Abstract
Recent studies suggest a potential role of lipid lowering drugs, fibrates and statins, in anticancer treatment. One candidate for tumor chemoprevention is fenofibrate, which is a potent agonist of peroxisome proliferator activated receptor alpha (PPARα). Our results demonstrate elevated expression of PPARα in the nuclei of neoplatic cells in 12 out of 13 cases of medulloblastoma, and of PPARc in six out of 13 cases. Further analysis demonstrated that aggressive mouse medulloblastoma cells, BsB8, express PPARα in the absence PPARγ, and human medulloblastoma cells, D384 and Daoy, express both PPARα and PPARγ. Mouse and human cells responded to fenofibrate by a significant increase of PPAR-mediated transcriptional activity, and by a gradual accumulation of cells in G1 and G2/M phase of the cell cycle, leading to the inhibition of cell proliferation and elevated apoptosis. Preincubation of BsB8 cells with fenofibrate attenuated IGF-I-induced IRS-1, Akt, ERKs and GSK3β phosphorylation, and inhibited clonogenic growth. In Daoy and D384 cells, fenofibrate also inhibited IGF-I-mediated growth responses, and simultaneous delivery of fenofibrate with low dose of the IGF-IR inhibitor, NVP-AEW541, completely abolished their clonogenic growth and survival. These results indicate a strong supportive role of fenofibrate in chemoprevention against IGF-I-induced growth responses in medulloblastoma.
Keywords: PPARα, fenofibrate, IGF-I, medulloblastoma
Medulloblastomas are highly malignant cerebellar tumors of the childhood, which originate from the external granule layer of the cerebellum and have an inherent tendency to spread in the CNS via cerebrospinal fluid. Medulloblastomas are characterized by a large number of genetic and epigenetic aberrations.1 Among them, overexpression of insulin-like growth factor receptor (IGF-IR), and insulin receptor substrate 1 (IRS-1) are frequently seen in these tumors.1–5 The IGF-IR is a membrane-associated tyrosine kinase capable of mediating a variety of biological responses including cell survival and cell proliferation.6,7 In the cerebellum, the IGF-IR has been shown in the granule cell layer and in Purkinje cells, and IGF-I protected cultures of cerebellar neurons from low potassium induced apoptosis.8,9 In medulloblastoma, the IGF-IR signaling system has been investigated quite intensively.4,10 Resent results from our laboratory demonstrate that medulloblastoma cell lines and medulloblastoma biopsies are characterized by an abundant presence of the IGF-IR, and its major signaling molecule, IRS-1.2,3 Importantly, we have detected the phosphorylated form of the IGF-IR (active) in the majority of medulloblastoma clinical samples examined.2 In addition, growth and survival of medulloblastoma cell lines cultured in suspension was strongly dependent on the presence of exogenous IGF-I.2,3 The low-molecular-weight inhibitor of the IGF-IR, NVP-AEW541, is one of the most effective inhibitors of the growth of medulloblastoma cells and induces massive apoptosis in suspension cultures of mouse and human medulloblastoma cell lines.11 We have therefore searched for an additional therapeutic target, which when combined with NVP-AEW541 could enhance the efficacy and decries toxicity the chemotherapy against medulloblastoma. Previous work in our laboratory indicate that PPARs agonists might serve as the additional agents, because they are known to affect both insulin receptor and IGF-I receptor signaling pathways,12,13 and are characterized by relatively low toxicity.14,15 PPARs are nuclear receptors, which belong to the superfamily of steroid hormone receptors.16 Their transcriptional activity after ligand activation is associated with the formation of heterodimers with the retinoid X receptors and binding to the specific DNA sequences.17 To date, three types of PPARs have been identified: PPARα, PPARβ and PPARγ. In the cerebellum all three cellular layers: the molecular, Purkinje cells and granule neurons, express PPARs.18,19 Recent reports indicate that activation of PPARα by fenofibrate attenuates clonogenic growth and migration of melanoma cell lines.14,15 Interestingly, fenofibrate has been widely used to lower plasma levels of triglycerides and cholesterol, to improve LDL:HDL ratio, and to prevent the development of arteriosclerosis mainly through regulation of apolipoprotein expression.20 Fenofibrate is also a potent ligand for PPARα, which has been originally discovered as a regulator of glucose and lipid metabolism, with potential anticancer properties.14,15 Since PPARs have not been studied in medulloblastomas, we attempted to ask whether their activation could repress malignant growth of these cerebellar tumors. Our results show elevated levels of PPARα and PPARγ in medulloblastoma clinical samples (Fig. 1), and in mouse and human medulloblastoma cell lines (Fig. 2). We have determined also that BsB8 mouse medulloblastoma cells express exclusively PPARα, and that D384 and Daoy human medulloblastoma cells express both PPARα and PPARγ. Mouse and human medulloblastoma cells responded to fenofibrate by a gradual withdrawal from the cell cycle at early time points after the treatment, which resulted in almost complete inhibition of cell proliferation and induction of apoptosis at later time points (after 72 hr). Fenofibrate attenuated IGF-I-induced phosphorylation events, which was accompanied by a severe retardation of their clonogenic growth. Although in human medulloblastoma cell lines fenofibrate was much less effective in triggering apoptosis, simultaneous delivery of fenofibrate with low doses of NVP-AEW541 (0.5 µM) resulted in a complete inhibition of their clonogenic growth. These results indicate that activation of PPARα by fenofibrate inhibits cell cycle progression in the first 48 hr, and after prolonged treatment (over 72 hr) it can induce apoptosis. Because of the low toxicity of fenofibrate,14,15 it could be considered as a supplementary treatment against IGF-I-mediated growth responses in medulloblastoma.
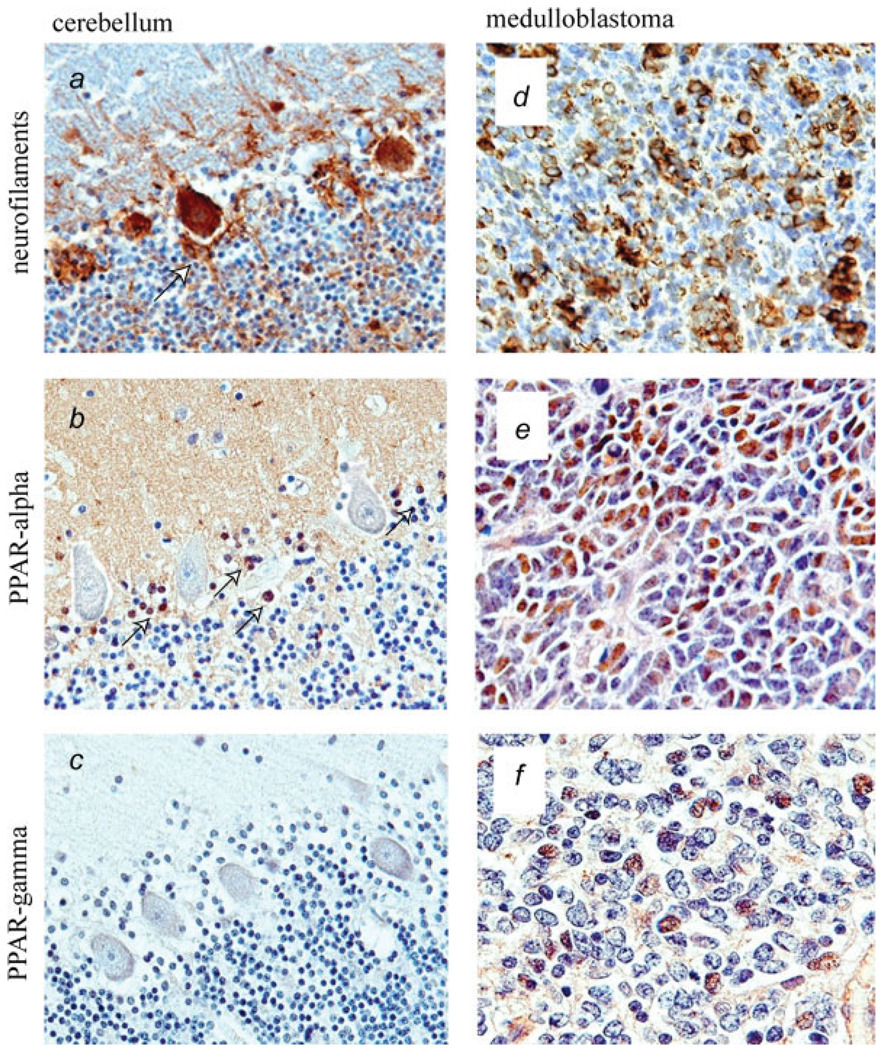
FIGURE 1.
