Limited information is available regarding eukaryotic H/ACA RNPs. This paper reports the first in vitro reconstitution of active eukaryotic H/ACA RNPs. Using this system, Li et al. also report the crystal structure of the yeast Cbf5–Nop10–Gar1 ternary complex, providing insight into the functional organization of eukaryotic H/ACA RNPs and into major differences with its archaeal counterparts.
Keywords: pseudouridylation, H/ACA RNA–protein complex, crystal structure, dyskeratosis congenita
Abstract
Box H/ACA ribonucleoprotein particles (RNPs) mediate pseudouridine synthesis, ribosome formation, and telomere maintenance. The structure of eukaryotic H/ACA RNPs remains poorly understood. We reconstituted functional Saccharomyces cerevisiae H/ACA RNPs with recombinant proteins Cbf5, Nop10, Gar1, and Nhp2 and a two-hairpin H/ACA RNA; determined the crystal structure of a Cbf5, Nop10, and Gar1 ternary complex at 1.9 Å resolution; and analyzed the structure–function relationship of the yeast complex. Although eukaryotic H/ACA RNAs have a conserved two-hairpin structure, isolated single-hairpin RNAs are also active in guiding pseudouridylation. Nhp2, unlike its archaeal counterpart, is largely dispensable for the activity, reflecting a functional adaptation of eukaryotic H/ACA RNPs to the variable RNA structure that Nhp2 binds. The N-terminal extension of Cbf5, a hot spot for dyskeratosis congenita mutation, forms an extra structural layer on the PUA domain. Gar1 is distinguished from the assembly factor Naf1 by containing a C-terminal extension that controls substrate turnover and the Gar1–Naf1 exchange during H/ACA RNP maturation. Our results reveal significant novel features of eukaryotic H/ACA RNPs.
Box H/ACA ribonucleoprotein particles (RNPs) are ancient molecular machines conserved in archaea and eukaryotes that function mainly to convert uridine (U) into pseudouridine (Ψ) post-transcriptionally at specific sites of rRNAs and snRNAs (Meier 2005; Matera et al. 2007; Kiss et al. 2010). Some special eukaryotic H/ACA RNPs assume functions other than modification. Conserved U17/snR30 RNPs mediate pre-rRNA processing and ribosome assembly (Morrissey and Tollervey 1993; Atzorn et al. 2004; Fayet-Lebaron et al. 2009), and vertebrate telomerases contain an H/ACA RNP domain that is required for their biogenesis and in vivo function (Mitchell et al. 1999).
Mature H/ACA RNPs are composed of a distinct H/ACA RNA and a set of highly conserved proteins: the catalytic subunit Cbf5 (dyskerin in humans), Nop10, Gar1, and Nhp2 (L7Ae in archaea). Eukaryotic H/ACA RNAs contain a 5′ hairpin followed by a single-stranded H box (ANANNA, where N is any nucleotide), and a 3′ hairpin followed by a related ACA box (Balakin et al. 1996; Ganot et al. 1997b). Each functional hairpin contains a large internal loop, called the pseudouridylation pocket, which is able to form a bipartite RNA duplex around the target uridine in substrates, hence determining the modification specificity (Ganot et al. 1997a). Archaeal H/ACA RNAs possess one to three copies of the hairpin unit that includes a downstream ACA motif and a kink-turn (K-turn) motif in the upper stem (Tang et al. 2002; Rozhdestvensky et al. 2003).
Much of our understanding of H/ACA RNP structure and mechanism of action comes from studies on archaeal complexes. Catalytically active archaeal H/ACA RNPs have been reconstituted with recombinant proteins (Baker et al. 2005; Charpentier et al. 2005) and are amenable for crystallographic analysis (Hamma et al. 2005; Li and Ye 2006; Manival et al. 2006; Rashid et al. 2006; Liang et al. 2007, 2009; Ye 2007; Duan et al. 2009). The crystal structure of Pyrococcus furiosus (Pf) H/ACA RNP assembled with a single-hairpin H/ACA RNA demonstrates that the upper stem of the guide RNA is bound sequentially by L7Ae, Nop10, and the catalytic domain of Cbf5, and the lower stem and the ACA motif are anchored at the PUA domain of Cbf5 (Li and Ye 2006). The substrate is recruited through base-pairing with the bipartite guides and extensive protein interactions, precisely placing the target uridine at the active site (Duan et al. 2009; Liang et al. 2009). Gar1 regulates substrate turnover through controlling the conformation of the thumb loop of Cbf5, which is essential for substrate loading (Li and Ye 2006; Duan et al. 2009).
Eukaryotic H/ACA RNPs are characterized by several features distinct from those of archaeal RNPs. Eukaryotic H/ACA RNAs are almost exclusively composed of two hairpins and lack the K-turn motif present in archaeal H/ACA RNAs, which is specifically recognized by L7Ae (Rozhdestvensky et al. 2003). Nhp2 is structurally similar to L7Ae (Koo et al. 2011) but, unlike L7Ae, can stably associate with Cbf5 and Nop10 (Wang and Meier 2004). Eukaryotic Nop10 also lacks the cysteine residues that coordinate a zinc ion in archaeal Nop10 (Hamma et al. 2005; Khanna et al. 2006).
Dyskeratosis congenita (DC) is an inheritable bone marrow failure syndrome, and causal mutations have been identified in the genes encoding telomerase components dyskerin, Nhp2, Nop10, hTR, and hTERT or the telomere-stabilizing protein TIN2 (Walne and Dokal 2009). Because of the high-sequence similarity between yeast and human H/ACA RNP proteins, the yeast H/ACA RNP structure will provide a better model than archaeal structures in understanding the effect of DC mutations. In particular, many DC mutations are present at the N- and C-terminal extensions (NTE and CTE) of Cbf5 that are conserved between yeast and human but that are missing in archaea.
The structure of eukaryotic H/ACA RNPs remains poorly understood, in large part, because of the lack of a good in vitro reconstitution system. The pseudouridylation activity of eukaryotic H/ACA RNPs has been reconstituted by assembling H/ACA RNAs with mammalian cell extracts or with purified yeast H/ACA RNPs in which endogenous RNAs have been digested by micrococcal nuclease (Wang et al. 2002; Wang and Meier 2004; Xiao et al. 2009). These systems depend on in vivo preassembled protein complexes and have limited utility in structural analysis. Moreover, the in vivo assembly of eukaryotic H/ACA RNPs requires at least two specific assembly factors: Naf1 and Shq1 (Dez et al. 2002; Fatica et al. 2002; Yang et al. 2002). It remains unclear whether eukaryotic H/ACA RNPs also require these factors for in vitro assembly.
In this study, we reconstitute active yeast H/ACA RNPs using bacteria-expressed recombinant proteins. We solve the structure of a Cbf5–Nop10–Gar1 ternary protein complex and analyze the structure–function relationship of yeast H/ACA RNPs. These results provide novel insights into the functional organization of eukaryotic H/ACA RNPs.
Results
Purification of recombinant yeast H/ACA RNP proteins
Yeast Cbf5 contains a core region (residues 1–394, hereafter Cbf5C) conserved in eukaryotes and a KKE/D-rich low-complexity tail at the C terminus, which nevertheless is functionally important in vivo (Jiang et al. 1993). Yeast Gar1 is composed of a central core domain (residues 32–124, hereafter Gar1C) flanked by two Gly/Arg-rich (GAR) domains. The core domain of Gar1 is sufficient for its function in yeast and human cells (Girard et al. 1994; Pogacic et al. 2000). We mainly used the core domain of Cbf5 and Gar1 for biochemical and structural study.
