Abstract
The initial assembly product of bacteriophage φ6, the procapsid, undergoes major structural transformation during the sequential packaging of its three segments of single-stranded RNA. The procapsid, a compact icosahedrally symmetric particle with deeply recessed vertices, expands to the spherical mature capsid, increasing the volume available to accommodate the genome by 2.5-fold. It has been proposed that expansion and packaging are linked, with each stage in expansion presenting a binding site for a particular RNA segment. To investigate procapsid transformation, we induced expansion by acidification, heating, and elevated salt concentration. Cryo-EM reconstructions for all three treatments produced the same partially expanded particle. Analysis by cryo-electron tomography showed that all vertices of a given capsid were either compact or expanded, indicating a highly cooperative transition. To benchmark the mature capsid, we analyzed filled (in vivo-packaged) capsids. When these particles were induced to release their RNA, they reverted to the same intermediate state as expanded procapsids (intermediate 1) or to a second, further expanded state (intermediate 2). This partial reversibility of expansion suggests that the mature spherical capsid conformation is obtained only when sufficient outwards pressure is exerted by packaged RNA. The observation of two intermediates is consistent with the proposed three-step packaging process. The model is further supported by the observation that a mutant capable of packaging the second RNA segment without previously packaging the first segment has enhanced susceptibility for switching spontaneously from the procapsid to the first intermediate state.
Keywords: cryo-electron microscopy, Cystoviridae, virus, capsid, segmented genome
Introduction
Bacteriophage φ6 offers an attractive system for studying encapsidation of the segmented genomes of double-stranded RNA viruses, due to its relative simplicity.1 The genome is packaged into a pre-assembled procapsid, as opposed to the co-assembly of the genome and capsid proposed for reoviruses.2 The selection of segments and the order in which they are packaged is also well understood, which is not the case for other segmented genome viruses in general.3, 4
φ6 replication starts with an initial assembly product, the procapsid, consisting of four proteins, P1 (capsid shell5), P2 (RNA-dependent RNA polymerase6), P4 (packaging motor7, 8), and P7 (packaging facilitator9). The procapsid shell is an icosahedral assembly of 120 P1 subunits, with P4 hexamers bound over the recessed vertices, and P2 monomers nestled between the tips of the vertices on the inside. Then the three segments of single-stranded RNA - s, m and l - are packaged, in that order.10 Once all three segments are packaged, minus-strand synthesis takes place within the capsid, followed by transcription.11
According to a model that has been proposed for regulating the order of packaging1, 10 the pac sequence of the s segment binds to the outer surface of the procapsid, which is then packaged by the P4 motor, leading to a structural transition that exposes the binding site for the m segment. The m segment is then packaged, leading to further expansion that allows the l segment to bind and be packaged. This model suggests the existence of at least two intermediate states between the compact procapsid and the spherical morphology of the mature fully packaged capsid.12 A candidate intermediate structure was reported by Butcher et al.13 as present in populations of isolated P14 procapsids (i.e. containing only P1 and P4). They also observed a similar partially expanded particle in preparations obtained by extracting complete virions with salt and EDTA.
The goal of the present study was to investigate structural transformations of the φ6 procapsid by cryo-electron microscopy in the context of the three-step packaging hypothesis. However, to date, in vitro packaging of φ6 procapsids has been hampered by relatively low efficiency (2–5%).14, 15 Noting that in some other viral systems, it has been possible to induce procapsid maturation by subjecting them to appropriate physical or chemical perturbations,16, 17, 18 we decided to follow this approach. Thus we investigated the effects of varying several environmental parameters (pH, ionic strength, and temperature) on φ6 procapsids. We also examined procapsids with a point mutation in P1 that is capable of packaging the m segment directly,19 in order to ascertain whether this property has some structural correlate. To compare the conformations observed with the mature surface lattice, we studied in vivo-packaged mature capsids, both intact and after causing them to release their RNA at elevated salt concentration.
Results
Conditions for the expansion of procapsids
In an earlier study in which P1’247 mutant procapsids were imaged,20 we observed a subpopulation of what appeared to be spontaneously expanded particles. This mutant, which harbors the E390A substitution in P1, is able to bind and package the m segment without previously packaging the s segment.19 The proportion of expanded particles was somewhat variable, ranging from 10–20%, but substantially greater than for wild-type (P1247) procapsids with a similar preparatory history (< 5%).
This motivated us to seek conditions under which both the mutant and wild type procapsids expand. Recalling that decreasing the pH promotes maturation of the HK97 procapsid,17 we decided to examine its effect on the φ6 procapsid. The acidification of φ6 procapsids led to larger fractions of expanded particles for both the wild type P1247 (~20% at pH 5.1) and the mutant P1’247 (~50% at pH 5.5 and ~95% at pH 5.0). However, massive aggregation took place at pH < 6 and there were indications of incipient disintegration at pH 5.0 (Figure 2A).
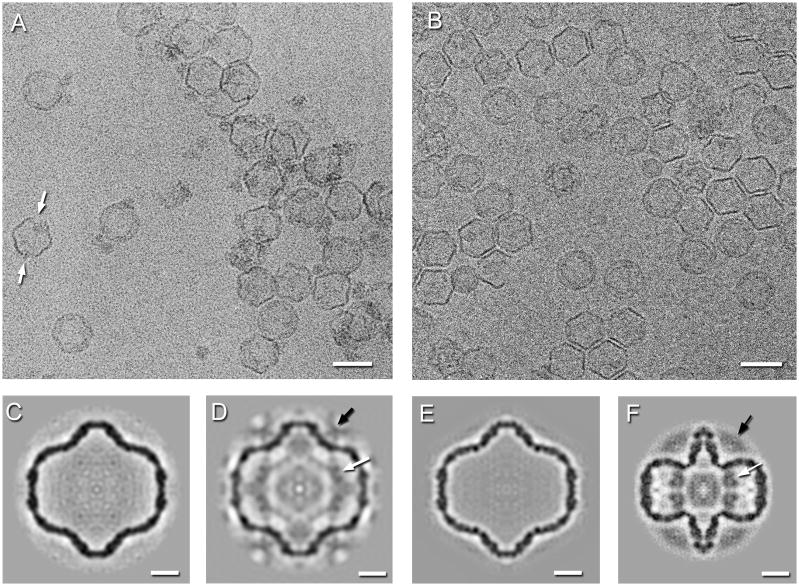
Figure 2.