Immunohistochemical detection of PPARs in medulloblastoma clinical samples. Evaluation of PPARα and PPARγ protein levels in medulloblastomas in comparison to normal cerebellar tissue. (a–c) View of the normal cerebellum where the classic layers, molecular, Purkinje cell and granular, can be identified. (a) Immunohistochemistry for neurofilaments highlights the Purkinje neurons and their adjacent Basket cells (arrow). (b) PPAR-α is absent in the majority of cells in normal cerebellum with exception of Basket cells, which express robust nuclear PPARα (arrow). (c) No expression of PPARγ was detected in normal cerebellar tissues. (d) Class III β-tubulin reveals the primitive neuronal phenotype of the tumors cells in a representative case of classic medulloblastoma. (e) PPARα is expressed in the nuclei of neoplastic cells in a representative case of medulloblastoma. (f) PPAR-γ is shown in the nuclei of some tumor cells in PPARγ positive case of medulloblastoma.
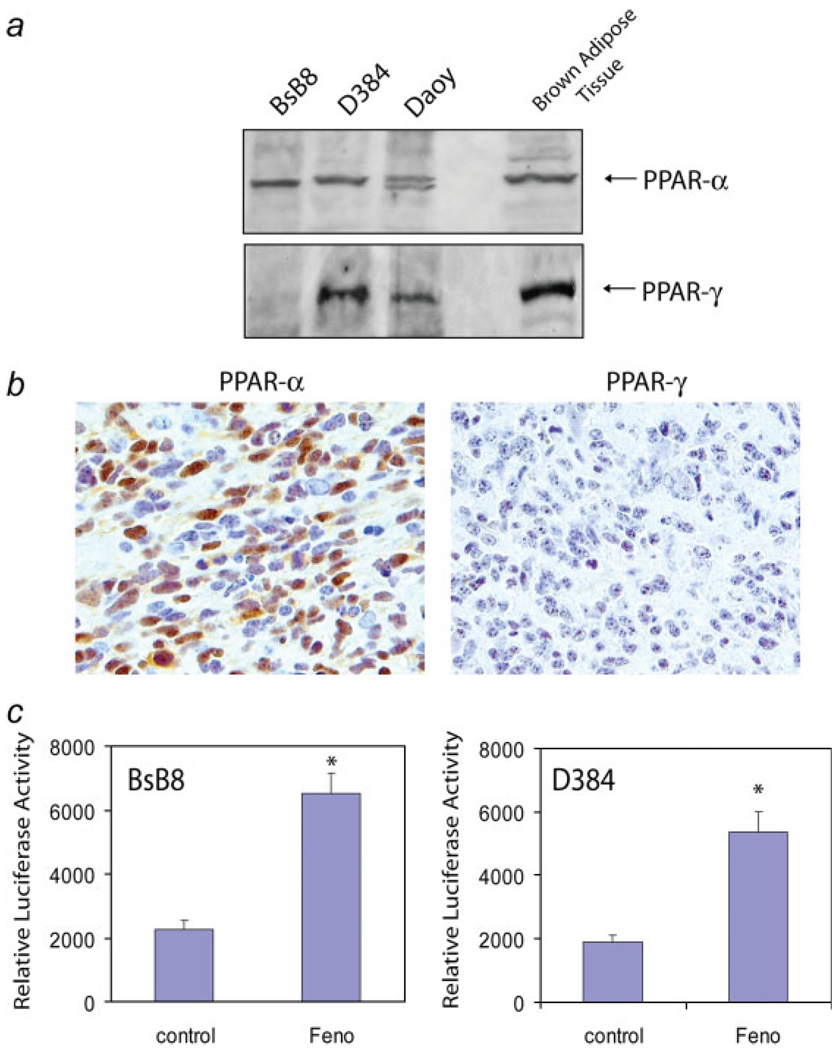
FIGURE 2.
Peroxisome proliferator-activated receptors (PPARs) in medulloblastoma cell lines. (a) Western blot showing PPARα and PPARα in two human (D384, Daoy) and one mouse (BsB8) medulloblastoma cell lines. Brown adipose tissue was used as a positive control. Note that human medulloblastoma cell lines express both PPARα and PPARγ and mouse medulloblastoma express PPARα in the absence of PPARγ. (b) Histological demonstration of a primitive neuroectodermal tumor (PNET) in mouse medulloblastoma model from which BsB8 cells have been developed. Note that mouse neoplastic cells express nuclear PPARα, but are negative for PPARγ. (c) PPARs transcriptional activity in BsB8 and D384 cells. The reporter plasmid contains luciferase gene driven by PPAR responsive element (PPRE), which consist of three copies of the J site from apo-AII gene promoter. The activation of PPAR elements was evaluated by a dual-Firefly/Renilla luciferase reporter system (Promega). Data are presented as mean ± SD calculated from two experiments in triplicates (n = 6). * indicates values statistically significantly different (p ≤ 0.05) from control (cells treated with vehicle only). Statistical significance between two measurements was determined with the two-tailed Student’s t test.
Material and methods
Immunohistochemistry
A total of 13 cases of human medulloblastoma were acquired from the pathology archives of the University Health Network, University of Toronto, Ontario, Canada. The formalin-fixed, paraffin-embedded samples were sectioned at 4 microns in thickness, stained with Hematoxilin and Eosin and histologically classified according to the World Health Organization Classification of Tumors of the Nervous System. Immunohistochemistry was performed using the avidin–biotin–peroxidase complex system, according to the manufacturer’s instructions (Vectastain Elite ABC Peroxidase Kit; Vector Laboratories, Burlingame, CA). Briefly, sections were deparaffinized in xylene and rehydrated through descending alcohols to water. For nonenzymatic antigen retrieval sections were heated in 0.01 M sodium citrate buffer (pH 6.0) to 95°C for 40 min and allowed to cool for 20 min at room temperature. To quench endogenous peroxidase, slides were then rinsed with PBS and incubated in MeOH/3% H2O2 for 20 min. Sections were then washed with PBS and blocked in PBS/0.1% BSA containing 5% normal horse or goat serum for 2 hr at room temperature, and incubated overnight at room temperature with primary antibodies. The primary antibodies used in our study include: a mouse monoclonal anti-PPARα antibody (1:100 dilution; Chemicon, Temecula, CA), a rabbit polyclonal anti-PPARγ (E-8, 1:300 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), a mouse monoclonal anti-class IIIβ-tubulin (TUJ1, 1:500 dilution; Covance, Berkeley, CA) and a mouse monoclonal cocktail for different molecular weight neurofilaments (SMI-312, 1:2000 dilution; Sternbeger Monoclonals, Lutherville, MA). Secondary antibodies used were biotin-conjugated horse anti-mouse and goat anti-rabbit IgGs (Vector). Sections were developed with a diaminobenzidine substrate (Sigma, St. Louis, MO), counterstained with hematoxylin and mounted with Permont.
Cell culture conditions
The following cell lines were used in our study: a mouse medulloblastoma, BsB8, isolated from a cerebellar tumor of a transgenic mouse expressing the early genome of human polyomavirus JC3,21; and two human medulloblastoma cell lines, D384 Med and Daoy. D384 are metastatic medulloblastomas isolated from peritoneal ascietes of a child with medulloblastoma,22 and Daoy are derived from a tumor in the posterior fossa of a 4-years-old boy (ATCC no. HTB186). BsB8 cells are very aggressive, capable of forming large tumors in subcutaneous tissue, as well as intracranial tumors after stereotactic injection of a small number of cells in to the brain, and have the ability of spreading into cerebellum. 1,23 BsB8 and Daoy were maintained as monolayer cultures in Dulbecco’s modified Eagle medium (DMEM) (GIBCO-BRL, Grand Island, NY) containing 10% fetal bovine serum (FBS), at 37°C in a 9.6% CO2 atmosphere. D384 were cultured in suspension, in the Eagle’s MEM supplemented with nonessential amino acids (GIBCO-BRL), 2 mM l-glutamine, 1 mM sodium pyruvate, and 20% FBS. The cells were made partially quiescent by 48 hr incubation in serum-free medium (SFM) (DMEM supplemented with 0.1% bovine serum albumin). Cellular growth and signaling responses were tested by stimulating serum starved cells with 50 ng/ml of recombinant IGF-I (GIBCO-BRL).
