Abstract
Accurate electrostatic descriptions of aqueous solvent are critical for simulation studies of bio-molecules, but the computational cost of explicit treatment of solvent is very high. A computationally more feasible alternative is a generalized Born implicit solvent description which models polar solvent as a dielectric continuum. Unfortunately, the attainable simulation speedup does not transfer to the massive parallel computers often employed for simulation of large structures. Longer cutoff distances, spatially heterogenous distribution of atoms and the necessary three-fold iteration over atom-pairs in each timestep combine to challenge efficient parallel performance of generalized Born implicit solvent algorithms. Here we report how NAMD, a parallel molecular dynamics program, meets the challenge through a unique parallelization strategy. NAMD now permits efficient simulation of large systems whose slow conformational motions benefit most from implicit solvent descriptions due to the inherent low viscosity. NAMD’s implicit solvent performance is benchmarked and then illustrated in simulating the ratcheting Escherichia coli ribosome involving ~250,000 atoms.
Introduction
Molecular dynamics (MD) is a computational method1 employed for studying the dynamics of nanoscale biological systems on nanosecond to microsecond timescales.2 Using MD, researchers can utilize experimental data from crystallography and cryo-electron microscopy (cryo-EM) to explore the functional dynamics of biological systems.3
Because biological processes take place in the aqueous environment of the cell, a critical component of any biological MD simulation is the solvent model employed.4,5 An accurate solvent model must reproduce water’s effect on solutes such as the free energy of solvation, dielectric screening of solute electrostatic interactions, hydrogen bonding and van der Waals interactions with solute. For typical biological MD simulations, solute is comprised of proteins, nucleic acids, lipids or other small molecules.
Two main categories of solvent models are explicit and implicit solvents. Explicit solvents, such as SPC6 and TIP3P,7 represent water molecules explicitly as a collection of charged interacting atoms and calculate a simple potential function, such as Coulomb electrostatics, between solvent and solute atoms. Implicit solvent models, instead, ignore atomic details of solvent and represent the presence of water indirectly through complex interatomic potentials between solute atoms only.8–10 There are advantages and disadvantages of each solvent model.
Simulation of explicit water is both accurate and natural for MD, but often computationally too demanding, not only since the inclusion of explicit water atoms increases a simulation’s computational cost through the higher atom count, but also because water slows down association and disassociation processes due to the relatively long relaxation times of interstitial water.11 The viscous drag of explicit water also retards large conformation changes of macromolecules.12
An alternative representation of water is furnished by implicit solvent descriptions which eliminate the need for explicit solvent molecules. Implicit water remains always equilibrated to the solute. The absence of explicit water molecules also eliminates the viscosity imposed on simulated solutes, allowing faster equilibration of solute conformations and better conformational sampling. Examples of popular implicit solvent models are Poisson-Boltzmann electrostatics,13,14 screened Coulomb potential,9,15 analytical continuum electrostatics16 and generalized Born implicit solvent.17
The generalized Born implicit solvent (GBIS) model, used by MD programs CHARMM,18,19 Gromacs,20,21 Amber22 and NAMD,23,24 furnishes a fast approximation for calculating the electrostatic interaction between atoms in a dielectric environment described by the Poisson-Boltzmann equation. The GBIS electrostatics calculation determines first the Born radius of each atom, which quantifies an atom’s exposure to solvent, and, therefore, its dielectric screening from other atoms. The solvent exposure represented by Born radii can be calculated with varying speeds and accuracies25 either by integration over the molecule’s interior volume26,27 or by pairwise overlap of atomic surface areas.17 GBIS calculations then determine the electrostatic interaction between atoms based on their separation and Born radii.
GBIS has benefited MD simulations of small molecules.28 For the case of large systems, whose large conformational motions29 may benefit most from an implicit solvent description, but which must be simulated on large parallel computers,30 the challenge to develop efficient parallel GBIS algorithms remains. In the following, we outline how NAMD addresses the computational challenges of parallel GBIS calculations and efficiently simulates large systems, demonstrated through benchmarks as well as simulations of the Escherichia coli ribosome, a RNA-protein complex involving ~250,000 atoms.
Methods
In order to characterize the challenges of parallel generalized Born implicit solvent (GBIS) simulations, we first introduce the key equations employed. We then outline the specific challenges that GBIS calculations pose for efficient parallel performance as well as how NAMD’s implementation of GBIS achieves highly efficient parallel performance. GBIS benchmark simulations, which demonstrate NAMD’s performance, as well as the ribosome simulations, are then described.
Generalized Born Implicit Solvent Model
The GBIS model8 represents polar solvent as a dielectric continuum and, accordingly, screens electrostatic interactions between solute atoms. GBIS treats solute atoms as spheres of low protein dielectric (εp = 1), whose radius is the Bondi31 van der Waals radius, in a continuum of high solvent dielectric (εs = 80).
The total electrostatic energy for atoms in a dielectric solvent is modeled as the sum of Coulomb and generalized Born (GB) energies,8
| (1) |
The total Coulomb energy for the system of atoms is the sum over pairwise Coulomb energies,
| (2) |
where the double summation represents all unique pairs of atoms within the interaction cutoff; the interaction cutoff for GBIS simulations is generally in the range 16–20 Å, i.e., longer than for explicit solvent simulations, where it is typically 8–12 Å. The reason for the wider cutoff is that particle-mesh Ewald summations, used to describe long-range Coulomb forces, cannot be employed for treatment of long-range GBIS electrostatics.