Cryo-electron micrographs of the P1’247 mutant procapsids at pH 5.0 (A) and of the P1247 wildtype procapsids heated to 60°C in 0.3 M KCl (B). Almost all particles appear expanded, however at the lower pH, some exhibit signs of disassembly at the 2-fold axes (white arrows). Scale bar: 500 Å. Single particle reconstructions of procapsids. (C) The spontaneously expanded P1’247 particle at pH 7.5. (D) The acid-induced expanded P1’247 particle at pH 5.5. (E) The expanded P1247 particle at 60°C and 0.3M KCl. (F) The compact P1’247 procapsid at pH 7.5. The arrows indicate densities associated with the RNA-dependent RNA polymerase, P2 (white), and packaging NTPase, P4 (black). Scale bars: 100 Å.
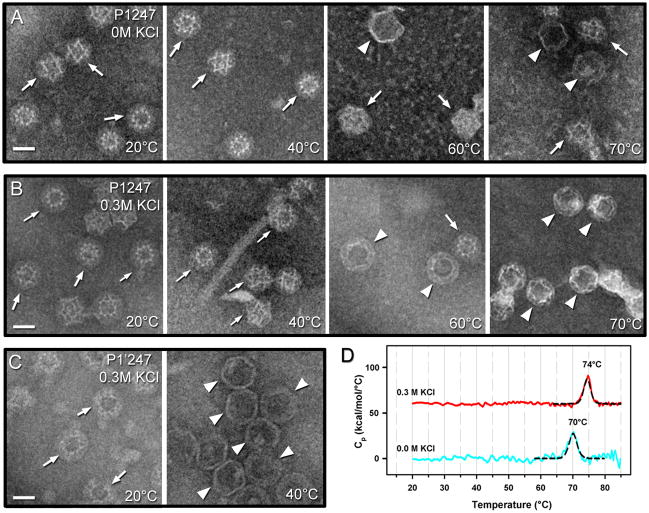
We next tested the effects of elevated temperature and ionic strength on procapsid expansion. To facilitate sampling multiple conditions, negative-staining electron microscopy was used. Preparations of both P1247 and P1’247 procapsids showed increased levels of expanded particles after incubation for 10 minutes at temperatures in the range, 20–70°C (micrographs from selected temperature experiments are presented in Figure 1A–C and Figure 2B). In general, the percentage of expanded particles increased with rising temperature, although the data had considerable scatter, probably as a result of aggregation removing ill-defined fractions of capsids from the suspension. Without any added salt, only about 10% of wild type procapsids expanded at 50 C, increasing to ~30% above 60 C (Figure 1A); no particles of any kind were observed at 80 C. At 0.3 M KCl, the expanded particles increased to ~50% at 50 C, and up to 90% above 60°C (Figure 1B), again, none were observed at 80 C. For the P1’247 mutant in 0.3 M KCl, most particles (>90%) were already expanded at 40 C (Figure 1C). We did not see any evidence of reversibility of procapsid expansion in a sample observed after cooling it down to 20 C, following incubation at 65 (data not shown).
Figure 1.
Negative-stain micrographs of P1247 procapsids in (A) low-salt and (B) high-salt at different temperatures within 20–70°C as indicated. Significantly more expanded (white arrowheads) than compact (white arrows) particles were observed in the high-salt conditions. (C) Negative-stain micrographs of P1’247 procapsids in 0.3M KCl and at 20° and 40°. All particles were already expanded at 40°C and no intact particles were detected at 60°C or higher temperatures. Scale bars: 300 Å. (D) Differential scanning calorimetry of P1247 procapsids in low salt (blue curve) and 0.3 M KCl (red curve) buffers. The endothermic peaks were fitted with a non-two state model (black curves).
Differential scanning calorimetry
In order to probe procapsid stability and to ascertain whether an energetic transition that could be correlated with expansion might be observed,21 we performed differential scanning calorimetry (DSC) (Figure 1D). With wild-type procapsids at concentrations above 0.2 mg/mL protein, the thermograms were compromised by large exothermic excursions, indicative of severe aggregation effects (not shown). This problem was solved by limiting runs to lower concentrations (0.1 mg/mL) but at the cost of lower signal-to-noise ratios. Procapsids in low-salt conditions, which poorly promote expansion in a heated sample (Figure 1A), exhibited an endothermic event at 70 C that we take to represent thermal denaturation of the procapsid. In 0.3 M salt, this event shifted to 74 C and we interpret it as denaturation of the expanded particle. We observed no event at lower temperature that could potentially represent expansion. However, as transitions other than denaturation tend to involve enthalpy exchanges of less than 10% of that of capsid denaturation (~5% in the case of P22,21 2–10% in the case of HK9722), the prospects to observe such an event were low on account of both the weak signal at low protein concentration and expansion being spread over a wide temperature interval.
Reconstructions of expanded procapsids
We performed 3D reconstructions of spontaneously expanded P1’247 procapsids (Figure 2C), P1’247 procapsids subjected to acidic pH (Figure 2D), P1247 procapsids heated in high salt (Figure 2E), and several other conditions (see Table 1 for details). The damage to and aggregation of P1’247 procapsids subjected to low pH treatment limited the numbers of particles in micrographs that were suitable for reconstruction and hence the resolution of the reconstructions (Figure 2D, Table 1). All the reconstructions of expanded particles are indistinguishable (Table 2 and Supplementary Figure 2), suggesting that the same conformation of the P1 shell is reached through each of the expansion-inducing treatments.