Luciferase assay
The PPARs transcriptional activity in BsB8 and D384 cells was determined by utilizing the JsTkpGL3 reporter plasmid, which contains the luciferase gene driven by PPAR responsive element (PPRE), which consists of three copies of the J site from apo-AII gene promoter.24 The activation of PPAR elements was evaluated by a dual-Firefly/Renilla luciferase reporter system (Promega, Madison, WI), using Femtomaster FB12 chemiluminometer (Zylux Corp., Oak Ridge, TN).
Western blot analysis
To evaluate phosphorylation levels of the selected IGF-IR signaling molecules, semiquiescent monolayer cultures were stimulated with IGF-I and total protein extracts collected at the indicated time points. The cells were lysed for 5 min on ice with 400 µl of lysis buffer A [50 mM HEPES, pH 7.5; 150 mM NaCl; 1.5 mM MgCl2; 1mM EGTA; 10% glycerol; 1% Triton X-100; 1 µM phenylmethylsulfonyl fluoride (PMSF); 0.2 mM Na-ortho-vanadate; 10 µg/ml aprotinin]. Total protein extracts (50 µg) were separated on a 4–15% gradient SDS-PAGE (BioRad, Hercules, CA) and transferred to nitrocellulose filters. The following primary anti-phospho antibodies were utilized: anti-pY612IRS-1 rabbit polyclonal (BioSource, Camarillo, CA); anti-pS473Akt, anti-pT202/Y204Erk1/2, and anti-pS21/9GSK3α/β (Cell Signaling Technology, Beverly, MA). In addition, anti-PPARα mouse monoclonal antibody (Chemicon) and anti-PPARγ rabbit polyclonal antibody (Santa Cruz Biotechnology) were utilized. To monitor loading conditions we used anti-IRS-1 (Upstate USA, Charlottesville, VA), anti-GSK3β, anti-Akt, anti-Erks antibodies (Cell Signaling, Danvers, MA) and anti-Grb-2 antibodies.
Cell cycle analysis and apoptosis
Aliquots of cells, 1 × 106/ml, were fixed in 70% ethanol for 30 min at 4°C. Cells were centrifuged at 1,600 rpm and the resulting pellets were resuspended in 1 ml of freshly prepared propidium iodide/RNaseA solution. Cell cycle distribution was analyzed with GuavaEasy Cyte system by using Guava CytoSoft cell cycle program according to the manufacturer recommendations (Guava Technologies, Hayward, CA). Based on differences in PI intensity of fluorescence, cells in G1, S, G2/M and subG1 were separated and counted. Each cell cycle experiment was repeated at least three times. Cells in subG1 are characterized by DNA content, which is below the DNA content in G1 (single copy of the genome), and are considered as a mixture of cells dying by apoptosis and/or necrosis. To monitor apoptosis, cells were additionally labeled by in situ terminal dUTP nick-end labeling (TUNEL) assay. We have utilized the Guava TUNEL kit and the Guava TUNEL EasyCyte program to determine the percentage of medulloblastoma cells undergoing apoptosis at different time points after fenofibrate treatment.
Cell growth in monolayer
BsB8 cells were seeded at a concentration of 2 × 104 cells/35 mm dish, in DMEM containing 10% FBS and 50ng/ml IGF-I. 24 hr after plating cells were either treated with 25 µM fenofibrate, or were left untreated. At 48 and 72 hr time points, the cells were collected by trypsinization and counted in a Bright-line Hemocytometer in the presence of Trypan blue. T0 is the cell number determined 16 hr after the initial plating of the cells, which reflects plating efficiency.
Clonogenic assay
Cells were plated at 1 × 103 on 6-well culture plates (Costar, Corning, NY) in DMEM supplemented with 10% FBS and 50 ng/ml of IGF-I. After attachment, cells were treated with fenofibrate, NVP-AEW541 (kindly provided by Novartis Pharma, Basel, Switzerland), or with combination of fenofibrate and NVP-AEW541 at the indicated concentration. Clonogenic potential was evaluated 10 days after the treatment by fixing and staining corresponding cell cultures with 0.25% Brilliant Blue in methanol, which was followed by macroscopic count of the clones.
Colony formation in soft agar
To assess anchorage-independent growth of D384, cells were plated at 1 × 104 per 35 mm/dish in DMEM containing 10% FBS, 50 ng/ml of IGF-I and 0.2% agarose, with 0.4% agarose underlay. The growth medium was additionally supplemented with 25 µM fenofibrate or with a combination of 25 µM fenofibrate and 0.5 µM NVP-AEW541. Clones larger that 125 µm in diameter were scored after continuous growth of the cells for 2 weeks.
Results
Detection of PPARs in medulloblastoma clinical samples
The results in Figure 1 demonstrate nuclear expression of PPARα and PPARγ in a significant number of tumor cells from two representative medulloblastoma biopsies (Figs. 1e and 1f). In normal human cerebellum (Fig. 1a–1c), PPARα was detected only in proximity to Purkinje cells, where a population of neurofilament positive Basket cells (Fig. 1a; arrow) was strongly positive for PPARα (Fig. 1b; arrows). In contrast, immunolabeling with anti-PPARγ antibody of normal human cerebellum (including molecular layer, granular layer, Purkinje cells and Basket cells) was negative (Fig. 1c). We have analyzed a total of 13 medulloblastoma clinical samples, and detected PPARα in 92%, and PPARγ in 46% of all medulloblastoma cases examined.
Fenofibrate -mediated activation of PPARα in medulloblastoma cell lines
We have previously reported strong antitumoral properties of a synthetic PPARα agonist, fenofibrate, which in melanoma cell lines attenuated constitutive phosphorylation of Akt.15 To investigate whether fenofibrate can affect IGF-I-mediated growth responses in medulloblastomas, we selected the aggressive mouse medulloblastoma cell line, BsB8, and two human medulloblastoma cell lines Daoy and D384, which are strongly responsive to the IGF-I stimulation.3,4,11 The western blot analysis depicted in Fig. 2a shows that BsB8, Daoy and D384 cells express PPARα protein at levels comparable to the control brown adipose tissue. In contrast to human medulloblastoma cell lines, which express both PPARα and PPARγ, BsB8 cells were negative for PPARγ. Our finding was additionally confirmed in JCV T-antigen transgenic mice (Fig. 2b), which develop cerebellar primitive neuroectodermal tumors, from which BsB8 were originally isolated.3,21 The luciferase based transcriptional activity assay demonstrated that the fenofibrate treatment resulted in three-fold and four-fold increase in the activity of PPREs in BsB8 and D384 cells, respectively (Fig. 2c), further confirming PPARα-mediated transcriptional activity in mouse and human medulloblastoma cell lines.