We observed that Cbf5C formed aggregates when expressed alone in Escherichia coli, but can be coexpressed and copurified with Nop10 in a soluble form. When purified alone, Gar1C was capable of reconstituting pseudouridylation activity together with other H/ACA RNP components (data not shown) but was barely bound to Cbf5C–Nop10, suggesting that only a small amount of such preparation was properly folded and functional. To overcome this problem, we coexpressed and copurified Gar1C with Cbf5C and Nop10. The obtained ternary complex of Cbf5C–Nop10–Gar1C was more stable and soluble than the Cbf5C–Nop10 binary complex and has been crystallized. In addition, full-length Cbf5 and full-length Gar1 can also be purified using the similar coexpression approach, although full-length Gar1 was prone to degradation (Fig. 1A). Nhp2 can be purified either alone or together with Cbf5 complexes. The copurified Cbf5 complexes contained stoichiometric amounts of each component (Fig. 1A). It should be noted that Nhp2 tended to dissociate from Cbf5 complexes during storage, likely because of a weak interaction (Henras et al. 2004). The availability of large amounts of soluble and pure Cbf5 complexes in various compositions allowed us to analyze the assembly, activity, and structure of yeast H/ACA RNP in detail.
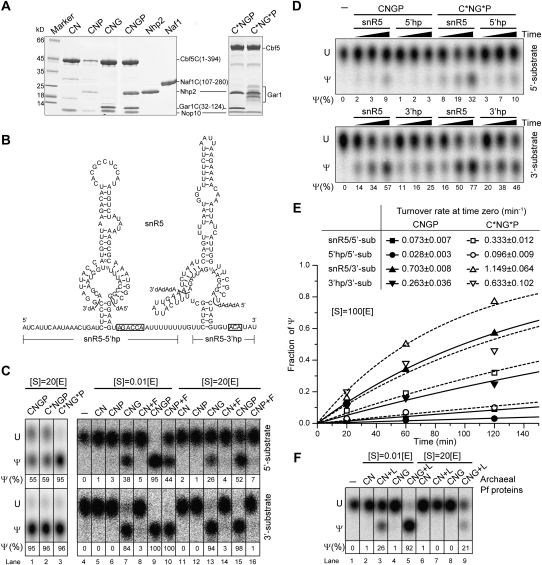
Figure 1.
Pseudouridylation activity of reconstituted yeast H/ACA RNPs. (A) Coomassie blue-stained SDS-PAGE gel of the purified yeast H/ACA RNP proteins. The migration positions of each protein and molecular marker are labeled. The boundaries of protein fragment constructs are shown in parentheses. Full-length Gar1 was partially degraded. (C) Cbf5C (1–394); (C*) full-length Cbf5; (N) Nop10; (P) Nhp2; (G) Gar1C (32–124); (G*) full-length Gar1; (F) Naf1C (107–280). Copurified complexes are denoted by concatenated words; for example, CNGP stands for the Cbf5C–Nop10–Gar1C–Nhp2 complex. (B) Sequence and secondary structure of snR5 H/ACA RNA with two cognate substrates bound at the pseudouridylation pockets. The target site is shown as pseudouridine (Ψ). The boundaries of single-hairpin constructs snR5-5′hp and snR5-3′hp are marked. Boxes H and ACA are boxed. (C) Pseudouridylation activity of snR5 H/ACA RNPs. SnR5 H/ACA RNPs (1 μM) with indicated protein compositions were incubated with 5′ or 3′ substrates of ∼0.01 or 20 μM for 1 h at 30°C. The substrates with a singly labeled 32P at the 3′ side of the target uridine were extracted and digested by RNase A/S7 nuclease into 3′-P mononucleotides, which were analyzed by thin-layer chromatography and autoradiograph. The fractions of converted Ψ are indicated, and the estimated error is ∼10%. Protein added separately is preceded by a plus symbol. (S) Substrate; (E) enzyme. (D) Time course of pseudouridylation. Yeast H/ACA RNPs (0.2 μM) composed of CNGP or C*NG*P and snR5, snR5-5′hp, or snR5-3′hp were incubated with 20 μM 5′ or 3′ substrate for 20, 60, and 120 min. (E) The time course data were fit to a single-exponential equation, and the derived substrate turnover rates at time 0 are indicated. (F) Pseudouridylation activity of Pf H/ACA RNPs. Pf H/ACA RNPs (1 μM) of indicated protein compositions were incubated with ∼0.01 or 20 μM substrate for 1 h at 37°C. (L) L7Ae.
Reconstituted H/ACA RNPs are active in site-specific pseudouridylation
The yeast snR5 H/ACA RNA has a 5′ and a 3′ hairpin that guides the formation of pseudouridine at positions 1124 and 1004 of 25S rRNA, respectively (Fig. 1B; Ganot et al. 1997a). We reconstituted H/ACA RNPs with snR5 and examined their activity in modifying substrates that are complementary to the 5′ or 3′ hairpin. These substrates were singly 32P-labeled at the 3′ side of the target uridine and digested into mononucleotides following the modification (Wang et al. 2002). Uridine and pseudouridine were then separated by thin-layer chromatography. To examine the basic catalytic activity of the enzyme, the modification was measured under single-turnover conditions in which the enzyme is ∼100-fold excess over the substrate. The modification was also performed with excess substrate to assess the substrate turnover rate.
The snR5 H/ACA RNP assembled with the core domain of Cbf5, and Gar1, Nop10, and Nhp2 were active in modification of the 5′ and 3′ substrates in a multiple-turnover manner, indicating that both hairpins of snR5 are functional in guiding modification (Fig. 1C, lane 1). Similar to archaeal enzymes (Duan et al. 2009), the yeast enzyme possesses the intrinsic ability to turn over the substrate at physiological temperature and in the absence of any RNA helicase.
Assembly with full-length Cbf5 did not appreciably change the reaction rate, suggesting that the KKE/D tail of Cbf5 is not involved in the modification (Fig. 1C, lane 2). In contrast, incorporation of full-length Gar1 (partially degraded) significantly increased the reaction rate for the 5′ substrate (Fig. 1C, lane 3). The effect on the 3′ substrate cannot be assessed under the experimental conditions, as it was modified to near completion by H/ACA RNPs with Gar1C or full-length Gar1. To quantify the reaction rate, we also measured the time course of modification using a substrate-to-enzyme ratio of 100 (Fig. 1D,E). The presence of GAR domain increased the substrate turnover rate at time 0 by 4.5-fold and 1.6-fold for the 5′ and 3′ substrates, respectively. Despite being genetically dispensable (Girard et al. 1994; Pogacic et al. 2000), the highly basic GAR domain has a role in substrate turnover, likely through transitive interactions with substrate RNAs.
Interestingly, the 3′ substrate was modified consistently faster (7.2 times on average) than the 5′ substrate by H/ACA RNPs of various compositions (Fig. 1C–E). The substrate turnover rate is probably inversely proportional to the interaction strength between the substrate and guide RNA: The 3′ substrate, which forms four G–C and eight A–U base pairs with the guide, is bound more weakly than the 5′ substrate, which forms five G–C, eight A/G–U, and possibly additional base pairs with the guide (Fig. 1B). In addition, a stem–loop insertion in the 3′ pseudouridylation pocket may affect the reaction kinetics.