Table 1.
Statistics of reconstructed maps of bacteriophage φ6 procapsids. All are in P buffer (10mM phosphate, 1mM MgCl2, pH 7.5 or 6.5) or T buffer (20mM Tris, 1mM MgCl2, pH 8.0) at room temperature with modifications
| Particle/Sample | State | pH | Salt † (M) | Temperature(°C) | Number of particles | Resolution (Å, FSC0.3) | 5-fold radius (Å) # |
|---|---|---|---|---|---|---|---|
| P1’247 | Compact§ (Fig. 2D) | 7.5 | - | 22 | 902 | 19 | 107 |
| Expanded (Fig. 2A) | 7.5 | - | 22 | 342 | 23 | 194 | |
| Expanded | 5.5 * | - | 22 | 89 | 31 | 195 | |
| Expanded (Fig. 2B) | 5.0 * | - | 22 | 412 | 37 | 193 | |
| Expanded | 6.5 | 0.5(K) | 22 | 715 | 23 | 193 | |
| P1247 | Compact§ | 7.5 | - | 22 | 3150 | 14 | 117 |
| Expanded (Fig. 2C) | 6.5 | 0.3(K) | 60 | 1474 | 18 | 186 | |
| Packaged P1247 | RNA-filled (Fig. 3C) | 8.0 | 0.1(N) | 22 | 2659 | 18 | 220 |
| RNA-released 1 (Fig. 3D) | 8.0 | 0.2(N) | 22 | 496 | 20 | 208 | |
| RNA-released 2 (Fig. 3E) | 8.0 | 0.2(N) | 22 | 558 | 20 | 211 | |
| RNA-released 3 (Fig. 3F) | 8.0 | 0.2(N) | 22 | 2026 | 19 | 197 |
Adjusted with 100 mM acetic acid buffer at the indicated pH.
Actual salt concentrations may be higher at the time of plunge-freezing due to rapid evaporation of solvent during specimen blotting; K: KCl; N: NaCl.
From Sen et al., 2008
The uncertainty in measuring the radius due to local structure is ~5Å.
Table 2.
Comparisons of the expansion intermediate reconstructions by Fourier shell correlation (0.3 cutoff): 1–6: first expansion intermediate; 7–8: second expansion intermediate. (The gray cells present the most similar comparisons)
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|
| 1 | 28 | 32* | 19 | 22 | 19 | 30 | 30 |
| 2 | 31* | 29 | 28 | 30 | 32 | 49 | |
| 3 | 32* | 30* | 32* | 51 | 63 | ||
| 4 | 18 | 19 | 30 | 30 | |||
| 5 | 21 | 30 | 30 | ||||
| 6 | 19 | 21 | |||||
| 7 | 18 |
1 P1’247 Expanded pH 7.5 (Fig. 2A)
2 P1’247 Expanded pH 5.5
3 P1’247 Expanded pH 5.0 (Fig. 2B)
4 P1’247 Expanded pH 6.5, 0.5 M KCl
5 P1247 Expanded pH 6.5, 0.3 M KCl (Fig. 2C)
6 P1247 RNA-released 3 (Fig. 3F)
7 P1247 RNA released 2 (Fig. 3E)
8 P1247 RNA released 1 (Fig. 3D)
Comparisons much better than the single particle resolution for the pH 5.0 expanded procapsid
Despite the relatively low resolution of the reconstruction of P1’247 procapsids at low pH, the densities associated with P2 and P4 are present. This indicates that at least some of these proteins remain attached during acidification. In contrast, all reconstructions from particles expanded spontaneously or by heat and salt (such as those in Figure 2A and C and others not shown) exhibit no P2-related densities on the inside, and very little P4-related density above the vertices. Prior experience has been that retention levels of P4 are variable,5, 20, 23 implying that P4 is dislodged relatively easily (a property used to prepare P4 from nucleocapsids8). Although we observed detachment of P4 molecules from P1247 procapsids after storage at 4°C for two weeks (Supplementary Figure 1), there is no reason to believe that expansion per se causes detachment of P4, as the protein is well represented on mature filled particles (see below). This point is considered further in the Discussion section. The dearth of P2 densities in these expanded particles may be attributed to their detachment from the fixed sites that they occupy in the procapsid. In tomograms, internal densities of appropriate size for P2 molecules are seen (Supplementary Figure 3), but not arranged in consistent positions and consequently smeared to invisibility in icosahedrally symmetrized reconstructions.
Salt-induced release of RNA from packaged mature capsids
To obtain a representation of the fully expanded capsid for comparison with in vitro-transformed procapsids, we isolated particles that had been packaged in vivo and were filled with double-stranded RNA and analyzed them by cryo-EM. Micrographs of this specimen in 0.1M salt showed mostly filled capsids but about 20% were empty (Figure 3A). At higher salt (0.2M), the fraction of empty particles increased to ~ 80% (Figure 3B). A reconstruction of the full particles showed nearly full occupancy of P4 at the vertex sites as well as several shells of RNA-associated density inside (Figure 3C). This is not to say that the RNA is strictly stratified but rather that the predominant inter-duplex spacing is 33 Å, the spacing between adjacent shells in the reconstruction.
Figure 3.