PPARα-mediated inhibition of growth responses
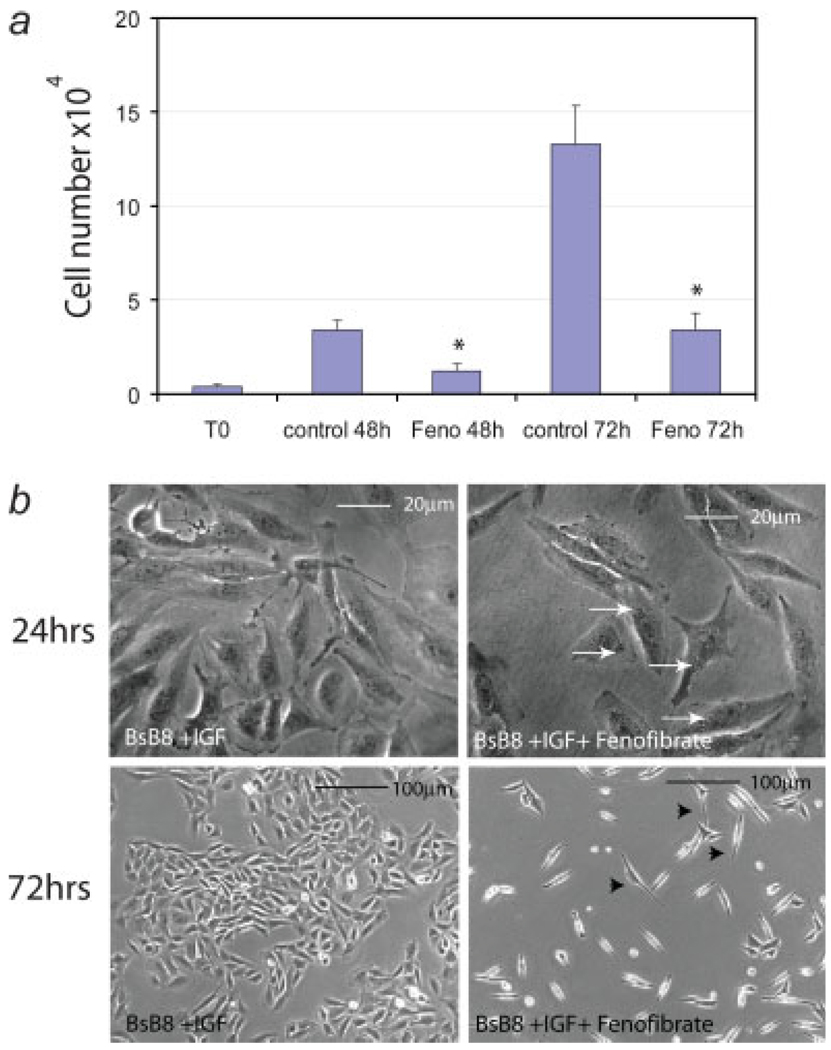
We than evaluated the monolayer growth of BsB8 cells, the condition in which medulloblastoma cells are quite resistant to the treatment against the IGF-IR function.11 The cells were plated in 10%FBS in the presence (Feno) or absence (control) of 25 µM fenofibrate.15 The cell number was determined at time zero (T0; 16 hr after initial plating), and at 48 and 72 hr after the fenofibrate treatment. The results in Figure 3a show that in comparison to BsB8 cells growing in the presence of 10%FBS, fenofibrate-treated cells demonstrated a 4-fold lower rate of cell proliferation. This growth inhibition was accompanied by morphological changes (Fig. 3b), including accumulation of perinuclear structures within first 24 hr after the treatment (arrows), which likely represents increased peroxisome proliferation,25 the appearance of spindle shaped large nondividing cells, between 48 and 72 hr (arrowheads), and apoptotic cells in later time points (Fig. 4b; subG1 cell population).
FIGURE 3.
Inhibition of growth responses of BsB8 cells after PPARα activation. (a) BsB8 mouse medulloblastomas were plated in the presence of 10% FBS at 2 × 104/35 mm dish. Attached cells were treated with 25 µM fenofibrate (Feno) or were left without treatment (control). The cell number was evaluated at T0 (16 hr after initial plating) and at 48 and 72 hr after the fenofibrate treatment. Data are presented as mean ± SD calculated from three experiments in triplicates (n = 9). * indicates values statistically significantly different (p ≤ 0.05) from control values (cells treated with vehicle only). Statistical significance between two measurements was determined with the two-tailed Student’s t test. (b) Photographic documentation of fenofibrate-mediated morphological changes and growth inhibition. Phase contrast images were taken from control (BsB8 + IGF-I) and fenofibrate-treated cultures (BsB8 + IGF-I + fenofibrate) at 24 hr (large magnification— to visualize accumulation of perinuclear vacuoles) and at 72 hr (small magnification—to visualize growth inhibition). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
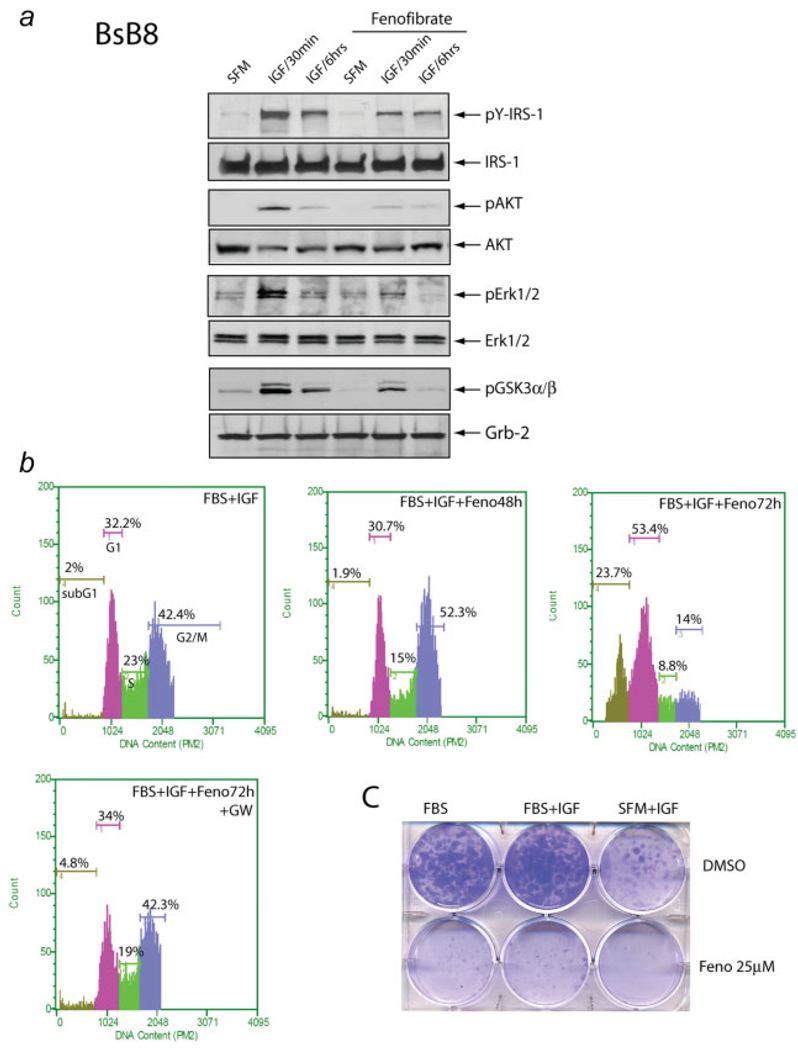
FIGURE 4.
Effects of fenofibrate on IGF-I signaling pathways. (a) After 24 hr incubation in serum-free medium (SFM), quiescent BSB8 were cultured in the presence or absence of fenofibrate for additional 24 hr. Subsequently, both cell groups were treated with IGF-I (50 ng/ml) for 30 min, 6 hr or were left without IGF-I treatment (SFM). Western blots were prepared with corresponding protein extracts (50 µg) separated on a 4–15% gradient SDS-PAGE, and were probed with the following primary antibodies: anti-(pSer473)Akt, anti-(pT202/Y204)Erk1/2, anti-(pS21/9) GSK3α/β and anti-(pY612) IRS-1. Anti-Grg-2, anti-IRS-1, anti-Akt and anti-Erk1/2 antibodies were used as loading controls. (b) Evaluation of cell cycle distribution after fenofibrate treatment. Exponentially growing monolayer cultures of BsB8 cells were treated with 25 µM fenofibrate (FBS + IGF + Feno) for either 48 of 72 hr, or were left without the fenofibrate (10% FBS + IGF). In one of the control experiments, the fenofibrate treatment was performed in the presence of PPARα synthetic antagonist GW-9662 (10 µM). Subsequently, cells were tripsinized, fixed in ethanol and the DNA content was determined by propidium iodide (PI) DNA labeling. Cell cycle distribution was evaluated by the Guava EayCyte flow cytometer. Note that 48 hr incubation of BsB8 cells in the presence of fenofibrate resulted in only a partial G2/M arrest. At 72 hr after the fenofibrate treatment, however, there is a significant increase of cells in sub-G1 (cell death), proportional decrease of cells in S and G2/M and accumulation in G1. (c) IGF-I and serum stimulated clonogenic growth of BsB8 cells is severely impaired by 25 µM fenofibrate. In control experiment, cells were treated with DMSO in which fenofibrate was diluted. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Effects of fenofibrate on IGF-I signaling pathways
Since fenofibrate modulates insulin-mediated cellular responses, and the IGF-IR signaling system is highly active in medulloblastomas,1 we asked whether the observed growth inhibitory effects of fenofibrate are associated with the inhibition of IGF-IR signaling pathway(s). After 24 hr incubation in SFM, semiquiescent BsB8 cells were cultured in the presence or absence of 25 µM fenofibrate for an additional 24 hrs. Subsequently, the cells were treated with IGF-I (50 ng/ml) for 30 min, 6 hr, or were left without the IGF-I treatment (SFM). The results in Figure 4a demonstrate a strong increase in the phosphorylation of IRS-1, Akt, Erks1/2 and GSK-3β during first 30 min after IGF-stimulation, and an expected desensitization of the signal at the 6 hr time point. Preincubation of the cells with 25 µM fenofibrate attenuated IGF-I-mediated phosphorylation events (Fig. 4a). Cell cycle analysis shown in Fig. 4b revealed accumulation of cells in G2/M phase, and decrease in the S phase of the cell cycle at the 48 hr time point, which was followed by the accumulation of cells in subG1 phase 72 hr after the treatment. This subG1 cells fraction (23.7%) was most likely associated with the loss of DNA content in cells undergoing apoptosis. Indeed, TUNEL assay demonstrated that 31% +/− 6 of BsB8 cells underwent apoptosis 72 hr after the fenofibrate treatment. These inhibitory effects of fenofibrate were almost completely abolished by GW-9662, which at 10 µM concentration is considered as a strong PPARα and PPARγ antagonist.15 In a similar manner, pre-incubation of BsB8 cells with 25 εM fenofibrate inhibited IGF-I -stimulated clonogenic growth (Fig. 4c).