The pairwise Coulomb energy, in eq. 2, is
| (3) |
where ke = 332 (kcal/mol)Å/e2 is the Coulomb constant, qi is the charge on atom i, and ri j is the distance between atoms i and j. The total GB energy for the system of atoms is the sum over pairwise GB energies and self-energies given by the expression
| (4) |
where the pair-energies and self-energies are defined as8
| (5) |
Here, Di j is the pairwise dielectric term,32 which contains the contribution of an implicit ion concentration to the dielectric screening, and is expressed as
| (6) |
where κ−1 is the Debye screening length which represents the length scale over which mobile solvent ions screen electrostatics. For an ion concentration of 0.2 M, room temperature water has a Debye screening length of κ−1 = ~7 Å. is8
| (7) |
The form of the pairwise GB energy in eq. 5 is similar to the form of the pairwise Coulomb energy in eq. 3, but is of opposite sign and replaces the 1/ri j distance dependence by . The GB energy bears a negative sign because the electrostatic screening counteracts the Coulomb interaction. The use of , instead of ri j, in eq. 5 heavily screens the electrostatic interaction between atoms which are either far apart or highly exposed to solvent. The more exposed an atom is to high solvent dielectric, the more it is screened electrostatically, represented by a smaller Born radius, αi.
Accurately calculating the Born radius is central to a GBIS model as the use of perfect Born radii allows the GBIS model to reproduce, with high accuracy, the electrostatics and solvation energies described by the Poisson-Boltzmann equation,33 and does it much faster than a Poisson-Boltzmann or explicit solvent treatment.34 Different GBIS models vary in how the Born radius is calculated; models seek to suggest computationally less expensive algorithms without undue sacrifice in accuracy. Many GBIS models35 calculate the Born radius by assuming atoms are spheres whose radius is the Bondi31 van der Waals radius and determine an atom’s exposure to solute through the sum of overlapping surface areas with neighboring spheres.36 The more recent GBIS model of Onufriev, Bashford and Case (GBOBC), applied successfully to MD of macromolecules37,38 and adopted in NAMD, calculates the Born radius as
| (8) |
where ψi, the sum of surface area overlap with neighboring spheres, is calculated through
| (9) |
As explained in prior studies,35,36,38 H(ri j,ρi,ρj) is the surface area overlap of two spheres based on their relative separation, ri j, and radii, ρi and ρi0; the parameters δ, β and γ in eq. 8 have been calculated to maximize agreement between Born radii described by eq. 8 and those derived from Poisson-Boltzmann electrostatics.38 ρi and ρj are the Bondi31 van der Waals radii of atoms i and j, respectively, while ρi0 is the reduced radius, ρi0 = ρi − 0.09 Å, as required by GBOBC.38
The total electrostatic force acting on an atom is the sum of Coulomb and GB forces; the net Coulomb force on an atom is given by
| (10) |
whose derivative ( ) is inexpensive to calculate. The required derivatives ( ) for the GB force, however, are much more expensive to calculate because depends on inter-atomic distances, ri j, both directly (c.f. eqs. 5 and 7) and indirectly through the Born radius (c.f. eqs. 5, 7, 8 and 9). The net GB force on an atom is given by
| (11) |
with r⃗i j = r⃗j − r⃗i. The required partial derivative of with respect to a Born radius, αk, is
| (12) |
The summations in eqs. 9, 11 and 12, require three successive iterations over all pairs of atoms for each GBIS force calculation, whereas calculating Coulomb forces for an explicit solvent simulation requires only one such iteration over atom-pairs. Also, because of the computational complexity of the above GBIS equations, the total cost of calculating the pairwise GBIS force between pairs of atoms is ~7× higher than the cost for the pairwise Coulomb force. For large systems and long cutoffs, the computational expense of implicit solvent simulations can exceed that of explicit solvent simulations; however, in this case, an effective speed-up over explicit solvent still arises due to faster conformational exploration as will be illustrated below. The trade-off between the speedup of implicit solvent models and the higher accuracy of explicit solvent models is still under investigation.34 Differences between GBIS and Coulomb force calculations create challenges for parallel GBIS simulations that do not arise in explicit solvent simulations.
Challenges in Parallel Calculation of GBIS Forces
Running a MD simulation in parallel requires a scheme to decompose the simulation calculation into independent work units that can be executed simultaneously on parallel processors; the scheme employed for decomposition strongly determines how many processors the simulation can efficiently utilize and, therefore, how fast the simulation will be. For example, a common decomposition scheme, known as spatial or domain decomposition, divides the simulated system into a three-dimensional grid of spatial domains whose side length is the interaction cutoff distance. Because the atoms within each spatial domain are simulated on a single processor, the number of processors utilized equals the number of domains.
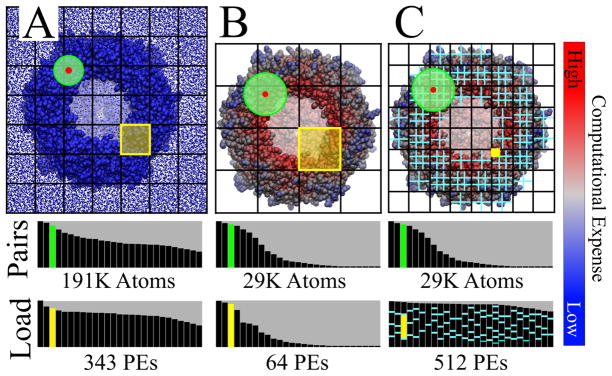
Although explicit solvent MD simulations perform efficiently in parallel, even for simple schemes such as naive domain decomposition, the GBIS model poses unique challenges for simulating large systems on parallel computers. We outline here the three challenges arising in parallel GBIS calculations and how NAMD addresses them. For the sake of concreteness, we use the SEp22 dodecamer (PDB ID 3AK8) as an example as shown in Figure 1.
Figure 1.