Role of Gar1 and Nhp2 in modification
The omission of Gar1 in the yeast RNP abolished most of the activity even under single-turnover conditions (Fig. 1C, lanes 6,12). The Gar1-minus complex sometimes displayed basal activity (<10% yield) under single-turnover conditions in other experiments (data not shown). In contrast, the archaeal Pf H/ACA RNP without Gar1 is more active under single-turnover conditions (Fig. 1F, lane 3; Baker et al. 2005; Charpentier et al. 2005). This suggests that Gar1 plays a more important role in yeast RNPs than in Pf RNPs in terms of facilitating the substrate to adopt a catalysis-competent conformation. Nevertheless, both the archaeal and eukaryotic enzymes require Gar1 for substrate turnover.
Yeast H/ACA RNPs lacking Nhp2 still displayed a pronounced, although reduced, activity in multiple-turnover reactions (Fig. 1C, lane 13). This is surprising, given that the archaeal counterpart L7Ae is critical even for basic catalytic activity (Fig. 1F, lane 4). Nhp2 and L7Ae apparently play different roles in eukaryotic and archaeal H/ACA RNPs, respectively.
The assembly factor Naf1 is inactive in substrate turnover
As the assembly factor Naf1 is not present in mature H/ACA RNPs, presumably, the H/ACA RNPs with Naf1 should not be active. We tested this idea by assembling H/ACA RNP with the core domain of Naf1 (residues 107–280, hereafter Naf1C), which is homologous to the core domain of Gar1 (Leulliot et al. 2007). The Naf1C RNP exhibited some activity in single-turnover reactions, which was dependent on the presence of Nhp2, but was incapable of substrate turnover (Fig. 1C, lanes 8,10,14,16). Hence, despite being a structural homolog, Naf1 cannot substitute Gar1 in the enzymatic reaction, which is consistent with its role as an assembly factor.
Single-hairpin H/ACA RNPs are also active
Eukaryotic H/ACA RNAs are characterized by a conserved two-hairpin structure. To investigate whether the enzymatic function of reconstituted yeast H/ACA RNP requires both hairpins in H/ACA RNA, we constructed snR5 derivatives snR5-5′hp and snR5-3′hp, which contained isolated 5′ and 3′ hairpins followed by box H and ACA (Fig. 1B), respectively. The assembled single-hairpin RNPs were also active in modifying cognate substrates in a multiple-turnover fashion (Fig. 1D,E). However, single-hairpin RNPs were 2.6-fold slower on average in substrate turnover than two-hairpin RNPs.
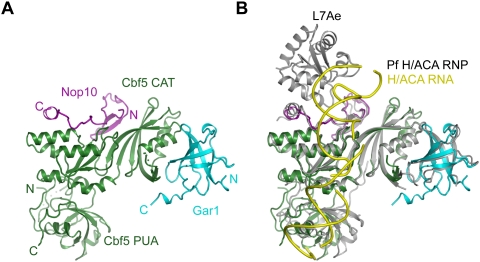
Crystal structure of the Cbf5C–Nop10–Gar1C complex
To understand the specific features of yeast H/ACA RNP, we determined the crystal structure of a ternary complex composed of Cbf5C, Nop10, and Gar1C at 1.9 Å resolution (Fig. 2A). The structure was solved by molecular replacement using the archaeal structure as a search model and refined to an Rwork/Rfree value of 0.201/0.231 (Table 1).
Figure 2.
Crystal structure of the yeast Cbf5C–Nop10–Gar1C complex. (A) Ribbon representation of the structure. Cbf5 is green, Nop10 is magenta, and Gar1 is cyan. The N and C termini of each protein are indicated. (CAT) The catalytic domain. (B) Structural alignment of the yeast Cbf5C–Nop10–Gar1C complex with the Pf H/ACA RNP. Pf H/ACA RNP structure is colored in gray for proteins and in yellow for RNA.
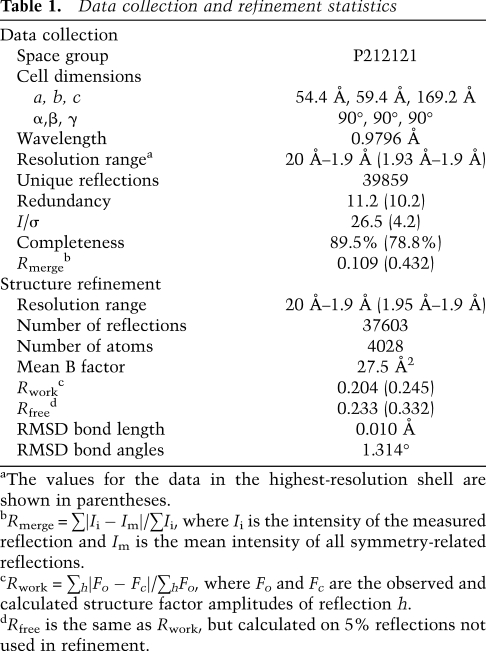
Table 1.
Data collection and refinement statistics
The structure of individual subunits, overall organization, and protein–protein interaction of the yeast complex are generally similar to those of archaeal structure (Fig. 2B; Hamma et al. 2005; Li and Ye 2006; Manival et al. 2006; Rashid et al. 2006). Nop10 and Gar1 bind at two orthogonal faces of the catalytic domain of Cbf5. Alignment of the individual domain of the yeast and Pf structures yields a root-mean-square deviation (RMSD) of 0.929 Å for the Cbf5 catalytic domain (178 Cα pairs), 0.62 Å for the Cbf5 PUA domain (71 Cα pairs), 0.813 Å for Gar1 (47 Cα pairs), and 1.56 Å for Nop10 (31 Cα pairs). Except for the PUA domain that rotates ∼20° relative to the catalytic domain and the interface between Cbf5 and the C-terminal helix of Nop10 (see below), domain movement is minor in the rest of the structure. The junction between the PUA domain and catalytic domain has been shown to be flexible in archaeal Cbf5 (Duan et al. 2009). The structure also reveals specific features in the three yeast proteins.
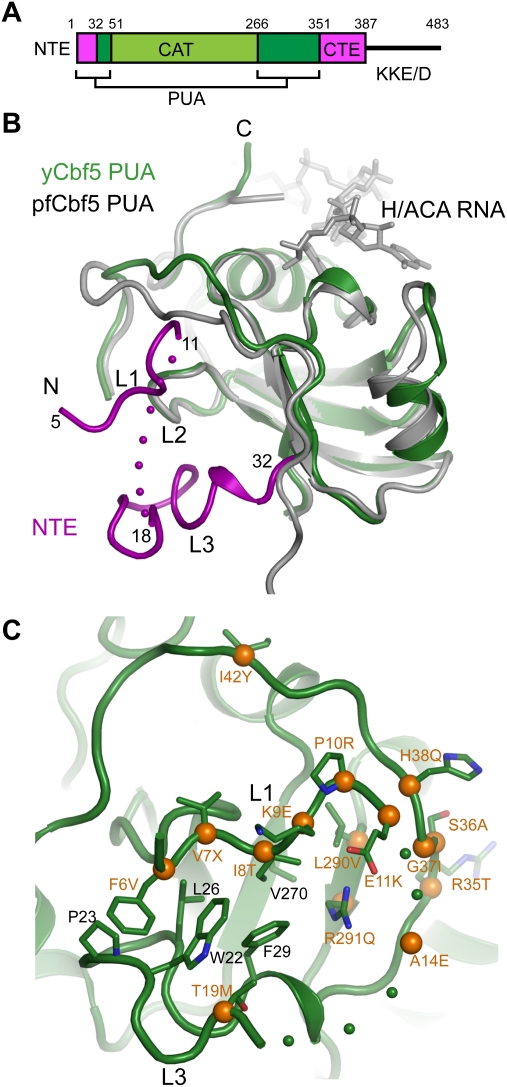
The PUA domain of yeast Cbf5 contains a new structural layer harboring many DC mutations
Eukaryotic Cbf5 is longer than archaeal Cbf5, with an NTE (residues 1–32 in yeast Cbf5), a CTE (residues 351–387), and a long C-terminal tail of KKE/D repeats (Fig. 3A). These extensions are particularly interesting, considering that 15 DC mutation sites were found in these regions and cannot be modeled from the archaeal Cbf5 structure (Marrone et al. 2005).