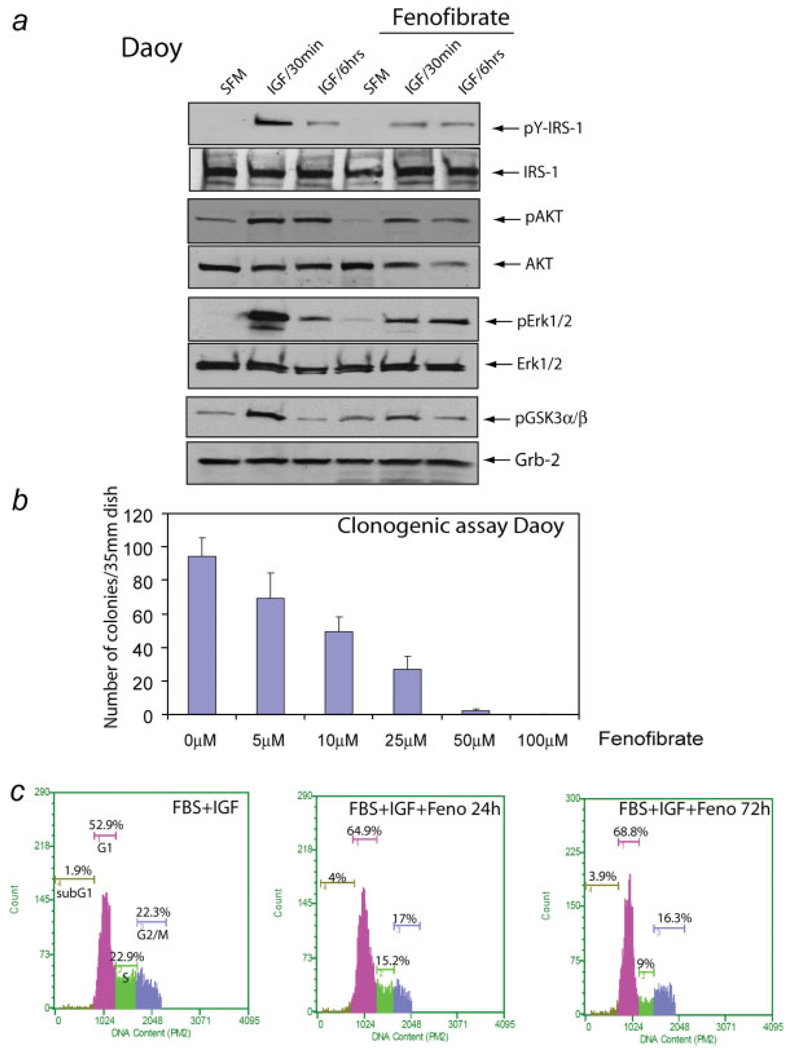
Since the human medulloblastoma cell lines, Daoy and D384, express both PPARα and PPARγ, we asked whether fenofibrate could inhibit their growth responses to IGF-I in a similar manner to BsB8 cells. The results in Figure 5 illustrate that fenofibrate partially downregulated IGF-I-mediated phosphorylation of IRS-1, Akt, ERKs and Gsk-3β, which was associated with dose dependent inhibition of the clonogenic growth of Daoy cells by fenofibrate (Fig. 5b). Although fenofibrate-mediated inhibition of growth was quite effective (Fig. 5b), expected apoptotic cell death was less apparent than in BsB8 cells (compare Figs. 4b and 5c). Also, in contrast to BsB8 cells in which G2/M arrest predominated, fenofibrate induced partial G1 arrest in Daoy cells, which was accompanied by only slightly elevated apoptosis (from 2% in control samples to 4% in fenofibrate treated samples) (Fig. 5c).
FIGURE 5.
Effects of fenofibrate on IGF-I -mediated cell signaling responses in Daoy cells. (a) After 48 hr incubation in serum-free medium (SFM), quiescent Daoy were cultured in the presence or absence of fenofibrate for additional 24 hr. The cells were subsequently stimulated with IGF-I (50 ng/ml) for 30 min and 6 hr, or were left without IGF-I treatment (SFM). Western blots were prepared with corresponding total protein extracts (50 µg), and were probed with the following primary antibodies: anti (pY612)IRS-1, anti-(pSer473)Akt, total Akt, anti-(pT202/Y204)Erk1/2, total Erk1/2 and anti-(pS21/9)GSK3α/β. Anti-Grb-2, anti-IRS-1, anti-Akt and anti-Erk1/2 antibodies were used as loading controls. (b) Clonogenic assay with Daoy cells in response to different concentrations of fenofibrate ranging from 0 to 100 µM. The cells were plated at clonal density (1 × 103 per 35 mm dish) and the number of clones was evaluated 10 days later. (c) Evaluation of cell cycle distribution after fenofibrate treatment. Exponentially growing monolayer cultures of Daoy cells were treated with 25 µM fenofibrate (Feno) for 24 or 72 hr, or were left without the fenofibrate treatment (FBS + IGF-I). After tripsinization, the cells were fixed in ethanol, and DNA content was determined by propidium iodide (PI) DNA labeling. Cell cycle distribution was evaluated by the Guava EayCyte flowcytometer. Note that 72 hr incubation of Daoy cells with fenofibrate resulted in a partial G1 arrest, slightly elevated fraction of cells in sub-G1 and a significant decrease of cells in S phase. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
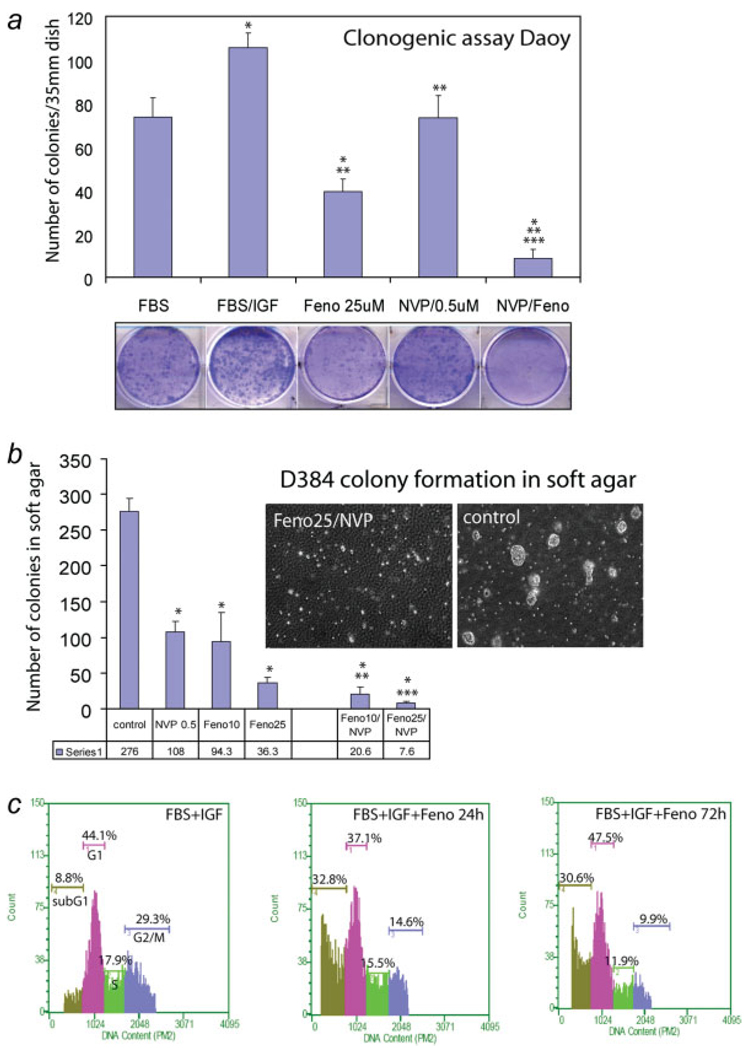
We attempted next to sensitize human medulloblastoma cell lines to fenofibrate by utilizing low concentrations of a specific IGF-IR inhibitor, NVP-AEW541 (Novartis). We have previously shown that medulloblastomas are sensitive to 1 µM NVP-AEW541 treatment when kept in suspension cultures, and were much more resistant to the same treatment when cultured in monolayer.11 The results depicted in Fig. 6 indicate that simultaneous treatment of Daoy and D384 cells with fenofibrate (25 µM) and NVP-AEW541 (0.5 µM) caused almost complete inhibition of the clonogenic growth of Daoy cells (Fig. 6a), and inhibited colony formation of D384 cells in soft agar (Fig. 6b). We have obtained 97.3% inhibition of the colony formation in soft agar in the presence of 0.5 µM NVP AEW541 (ref.) and 25 µM fenofibrate (Fig. 6b). Note also that D384 cells were partially resistant to 1 µM NVP AEW541 in soft agar assay.11 Results in Figure 6C revealed that cell cycle distribution of D384 cell growing in suspension was quite different from the cell cycle obtained from BsB8 and Daoy cells, which grew as monolayer cultures (compare Figs. 4b, 5c, and 6c). The major difference was associated with relatively high level of subG1 fraction of the cells in optimal growth conditions (8.8%). Fenofibrate treatment increased the accumulation of cells in subG1 phase from 8.8% to 32.8% during first 24 hr of the treatment, and this high rate of apoptosis was also detected in 72 hr time point. At this late time point the percentage of cells in S and G2/M phase of the cell cycle also decreased, indicating that both cell death and attenuation of cell cycle progression contribute to the inhibitory action of fenofibrate on D384 cells.