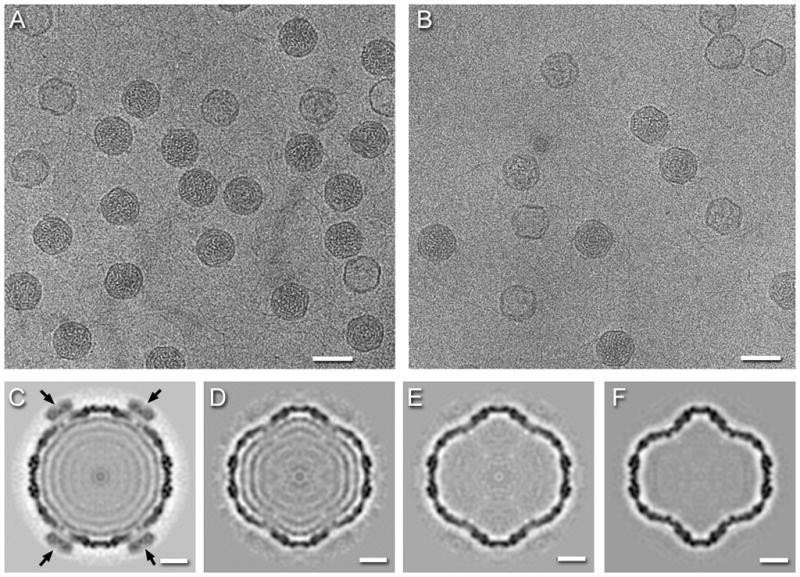
Cryo-electron micrographs of RNA-packaged P1247 capsids in (A) 0.1 M NaCl, and (B) 0.2 M NaCl. Scale bars: 500 Å. Reconstructions of the capsid in (C) 0.1 M NaCl and (D–E) three forms in 0.2 M NaCl showing partial or complete RNA-release. The black arrows indicate high densities of P4 subunits in the packaged capsid (C). Scale bars: 100 Å.
At higher salt concentration, most particles were completely empty, while some retained varying amounts of RNA. By visual inspection, there appeared to be some structural variability in the emptied capsids. To pursue this eventuality more quantitatively, we applied classification techniques (see Methods) to probe for multiple capsid species. This analysis yielded three stable reconstructions with similar resolutions (Figure 3D–F). None of them show either P4 or P2, the former probably dissociated due to the increased salt concentration, and the latter likely due to random distribution in the packaged and expanded particles. The coincidence of the loss of P4 and RNA hints at a role of P4 in the retention of dsRNA.
The first of these reconstructions (Figure 3D), accounting for 16% of the data, showed considerable amounts of RNA remaining. It is very similar in terms of the RNA organization and surface shell structure to the fully packaged particle, but with the spacing between the second and third shells increased to 36Å. The vertices are also slightly indented (by ~9Å) relative to those in Figure 3C (Supplementary Figure 4). The second reconstruction (Figure 3E), from 18% of the data, showed no traces of RNA, and a P1 shell that is indistinguishable from that in Figure 3D (Table 2). The majority particle in this sample (66% of the data, Figure 3F) was empty, with more deeply indented vertices (by ~23Å relative to the fully packaged capsid) and indistinguishable from the acid- and heat-expanded procapsids (Table 2 and Supplementary Figure 2).
Classification of individual vertices of compact and expanded capsids
To visualize single features of individual procapsids (rather than averaged features of reconstructions), we generated cryo-electron tomograms of P1247 procapsids captured at 60°C in 10 mM Tris, 1 mM MgCl2 and 0.3 M KCl at pH 6.5. The expected two kinds of particles were observed: (i) compact procapsids with recessed vertices and characteristic pentagram profiles in the 5-fold views; and (ii) expanded intermediates with rounder shells and more spacious interiors (Figure 4A). Averages of aligned subtomograms appeared to be the compact procapsid and the expansion intermediate (Figures 4B–E), but the resolutions obtained (~46 Å) were much lower than was previously determined for the compact procapsid at room temperature.24 Some expanded particles showed vertices at slightly different levels of expansion. This raised the question of whether the expansion states of vertices are uniform or can be mixed in each particle, and if the latter, whether there is any pattern to expansion.
Figure 4.
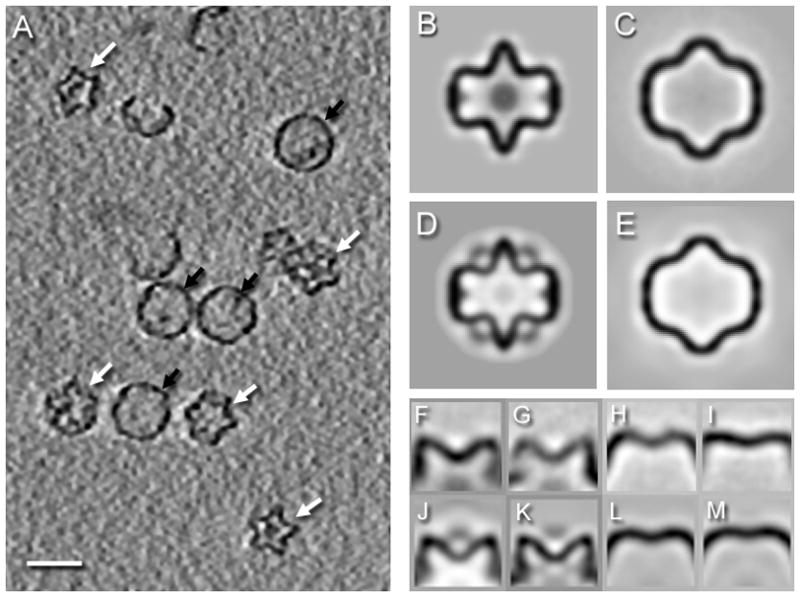
(A) Section through a denoised tomogram of P1247 procapsids at 60 °C in 0.3 M KCl, showing both compact (white arrows) and expanded (black arrows) particles. Scale bar: 500 Å. Central sections of icosahedral maps of the compact procapsid (B) and the expanded particle (C) of P1247 procapsids averaged from 120 and 174 non-denoised subtomograms, and with resolution estimates of 44 Å and 46 Å, respectively (FSC0.3). As a comparison, central sections through suitably resolution-limited single particle reconstructions of (D) the compact procapsid and (E) the first expansion intermediate are shown. Side views of 5-fold averaged vertex classes from subtomograms of compact procapsids (F,G) and expanded particles at 60°C (H,I). For comparison, average vertices from a single particle reconstruction (J) and a subtomogram average (K) of a compact P1247 procapsid at 22°C{Nemecek, 2010 #12573} and average vertices from single particle reconstructions of expansion intermediates 1 (K) and 2 (L) obtained from RNA-packaged procapsids after release of RNA.