Work decomposition for implicit solvent and explicit solvent simulations. A SEp22 do-decamer is shown (front removed to show interior) with overlaid black grid illustrating domain decomposition for explicit solvent (A), implicit solvent (B) and NAMD’s highly parallel implicit solvent (C). Atoms are colored according to relative work required to calculate the net force with blue being least expensive and red being most expensive. The number of neighbor-pairs within the interaction cutoff (green circle, Pairs) for an atom (red circle) varies more in implicit solvent than explicit solvent, as does the number of atoms within a spatial domain (yellow box, Load), each domain being assigned to a single processor (PE). (A) Because explicit solvent has a spatially homogeneous distribution of atoms, it has a balanced work load among processors using simple domain decomposition. (B) Domain decomposition with implicit solvent suffers from the spatially heterogenous atom distribution; the work load on each processor varies highly. (C) NAMD’s implicit solvent model (cyan grid representing force decomposition and partitioning), despite having varying number of atoms per domain and varying computational cost per atom, still achieves a balanced workload among processors.
Challenge 1: Dividing workload among processors
With a 12 Å cutoff, traditional domain decomposition divides the SEp22 dodecamer explicit solvent simulation (190,000 protein and solvent atoms) into 7 × 7 × 7 = 343 domains which efficiently utilize 343 processors (see Figure 1A). Unfortunately, with a 16 Å cutoff for the implicit solvent treatment, the same decomposition scheme divides the system (30,000 protein atoms) into 4 × 4 × 4 = 64 domains which can only utilize 64 processors (see Figure 1B). An efficient parallel GBIS implementation must employ a decomposition scheme which can divide the computational work among many (hundreds or thousands) processors (see Figure 1C).
Challenge 2: Workload imbalance from spatially heterogenous atom distribution
Due to the lack of explicit water atoms, the spatial distribution of atoms in a GBIS simulation (see Figure 2) is not uniform as it is for an explicit solvent simulation (see Figure 1A); some domains contain densely packed atoms while others are empty (see Figure 1B). Because the number of atoms varies highly among implicit solvent domains, the workload assigned to each processor also varies highly. The highly varying workload among processors for domain decomposition causes the naive decomposition scheme to be inefficient and, therefore, slow. An efficient parallel GBIS implementation must assign and maintain an equal workload on each processor (see Figure 1C).
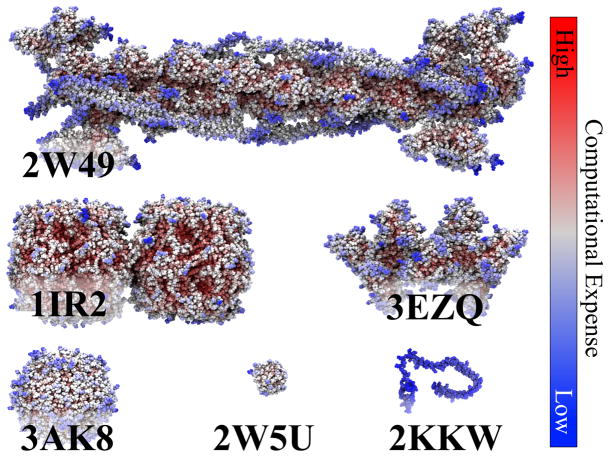
Figure 2.
Biomolecular systems in benchmark. The performance of NAMD’s parallel GBIS implementation was tested on six structures (Protein Data Bank IDs shown); the number of atoms and interactions are listed in Table 2. To illustrate the spatially heterogeneous distribution of work, each atom is colored by the relative time required to compute its net force with blue being fastest and red being slowest.
Challenge 3: Three iterations over atom-pairs per timestep
Instead of requiring one iteration over atom-pairs to calculate electrostatic forces, GBIS requires three independent iterations over atom-pairs (c.f. eqs. 9, 11 and 12). Because each of these iterations depends on the previous iteration, the cost associated with communication and synchronization is tripled. An efficient parallel GBIS implementation must schedule communication and computation on each processor as to maximize efficiency.
Parallelization Strategy
NAMD’s unique strategy39 for fast parallel MD simulations23 enables it to overcome the three challenges of parallel GBIS simulations. NAMD divides GBIS calculations into many small work units using a three-tier decomposition scheme, assigns a balanced load of work units to processors, and schedules work units on each processor to maximize efficiency.
NAMD’s three-tier work decomposition scheme40 directly addresses Challenge 1 of parallel GBIS calculations. NAMD first employs domain decomposition to initially divide the system into a three-dimensional grid of spatial domains. Second, NAMD assigns a force work unit to calculate pairwise forces within each domain and between each pair of adjacent domains. Third, each force work unit is further partitioned into up to ten separate work units, where each partition calculates only one tenth of the atom-pairs associated with the force work unit. Dividing force work units into partitions based on computational expense avoids the unnecessary communication overhead arising from further partitioning already inexpensive force work units belonging to under populated domains. Adaptively partitioning the force work units based on computational expense improves NAMD’s parallel performance even for non-implicit solvent simulations. NAMD’s decomposition scheme is able to finely divide simulations into many (~40,000 for SEp22 dodecamer) small work units and, thereby, utilize thousands of processors.
NAMD’s load balancer, a tool employed to ensure each processor carries an equivalent workload, initially distributes work units evenly across processors, thus partially overcoming Challenge 2 of parallel GBIS calculations. However, as atoms move during a simulation, the number of atoms in each domain fluctuates (more so than for the explicit solvent case) which causes the computational workload on each processor to change. NAMD employs a measurement based load balancer to maintain a uniform workload across processors during a simulation; periodically, NAMD measures the computational cost associated with each work unit and redistributes work units to new processors as required to maintain a balanced workload among processors. By continually balancing the workload, NAMD is capable of highly efficient simulations despite spatially heterogeneous atom distributions common to implicit solvent descriptions.