FIGURE 6.
Growth inhibition of human medulloblastoma cell lines by fenofibrate and NVP-AEW541. (a) Clonogenic growth of Daoy cells. The cells were plated at clonal density (1 × 103 per 35 mm dish), in the presence of 10% FBS, FBS + IGF-I (50 ng/ml) or in FBS + IGF-I supplemented by a single dose of 0.5 µM NVP-AEW541 (NVP/0.5 µM); a single dose of 25 µM fenofibrate (feno/25 µM); or a combination of 0.5 µM NVP-AEW541 and 25 µM fenofibrate (NVP/Feno). At day 15, cells were stained with crystal violet and the clones were counted macroscopically. Data are presented as mean ± SD calculated from two experiments in triplicates (n = 6). * indicates values statistically significantly different (p ≤ 0.05) from FBS. ** indicates values statistically significantly different (p ≤ 0.05) from FBS + IGF. *** indicates a value statistically significantly different (p ≤ 0.05) from feno/10 µM, or from NVP/0.5 µM. Statistical significance between two measurements was determined with the two-tailed Student’s t test. Note a complete inhibition of clonogenic growth of Daoy cells by a combination of fenofibrate and NVP-AEW541 used at relatively low concentrations. Inset: representative examples of the clonogenic assay in five culture conditions described above. (b) Soft agar assay for D384 cells. In basic growth condition, D384 cells were plated in DMEM containing 10% FBS and 50 ng/ml IGF-I (control). The growth medium was additionally supplemented with 25 µM fenofibrate (Feno) or with a combination of 25 µM fenofibrate and 0.5 µM NVP-AEW541 (Feno/NVP). Clones larger that 125 lm in diameter were scored after 2 weeks of continuous growth. Data are presented as mean ± SD calculated from three experiments in triplicates (n=9). * indicates values statistically significantly different (p ≤ 0.05) from the control (untreated cells cultured in FBS supplemented with 50 ng/ml of IGF-I). ** indicates values statistically significantly different (p ≤ 0.05) from 10µM fenofibrate and from 0.5 µM NVP AEW541. *** indicates values statistically significantly different (p ≤ 0.05) from 25 µM fenofibrate and from 0.5 µM NVP AEW541. Inset: representative examples of the soft agar assay. (c) Evaluation of cell cycle distribution after fenofibrate treatment. Exponentially growing suspension cultures of D384 cells were treated with 25 µM fenofibrate (FBS + IGF + Feno) for either 24 of 72 hr, or were left without the fenofibrate treatment (10% FBS + IGF). Subsequently, cells were tripsinized, fixed in ethanol and the DNA content was determined by propidium iodide (PI) DNA labeling. Cell cycle distribution was evaluated by the Guava EayCyte flow cytometer. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Discussion
The presented results demonstrate for the first time the nuclear presence of PPARα and PPARγ proteins in medulloblastoma cell lines and in medulloblastoma biopsies. Mouse and human tumor cells responded to fenofibrate treatment with a significant increase of PPAR-mediated transcriptional activity, and an accumulation of cells in G2/M phase (BsB8 mouse cells) and G1 phase (Daoy and D384 human cells) of the cell cycle. Pre-incubation of BsB8 cells with 25 µM fenofibrate for 24 hr attenuated IGF-I-induced IRS-1, Akt, ERKs and GSK3β phosphorylation, which resulted in a severe retardation of their clonogenic growth. In Daoy and D384 human cells, preincubation with fenofibrate partially inhibited IGF-I-mediated phosphorylation and growth responses; and together with a low dose of the IGF-IR inhibitor, NVP-AEW541, fenofibrate completely inhibited clonogenic growth and colony formation in soft agar. This indicates an important role of fenofibrate in chemoprevention against IGF-I-induced growth responses in medulloblastoma.
Growth and survival of medulloblastoma cells in vitro strongly depends on the activation of IGF-IR.11,26 In medulloblastoma clinical samples the IGF-IR and its major signaling molecule, IRS-1, are strongly upregulated and active (tyrosine phosphorylated).2 Different molecular and pharmacological manipulations directed against the IGF-IR were effective in attenuating medulloblastoma growth and survival in vitro,3,11 and in experimental animals.1,23 Our results demonstrate that 24 hr pre-incubation of mouse and human medulloblastoma cell lines with fenofibrate inhibited IGFI-mediated phosphorylation events (Figs. 4a and 5a). This result could be seen as a surprise since fenofibrate has been found to increase insulin-mediated signaling responses including enhanced glucose uptake, mitochondrial glucose oxidation and reduced adiposity, 13 and is frequently used to counteract metabolic abnormalities associated with insulin resistance in type-2 diabetes.27–29 However, fenofibrate treatment inhibited VEGF- and bFGF-induced endothelial cells migration, which was accompanied by a decrease in Akt phosphorylation.30,31 In melanoma cells, fenofibrate attenuated both cell invasiveness and clonogenic growth by inhibiting constitutively active in these tumors Akt.14,15 In contrast, some authors report a rapid but transient increase of Erk1/2 and Akt phosphorylation caused by PPARα and PPARγ agonists.32–34 In those studies, changes in the phosphorylation were detected shortly after fenofibrate treatment (in minutes), and therefore, were not dependent on the canonical activity of PPARα as a transcription factor.34,35 In our experiments, fenofibrate induced PPARs transcriptional activity and the biological consequences of its action accumulated over time, leading to cell cycle arrest, and to apoptotic cell death in later time points after the treatment (Fig. 4b). These long lasting cytostatic and cytotoxic responses to fenofibrate and the expression of PPARα in medulloblastoma clinical samples (Fig. 1) indicate a potential therapeutic application of fenofibrate against medulloblastoma.