To analyze the expansion level of individual vertices within single procapsids, we extracted and realigned subtomograms of the vertices from aligned non-denoised particles. The box size of extracted vertices was chosen so that each extracted subtomogram contained the 5-fold vertex and the neighboring 3-fold vertices. Classification of all vertices extracted from both compact and expanded procapsids from the same tomograms converged to four classes of vertices (Figure 4F–I). Two classes (Figure 4F,G) are similar to the compact procapsid vertex (Figure 4J,K) and the other two classes (Figure 4H,I) resemble vertices from the two expansion intermediates (Figure 4L,M). While the compact-expanded distinction is very clear, the differences within these sets are less certain. The subtle differences between the two compact vertices, or between the two expanded vertices, can in part be ascribed to the anisotropic resolution in the tomograms, and in part to apparent distortions of the P1 shell. We therefore can only classify the vertices reliably as compact or expanded.
In cataloguing the distribution of compact and expanded vertices within individual procapsids, we found only one class of vertex (either compact or expanded) in every particle, with a few exceptions. All of the exceptions were examined and found to be ambiguities in the data, such as missing or poorly represented density. We therefore have no evidence for a mixture of vertex classes in the same particle.
Discussion
Packaging of the φ6 genome follows a specific program, whereby the three segments are packaged in the order: s → m → l.10 During packaging and minus-strand synthesis, the procapsid expands by 150% in internal volume, enabling it to accommodate the complete genome (Figure 5). A current model holds that the pac site of each segment recognizes a particular conformational state of the P1 shell, and switching between these states specifies the order of packaging.1 This model predicts that there should be at least four conformational states of the capsid: the procapsid; and maturing particles respectively packaged with the s, s+m, and s+m+l segments.12 Throughout the packaging process, the P4 NTPase remains bound to the outer surface at the fivefold vertices, while P2, the internalized polymerase, replicates the genome during or after the last packaging step. Here we endeavored to shed light on the structural transitions of which the procapsid is capable.
Figure 5.
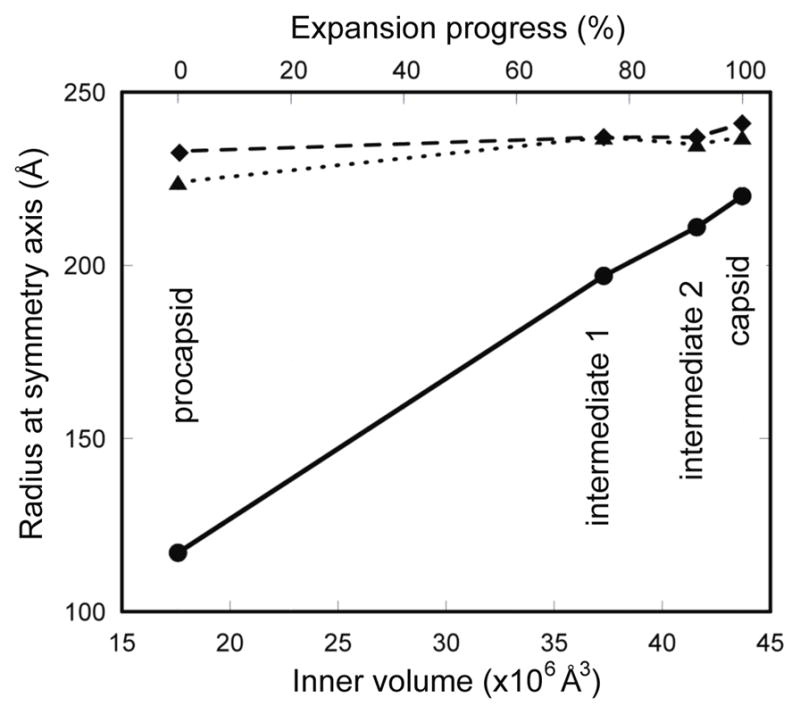
(A) The procapsid expansion is largely an outward movement of the five-fold vertices, showing a linear relationship with the internal volume. The radius was measured at the three symmetry axes: 5-fold (solid line), 3-fold (dotted line), 2-fold (dashed line). Procapsid expansion is also shown in Supplementary movies 1 and 2.
P2 detaches from the inner surface during expansion
P2 binds to the inner surface of the procapsid near the three-fold axes of symmetry.20, 24 In most reconstructions of expanded procapsids and all reconstructions of RNA-filled and emptied capsids, we could find no trace of P2-related density (Figures 2A,C, 3C–F). However, tomography revealed numerous randomly distributed P2-sized densities inside expanded procapsids (Figure 4A and Supplementary Figure 3). It follows that most if not all P2 molecules detach from their initial binding sites when the procapsid expands. Although the distribution of P2 in expanded particles is unclear, there is evidence that the binding of host proteins to P1 alters the activity of P2, indicating that P1 and P2 continue to interact. 25
Two intermediate states of the P1 shell
We found that treatment of procapsids with acid pH (5.0 to 6.5), salt (0.3 – 0.5M), and/or heat (up to 60°C) produced the same altered conformation of the P1 shell. This conformation was also represented in spontaneously transformed mutant P1’247 procapsids and in capsids that had been filled with RNA in vivo and subsequently emptied in vitro. This appears to be the same state as was reported by Butcher et al.,13 either starting from in vitro-assembled P14 procapsids or with capsids extracted from mature virions. In these particles, the vertices have moved outwards by ~80Å, 78% of the way towards their final positions and the internal volume is increased by 112% (Figure 5, Supplementary movies 1 and 2).