Figure 3.
The yeast PUA domain and DC mutations. (A) Domain organization of yeast Cbf5. The NTE, catalytic domain (CAT), PUA domain, CTE, and KKE/D repeat are labeled. (B) Structural alignment of the PUA domain of yeast and Pf Cbf5. The NTE of yeast Cbf5, colored in magenta, forms an extra structural layer on the PUA domain and is divided into loops L1, L2, and L3. The ACA motif of H/ACA RNA bound to the Pf PUA domain is shown as sticks. Dots denote disordered residues. (C) DC mutations surrounding the NTE. DC mutation sites mapped to the yeast Cbf5 structure are marked by Cα spheres (orange) and yeast residue names, followed by the amino acids found in DC.
The Cbf5 CTE was not observed in crystal, likely due to protein degradation and intrinsic disorder (Supplemental Fig. S1). The NTE is partially structured and forms a new structural layer that covers the β barrel of the PUA domain on the side opposite to the archaeal RNA-binding surface (Fig. 3B). For the purpose of this discussion, we divide the NTE into three parts: loop L1 (residues 5–11), loop L2 (residues 12–17), and loop L3 (residues 18–33). Loop L2 was not modeled because of its missing electron density. Loop L1, in an extended conformation, is encircled by loop L3 and the rest of N-terminal residues of the PUA domain. Loop L1 packs against loop L3 and the surface of the β barrel by hydrophobic interactions involving Phe6, Ile8, Trp22, Pro23, Leu26, Phe29, and Val270. Notable, loop L1 displayed high temperature factors in this reported structure and was invisible in two other crystals analyzed. The N-terminal region of Cbf5C was partially degraded in crystal, particularly in the disordered L2 loop, which may lead to a low occupancy of L1 (Supplemental Fig. S1).
Notably, a large number of DC mutation sites are clustered around the new surface formed by the NTE (Fig. 3C). Specifically, each residue in loop L1 was found to be mutated in DC patients (Marrone et al. 2005). Mutations F6V, I8T, and P10R; a Val7 deletion in loop L1; and mutations L290V and R291Q in the bottom floor that supports loop L1 may directly disturb the packing of loop L1. These disease mutations suggest that the NTE is important to the structure and/or function of dyskerin.
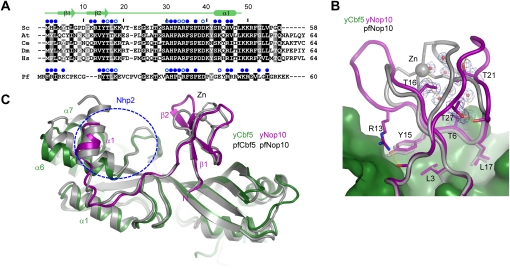
Structure of yeast Nop10
In the yeast complex, Nop10 adopts an elongated structure composed of an N-terminal ribbon domain, an extended linker, and a C-terminal α helix and contacts extensively with the catalytic domain of Cbf5. The ribbon domain of Pf Nop10 encapsulates a zinc ion with four cysteine residues, whereas the ribbon domain of yeast Nop10 lacks the zinc-binding cysteine residues and does not bind a zinc ion (Fig. 4A). Despite this difference, the two ribbon domains share a similar fold and a similar interface with Cbf5 (Fig. 4B,C). In the absence of zinc binding, the space between the two halves of the ribbon domain of yeast Nop10 is half-opened and filled with water molecules. Except for four packed methyl groups from Thr6, Thr16, Thr21, and Thr27, the yeast ribbon domain contains no hydrophobic core that stabilizes its tertiary fold (Fig. 4B). In fact, yeast Nop10 alone is largely unstructured, with only the first β hairpin persistent in solution (Hamma et al. 2005; Khanna et al. 2006; Reichow and Varani 2008). In the absence of zinc binding, the interaction with Cbf5 becomes critical to the maintenance of the ribbon structure.
Figure 4.
Structure comparison of yeast and Pf Nop10. (A) Sequence alignment of Nop10 proteins from Saccharomyces cerevisiae (Sc), Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), and P. furiosus (Pf). Residues conserved in 100% and 80% of eukaryotic Nop10 sequences are shaded black and gray, respectively. Residues of Pf Nop10 that match the conserved residues in eukaryotic Nop10 are shaded. Residues in yeast and Pf Nop10 whose solvent-accessible surfaces are buried by at least 30 Å2 and 10 Å2 due to Cbf5 association are marked by solid and open blue circles, respectively. (B) Comparison of the ribbon domain of yeast and Pf Nop10. Residues that interact with Cbf5 and maintain the fold are shown as sticks. Yeast Nop10 residues are labeled. The 2fo–fc electron density map contoured at 1.5 σ level is shown for the interior of the ribbon domain. (C) Structural alignment of yeast Cbf5–Nop10 with Pf H/ACA RNP. The blue circle indicates the putative Nhp2-binding site.
The C-terminal 10 residues of Nop10 were invisible in crystal, likely due to protein degradation (Supplemental Fig. S1). As in the archaeal Cbf5–Nop10 structure, the C-terminal helix α1 of yeast Nop10 is packed against helix α6 of Cbf5 at a right angle. However, the two interacting α helices slide along each other, leading to a different interface in the yeast complex (Fig. 4C). Specifically, helix α6 of Cbf5 moves in a direction perpendicular to its helical axis toward helix α1 of Cbf5, while helix α1 of Nop10 shifts in the opposite direction along its helical axis. Nhp2 has been shown to stably associate Cbf5–Nop10 in the absence of RNA (Wang and Meier 2004) and may bind Nop10 in a different manner than L7Ae, which cannot form a stable protein complex with Cbf5–Nop10 (Baker et al. 2005; Charpentier et al. 2005). The structural change at the C-terminal region of Nop10 may generate a new interface for Nhp2.
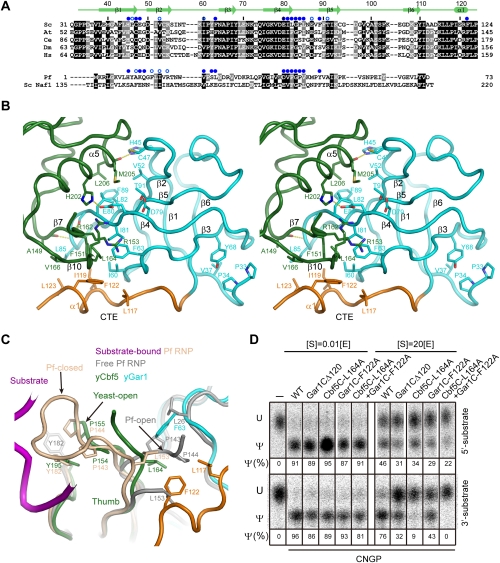
The core domain of yeast Gar1 contains a novel CTE interacting with the thumb
The core domain of yeast Gar1 adopts a six-stranded β barrel as archaeal Gar1 does, but has an NTE and CTE (Figs. 2B, 5A). In the NTE (residues 32–38), Pro33 and Pro34 are highly conserved in eukaryotic Gar1. Removal of the two equivalent proline or more residues in human Gar1 was shown to cause protein aggregation and a loss of nucleolar and Cajal body localization (Pogacic et al. 2000). The structure demonstrates that the two prolines are part of a hydrophobic core and should be important for structural integrity (Fig. 5B). The CTE (residues 113–124) contains a short α helix, α1, that makes hydrophobic interactions with the thumb of Cbf5.