Though the three iterations over atom-pairs hurt parallel efficiency by requiring additional (compared to the explicit solvent case) communication and synchronization during each timestep, NAMD’s unique communication scheme is able to maintain parallel efficiency. Unlike most MD programs, NAMD is capable of scheduling work units on each processor in an order which overlaps communication and computation, thus maximizing efficiency. NAMD’s overall parallel strategy of work decomposition, workload balancing and work unit scheduling permits fast and efficient parallel GBIS simulations of even very large systems.
Performance Benchmark
To demonstrate the success of NAMD’s parallel GBIS strategy, protein systems of varying sizes and configurations were simulated on 2–2048 processor cores using NAMD version 2.8. We also compare against an implementation of domain decomposition taken from Amber’s PMEMD version 9,22 which also contains the original implementation of the GBOBC implicit solvent model.38 The benchmark consists of six systems, listed in Table 2 and displayed in Figure 2, chosen to represent small (2,000 atoms), medium (30,000 atoms) and large (150,000 atoms) systems.
Table 2.
NAMD and domain decomposition benchmark data. Speed, in units seconds/step, for both NAMD and the domain decomposition algorithm for the six test systems (see Figure 2) on 2–2048 processors (procs). Also listed are the number of atoms and pairs of atoms (in millions, M) within the cutoff (16 Å for implicit solvent and 12 Å for explicit solvent) in the initial structure. Data is not presented for higher processor counts with slower simulation speeds. An asterisk marks highest processor count for which doubling the number of processors increased simulation speed by at least 50%. 3AK8-E uses explicit solvent. Each simulated system demonstrates that NAMD can efficiently utilize at least twice the number of processors as domain decomposition and, thereby, achieves simulation speeds much faster than for domain decomposition.
| PDB ID | 2KKW | 2W5U | 3EZQ | 3AK8 | 3AK8-E | 2W49 | 1IR2 |
|---|---|---|---|---|---|---|---|
| atoms | 2,016 | 2,412 | 27,600 | 29,479 | 191,686 | 138,136 | 149,860 |
| pairs | 0.25 M | 1 M | 13.2 M | 15.7 M | 63.8 M | 65.5 M | 99.5 M |
| # procs | NAMD sec/step | ||||||
|---|---|---|---|---|---|---|---|
| 2 | 0.0440 | 0.167 | 2.01 | 2.87 | 2.25 | 10.4 | 16.3 |
| 4 | 0.0225 | 0.0853 | 1.00 | 1.44 | 1.12 | 5.47 | 8.70 |
| 8 | 0.0122 | 0.0456 | 0.505 | 0.726 | 0.568 | 2.61 | 4.09 |
| 16 | 0.00664 | 0.0228 | 0.260 | 0.371 | 0.286 | 1.31 | 2.05 |
| 32 | 0.00412* | 0.0126 | 0.136 | 0.191 | 0.146 | 0.661 | 1.03 |
| 64 | - | 0.00736* | 0.0700 | 0.105 | 0.0868 | 0.333 | 0.520 |
| 128 | - | - | 0.0447 | 0.0575 | 0.0523 | 0.169 | 0.267 |
| 256 | - | - | 0.0321 | 0.0340 | 0.0288* | 0.0935 | 0.148 |
| 512 | - | - | 0.0224* | 0.0171* | - | 0.0618 | 0.0806 |
| 1024 | - | - | - | - | - | 0.0461* | 0.0563* |
| 2048 | - | - | - | - | - | 0.0326 | 0.0486 |
| # procs | Domain Decomposition sec/step | ||||||
|---|---|---|---|---|---|---|---|
| 2 | 0.0613 | 0.208 | 5.57 | 7.15 | - | 306 | 373 |
| 4 | 0.0312 | 0.104 | 2.83 | 3.67 | - | 169 | 203 |
| 8 | 0.0163 | 0.0535 | 1.43 | 1.84 | - | 92.5 | 111 |
| 16 | 0.0090* | 0.0277 | 0.727 | 0.934 | - | 51.6 | 62.0 |
| 32 | 0.0070 | 0.0162* | 0.391 | 0.506 | - | 25.9 | 31.3 |
| 64 | - | 0.013 | 0.254* | 0.307* | - | 13.1 | 15.7 |
| 128 | - | - | 0.191 | 0.220 | - | 6.74 | 8.08 |
| 256 | - | - | - | - | - | 3.63 | 4.28 |
| 512 | - | - | - | - | - | 2.16* | 2.66* |
| 1024 | - | - | - | - | - | 1.95 | 2.07 |
The following simulation parameters were employed for the benchmark simulations. A value of 16 Å was used for both nonbonded interaction cutoff and the Born radius calculation cutoff. An implicit ion concentration of 0.3 M was assumed. A time step of 1 fs was employed with all forces being evaluated every step. System coordinates were not written to a trajectory file. For the explicit solvent simulation (Table 2: 3AK8-E), nonbonded interactions were cutoff and smoothed between 10 and 12 Å, with PME41 electrostatics, which require periodic boundary conditions, being evaluated every four steps.
Simulations were run on 2.3 GHz processors with 1 GB/s network interconnect for 600 steps. NAMD’s speed is reported during simulation and was averaged over the last 100 steps (first 500 steps are dedicated to initial load balancing). The speed of the domain decomposition implementation, in units seconds per timestep, was calculated as (“Master NonSetup CPU time”)/(total steps); simulating up to 10,000 steps did not return noticeably faster speeds. Table 2 reports the simulation speeds in seconds/step for each benchmark simulation; Figure 3 presents simulation speeds scaled by system size in terms of the number of pairwise interactions per second (pips) calculated.
