Abstract
Here, a comprehensive proteomic analysis of the chromoplasts purified from sweet orange using Nycodenz density gradient centrifugation is reported. A GeLC-MS/MS shotgun approach was used to identify the proteins of pooled chromoplast samples. A total of 493 proteins were identified from purified chromoplasts, of which 418 are putative plastid proteins based on in silico sequence homology and functional analyses. Based on the predicted functions of these identified plastid proteins, a large proportion (∼60%) of the chromoplast proteome of sweet orange is constituted by proteins involved in carbohydrate metabolism, amino acid/protein synthesis, and secondary metabolism. Of note, HDS (hydroxymethylbutenyl 4-diphosphate synthase), PAP (plastid-lipid-associated protein), and psHSPs (plastid small heat shock proteins) involved in the synthesis or storage of carotenoid and stress response are among the most abundant proteins identified. A comparison of chromoplast proteomes between sweet orange and tomato suggested a high level of conservation in a broad range of metabolic pathways. However, the citrus chromoplast was characterized by more extensive carotenoid synthesis, extensive amino acid synthesis without nitrogen assimilation, and evidence for lipid metabolism concerning jasmonic acid synthesis. In conclusion, this study provides an insight into the major metabolic pathways as well as some unique characteristics of the sweet orange chromoplasts at the whole proteome level.
Keywords: Chromoplast, citrus, metabolism, proteome, ripening
Introduction
Fruit development and ripening of plants involve a series of complex biochemical and physiological changes such as an increase in sugar content, reduction in acid levels, and accumulation of carotenoids. Carotenoid biosynthesis is believed to occur in chromoplasts, which are non-photosynthetic plastids often present in flowers and fruits. Chromoplasts are responsible for the colour of flowers and fruits, and they could promote plant reproduction by attracting insects to pollinate flowers and animals to spread seeds.
Despite the importance of chromoplasts in enhancing the sensory and nutritional quality of fruits, the physiological role of chromoplasts during fruit ripening is not clear; whether the formation of chromoplasts is the cause or the consequence of the fruit ripening process remains to be clarified. It has been reported that, besides being the site in which carotenoid biosynthesis takes place, chromoplasts seem to encompass multiple essential metabolism pathways such as the synthesis of starch, fatty acids, and amino acids, and the oxidative pentose phosphate pathway (Neuhaus and Emes, 2000; Lopez-Juez and Pyke, 2005). However, experimental evidence is needed to test if this notion is true for a wide range of plant species. It has been suggested that >95% of the ∼2700 plastid proteins detected are imported rather than synthesized in situ (Millar et al., 2006). Consistent with this estimation, the chloroplast genome of citrus contains only 133 genes (Bausher et al., 2006), which alone apparently could not explain the various metabolic activities in the chromoplast, thus implying extensive nuclear protein import into the chromoplast. Recently, proteomics has become an efficient tool to study the protein composition and cellular functions of subcellular organelles such as chloroplasts (Kleffmann et al., 2004; Zybailov et al., 2009). Many studies have been focused on subcellular localization/targeting of chloroplast proteins to the thylakoid and lumen (Peltier et al., 2002; Schubert et al., 2002), the envelope (Ferro et al., 2003), the stroma (Peltier et al., 2006), and the plastoglobules (Ytterberg et al., 2006). However, the presence of highly abundant photosynthetic proteins in the chloroplast makes detection of novel chloroplast proteins increasingly difficult. Proteomic analyses with non-photosynthetic plastids may overcome this constraint and increase the coverage of the plastid proteomes. Such studies have been reported on rice etioplasts (von Zychlinski et al., 2005), tobacco proplastid (Baginsky et al., 2004), wheat amyloplast (Andon et al., 2002; Balmer et al., 2006), bell pepper chromoplast (Siddique et al., 2006), and tomato chromoplast (Barsan et al., 2010).