Figure 5.
Interaction between yeast Gar1 and Cbf5. (A) Sequence alignment of the Gar1 core domain from S. cerevisiae (Sc), A. thaliana (At), C. elegans (Ce), D. melanogaster (Dm), and H. sapiens (Hs). Residues that are conserved in 100% and 80% of the five eukaryotic sequences are shaded black and gray, respectively. Pf Gar1 and Sc Naf1 are aligned based on structural homology with yeast Gar1. Their residues are shaded if they match the conserved residues of eukaryotic Gar1. Yeast and Pf Gar1 residues whose solvent-accessible surfaces are buried by at least 30 Å2 and 10 Å2 due to Cbf5 association are marked by solid and open blue circles, respectively. (B) The Cbf5–Gar1 interface in cross-eye stereo view. The relevant residues are shown as sticks, with oxygen atoms in red and nitrogen atoms in blue. The CTE of Gar1 is colored orange. (C) The thumb in the yeast (y) Cbf5C–Nop10–Gar1C complex compared with the thumb of Pf H/ACA RNP in the closed and open conformation. Interacting residues are indicated. (D) The interaction between the Gar1 CTE and the Cbf5 thumb is important for substrate turnover. The snR5 CNGP complexes (1 μM) containing wild-type proteins or mutant Gar1CΔ120 (residues 32–120), Cbf5C L164A, and Gar1C F122A were incubated with 5′ or 3′ substrates of ∼0.01 or 20 μM for 1 h at 30°C.
Gar1 binds Cbf5 with two interfaces through many hydrophobic contacts and a few hydrogen bonds (Fig. 5B). The first interface, similar to that in the archaeal complex (Rashid et al. 2006), involves a Gar1 surface spanning strands β2, β5, and β4 that contacts helix α5 of Cbf5. The second interface is very different from its archaeal counterpart and constitutes strand β4, loop β4–β5, loop β2–β3, and the eukaryote-specific helix α1 of Gar1 that contact the base of the Cbf5 thumb.
Studies on archaeal H/ACA RNPs have shown that the highly conserved thumb is flexible and critical for substrate recruitment and turnover (Li and Ye 2006; Duan et al. 2009). In the substrate-loaded archaeal H/ACA RNP structures, it adopts a closed conformation, pinching the substrate at the active site (Duan et al. 2009; Liang et al. 2009), while in the substrate-free state or an inactive substrate complex, the thumb is partially disordered and docks at Gar1 in an open conformation (Fig. 5C; Li and Ye 2006; Liang et al. 2007). Breaking of the extensive interactions between the substrate and the thumb is expected to occur prior to product release after modification. Gar1 may facilitate this process by providing a low-energy docking site for the thumb.
In the yeast protein complex, the thumb is partially disordered, and the structured region adopts a conformation resembling, but not identical to, the closed conformation seen in the substrate-loaded archaeal structure (Fig. 5C). The thumb cannot adopt a Pf-like open conformation because the analogous Gar1 docking site is physically blocked by the side chain of Phe63, Leu117, and Phe122 in yeast Gar1. In the yeast “open” conformation, Pro154 and Pro155 are docked favorably over the aromatic ring of Try195. Phe151 and Leu164 of the thumb are inserted into a hydrophobic pocket of Gar1 that is composed of Ile60, Phe63, Ile81, Leu85, I119, and Phe122. The guanidium group of Cbf5 Arg153 stacks over the aromatic ring of Gar1 Phe63 (Fig. 5B,C).
Compared with the Pf closed conformation, the “open” yeast thumb is closer to Gar1 (Fig. 5C). The yeast thumb may undergo only subtle conformational changes during the substrate loading and release cycle. For example, Leu164 might need to be released from its binding pocket in Gar1 upon substrate loading.
The CTE of Gar1 regulates substrate turnover
Although the yeast thumb adopts a different open conformation than its archaeal counterpart, the new thumb–Gar1 interface may also regulate substrate turnover. To test this idea, we made mutagenesis to interfere with the thumb–Gar1 CTE interaction and measured the activity of mutant snR5 RNPs under single- and multiple-turnover conditions. Specially, we truncated the C termini of Gar1C to residue 120 (Gar1CΔ120) or made alanine substitutions of Phe122 in the CTE of Gar1C and Leu164 in the thumb of Cbf5C, two interacting hydrophobic residues. The mutant enzymes all retained basic catalytic activity in single-turnover reactions, but were impaired in substrate turnover to various degrees in multiple-turnover reactions (Fig. 5D). The mutational effect was additive for the double mutation F122A/L164A. The 3′ substrate, which has a faster turnover rate, was more sensitive to the mutations than the 5′ substrate. These results suggest that the CTE–thumb interaction regulates substrate turnover. The assembly factor Naf1 does not contain an equivalent CTE (Fig. 5A), which at least partially accounts for its inability in substrate turnover (Fig. 1C).
The CTE renders Gar1 to bind Cbf5 tighter than Naf1
The CTE has been shown to be critical for human Gar1 to localize in the nucleolus and associate with U17 H/ACA RNA (Pogacic et al. 2000). During the in vivo assembly pathway of H/ACA RNPs, Naf1 is first assembled with Cbf5 at the transcription site of H/ACA RNAs and then replaced by Gar1 in mature RNPs in nucleoli and Cajal bodies (Ballarino et al. 2005; Yang et al. 2005; Darzacq et al. 2006). The mechanism by which Gar1 replaces Naf1 remains unknown. One possibility is that Gar1 has a higher affinity for Cbf5 than Naf1. In addition to regulating substrate turnover, the observed Gar1 CTE–Cbf5 interaction, which is absent in Naf1, may strengthen the binding of Gar1 to Cbf5 and facilitate the displacement of Naf1.
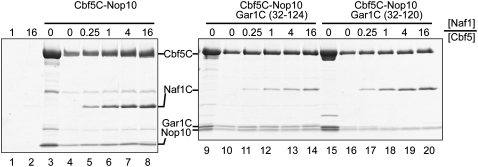
To test this notion, we assembled Naf1C with Cbf5C complexes and used Ni beads to pull down His-tagged Cbf5C and any associated proteins (Fig. 6). Naf1C was retained insignificantly on Ni beads by itself (Fig. 6, lanes 1,2) and readily by Cbf5C–Nop10 in a stoichiometric manner (Fig. 6, lanes 3–8), indicating a specific interaction between them. A small amount of Naf1C was retained by a preassembled Cbf5C–Nop10–Gar1C complex, even when Naf1C was 16-fold excess. The results indicate that Naf1C competes weakly with Gar1C to bind Cbf5. However, truncation of the C-terminal four residues of the CTE in Gar1C (Gar1CΔ120) dramatically increased the ability of Naf1C to displace Gar1C. Gar1CΔ120, but not Gar1C with an intact CTE, was completely displaced in the presence of one molar equivalent of Naf1C (Fig. 6, cf. lanes 12 and 18). Moreover, a fraction of Gar1CΔ120 was dissociated from Cbf5C–Nop10 even in the absence of Naf1C during the pull-down process, further supporting that the Cbf5–Gar1CΔ120 interaction was weakened (Fig. 6, cf. lanes 15 and 16). We were not able to perform the reciprocal competition experiment (namely, using Gar1C to bind Cbf5C–Nop10–Naf1C) because free recombinant Gar1C is incapable of binding Cbf5C in our hands. We can conclude that the CTE contributes critically to the Gar1–Cbf5 interaction and could allow Gar1 to dislodge Naf1 from Cbf5 during H/ACA RNP maturation.