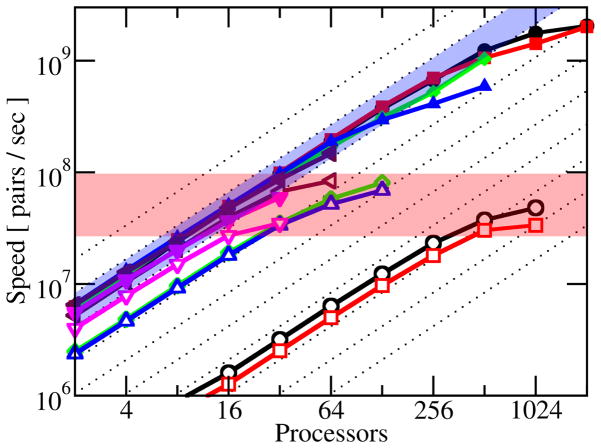
Figure 3.
Parallel performance of NAMD and domain decomposition implicit solvent. Computational speed (pairwise interactions per second) for six biomolecular systems (see Figure 2): 1IR2 (black circle), 2W49 (red square), 3AK8 (green diamond), 3EZQ (blue up triangle), 2W5U (maroon left triangle), and 2KKW (magenta down triangle). NAMD’s parallel implicit solvent implementation (solid shapes) performs extremely well in parallel as seen by performance increasing linearly (blue highlight) with number of processors and is independent of system size. Performance of domain decomposition implicit solvent, however, suffers in parallel (empty shapes). Not only does there appear to be a maximum speed of 108 pairs/sec (red highlight) regardless of processor count, but the large systems (1IR2, 2W49) also perform at significantly lower efficiency than the small systems (2E5U, 2KKW). Diagonal dotted lines represent perfect speedup. See also Table 2.
Implementation Validation
The correctness of our GBIS implementation was validated by comparison to the method’s38 original implementation in Amber.22 Comparing the total electrostatic energy (see eq. 1) of the six test systems as calculated by NAMD and Amber demonstrates their close agreement. Indeed, Table 1 shows that the relative error, defined through
Table 1.
Total electrostatic energy of benchmark systems. To validate NAMD’s implicit solvent implementation, the total electrostatic energy (in units kcal/mol) of the six benchmark systems was calculated by NAMD and Amber implementations of the GBOBC implicit solvent;38 error is calculated through eq. 13.
| NAMD | Amber | error | |
|---|---|---|---|
| 2KKW | −5,271.42 | −5,271.23 | 3.6E-05 |
| 2W5U | −6,246.10 | −6,245.88 | 3.5E-05 |
| 3EZQ | −94,291.43 | −94,288.17 | 3.4E-05 |
| 3AK8 | −89,322.23 | −89,319.13 | 3.4E-05 |
| 2W49 | −396,955.31 | −396,941.54 | 3.4E-05 |
| 1IR2 | −426,270.23 | −426,255.45 | 3.4E-05 |
| (13) |
is less than 4 × 10−5 for all structures in Figure 2.
Molecular Dynamics Flexible Fitting of Ribosome
To illustrate the utility of NAMD’s parallel GBIS implementation, we simulate the Escherichia coli ribosome. The ribosome is the cellular machine that translates genetic information on mRNA into protein chains.
During translation, tRNAs, with their anti-codon loops to be matched to the genetic code on mRNA, carry amino acids to the ribosome. The synthesized protein chain is elongated by one amino acid each time a cognate tRNA (with its anti-codon loop complementary to the next mRNA codon) brings an amino acid to the ribosome; a peptide bond is formed between the new amino acid and the existing protein chain. During protein synthesis, the ribosome complex fluctuates between two conformational states, namely the so-called classical and ratcheted state.42
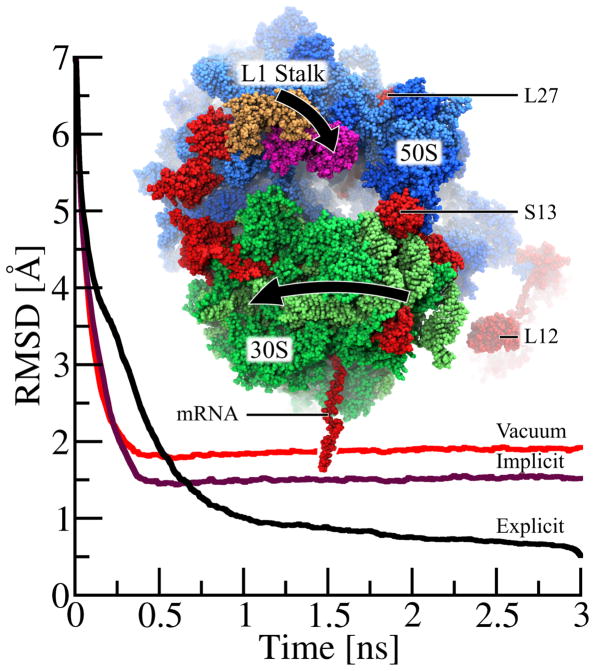
During transition from classical to ratcheted state, the ribosome undergoes multiple, large conformational changes, including an inter-subunit rotation between its 50S and 30S subunits42 and the closing of its L1 stalk in the 50S subunit43 (see Figure 4). The large conformational changes during the transition from classical to ratcheted state are essential for translation44 as suggested by previous cryo-EM data.45 To demonstrate the benefits of NAMD GBIS, we simulate the large conformational changes during ratcheting of the ~250,000-atom ribosome using molecular dynamics flexible fitting.
Figure 4.