An alternative concept of PPAR α antitumoral action is associated with cancer cell energy metabolism. This notion has been initiated by Otto Warburg, who indicated a distinctive dependence of tumor cell metabolism from glycolysis, even when there is sufficient amount of oxygen available for much more effective oxidative phosphorylation.36,37 Only recently, it has been established that the inclination of tumor cells for glycolysis is mainly driven by mitochondrial dysfunction.38,39 PPARα, which is a transcriptional activator of fatty acid β-oxidation machinery (e.g. acyl-CoA oxidase, acyl-CoA synthetase, carnitine palmitoyl transferase, fatty acid binding protein, fatty acid transporter), can switch energy metabolism towards fatty acid degradation, and decrease glucose uptake by repressing glucose transporter GLUT4.40,41 Interestingly, PPARα acts as a direct sensor for fatty acids, which are considered as natural ligands for this nuclear receptor.42,43 According to fatty acid–glucose cycle paradigm, increased rate of fatty acid and ketone bodies oxidation forces the decline in glucose utilization through the inhibition of glycolytic enzymes.44,45. This concept was supported by the results of animal studies, showing that during fasting activated PPARα can divert energy metabolism from the glucose to fatty acid utilization as a primary source of energy. In addition, loss-of-function mutations of genes encoding the Krebs cycle enzymes (such as succinate dehydrogenase and fumarate hydratase) are observed in uterine leiomyomas, renal carcinomas, paragangliomas and phaeochromocytomas.46 Therefore, glycolysis-promoting metabolism of the cancer cell could relieve the selection pressure and support growth and survival of the cells with defective mitochondrial system. Note, however, that such cells could be brought to the verge of metabolic catastrophe in the condition of limited glucose availability, or when the oxidative metabolism is forced pharmacologically. This opens an opportunity for the use of PPARα ligands, including fenofibrate, as they should be selectively toxic for cancer cells and neutral for normal cells.
In mouse medulloblastoma cells, the treatment with 25 µM fenofibrate was sufficient to stop cell proliferation, and to induce massive apoptosis (Fig. 4b). The concentration of fenofibrate was comparable to plasma concentrations of fenofibric acid, an active metabolite of fenofibrate, detected in patients during standard hyperlipidemia treatment (300 mg regular or 250 mg slow release capsules daily). In such patients, the plateau phase plasma concentration of fenofibric acid is approximately 10–12 µg/ml (28–33 µM).47 Therefore, 25 µM fenofibrate holds promise for low toxicity systemic chemotherapy against medulloblastoma in comparison to current regimens many of which are highly toxic.48,49 In human medulloblastoma cells, Daoy and D384, which express PPARα and PPARγ, 25 µM fenofibrate attenuated IGF-I-mediated signaling events including IRS-1, Akt, ERKs and GSK-3β phosphorylation (Fig. 5a), and triggered cell cycle arrest (Fig. 5b). However, there was induction of apoptosis than in BsB8 cells (Fig. 4b). Although increasing fenofibrate concentration to 50 µM induced apoptosis in Daoy cells (not shown), this higher concentration could be systemically toxic. We therefore tested the effects of 25 µM fenofibrate in combination with low doses (up to 0.5 µM) of the IGF-IR inhibitor NVP-AEW541. This low-molecular-weight inhibitor of the IGF-IR kinase activity has been previously shown to limit the growth and survival of medulloblastoma cell lines in anchorage-independent culture conditions.11 Interestingly, medulloblastoma cells in monolayer cultures were quite resistant to the treatment with 1 µM NVP-AEW541.11 In our study, lower dose of NVP-AEW541 (0.5 µM) used in combination with 25 µM fenofibrate completely inhibited clonogenic growth of Daoy cells (Fig. 6a), and efficiently inhibited colony formation of D384 cells in soft agar (Fig. 6b).
Although the mechanism by which fenofibrate attenuates IGF-I signaling responses is still under investigation, it has been recently reported that fenofibrate increases plasma membrane rigidity in a manner similar to the elevated cholesterol content in cell membranes.50 In that report, fenofibrate did not change the membrane content of cholesterol, but increased plasma membrane rigidity, altering activities of integral membrane proteins such as sarco (endo)plasmic reticulum Ca2+-ATPase and β-secretase-mediated cleavage of APP.50 Further experiments are required to determine whether similar fenofibrate-mediated changes in the fluidity of plasma membrane are indeed responsible for the attenuation of ligand-induced clusterization of the IGF-IR molecules—a critical step in autophosphorylation of the receptor molecules and the initiation of growth promoting signaling cascades.
Acknowledgements
We thank Dr. Shohreh Amini for her editorial help.
Grant sponsor: NIH; Grant numbers: RO1CA095518-01 and PO1 NS36466-06.
References
- 1.Reiss K. Insulin-like growth factor-I receptor—a potential therapeutic target in medulloblastomas. Expert Opin Ther Targets. 2002;6:539–544. doi: 10.1517/14728222.6.5.539. [DOI] [PubMed] [Google Scholar]
- 2.Del Valle L, Enam S, Lassak A, Wang JY, Croul S, Khalili K, Reiss K. Insulin-like growth factor i receptor activity in human medulloblastomas. Clin Cancer Res. 2002;8:1822–1830. [PubMed] [Google Scholar]
- 3.Wang JY, Del Valle L, Gordon J, Rubini M, Romano G, Croul S, Peruzzi F, Khalili K, Reiss K. Activation of the IGF-IR system contributes to malignant growth of human and mouse medulloblastomas. Oncogene. 2001;20:3857–3868. doi: 10.1038/sj.onc.1204532. [DOI] [PubMed] [Google Scholar]
- 4.Patti R, Reddy CD, Geoerger B, Grotzer MA, Raghunath M, Sutton LN, Phillips PC. Autocrine secreted insulin-like growth factor-I stimulates MAP kinase-dependent mitogenic effects in human primitive neuroectodermal tumor/medulloblastoma. Int J Oncol. 2000;16:577–584. doi: 10.3892/ijo.16.3.577. [DOI] [PubMed] [Google Scholar]
- 5.Rao G, Pedone CA, Valle LD, Reiss K, Holland EC, Fults DW. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene. 2004;23:6156–6162. doi: 10.1038/sj.onc.1207818. [DOI] [PubMed] [Google Scholar]
- 6.Baserga R, Sell C, Porcu P, Rubini M. The role of the IGF-I receptor in the growth and transformation of mammalian cells. Cell Prolif. 1994;27:63–71. doi: 10.1111/j.1365-2184.1994.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 7.Baserga R, Peruzzi F, Reiss K. The IGF-1 receptor in cancer biology. Int J Cancer. 2003;107:873–877. doi: 10.1002/ijc.11487. [DOI] [PubMed] [Google Scholar]
- 8.D’Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci USA. 1993;90:10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui H, Meng Y, Bulleit RF. Inhibition of glycogen synthase kinase 3beta activity regulates proliferation of cultured cerebellar granule cells. Brain Res Dev Brain Res. 1998;111:177–188. doi: 10.1016/s0165-3806(98)00136-9. [DOI] [PubMed] [Google Scholar]
- 10.Kurihara M, Tokunaga Y, Ochi A, Kawaguchi T, Tsutsumi K, Shigematsu K, Niwa M, Mori K. [Expression of insulin-like growth factor I receptors in human brain tumors: comparison with epidermal growth factor receptor by using quantitative autoradiography] No To Shinkei. 1989;41:719–725. [PubMed] [Google Scholar]
- 11.Urbanska K, Trojanek J, Del Valle L, Eldeen MB, Hofmann F, Garcia-Echeverria C, Khalili K, Reiss K. Inhibition of IGF-I receptor in anchorage-independence attenuates GSK-3beta constitutive phosphorylation and compromises growth and survival of medulloblastoma cell lines. Oncogene. 2007;26:2308–2317. doi: 10.1038/sj.onc.1210018. [DOI] [PubMed] [Google Scholar]
