Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25: 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A 2009. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 37: W317–W322 doi: 10.1093/nar/gkp416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Lis JT 1991. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol 11: 5285–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR 2010. Active RNA polymerases: Mobile or immobile molecular machines? PLoS Biol 8: e1000419 doi: 10.1371/journal.pbio.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MM 1978. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J Cell Biol 79: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: Dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol 28: 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65 doi: 10.1371/journal.pbio.0060065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT 1978. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17: 5524–5531 [DOI] [PubMed] [Google Scholar]
- Weber CM, Henikoff JG, Henikoff S 2010. H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol 17: 1500–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yao J, Chen R, Li W, He X 2011. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res 21: 718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF 2009. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci 106: 5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF 2006. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev 20: 2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Victor JM 2009. How are nucleosomes disrupted during transcription elongation? HFSP J 3: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]