Citrus is one of the most important fruit crops in the world. Different from the model fruit tomato which represents climacteric fruit, citrus has a non-climacteric fruit ripening behaviour. Additionally, citrus fruits exhibit a unique anatomical fruit structure, being made up of two major sections, the pericarp and the edible endocarp. The unique anatomical features of citrus fruits plus their unique biochemical composition suggest that the citrus fruit ripening processes may involve some developmental programmes or metabolic pathways specific to citrus. For example, unlike most climacteric fruits, the conversion of chloroplasts to chromoplasts in citrus pericarp is reversible even with fully differentiated chromoplasts (Goldschmidt, 1988). In the past several decades, studies on citrus metabolism in relation to fruit ripening have been mainly focused on physiological aspects. The recent development of genome-wide expressed sequence tag (EST) and large-scale proteomics projects have promoted the understanding of citrus fruit metabolism during ripening at the molecular level (Chen et al., 2006; Katz et al., 2007, 2010; Muccilli et al., 2009; Pan et al., 2009; Yun et al., 2010). However, little attention has been paid to the characterization of organelles and organelle-specific proteins at the subcellular level during citrus fruit ripening.
In this study, a comprehensive proteomic analysis was initiated to identify proteins in citrus chromoplasts that may be involved in the ripening processes of citrus fruits. By using purified chromoplasts prepared from the edible endocarp of fruits of a red-fleshed sweet orange cultivar ‘Hong Anliu’ (Liu et al., 2007), 418 putative plastid proteins were identified that are predicted to be involved in multiple common and/or some potentially citrus chromoplast-specific metabolic pathways contributing to citrus fruit ripening and/or stress responses to environmental changes. The results represent an important first step toward understanding the metabolic activities within and differentiation of citrus chromoplasts during fruit ripening at the whole proteome level.
Materials and methods
Plant material
Fruits of the red-flesh mutant ‘Hong Anliu’ (Citrus sinensis [L.] Osbeck), which were grown at the Institute of Citrus Research located in Guilin, Guangxi Province, China, were harvested and stored at 4 °C until use. The fruits at 220 days after flowering (DAF) were collected from five trees, with 10 representative fruits from each tree; thus a total of 50 fruits were sampled. Representative pulp from these 50 fruits was pooled together for chromoplast isolation.
Isolation and purification of chromoplasts
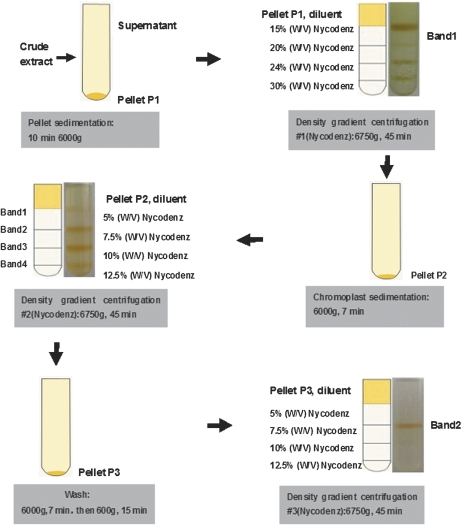
Chromoplasts were isolated by using a protocol involving a combination of three consecutive Nycodenz (Axis-Shield PoC AS, Oslo, Norway) density gradient centrifugations (Fig. 1). All steps of the isolation procedure were carried out at 4 °C. Prior to isolation of chromoplasts, the peel and seeds of the oranges were eliminated and the pulp (∼80 g) was cut into small pieces and homogenized in 300 ml of ice-cold GR buffer [1 mM NaP2O7, 50 mM HEPES, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 2 mM MnCl2, 2 mM dithiothreitol (DTT), pH 6.8] with a cold Waring blender (Hadjeb et al., 1988) at low speed for 8–10s. The homogenate was filtered through two layers of Micracloth® (Calbiochem) and the pooled filtrates were subsequently centrifuged for 5min at 400 g to remove cellular debris. The supernatant was recovered and centrifuged for 10 min at 6000 g, resulting in Pellet P1 in Fig. 1. The pellets were gently resuspended in 10 ml of GR buffer and subject to the first Nycodenz density gradient centrifugation. A 30% (w/v) Nycodenz stock solution (1 mM NaP2O7, 50 mM HEPES, 2 mM EDTA, 1 mM MgCl2, 2 mM DTT, pH 6.8) was diluted with GR buffer to the required Nycodenz concentration. The crude fraction was separated on a Nycodenz step gradient at 15, 20, 24, and 30% (w/v) Nycodenz (1:1:1:1) in GR buffer with centrifugation at 6750 g for 45 min. Four zones were separated by Nycodenz gradient centrifugation, termed bands 1–4. Each band was carefully recovered with a pipettor and examined by light microscopy using an Olympus/Cover-018 microscope. Compared with other bands, only band 1 contained a few chromoplasts, thus it was pooled. The pooled solution of band 