The second intermediate is distinguished from the first one by the vertices having moved further outwards by another ~14 Å, now ~ 91% of the way towards their positions in the mature capsid and with a further increase in internal volume, now 136% more than in the procapsid (see the difference map in Supplementary Figure 5). To date, this state has been observed only in RNA-voided preparations of filled capsids and it remains an assumption that it is also a staging-post on the expansion pathway. This assumption is based on its conformation being midway between the first intermediate and the mature capsid (Figure 5).
Implications for the 3-step packaging model
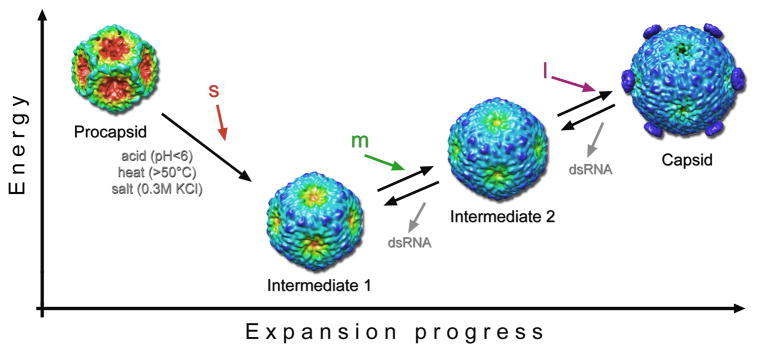
Figure 6 summarizes an updated model for RNA packaging in the φ6 system. The first intermediate appears to be a low-energy state as we observed no tendency of these particles to revert to the procapsid conformation when the stimuli that provoked their formation (heat, low pH, elevated salt) were removed. Huiskonen et al.26 suggested that the trigger for the first expansion step may be provided by NTP hydrolysis by P4. However, our experiments did not include NTP’s and the correlation of loss of P4 with expansion during mild environmental manipulation indicates a different driving force. The energy barrier for the initial expansion may be relatively low, and during packaging of the s segment, moderate pressure exerted on the P1 shell may be enough to switch it to the first expansion intermediate. Although the inner volume of the compact procapsid is 17.6 × 106 Å3, packaged RNA would affect the recessed five-fold vertices already when its size exceeds the central spherical volume of the procapsid. Yoffe et al.27 characterized the size of ssRNA strands by the longest path across the predicted RNA secondary structure and derived a simple approximation for the radius of gyration as a function of the number of nucleotides. The estimated radius of gyration for the s segment is ~150Å, somewhat larger than the 107 Å radius of the tips of the recessed five-fold vertices. The s segment may therefore exert enough pressure on the P1 shell to initiate the expansion, particularly if the inner surface of the procapsid was negatively charged and the expansion triggered by electrostatic repulsion. An alternative trigger for procapsid expansion can be interaction of the s segment with the P1 shell, either through its pac sequence on the outside, or with an internal surface after packaging. Consistent with idea that this transition should be highly cooperative so as to avoid packaging more than one s segment,26 we only observed particles with all their vertices in either the compact or the expanded state, never a mixture.
Figure 6.
Sequential steps of procapsid expansion. Here we tentatively correlate the structures produced by perturbing the procapsid or the RNA-filled capsid in vitro by environmental factors (acid, heat and/or salt) with packaging intermediates that it has not yet been possible to observe directly. The procapsid transforms in a cooperative manner to the intermediate 1 state in vitro in response to any of several treatments or, we hypothesize, by packaging the s segment. Further transformation to the intermediate 2 state would then be driven by packaging the m segment. Finally, packaging of the l segment and RNA replication yield the fully expanded capsid. To date, intermediate 2 has only been observed on perturbing the mature capsid and not on driving expansion further past the intermediate 1 state. This observation suggests that the lowest free energy state of the P1 shell in the absence of packaged RNA is intermediate 1. The reconstructions are color-coded according to the radial distance from the particle center from red to blue to emphasize the main conformational change: the outward movement of the five-fold vertices. Vertex-mounted P4 hexamers are present on the capsid reconstruction and are inferred to have been shed from the particles used to calculate the intermediate 1 and 2 reconstructions (see Discussion).
The observed reversion of emptied capsids to the two expansion intermediate conformations implies that pressure from internalized RNA is needed to push the procapsid into its fully mature conformation, and possibly also the second intermediate. In this property, the φ6 capsid differs from the capsids of dsDNA viruses such as HK9728 or HSV29 in which only slight differences are observed between filled capsids and empty but nominally mature capsids. The volumetric increase in the first intermediate of 112% leaves only a small further increase to the 150% of the fully packaged capsid over the procapsid (Figure 5). Within this range lies the second intermediate we propose corresponds to the capsid packaged with the s and m segments, as well as the fully packaged capsid before replication (i.e., with ssRNA as opposed to the dsRNA in the particle shown in Figure 3C) that we have yet to observe.
In conclusion we propose that the first expansion intermediate may be equated with the conformation putatively achieved after packaging the s segment, in which the particle is competent to package the m segment. This is strongly supported by the propensity of the P1’247 mutant to expand and its ability to bind and package the m segment in the absence of the s segment.30 In terms of this scenario, we envisage a mixed population of procapsids with some still unexpanded and packaging the s segment, and others already expanded and packaging the m segment.
Materials and Methods
Preparation of φ6 procapsids and packaged mature capsids
Procapsids were produced in Escherichia coli strain JM109, using either the plasmid pLM687 to coexpress wildtype P1, P2, P4 and P7 subunits (P1247) or the plasmid pLM2541 to coexpress the E390A mutant of P1 and wildtype P2, P4, and P7 subunits (P1’247).19 The RNA-packaged mature capsids were prepared from the strain LM4383 and contained only segments L (6.4 kb) and S (4.0 kb, modified to contain a kanamycin resistance gene).25, 31 The capsids were purified as described previously,20 and stored at 4°C in buffer P (10 mM potassium phosphate, 1 mM MgCl2, 50% sucrose, pH 7.5). Prior to experiments, the samples were buffer exchanged into buffer T (20 mM Tris, 1 mM MgCl2, pH 6.5) with additional salts as indicated, using Zeba-midi buffer exchange columns with a 7-kDa cutoff (Thermo Scientific, Rockford, IL).