- 12.Lecka-Czernik B, Ackert-Bicknell C, Adamo ML, Marmolejos V, Churchill GA, Shockley KR, Reid IR, Grey A, Rosen CJ. Activation of peroxisome proliferator-activated receptor gamma (PPARgamma) by rosiglitazone suppresses components of the insulin-like growth factor regulatory system in vitro and in vivo. Endocrinology. 2007;148:903–911. doi: 10.1210/en.2006-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerre-Millo M, Gervois P, Raspe E, Madsen L, Poulain P, Derudas B, Herbert JM, Winegar DA, Willson TM, Fruchart JC, Berge RK, Staels B. Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J Biol Chem. 2000;275:16638–16642. doi: 10.1074/jbc.275.22.16638. [DOI] [PubMed] [Google Scholar]
- 14.Grabacka M, Placha W, Plonka PM, Pajak S, Urbanska K, Laidler P, Slominski A. Inhibition of melanoma metastases by fenofibrate. Arch Dermatol Res. 2004;296:54–58. doi: 10.1007/s00403-004-0479-y. [DOI] [PubMed] [Google Scholar]
- 15.Grabacka M, Plonka PM, Urbanska K, Reiss K. Peroxisome proliferator-activated receptor alpha activation decreases metastatic potential of melanoma cells in vitro via down-regulation of Akt. Clin Cancer Res. 2006;12:3028–3036. doi: 10.1158/1078-0432.CCR-05-2556. [DOI] [PubMed] [Google Scholar]
- 16.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 17.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 19.Kainu T, Wikstrom AC, Gustafsson JA, Pelto-Huikko M. Localization of the peroxisome proliferator-activated receptor in the brain. Neuroreport. 1994;5:2481–2485. doi: 10.1097/00001756-199412000-00019. [DOI] [PubMed] [Google Scholar]
- 20.Staels B, Van Tol A, Fruchart JC, Auwerx J. Effects of hypolipidemic drugs on the expression of genes involved in high density lipoprotein metabolism in the rat. Isr J Med Sci. 1996;32:490–498. [PubMed] [Google Scholar]
- 21.Krynska B, Otte J, Franks R, Khalili K, Croul S. Human ubiquitous JCV(CY) T-antigen gene induces brain tumors in experimental animals. Oncogene. 1999;18:39–46. doi: 10.1038/sj.onc.1202278. [DOI] [PubMed] [Google Scholar]
- 22.He XM, Wikstrand CJ, Friedman HS, Bigner SH, Pleasure S, Trojanowski JQ, Bigner DD. Differentiation characteristics of newly established medulloblastoma cell lines (D384 Med, D425 Med, and D458 Med) and their transplantable xenografts. Lab Invest. 1991;64:833–843. [PubMed] [Google Scholar]
- 23.Wang JY, Del Valle L, Peruzzi F, Trojanek J, Giordano A, Khalili K, Reiss K. Polyomaviruses and cancer–interplay between viral proteins and signal transduction pathways. J Exp Clin Cancer Res. 2004;23:373–383. [PubMed] [Google Scholar]
- 24.Vu-Dac N, Schoonjans K, Kosykh V, Dallongeville J, Fruchart JC, Staels B, Auwerx J. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J Clin Invest. 1995;96:741–750. doi: 10.1172/JCI118118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lores Arnaiz S, Travacio M, Monserrat AJ, Cutrin JC, Llesuy S, Boveris A. Chemiluminescence and antioxidant levels during peroxisome proliferation by fenofibrate. Biochim Biophys Acta. 1997;1360:222–228. doi: 10.1016/s0925-4439(97)00004-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Zeng ZC, Bui TA, Sonoda E, Takata M, Takeda S, Iliakis G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene. 2001;20:2212–2224. doi: 10.1038/sj.onc.1204350. [DOI] [PubMed] [Google Scholar]
- 27.Bajaj M, Suraamornkul S, Hardies LJ, Glass L, Musi N, Defronzo RA. Effects of peroxisome proliferator-activated receptor (PPAR)-alpha and PPAR-gamma agonists on glucose and lipid metabolism in patients with type 2 diabetes mellitus. Diabetologia. 2007;8:1723–1731. doi: 10.1007/s00125-007-0698-9. [DOI] [PubMed] [Google Scholar]
- 28.Tan CE, Chew LS, Tai ES, Chio LF, Lim HS, Loh LM, Shepherd J. Benefits of micronised Fenofibrate in type 2 diabetes mellitus subjects with good glycemic control. Atherosclerosis. 2001;154:469–474. doi: 10.1016/s0021-9150(00)00497-4. [DOI] [PubMed] [Google Scholar]
- 29.Wu TJ, Ou HY, Chou CW, Hsiao SH, Lin CY, Kao PC. Decrease in inflammatory cardiovascular risk markers in hyperlipidemic diabetic patients treated with fenofibrate. Ann Clin Lab Sci. 2007;37:158–166. [PubMed] [Google Scholar]
- 30.Goetze S, Eilers F, Bungenstock A, Kintscher U, Stawowy P, Blaschke F, Graf K, Law RE, Fleck E, Grafe M. PPAR activators inhibit endothelial cell migration by targeting Akt. Biochem Biophys Res Commun. 2002;293:1431–1437. doi: 10.1016/S0006-291X(02)00385-6. [DOI] [PubMed] [Google Scholar]
- 31.Varet J, Vincent L, Mirshahi P, Pille JV, Legrand E, Opolon P, Mishal Z, Soria J, Li H, Soria C. Fenofibrate inhibits angiogenesis in vitro and in vivo. Cell Mol Life Sci. 2003;60:810–819. doi: 10.1007/s00018-003-2322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845–5853. doi: 10.1074/jbc.M208394200. [DOI] [PubMed] [Google Scholar]
- 33.Gardner OS, Dewar BJ, Earp HS, Samet JM, Graves LM. Dependence of peroxisome proliferator-activated receptor ligand-induced mitogen-activated protein kinase signaling on epidermal growth factor receptor transactivation. J Biol Chem. 2003;278:46261–46269. doi: 10.1074/jbc.M307827200. [DOI] [PubMed] [Google Scholar]
- 34.Takeda K, Ichiki T, Tokunou T, Iino N, Takeshita A. 15-Deoxy-delta 12,14-prostaglandin J2 and thiazolidinediones activate the MEK/ERK pathway through phosphatidylinositol 3-kinase in vascular smooth muscle cells. J Biol Chem. 2001;276:48950–48955. doi: 10.1074/jbc.M108722200. [DOI] [PubMed] [Google Scholar]
- 35.Pauley CJ, Ledwith BJ, Kaplanski C. Peroxisome proliferators activate growth regulatory pathways largely via peroxisome proliferator-activated receptor alpha-independent mechanisms. Cell Signal. 2002;14:351–358. doi: 10.1016/s0898-6568(01)00260-1. [DOI] [PubMed] [Google Scholar]
- 36.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 37.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 38.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed W, Ziouzenkova O, Brown J, Devchand P, Francis S, Kadakia M, Kanda T, Orasanu G, Sharlach M, Zandbergen F, Plutzky J. PPARs and their metabolic modulation: new mechanisms for transcriptional regulation? J Intern Med. 2007;262:184–198. doi: 10.1111/j.1365-2796.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 41.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol. 2002;34:1249–1257. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 42.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 44.Randle PJ. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev. 1998;14:263–283. doi: 10.1002/(sici)1099-0895(199812)14:4<263::aid-dmr233>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe RR. Metabolic interactions between glucose and fatty acids in humans. Am J Clin Nutr. 1998;67:519S–526S. doi: 10.1093/ajcn/67.3.519S. [DOI] [PubMed] [Google Scholar]
- 46.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 47.Doser K, Guserle R, Nitsche V, Arnold G. Comparative steady state study with 2 fenofibrate 250 mg slow release capsules. An example of bioequivalence assessment with a highly variable drug. Int J Clin Pharmacol Ther. 1996;34:345–348. [PubMed] [Google Scholar]
- 48.Corden BJ, Strauss LC, Killmond T, Carson BS, Wharam MD, Kumar AJ, Piantadosi S, Robb PA, Phillips PC. Cisplatin, ara-C and etoposide (PAE) in the treatment of recurrent childhood brain tumors. J Neurooncol. 1991;11:57–63. doi: 10.1007/BF00166998. [DOI] [PubMed] [Google Scholar]
- 49.Knight KR, Kraemer DF, Neuwelt EA. Ototoxicity in children receiving platinum chemotherapy: underestimating a commonly occurring toxicity that may influence academic and social development. J Clin Oncol. 2005;23:8588–8596. doi: 10.1200/JCO.2004.00.5355. [DOI] [PubMed] [Google Scholar]
- 50.Gamerdinger M, Clement AB, Behl C. Cholesterol-like effects of selective COX inhibitors and fibrates on cellular membranes and amyloid-{beta} production. Mol Pharmacol. 2007;72:141–151. doi: 10.1124/mol.107.034009. [DOI] [PubMed] [Google Scholar]