1 was diluted five times and then centrifuged at 6000 g for 7 min to obtain the ‘Pellet P2’ (Fig. 1). The Pellet P2 was further resuspended in 1.5 ml of GR buffer, and carefully transferred onto the top of the second density gradient consisting of 2 ml of 5%, 2.5 ml of 7.5%, 1.7 ml of 10%, and 1.3 ml of 12.5% (w/v) Nycodenz. After centrifugation at 6750 g for 45 min the yellowish-red band (band 2) formed at the interface of 5–7.5% (w/v) Nycodenz containing concentrated chromoplasts was recovered, diluted five times with GR buffer, and pelleted at 6000 g for 7 min The pellets were then resuspended in 10 ml of GR buffer and centrifugated at 600 g for 15 min resulting in the third plastid pellet (Pellet P3, Fig. 1). In order to obtain highly purified chromoplasts, the second density gradient centrifugation was repeated (Fig. 1). The last chromoplast layer (band 2) was recovered, diluted 10 times with GR buffer, and pelleted at 6000 g for 7 min to remove Nycodenz. The pellets were further resuspended in 20 ml of GR buffer and centrifuged at 2000 g for 10 min. The supernatant was discarded and the resulting pellet containing the chromoplast fraction was frozen with liquid nitrogen and stored at –80 °C.
Fig. 1.
Work flow of the isolation of chromoplasts from citrus pulp by Nycodenz density gradient centrifugation.
Protein extraction
Purified chromoplasts were treated with a lysis buffer (8 M urea, 2 M thiourea, 4% CHAPS, 2% pharmalyte 3–10, 13 mM DTT, complete EDTA-free protease inhibitor cocktail) and incubated for 20 min at room temperature with shaking. Proteins were collected after the insoluble debris was removed by centrifugation at 8000 g at room temperature for 15 min. The protein concentration was measured with the Bio-Rad protein assay kit based on the Lowry method using bovine serum albumin (BSA) as standard.
Western blot analysis
Western blot analysis was performed using polyclonal antibodies at the appropriate dilution against Rubisco large subunit (RubcL; 53 kDa, 1:3000), mitochondrial voltage-dependent anion-selective channel protein 1 (Vdac1; 29 kDa, 1:2000), vacuolar ATPase (V-ATPase; 26–31 kDa, 1:2000), and cytoplasmic UDP-glucose pyrophosphorylase (UDPase; 51.6 kDa, 1:2000) from Agrisera®. Total fruit pulp proteins were extracted from the ‘Hong Anliu’ fruits harvested at 220 DAF according to Pan et al. (2009). Total fruit pulp and chromoplast proteins (30 μg) were separated by 15% SDS–PAGE, electrotransferred to a PVDF membrane (Hybond ECL, GE Healthcare) using a Trans-Blot SD semi-dry Electrophoretic Transfer Cell (BioRad), treated with blocking TBST buffer [20 mM TRIS-HCl, 150 mM NaCl, 0.05% (v/v) Tween-20] and subsequently incubated for 2 h with polyclonal antibodies diluted as indicated above in TBST buffer. Antibody-bound proteins were detected using the Thermo Scientific® Kit (SuperSignal West Pico Chem iluminescent Substrate) after incubation with anti-rabbit secondary antibody [peroxidase-Conjugated immunoPure Goat Anti-Rabbit IgG (H+L) (Pierce®)], diluted 1:2000 in TBST. Two technical replicates were conducted in the western blot analysis.
Protein digestion
An appropriate volume of protein samples was re-dissolved in reducing solution (6 M guanidine-HCl, 100 mM TRIS, pH 8.3) at a concentration of ∼3 μg μl−1. Next, 1 M DTT (final concentration of 10 mM) was added to the protein sample followed by incubation at 37 °C for 2.5h. Afterwards, 1M indole acetic acid (IAA; final concentration of 50 mM) was added to the solution which was then kept at room temperature for 40 min in the dark. The protein solution was subjected to ultrafiltration (3K) with addition of 100 mM NH4HCO3, and then centrifuged at 12 000 rpm, 4 °C for 2–4h until there was no liquid on the surface of the ultrafiltration tube. The ultrafiltration tube was turned over, and the contents were transferred to a new tube, and then centrifuged at 12 000 rpm, 4 °C for 5 min, and ∼10 μl of solution was obtained. Appropriate volumes of 100mM NH4HCO3 were used to wash the ultrafiltration membrane. A 3 μg μl−1 sample was acquired and then transferred into a new tube. The protein solution was incubated with trypsin (trypsin:protein=1:50) at 37 °C overnight. The digested peptides were subjected to ultrafiltration (10K) and centrifuged (12000 rpm, 4 °C, 90 min) until there was no liquid on the surface of the ultrafiltration tube, and then lyophilized.