Figure 6.
The CTE allows Gar1 to bind Cbf5 tighter than Naf1. Copurified Cbf5C–Nop10 and Cbf5C–Nop10–Gar1C and Cbf5C–Nop10–Gar1CΔ120 complexes (200 pmol) were incubated with 0, 50, 200, 800, and 3200 pmol of Naf1C and pulled down by Ni beads. Only Cbf5C contains a His6 tag among these proteins. Lanes 1 and 2 show retention of 200 and 3200 pmol of Naf1C. Lanes 3, 9, and 15 are 60 pmol of Cbf5C complexes prior to pull down.
Nhp2 occupies a parallel position as L7Ae in H/ACA RNP
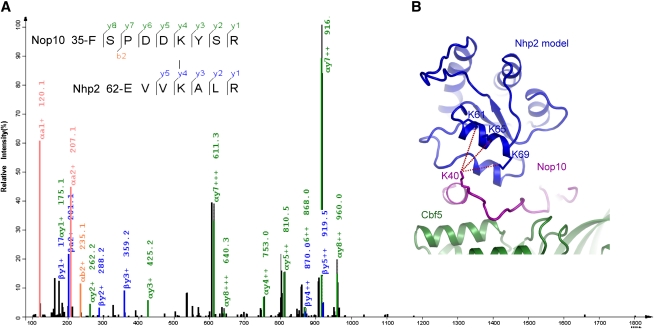
A previous study using in vitro translated mammalian proteins showed that Nhp2 interacts stably with the Cbf5 and Nop10 complex (Wang and Meier 2004). Our purification of the Cbf5C–Nop10–Nhp2 ternary complex confirms that this interaction also occurs in recombinant yeast proteins (Fig. 1A). Like L7Ae, Nhp2 possibly binds Nop10 near the upper stem of H/ACA RNA (Fig. 2B; Li and Ye 2006). To experimentally probe the location of Nhp2, we treated the copurified Cbf5C–Nop10–Gar1C–Nhp2 complex with a bivalant amine-reactive cross-linker, bis(sulfosuccinimidyl)suberate (BS3), and identified cross-linked peptides by mass spectroscopy (Fig. 7A). We identified three intermolecular cross-linked peptides that were formed between Nop10 Lys40 and Nhp2 Lys61, Lys65, and Lys69. Because BS3 has a spacer arm of 11.4 Å, the two cross-linked amine groups should be in close proximity, within a distance of 24 Å, given the flexibility and a 6 Å length of the lysine side chain. If Nhp2 is modeled as L7Ae in H/ACA RNP, the amine group of Nop10 Lys40 would be 9–13 Å away from that of the three Nhp2 lysines (Fig. 7B). The cross-linking results experimentally confirm that L7Ae and Nhp2 occupy a similar position in H/ACA RNP, despite their different modes of interaction with protein and RNA.
Figure 7.
Interaction between Nhp2 and Nop10 analyzed by chemical cross-linking and mass spectroscopy. (A) A representative fragmentation spectrum of a BS3 cross-linked peptide pair consisting of FSPDDKYSR (α peptide) from Nop10 and EVVKALR (β peptide) from Nhp2. The observed C-terminal fragments (y ions) resulting from a single event of peptide bond cleavage are highlighted in green if the cleavage occurs on the α peptide or blue if it occurs on the β peptide. The corresponding N-terminal fragments are called b ions, from which a further loss of carbonyl results in a ions. As is typical of collision-induced dissociation, y ions are the dominant species. Only b2+, a1+, and a2+ from the breakage of the α peptide are observed. (B) Three cross-linked lysine pairs between Nop10 and Nhp2 are shown in the model of yeast Nhp2 protein complex.
Discussion
We purified recombinant Cbf5 protein complexes in large amounts using a coexpression–copurification strategy, allowing for the detailed study of the structure–function relationship of yeast H/ACA RNPs. We show that the four yeast H/ACA RNP proteins Cbf5, Nop10, Nhp2, and Gar1 can form a protein-only complex in bacteria (Fig. 1A) and form a functional pseudouridine synthase with H/ACA RNAs in vitro (Fig. 1C). Therefore, yeast H/ACA RNPs, like their archaeal counterparts, also possess the intrinsic ability to self-assemble in the absence of assembly factors. The observed interaction patterns among the four proteins are consistent with previous results (Henras et al. 2004; Wang and Meier 2004).
The structure of the yeast Cbf5C–Gar1C–Nop10 ternary complex reveals a general similarity to the archaeal structure as well as many unique features that are likely conserved in eukaryotic RNPs. The small ribbon domain of yeast Nop10 is loosely packed in the absence of zinc coordination but still maintains a similar set of interactions with Cbf5. It appears that the interaction with Cbf5, rather than the ribbon domain structure itself, is under the most selective pressure in evolution. We observed structural changes in the interface between the C-terminal α helix of Nop10 and Cbf5, which may be related to Nhp2 that binds at this region (Fig. 7).
We observed a novel CTE in the core domain of Gar1 that contacts with the thumb of Cbf5. This interaction induces an open conformation of the thumb that is distinct from the open conformation observed for the Pf H/ACA RNP structure. We show that the thumb–CTE interaction is important for substrate turnover regulation. The additional interaction between the CTE of Gar1 and the thumb may lead to a better control on the pseudouridylation reaction that needs to occur in the moderate temperature range in which eukaryotes live.
The CTE appears to be a key feature that differentiates Gar1 from the assembly factor Naf1. Naf1 uses a Gar1 core-like domain to interact with Cbf5 and could even partially fulfill the role of Gar1 in maintaining a catalysis-competent conformation of the enzyme (Fig. 1C). However, Naf1, which lacks a corresponding CTE, is inactive in substrate turnover and binds Cbf5 more weakly than Gar1. Gar1 with a truncated CTE could be easily displaced from the Cbf5 complex by Naf1 (Fig. 6) and also lost localization in nucleoli and Cajal bodies (Pogacic et al. 2000). The CTE appears to provide a structural basis that allows Gar1 to replace Naf1 during H/ACA RNP biogenesis.
The NTE and CTE of Cbf5 are concentrated with many DC mutations, which could not be mapped to the previous archaeal Cbf5 structure. The yeast Cbf5 structure now provides an improved model for interpreting these mutations around the NTE that forms a new structural layer in the PUA domain. The CTE appears not to be part of the PUA domain and is instead involved in binding the assembly factor Shq1 (S Li, J Duan, and K Ye, in prep.).
Structural and functional changes in the upper stem region of yeast H/ACA RNPs
Eukaryotic and archaeal H/ACA RNPs display major structural differences in the region around the upper stem of H/ACA RNAs. Archaeal RNAs contain a K-turn motif in the upper stem that has a conserved distance from the pseudouridylation pocket and is bound specifically by L7Ae (Rozhdestvensky et al. 2003). L7Ae contacts Nop10 in the archaeal H/ACA RNP structure (Li and Ye 2006) but does not stably associate with Cbf5–Nop10 in the absence of RNA (Baker et al. 2005; Charpentier et al. 2005). In contrast, in eukaryotic complexes, no K-turn is present in the upper stem of H/ACA RNAs, and Nhp2 associates stably with the Cbf5–Nop10 protein complex (Wang and Meier 2004). Despite these differences, our chemical cross-linking data support the model that Nhp2 is located at a position in H/ACA RNPs similar to L7Ae and contacts Nop10 (Fig. 7). As Nhp2 binds to RNAs of various structures (Henras et al. 2001) and assumes a structure similar to L7Ae (Koo et al. 2011), it could potentially interact with the upper stem of H/ACA RNAs.