Differential Scanning Calorimetry
DSC measurements were performed using a VP-DSC calorimeter from MicroCal, LLC (Northampton, MA). The instrument was run without feedback with at least 60 min equilibration times prior to, and between, the 60°C/h scans. Samples, at concentrations over the range 0.1–1.0 mg/mL, were scanned three times from 15 to 90 °C with rapid cooling between scans. Samples at concentrations above 0.2 mg/mL exhibited deep and broad exothermic signal above ~50 °C due to precipitation of procapsids in the sample. DSC data were corrected for instrument baselines (determined by running the dialysis buffer in both reference and sample cells just prior to placing protein in the sample cell) and normalized for scan rate and protein concentration. Data conversion and analysis were performed with Origin software (OriginLab Corporation, Northampton, MA). The baseline under denaturation peaks was approximated with a polynomial function and the peaks fitted with a non-two-state model with no ΔCp. This model considered cooperativity in procapsid denaturation (ΔHcal ≠ ΔHvHoff) that had to be included to fit the denaturation peaks reliably.
Negative-stain electron microscopy
For negative staining, the sample was diluted to ~0.1 mg/mL and stabilized at a chosen temperature for ~10 min. A 5 μl aliquot was deposited on a 300-mesh copper grid with a continuous carbon layer. The sample was blotted, grid washed with Mili-Q ultrapure water and stained with methylamine tungstate (Nanoprobes, Yaphank, NY). Micrographs were collected on CM120 electron microscope using a CCD camera at a magnification of 22,000x. The fraction of expanded intermediates at each temperature was determined for 100–500 procapsids picked from several micrographs that were collected from different parts of the grid.
Cryo-electron microscopy
Samples (3.5 μl) at ~10 mg/mL protein concentration were equilibrated at the desired temperature and applied to grids with holey carbon films, blotted, and plunge-frozen in liquid ethane using a Vitrobot (FEI, Hillsboro, OR). The grid and sample chamber of the Vitrobot were preheated to the temperature of the sample to minimize temperature gradients. The vitrified specimens were imaged at 38,000x magnification with a CM200 FEG microscope (FEI, Eindhoven, The Netherlands) fitted with a Gatan 626 cryoholder and operating at an accelerating voltage of 120 keV. Micrographs were recorded at 12–15 e−/Å2 per exposure at 1.0–2.0 μm underfocus.
Single particle processing
Micrographs were digitized using a Nikon Super Coolscan 9000ED at 4000 dpi (1.63 Å/pixel). Image processing was done with the Bsoft package.32 For each micrograph, the contrast transfer function parameters were estimated in bshow. Single particles were picked and extracted in bshow and corrected to the contrast transfer function by phase-flipping using bctf. All of the micrographs and the picked particles were binned by a factor of 2 to accelerate image processing, giving a pixel size of 3.26 Å. Origins and orientations of single particles were determined by projection matching using programs borient and brefine. An angular step size of 1° for global search and 0.2° for local refinement within a 20–100 Å resolution range were used in final iterations. The reconstructions were done with breconstruct and the resolution determined by Fourier shell correlation and using a 0.3 cut-off (Table 1). Dimensions of each reconstruction were determined by several measurements of the outer diameter along the 2-fold, 3-fold and 5-fold axes.
Multi-reference classification
The classification was done as described in Heymann et al.33 Six starting reference maps were chosen to represent the known range of P1 shell conformations: The wild type P147 procapsid,20 the expanded mutant P1’247 at pH 5.5, 5.0, and 7.5, and initial single reconstructions of the packaged particle at 100 and 200 mM NaCl (obtained using as reference the nucleocapsid map from the EMDB: EMD-1207.34 After alignment and competitive classification, only the classes derived from the P1’247 references at pH 5.5 and 7.5 and the higher salt reference map had significant numbers of particles assigned to them, and the other three were discarded. This procedure was repeated 8 more times, each time discarding classes with very low numbers of particles, combining classes where the reconstructions were indistinguishable at the measured resolution, and generating new intermediate references by combining pairs of previous maps. The final classes of particles remained stable through the last three rounds of alignment and assignment. The final result yielded four classes, with one being the fully packaged particle and containing only 3% of the particle images. The other three were kept as representative of the data set of particles ranging from empty to some still containing RNA.
Internal volumes
Each map was segmented with bsegment to isolate and extract the P1 shell. This map was then low-pass filtered to 30 Å to close any openings in the shell. The threshold for 100% mass was determined and used to define a mask with an inner volume taken to represent the volume within the shell. Tests using the available two nucleocapsid maps in the EMDB (EMD-1207 and EMD-130134) suggest a precision of about 2%.
Cryo-electron tomography
Procapsid samples were mixed with 10-nm colloidal gold particles conjugated with BSA (AURION, Wageningen, The Netherlands), at room temperature or heated to 60 °C and vitrified using a Vitrobot (FEI) with the sample chamber prepared at the same temperature. A Tecnai-12 electron microscope (FEI) operating at 120 kV with a LaB6 source and equipped with an energy filter (GIF 2002; Gatan, Pleasanton, CA) was used to collect tilt series at 4 μm defocus, covering the range ±66° in 2° increments. The tilt series were acquired using the SerialEM package,35 for a total dose of ~70 e−/Å2 per series. Images were recorded on a 2048 × 2048-pixel CCD camera (Gatan, Pleasanton, CA) at 38,500x magnification (7.8 Å/pixel).