Protein identification
The separation and identification of the digested protein was conducted using a Finnigan LTQ mass spectrometer (ThermoQuest, San Jose, CA, USA) coupled with a Surveyor HPLC system (ThermoQuest). First, a Microcore RP column (C18 0.15 mm×120 mm; ThermoHypersil, San Jose, CA, USA) was used to separate the protein digests. Solvent A was 0.1% (v/v) formic acid, and solvent B was 0.1% (v/v) formic acid in 100% (v/v) acetonitrile. The gradient was held at 2% solvent B for 15 min, and increased linearly to 98% solvent B in 60 min. The peptides were eluted from the C18 microcapillary column at a flow rate of 150 μl min−1 and then electrosprayed directly into the LTQ mass spectrometer with the application of a spray voltage of 3.2 kV and a the capillary temperature of 200 °C. The full scan ranges from m/z 400 to 2000. Protein identification using tandem mass spectrometry (MS/MS) raw data was performed with SEQUEST software (University of Washington, licenced to Thermo Finnigan) in combination with citrus ESTs [CICLGI and CIGS (http://compbio.dfci.harvard.edu/index.html; download from May 2010)] and the NCBI green plant protein database (http://www.ncbi.nlm.nih.gov/; download from May 2010). A relative molecular mass of 57Da was added to the average molecular mass of cysteines in MS/MS data searching. Both b ions and y ions were also included in the database search. Protein identification results were filtered with the Xcorr (1+ ≥1.9, 2+ ≥2.2, 3+ ≥3.75) and DelCn (≥0.1).
Identified proteins were functionally classified according to MapMan (http://mapman.mpimp-golm.mpg.de/pageman/index.shtml) using annotations retrieved from databases. Homologies of the detected proteins were searched against (i) three plastidial databanks: Plprot (Kleffmann et al., 2006), SUBA (Heazlewood et al., 2007), and PPDB (Sun et al., 2008); and (ii) the Arabidopsis chloroplast (Zybailov et al., 2009), the tobacco proplastid (Baginsky et al., 2004), the rice etioplast (von Zychlinski et al., 2005), the wheat amyloplast (Balmer et al., 2006), the pepper chromoplast (Siddique et al., 2006), and the tomato chromoplast (Barsan et al., 2010) with a cut-off expectation value of e−20 of having significant homology. Predictions of subcellular localization were carried out using TargetP (http://www.cbs.dtu.dk/services/TargetP/), ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and WoLF PSORT (http://wolfpsort.seq.cbrc.jp/). Those proteins whose close homologues were experimentally demonstrated to localize exclusively outside plastids on the basis of either ‘direct assay’ or ‘traceable author statement’ (Richly and Leister, 2004) were considered as non-chromoplast contaminants. The criterion for being a ‘close homologue’ is any protein having at least 70% pair-wise identity with the query sequence and/or a cut-off expectation value of e−40 (Nair and Rost, 2002) following BLASTP searches against the NCBI non-redundant protein database (www.ncbi.nlm.nih.gov/BLAST/), Uniprot http://services.uniprot.org/blast), or The Arabidopsis Information Resource database (http://www.arabidopsis.org/). Metabolic pathway reconstruction was performed using the PlantCyc database (http://www.gramene.org/pathway/).
Results and Discussion
Isolation of highly purified citrus chromoplasts with Nycodenz density gradient centrifugations
Isolation of highly purified chromoplasts is the prerequisite for chromoplast proteomic analysis, since minor contamination with proteins from other cell organelles would result in misinterpretation of the protein data from mass spectrometry (Reumann et al., 2007). Published methods involving Percoll (Hadjeb et al., 1988; Siddique et al., 2006) and sucrose density gradient centrifugation (Barsan et al., 2010) were first used to try to isolate and purify citrus chromoplasts. It was found that chromoplasts produced by these methods were significantly contaminated with other cell organelles, and/or were broken due to strong osmotic stress (data not shown). A new protocol was thus developed involving three consecutive Nycodenz density gradient centrifugations to isolate and purify intact chromoplasts effectively from citrus pulp (for details, see the Materials and methods and below).