Molecular dynamics flexible fitting (MDFF) of ribosome with NAMD’s GBIS method. While matching the 250,000-atom classical ribosome structure into the EM map of a ratcheted ribosome, the 30S subunit (green) rotates relative to the 50S subunit (blue) and the L1 stalk moves 30 Å from its classical (tan) to its ratcheted (magenta) position. Highlighted (red) are regions where the implicit solvent structure agrees with the explicit solvent structure much more closely than does the in vacuo structure. The root-mean-squared deviation (RMSDsol,exp(t)) of the ribosome, defined in eq. 14, with the final fitted explicit solvent structure as reference, is plotted over time for explicit solvent (RMSDexp,exp(t) in black), implicit solvent (RMSDimp,exp(t) in purple) and in vacuo (RMSDvac,exp(t) in red) MDFF. While the explicit solvent MDFF calculation requires ~1.5–2 ns to converge to its final structure, both implicit solvent and vacuum MDFF calculation require only 0.5 ns to converge. As seen by the lower RMSD values for t > 0.5 ns, the structure derived from the implicit solvent fitting agrees more closely with the final explicit solvent structure than does the in vacuo structure. While this plot illustrates only the overall improvement of the implicit solvent structure over the in vacuo structure, the text discusses key examples of ribosomal proteins (L27, S13 and L12) whose structural quality is significantly improved by the use of implicit solvent.
The molecular dynamics flexible fitting (MDFF) method3,46,47 is a MD simulation method that matches crystallographic structures to an electron microscopy (EM) map; crystallographic structures often correspond to non-physiological states of biopolymers while EM maps correspond often to functional intermediates of biopolymers. MDFF-derived models of the classical and ratcheted state ribosome provide atomic-level details crucial to understanding protein elongation in the ribosome. The MDFF method adds to a conventional MD simulation an EM map-derived potential, thereby driving a crystallographic structure towards the conformational state represented by an EM map. MDFF was previously applied to successfully match crystallographic structures of the ribosome to ribosome functional states as seen in EM.48–52 Shortcomings of MDFF are largely due to the use of in vacuo simulations; such use was necessary hitherto as simulations in explicit solvent proved too cumbersome. Implicit solvent MDFF simulations promise a significant improvement of the MDFF method. We applied MDFF here, therefore, to fit an atomic model of a classical state ribosome into an EM map of a ratcheted state ribosome.45
The classical state in our simulations is an all-atom ribosome structure53 with 50S and 30S subunits taken from PDB IDs 2I2V and 2I2U, respectively,54 and the complex fitted to an 8.9 Å resolution classical state EM map.45 In the multistep protocol for fitting this classical state ribosome to a ratcheted state map,46 the actual ribosome is fitted first, followed by fitting the tRNAs. Since the fitting of the ribosome itself exhibits the largest conformational changes (inter-subunit rotation and L1-stalk closing), we limit our MDFF calculation here to the ribosome and do not include tRNAs.
Three MDFF simulations were performed using NAMD23 and analyzed using VMD.55 The MDFF simulations are carried out in explicit TIP3P7 solvent, in implicit solvent and in vacuo. All simulations were performed in the NVT ensemble with the AMBER99 force field,56 employing the SB57 and BSC058 corrections and accounting for modified ribonucleosides.59 The grid scaling parameter,3 which controls the balance between MD force field and the EM-map derived force field, was set to 0.3. Simulations were performed using a 1 fs timestep with nonbonded forces being evaluated every two steps. Born radii were calculated using a cutoff of 14 Å, while the nonbonded forces were smoothed and cut off between 15 and 16 Å. An implicit ion concentration of 0.1 M was assumed with protein and solvent dielectric set to 1 and 80, respectively. A Langevin thermostat with a damping coefficient of 5 ps−1 was employed to hold the temperature to 300 K. In the explicit solvent simulation, the ribosome was simulated in a periodic box of TIP3P water7 including an explicit ion concentration of 0.1 M, with nonbonded forces cut off at 10 Å and long-range electrostatics calculated by PME every four steps. The in vacuo simulation utilized the same parameters as explicit solvent, but without inclusion of solvent or bulk ions, and neither PME nor periodicity were employed.
Each system was minimized for 5000 steps before performing MDFF for 3 ns. For the explicit solvent simulation, an additional 0.5 ns equilibration of water and ions was performed, with protein and nucleic acids restrained, before applying MDFF.
To compare behavior of solvent models during the ribosome simulations, we calculate the root-mean-square deviation between models as
| (14) |
where r⃗i,sol(t) denotes the atomic coordinates at time t of the simulation corresponding to one of the three solvent models (exp, imp or vac) and r⃗i,ref denotes the atomic coordinates for the last time step (tf = 3 ns) of the simulation using the reference solvent model (exp, imp or vac) as specified below. Unless otherwise specified, the summation is over the N = 146,000 heavy atoms excluding the mRNA, L10 and L12 protein segments which are too flexible to be resolved by the cryo-EM method.
Results
Performance Benchmarks
The results of the GBIS benchmark simulations are listed in Table 2. Figure 3 illustrates the simulation speeds, scaled by system size, as the number of pairwise interactions per second (pips) calculated. For a perfectly efficient algorithm, pips would be independent of system size or configuration and would increase proportionally with processor count. NAMD’s excellent parallel GBIS performance is demonstrated as pips is nearly the same for all six systems and increases almost linearly with processor count as highlighted (in blue) in Figure 3.
The domain decomposition algorithm22 performs equally well for small systems, but its performance suffers significantly when system size and processor count are increased. The domain decomposition implementation also appears to be limited to a pips maximum of 108 pairs/sec across all system sizes, no matter how many processors are used, as highlighted (in red) in Figure 3. NAMD runs efficiently on twice the number of processors compared to domain decomposition and greatly outperforms it for the large systems tested. The SEp22 dodecamer timings for both implicit (3AK8) and explicit (3AK8-E) solvent reported in Table 2 demonstrate that NAMD’s parallel GBIS implementation is as efficient as its parallel explicit solvent capability. We note that the simulation speed for GBIS can be further increased, without significant loss of accuracy, by shortening either the interaction or Born radius calculation cutoff distance.