L7Ae is critical for the enzymatic activity of archaeal H/ACA RNPs. Removal of L7Ae significantly impairs even the basic catalytic activity (Baker et al. 2005; Charpentier et al. 2005) and also completely abolishes substrate turnover (Fig. 1F). In the structure of substrate-loaded archaeal H/ACA RNPs (Duan et al. 2009; Liang et al. 2009), the upper stem coaxially stacks with the substrate–guide duplex formed between the 3′ arm of the substrate and the 5′ guide. Proper anchoring of the upper stem by L7Ae would be important to correctly place the substrate at the active site. Accordingly, in a structure of H/ACA RNP lacking L7Ae, the substrate was not loaded in the active site (Liang et al. 2007).
In stark contrast to the critical role of L7Ae in assembling archaeal H/ACA RNAs and maintaining the catalytic-competent conformation of loaded substrates, we found that the yeast H/ACA RNP assembled without Nhp2 is highly active in both single- and multiple-turnover reactions (Fig. 1C). Hence, Nhp2 and its potential interaction with the upper stem of H/ACA RNA are largely dispensable to the enzymatic function of yeast H/ACA RNP, although Nhp2 is still required for optimal substrate turnover (∼50% reduction in reaction rate without Nhp2). We believe that the functional change between L7Ae and Nhp2 is related to the structural variability in the upper stem of eukaryotic H/ACA RNAs. In archaeal structure, the conserved K-turn motif and its interaction with L7Ae would precisely place the upper stem in the RNP structure. However, in eukaryotic H/ACA RNAs, the upper stem is highly heterogeneous, decorated by bulges and mismatches. The Nhp2–RNA interaction, if it exists, may also be variable among different H/ACA RNPs. It makes sense that eukaryotic H/ACA RNPs have evolved to not rely on a variable structure/interaction for the essential function of catalysis. An understanding of whether and how the upper stem coordinates the conformation of substrate RNA requires further study of eukaryotic H/ACA RNP structure.
Each single-hairpin domain of yeast H/ACA RNP is an independent structural and functional unit in vitro
Except in some early-diverging eukaryote Trypanosome brucei and Euglena gracilis whose H/ACA RNAs contain only a single hairpin (Liang et al. 2001; Russell et al. 2004), H/ACA RNAs from higher eukaryotic organisms universally contain two hairpins. The two hairpins likely associate with two sets of H/ACA proteins, forming a bipartite structure (Watkins et al. 1998). This evolutionary conservation implies that hairpin duality is functionally important. A previous in vivo study showed that the two hairpins are highly interdependent because disruption of either hairpin would inactivate the function of the other (Bortolin et al. 1999). Moreover, both hairpins of many H/ACA RNAs, except for the special U17 and hTR RNA, were shown to be required for RNP assembly in HeLa cell extracts (Dragon et al. 2000). The reason that eukaryotic H/ACA RNAs need two hairpins remains unknown. One possibility is that the assembly and activity depend on the presence of both hairpins. However, our results indicate that the isolated single hairpins of snR5 are also active in guiding pseudouridylation, suggesting that the single-hairpin domain is an independent structural and functional unit of yeast H/ACA RNP, at least in vitro. Two-hairpin RNPs appear more efficient in catalysis than single-hairpin RNPs, which might provide some functional advantage in vivo.
Materials and methods
Plasmids
The DNA sequences that encode full-length Cbf5 (residues 1–483) or Cbf5C (residues 1–394) and six C-terminal histidines were cloned into an ampicillin-resistant pETDuet-1 plasmid at the NcoI and EcoRI sites of the first multiple clone site (Novagen). The full-length Nop10 gene (residues 1–58) was cloned to the Cbf5-containing pETDuet-1 plasmid at the NdeI and XhoI sites of the second multiple clone site. The gene of full-length Gar1 (residues 1–205) or Gar1C (residues 32–124) was cloned into a kanamycin-resistant pET28a plasmid at the NdeI and EcoRI sites as a fusion to an N-terminal His6 tag and thrombin cleavage site. The full-length Nhp2 (residues 1–156) gene was cloned into the second multiple clone site of a pCDFDuet-1 plasmid without any affinity tag at the NdeI and EcoRI sites. The Nhp2 gene was also cloned into the second multiple clone site of a modified pETDuet-1 plasmid at the HindIII and EcoRI sites, in which Nhp2 was fused to an N-terminal His6 tag and PreScission cleavage site. Naf1C (residues 107–280) was cloned into a modified pCDFDuet-1 plasmid at the EcoRI and HindIII sites and was fused to an N-terminal His6 tag and PreScission cleavage site. Mutagenesis was created by QuikChange (Stratagene). All clones were validated by DNA sequencing. The generated plasmids and used primers are listed in Supplemental Tables S1 and S2.
Protein expression and purification
All proteins were expressed in E. coli Rosetta(DE3) or BL21(DE3) strains. The cells were resuspended in 50 mM Tris-Cl (pH 8.0), 1 M NaCl, and 5% glycerol and lysed by a high-pressure cell breaker. The Cbf5C–Nop10 complex was coexpressed from pETDuet-1 harboring both genes. The Cbf5C–Nop10–Gar1C complex was coexpressed from pETDuet-1 plasmid expressing Cbf5C and Nop10 and pET28a plasmid expressing Gar1C. For the purification of the Nhp2 complex, excessive bacteria expressing Nhp2 from pCDFDuet-1 were mixed with bacteria coexpressing Cbf5C–Nop10 or Cbf5C–Nop10–Gar1C before cell lysis. All Cbf5-containing complexes were purified similarly through HisTrap, heparin, and gel filtration chromatography (GE Healthcare). The His6 tag of Gar1 was cleaved by thrombin, if needed, before gel filtration chromatography. His-tagged Nhp2 alone was expressed from pETDuet-1 plasmid and purified through HisTrap, heparin, and gel filtration chromatography. The His6 tag on Nhp2 was cleaved by PreScission after HisTrap chromatography. Naf1C was purified by HisTrap chromatography, followed by His6 tag cleavage by PreScission and heparin chromatography. Protein complexes with full-length Cbf5 or full-length Gar1 were expressed and purified similarly. Protein concentrations were determined using absorbance at 280 nm and calculated molar extinction coefficiency. H/ACA RNAs were prepared by in vitro transcription using linearized plasmid DNA templates or PCR products.
Naf1 competition assay
Gar1C (residues 32–124) and Gar1CΔ120 (residues 32–120) were coexpressed and copurified with Cbf5C and Nop10. The N-terminal His6 tag of Gar1 was cleaved by thrombin before gel filtration chromatography. Cbf5C complexes (200 pmol) were incubated with 0, 50, 200, 800, and 3200 pmol of Naf1C in 100 μL of binding buffer containing 50 mM sodium phosphate (pH 7.6), 0.4 M KCl, and 5% glycerol for 1 h at room temperature. The reactions were added with 30 μL of Ni beads and an additional 400 μL of binding buffer and nutated for 1 h at 4°C. The Ni beads were washed once with 1 mL of 40 mM imidazole in binding buffer and eluted with 30 μL of 500 mM imidazole in binding buffer. Then, 10 μL of eluate was mixed with 2× SDS loading buffer and resolved in a 4%–12% gradient SDS-PAGE gel.