Tomogram reconstruction
Data were processed using the Bsoft package.36 Projections were aligned using gold particles as fiducial markers and tomograms were reconstructed. The tomograms were denoised by 100 iterations of anisotropic non-linear diffusion37 with the program bnad. Individual procapsids were extracted from tomograms in bshow and aligned to the procapsid map derived from single particle reconstruction with bfind and using an appropriate missing wedge mask. The orientations of the subtomograms were refined in two additional iterations with bfind. The in-plane resolution of the full tomograms was estimated to be ~70 Å by the NLOO-2D method38 with the 0.5 cutoff using btomres.
Classification and averaging of extracted vertices
Individual 5-fold vertices were extracted from aligned subtomograms using a box of 40×40×40 voxels positioned on the 5-fold axis at a radius of 175 Å. To generate an initial series of vertex template classes, two vertices extracted from icosahedrally symmetrized single particle reconstructions of the compact and expanded procapsid maps were used to produce 10 initial references gradually changing from the compact to expanded states. All individual vertices were aligned to each initial reference and classified according to the best cross-correlation coefficient. Members of each class were averaged and served as templates for further rounds of multi-reference alignments and averaging using the program bxb. Similar class averages were merged and classes with low population of particles were eliminated after each round of alignments and mutually exclusive classification. After several iterations, only four classes remained, with two classes each for the compact and expanded procapsids. Masking of the missing wedge during alignments had minimal effect on the results due to the small size of extracted vertices (40×40×40 voxels).
Supplementary Material
Acknowledgments
We thank Austin Luskin, a summer student at the NIH, for initial DSC measurements. This work was supported by the Intramural Research Program of NIAMS and by grant GM34352 to L.M. from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.McDonald SM, Patton JT. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011;19:136–44. doi: 10.1016/j.tim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20:114–20. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noda T, Kawaoka Y. Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Rev Med Virol. 2010;20:380–91. doi: 10.1002/rmv.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olkkonen VM, Bamford DH. The nucleocapsid of the lipid-containing double-stranded RNA bacteriophage phi 6 contains a protein skeleton consisting of a single polypeptide species. Journal of Virology. 1987;61:2362–7. doi: 10.1128/jvi.61.8.2362-2367.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin EV, Gorbalenya AE, Chumakov KM. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989;252:42–6. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- 7.Frilander M, Bamford DH. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage phi 6: the three segments modulate each other’s packaging efficiency. Journal of Molecular Biology. 1995;246:418–28. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- 8.Paatero AO, Syvaoja JE, Bamford DH. Double-stranded RNA bacteriophage phi 6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. Journal of Virology. 1995;69:6729–34. doi: 10.1128/jvi.69.11.6729-6734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juuti JT, Bamford DH. RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage phi 6. Journal of Molecular Biology. 1995;249:545–54. doi: 10.1006/jmbi.1995.0317. [DOI] [PubMed] [Google Scholar]
- 10.Qiao X, Qiao J, Mindich L. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage phi6. Proc Natl Acad Sci U S A. 1997;94:4074–9. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frilander M, Gottlieb P, Strassman J, Bamford DH, Mindich L. Dependence of minus-strand synthesis on complete genomic packaging in the double-stranded RNA bacteriophage phi 6. Journal of Virology. 1992;66:5013–7. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–60. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. Embo J. 1997;16:4477–87. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frilander M, Poranen M, Bamford DH. The large genome segment of dsRNA bacteriophage phi6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Poranen MM, Paatero AO, Tuma R, Bamford DH. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol Cell. 2001;7:845–54. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- 16.Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- 17.Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–63. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- 18.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao X, Qiao J, Mindich L. Analysis of specific binding involved in genomic packaging of the double-stranded-RNA bacteriophage phi6. J Bacteriol. 2003;185:6409–14. doi: 10.1128/JB.185.21.6409-6414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen A, Heymann JB, Cheng N, Qiao J, Mindich L, Steven AC. Initial location of the RNA-dependent RNA polymerase in the bacteriophage Phi6 procapsid determined by cryo-electron microscopy. J Biol Chem. 2008;283:12227–31. doi: 10.1074/jbc.M710508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galisteo ML, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys J. 1993;65:227–35. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 2005;24:1352–1363. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haas F, Paatero AO, Mindich L, Bamford DH, Fuller SD. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J Mol Biol. 1999;294:357–72. doi: 10.1006/jmbi.1999.3260. [DOI] [PubMed] [Google Scholar]
- 24.Nemecek D, Heymann JB, Qiao J, Mindich L, Steven AC. Cryo-electron tomography of bacteriophage phi6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J Struct Biol. 2010;171:389–96. doi: 10.1016/j.jsb.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao X, Sun Y, Qiao J, Mindich L. The role of host protein YajQ in the temporal control of transcription in bacteriophage Phi6. Proc Natl Acad Sci U S A. 2008;105:15956–60. doi: 10.1073/pnas.0807489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–48. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc Natl Acad Sci U S A. 2008;105:16153–8. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duda RL, Ross PD, Cheng N, Firek BA, Hendrix RW, Conway JF, Steven AC. Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol. 2009;391:471–83. doi: 10.1016/j.jmb.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26:479–89. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao J, Qiao X, Sun Y, Mindich L. Isolation and analysis of mutants of double-stranded-RNA bacteriophage phi6 with altered packaging specificity. J Bacteriol. 2003;185:4572–7. doi: 10.1128/JB.185.15.4572-4577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Qiao X, Mindich L. Construction of carrier state viruses with partial genomes of the segmented dsRNA bacteriophages. Virology. 2004;319:274–9. doi: 10.1016/j.virol.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Heymann JB, Conway JF, Steven AC. Molecular dynamics of protein complexes from four-dimensional cryo-electron microscopy. J Struct Biol. 2004;147:291–301. doi: 10.1016/j.jsb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM, Chiu W. EMDataBank. org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–64. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–42. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–50. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 38.Cardone G, Grünewald K, Steven AC. A resolution criterion for electron tomography based on cross-validation. J Struct Biol. 2005;151:117–29. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.