Ribosome Simulation
To demonstrate the benefit of NAMD’s GBIS capability for simulating large structures, a high-resolution classical state ribosome structure was fitted into a low-resolution ratcheted state EM map in an in vacuo MDFF simulation as well as MDFF simulations employing explicit and implicit solvent. During the MDFF simulation, the ribosome undergoes two major conformational changes: closing of the L1 stalk and rotation of the 30S subunit relative to the 50S subunit (see Figure 4).
To compare the rate of convergence and relative accuracy of solvent models, the RMSD values characterizing the three MDFF simulations were calculated using eq. 14. Figure 4 plots RMSDexp,exp(t), RMSDimp,exp(t) and RMSDvac,exp(t) that compare each MDFF simulation against the final structure reached in the explicit solvent case. We note that using the initial rather than final structure as the reference could yield a slightly different characterization of convergence,60 e.g., a slightly different convergence time. As manifested by RMSDimp,exp(t) and RMSDvac,exp(t), the implicit solvent and vacuum MDFF calculations converge to their respective final structures in 0.5 ns compared to ~1.5–2 ns for the explicit solvent case, i.e., for RMSDexp,exp(t).
The final structures obtained from the MDFF simulations are compared in Table 3 through the RMSDsol,ref(t) values for t = 3 ns. The ribosome structure from GBIS MDFF closely agrees with the one from explicit solvent MDFF as indicated by the value RMSDimp,exp(3ns) = 1.5 Å; the in vacuo MDFF ribosome structure, however, compares less favorably with the explicit solvent MDFF structure as suggested by the larger value RMSDvac,exp(3ns) = 1.9 Å. While the 0.4 Å improvement in RMSD of the GBIS MDFF, over in vacuo MDFF, structure implies an overall enhanced quality, certain regions of the ribosome are particularly improved.
Table 3.
Root-mean-square deviation (RMSDsol,ref(3 ns) in Å) between the three final ribosome structures matched using explicit solvent, GBIS, and in vacuo MDFF. The GBIS and explicit solvent MDFF structures closely agree as seen by RMSDimp,exp(3ns) = 1.5 Å, while the in vacuo MDFF structure deviates from the explicit solvent MDFF structure by RMSDvac,exp(3ns) = 1.9 Å. See also Figure 4.
| Reference | ||||
|---|---|---|---|---|
| exp | imp | vac | ||
| Solvent | exp | 0 | 1.5 | 1.9 |
| imp | 1.5 | 0 | 2.1 | |
| vac | 1.9 | 2.1 | 0 | |
The regions with the highest structural improvement (highlighted red in Figure 4) belong to segments at the exterior of the ribosome and to segments not resolved by and, therefore, not coupled to the EM map, i.e., not being directly shaped by MDFF. For proteins at the exterior of the ribosome, GBIS MDFF produces higher quality structures than in vacuo MDFF, because these proteins are highly exposed to solvent and, therefore, require a solvent description. The structural improvement for several exterior solvated proteins, calculated by RMSDvac,exp(3ns) – RMSDimp,exp(3ns), is 3.5 Å, 2.4 Å and 1.6 Å for ribosomal proteins S6, L27 and S13 (highlighted red in Figure 4), respectively. Accurate modeling of these proteins is critical for studying the translation process of the ribosome. The L27 protein, for example, not only facilitates the assembly of the 50S subunit, it also ensures proper positioning of the new amino acid for peptide bond formation.61 The S13 protein, located at the interface between subunits, is critical to the control of mRNA and tRNA translocation within the ribosome.62
The use of GBIS for MDFF also increases structural quality in regions where the EM map does not resolve the ribosome’s structure and, therefore, MDFF does not directly influence conformation; though it is most important that MDFF correctly models structural regions defined in the EM map, it is also desirable that it correctly describes regions of crystal structures not resolved by the EM map. The structural improvement, over in vacuo MDFF, of the unresolved segments is 8.3 Å for mRNA and 4.9 Å for L12 (highlighted red in Figure 4). The L12 segment is a highly mobile ribosomal protein in the 50S subunit that promotes binding of factors which stabilize the ratcheted conformation; L12 also promotes GTP hydrolysis which leads to mRNA translocation.63 As clearly demonstrated, the use of GBIS MDFF, instead of in vacuo MDFF, improves the MDFF method’s accuracy for matching crystallographic structures to EM maps, particularly for highly solvated or unresolved proteins.
To compare computational performance of the solvent models for MDFF, each ribosome simulation was benchmarked on 1020 processor cores (3.5 GHz processors with 5 GB/s network interconnect); the simulation speed for explicit solvent MDFF is 3.6 ns/day, for implicit solvent MDFF it is 5.2 ns/day and for vacuum MDFF it is 37 ns/day. GBIS MDFF performs 50% faster than does explicit solvent MDFF, but seven times slower than in vacuo MDFF. NAMD’s GBIS implementation is clearly able to achieve a more accurate MDFF match of the ribosome structure (see Table 3) than does an in vacuo MDFF calculation and does so at a lower computational cost than explicit solvent MDFF.
Conclusions
The generalized Born implicit solvent (GBIS) model has long been employed for molecular dynamics simulations of relatively small bio-molecules. NAMD’s unique GBIS implementation can also simulate very large systems, such as the entire ribosome, and does so efficiently on large parallel computers. The new GBIS capability of NAMD will be beneficial to accelerate in simulations the slow motions common to large systems by eliminating viscous drag from water.