Crystallization and structure determination
The Cbf5C–Nop10–Gar1C complex (20 mg/mL in 50 mM Tris-Cl at pH 8.0, 500 mM NaCl, 10% glycerol, 1 mM DTT) was mixed with a reservoir solution (0.2 M sodium malonate at pH 8.04, 7% [w/v] polyethylene glycol 3350), both at volumes of 1 μL, and was crystallized at 20°C by the hanging-drop vapor diffusion method. The crystal was cryoprotected in 0.2 M sodium malonate (pH 8.04), 25% (w/v) polyethylene glycol 3350, and 10% glycerol before freezing in liquid nitrogen.
The diffraction data were collected at the Beijing Synchrotron Radiation Facility beamline 3W1A and processed by Denzo and Scalepack (Otwinowski and Minor 1997). The diffraction data had an overlap problem and were ∼80% complete at the high-resolution shells. The structure was solved by molecular replacement in Phaser (McCoy et al. 2007) using the corresponding part of the Pf H/ACA RNP structure as a search model (Li and Ye 2006). The crystal has a space group of P212121 with cell dimensions of a = 54.4 Å, b = 59.4 Å, and c = 169.2 Å, and contains one trimer in the asymmetric unit. The structure was built in Coot (Emsley and Cowtan 2004) and refined in Refmac to 1.9 Å resolution (Murshudov et al. 1999). The final model contains Cbf5 (residues 5–11, 18–155 and 161–350), Nop10 (1–48), and Gar1 (33–124), and 280 water molecules. Analysis by RAMPAGE shows that 97.6% of the residues are in favored regions, 2.2% are in allowed regions, and 0.2% are in outlier regions (Lovell et al. 2003). The structural figures were prepared in PyMOL (DeLano 2002).
Pseudouridylation activity assay
The 32P-labeled 5′ substrate for snR5 was prepared by ligating a 5′ half RNA (5′-dAGGCCAUUUU-3′) and a 5′-32P-labeled 3′ half RNA (5′-UGGUAAGCAdA-3′) annealed to a bridging DNA (5′-TTGCTTACCAAAAATGGCCTT-3′). The 32P-labeled 3′ substrate for snR5 was generated by ligating a 5′ half RNA (5′-dAdAdAAGCGAAU-3′) and a 5′-32P-labeled 3′ half RNA (5′-GAUUAGAdAdAdA-3′) annealed to a bridging DNA (5′-TTTCTAATCATTCGCTTT-3′). Unlabeled 5′ and 3′ substrate have the sequences 5′-GGCCAUUUUUGGUAAGCA-3′ and 5′-AAGCGAAUGAUUAGA-3′, respectively. The guide RNA (RNA1) and cognate substrate for Pf H/ACA RNP were described previously (Duan et al. 2009).
The two-hairpin snR5 RNA and single-hairpin RNAs (snR5-5′hp, snR5-3′hp, and Pf RNA1) were assembled with two and one molar equivalents of proteins, respectively. The modification reactions contained 1 or 0.2 μM H/ACA RNP (referring to the guide RNA concentration), ∼0.01 μM 32P-labeled substrate, and 0 or 20 μM unlabeled substrate in a 10-μL volume of reaction buffer. The reaction buffer was composed of 1 M NaCl and 50 mM phosphate (pH 7.6) for Pf H/ACA RNPs, and 500 mM NaCl and 25 mM Tris-Cl (pH 8.0) for yeast H/ACA RNPs. The reactions were performed at 37°C for Pf RNPs and at 30°C for yeast RNPs. RNA extraction, digestion, and thin-layer chromatography were performed as previously described (Duan et al. 2009). The time course data were fit to the equation y = 1 − e−At, where y is a fraction of Ψ, t is time in minutes, and A is the rate constant. The substrate turnover rate at time 0 was calculated as A multiplied by the substrate-to-enzyme ratio.
Chemical cross-linking and mass spectroscopy
The copurified Cbf5C–Nop10–Gar1C–Nhp2 protein complex (1 μM) was incubated with 1 mM BS3 (Thermo Scientific) in 50 mM HEPES (pH 8.0) and 500 mM NaCl for 1 h at room temperature. The reaction was terminated by ammonium bicarbonate with a final concentration of 20 mM. The detailed protocol of sample preparation and mass spectroscopy analysis will be described elsewhere.
Accession code
The atomic coordinates and structure factors for the yeast Cbf5C–Nop10–Gar1C complex structure have been deposited in the Protein Data Bank under accession number 3U28.
Acknowledgments
We thank the staff at the Beijing Synchrotron Radiation Facility for assistance in data collection, and the NIBS protein facility for mass spectroscopy analysis. This research was supported by the Chinese Ministry of Science and Technology through 973 (2010CB835402) and 863 (2008AA022310) projects and the Beijing Municipal Government.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.175299.111.
References
- Atzorn V, Fragapane P, Kiss T 2004. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol 24: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP 2005. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 25: 5396–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Ganot P, Kiss T 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B, Muller S, Branlant C 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 33: 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL 2002. The PyMOL user's manual. Delano Scientific, San Carlos, CA [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol Cell Biol 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Pogacic V, Filipowicz W 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol 20: 3037–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fatica A, Dlakic M, Tollervey D 2002. Naf1p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514 [PMC free article] [PubMed] [Google Scholar]
- Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T 2009. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J 28: 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Girard JP, Bagni C, Caizergues-Ferrer M, Amalric F, Lapeyre B 1994. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J Biol Chem 269: 18499–18506 [PubMed] [Google Scholar]
- Hamma T, Reichow SL, Varani G, Ferre-D'Amare AR 2005. The Cbf5–Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol 12: 1101–1107 [DOI] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M 2001. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 29: 2733–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M 2004. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 10: 1704–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon HJ, Fouquet C, Carbon J 1993. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol Cell Biol 13: 4884–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M, Wu H, Johansson C, Caizergues-Ferrer M, Feigon J 2006. Structural study of the H/ACA snoRNP components Nop10p and the 3′ hairpin of U65 snoRNA. RNA 12: 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jady BE 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606 [DOI] [PubMed] [Google Scholar]
- Koo BK, Park CJ, Fernandez CF, Chim N, Ding Y, Chanfreau G, Feigon J 2011. Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol 411: 927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Godin KS, Hoareau-Aveilla C, Quevillon-Cheruel S, Varani G, Henry Y, Van Tilbeurgh H 2007. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J Mol Biol 371: 1338–1353 [DOI] [PubMed] [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu L, Michaeli S 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J Biol Chem 276: 40313–40318 [DOI] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H 2007. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 14: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16: 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC 2003. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50: 437–450 [DOI] [PubMed] [Google Scholar]
- Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C 2006. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5–aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res 34: 826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A, Walne A, Dokal I 2005. Dyskeratosis congenita: Telomerase, telomeres and anticipation. Curr Opin Genet Dev 15: 249–257 [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol 13: 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol 20: 9028–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H 2006. Crystal structure of a Cbf5–Nop10–Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 21: 249–260 [DOI] [PubMed] [Google Scholar]
- Reichow SL, Varani G 2008. Nop10 is a conserved H/ACA snoRNP molecular adaptor. Biochemistry 47: 6148–6156 [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Schnare MN, Gray MW 2004. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA 10: 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Dokal I 2009. Advances in the understanding of dyskeratosis congenita. Br J Haematol 145: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Meier UT 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 23: 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Query CC, Meier UT 2002. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol Cell Biol 22: 8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang C, Schattner P, Yu YT 2009. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 15: 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G 2002. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J Biol Chem 277: 45235–45242 [DOI] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 25: 3295–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K 2007. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol 17: 287–292 [DOI] [PubMed] [Google Scholar]