Acknowledgments
The authors thank Chris Harrison for helpful discussion as well as Laxmikant Kalé’s Parallel Programming Laboratory for parallel implementation advice. This work was supported by the National Science Foundation (NSF PHY0822613) and National Institutes of Health (NIH P41-RR005969) grants to K.S. and a Molecular Biophysics Training Grant fellowship to D.T. Computer time at the Texas Advanced Computing Center was provided through the National Resource Allocation Committee grant (NCSA MCA93S028) from the National Science Foundation.
References
- 1.Lee EH, Hsin J, Sotomayor M, Comellas G, Schulten K. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freddolino PL, Schulten K. Biophys J. 2009;97:2338–2347. doi: 10.1016/j.bpj.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daura X, Mark AE, van Gunsteren WF. Comput Phys Commun. 1999;123:97–102. [Google Scholar]
- 5.Daidone I, Ulmschneider MB, Nola AD, Amadei A, Smith JC. Proc Natl Acad Sci USA. 2007;104:15230–15235. doi: 10.1073/pnas.0701401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. D. Reidel Publishing Company; 1981. pp. 331–342. [Google Scholar]
- 7.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 8.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 9.Hassan SA, Mehler EL. Proteins: Struct, Func, Gen. 2002;47:45–61. doi: 10.1002/prot.10059. [DOI] [PubMed] [Google Scholar]
- 10.Holst M, Baker N, Wang F. J Comp Chem. 2000;21:1343–1352. [Google Scholar]
- 11.Nandi N, Bagchi B. J Phys Chem B. 1997;101:10954–10961. [Google Scholar]
- 12.Rhee YM, Pande VS. J Phys Chem B. 2008;112:6221–6227. doi: 10.1021/jp076301d. [DOI] [PubMed] [Google Scholar]
- 13.Lu B, Cheng X, Huang J, McCammon JA. Comput Phys Commun. 2010;181:1150–1160. doi: 10.1016/j.cpc.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker NA. Curr Opin Struct Biol. 2005;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Hassan SA, Mehler EL, Zhang D, Weinstein H. Proteins: Struct, Func, Gen. 2003;51:109–125. doi: 10.1002/prot.10330. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer M, Karplus M. J Phys Chem. 1996;100:1578–1599. [Google Scholar]
- 17.Qiu D, Shenkin PS, Hollinger FP, Still WC. J Phys Chem. 1997;101:3005–3014. [Google Scholar]
- 18.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 19.Brooks BR, et al. J Comp Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theor Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 21.Larsson P, Lindahl E. J Comp Chem. 2010;31:2593–2600. doi: 10.1002/jcc.21552. [DOI] [PubMed] [Google Scholar]
- 22.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, DeBolt S, Ferguson D, Seibel G, Kollman P. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 23.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner DE, Ma W, Chen Z, Schulten K. Biophys J. 2011;100:2548–2556. doi: 10.1016/j.bpj.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feig M, Onufreiv A, Lee MS, Im W, Case DA, Brooks CL. J Comp Chem. 2004;25:265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 26.Lee MS, Salsbury FR, Brooks CL. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 27.Im W, Lee MS, Brooks CL. J Comp Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 28.Shivakumar D, Deng Y, Roux B. J Chem Theor Comp. 2009;5:919–930. doi: 10.1021/ct800445x. [DOI] [PubMed] [Google Scholar]
- 29.Grant BJ, Gorfe AA, McCammon JA. Theoret Chim Acta. 2010;20:142–147. [Google Scholar]
- 30.Schulz R, Lindner B, Petridis L, Smith JC. J Chem Theor Comp. 2009;5:2798–2808. doi: 10.1021/ct900292r. [DOI] [PubMed] [Google Scholar]
- 31.Bondi A. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 32.Srinivasan J, Trevathan MW, Beroza P, Case DA. Theoret Chim Acta. 1999;101:426–434. [Google Scholar]
- 33.Onufriev L, Case DA, Bashford D. J Comp Chem. 2002;23:1297–1304. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- 34.Feig M, Brooks CL. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Hawkins GD, Cramer CJ, Truhlar DG. J Phys Chem. 1996;100:19824–19839. [Google Scholar]
- 36.Schaefer M, Froemmel C. J Mol Biol. 1990;216:1045–1066. doi: 10.1016/S0022-2836(99)80019-9. [DOI] [PubMed] [Google Scholar]
- 37.Onufriev A, Bashford D, Case DA. J Phys Chem. 2000;104:3712–3720. [Google Scholar]
- 38.Onufriev A, Bashford D, Case DA. Proteins: Struct, Func, Bioinf. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 39.Kalé LV, Krishnan S. Charm++Parallel Programming with Message-Driven Objects. In: Wilson GV, Lu P, editors. Parallel Programming using C++ MIT Press; 1996. pp. 175–213. [Google Scholar]
- 40.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comp Phys. 1999;151:283–312. [Google Scholar]
- 41.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornish P, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horan LH, Noller HF. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells DB, Abramkina V, Aksimentiev A. J Chem Phys. 2007;127:125101. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agirrezabala X, Scheiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. EMBO J. 2011;30:1497–1507. doi: 10.1038/emboj.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. J Mol Biol. 2010;402:741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berk V, Zhang W, Pai RD, Cate JHD. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 57.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aduri R, Psciuk BT, Saro P, Taniga H, Schlegel HB, SantaLucia J. J Chem Theor Comp. 2007;3:1464–1475. doi: 10.1021/ct600329w. [DOI] [PubMed] [Google Scholar]
- 60.Stella L, Melchionna S. J Chem Phys. 1998;109:10115–10117. [Google Scholar]
- 61.Wower IK, Wower J, Zimmermann RA. J Biol Chem. 1998;273:19847–19852. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 62.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 63.Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]