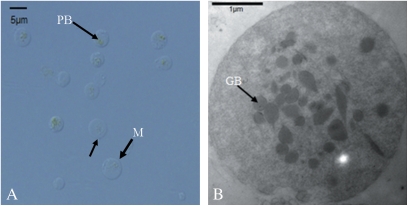
It should be pointed out that isolation of purified intact chromoplasts from oranges is exceptionally difficult, because (i) the abundance of chromoplasts in oranges is relatively low when compared with that in tomato and bell pepper; and (ii) oranges have a high sugar content which makes separation of organelles from cell debris more difficult due to a high level of viscosity of the crude extracts. Based on the authors’ experience, the first Nycodenz density gradient centrifugation was the most important step, because chromoplasts rose to the top of the solution after centrifugation and accumulated at the interface between 0% and 15% Nycodenz (Fig. 1, density gradient centrifugation #1, band 1). After band 1 was recovered from the next purification process, the remaining density gradient could be reused for chromoplast isolation to increase the yield while reducing the cost. The second consecutive Nycodenz density gradient centrifugation produced four visible yellow-reddish bands. Band 2 (in Fig. 1, density gradient centrifugation #2) contained abundant chromoplast-specific structures (prolamellar body and membrane) with a size of 2–4 μm measured under a light microscope (Fig. 2), whereas the other bands contained cell debris with few or no chromoplasts (data not shown). Therefore, band 2 was recovered, further purified by density gradient centrifugation #3, and enriched for subsequent protein preparation (Fig. 1).
Fig. 2.
(A) Light microscopy of purified chromoplasts. A representative micrograph of the third Nycodenz gradient band 2 that contains a clean fraction of chromoplasts ranging from 2 μm to 4 μm. Arrows indicate the chromoplast-specific prolamellar body (PB) and membrane (M). (B) An electron micrograph of purified chromoplasts after the second density gradient centrifugation shows the clear structure of chromoplasts. Arrows indicate the plastoglobulus (GB). (This figure is available in colour at JXB online.)
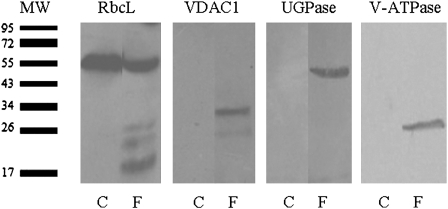
The purity of the chromoplast pellet was then assessed by western blot analysis using polyclonal antibodies against marker proteins for different cell compartments. As expected, the plastid marker protein RbcL was readily detected in the total proteins extracted from the isolated chromoplasts; in contrast, marker proteins predicted to localize to the mitochondria (VDAC1), cytosol (UGPase), and vacuole (V-ATPase) could not be detected (Fig. 3). As a positive control, all these marker proteins were detected in the total protein extracts of the ‘Hong Anliu’ fruits (Fig. 3). These results suggested that the chromoplasts prepared using the Nycodenz density gradient centrifugation method were of high purity and suitable for subsequent proteomic analysis.
Fig. 3.
Western blots for assessment of the purity of chromoplasts. Total proteins prepared from whole fruits (as control) or purified chromoplasts were subject to gel blotting using antibodies against the plastidial large Rubisco subunit (RbcL), mitochondrial voltage-dependent anion-selective channel protein 1 (VDAC1), cytosolic UGPase, and vacuolar ATPase. C, purified chromoplast; F, whole fruit; MW, the actual molecular weight of the marker proteins.
Protein identification and functional classification of the citrus chromoplast proteins
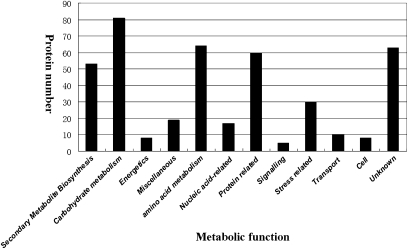
Total proteins were extracted from the purified chromoplasts, trypsin digested, and the resultant peptides were subject to GeLC-MS/MS shotgun analysis. A total of 493 chromoplast protein candidates were identified using the criteria described in the Materials and methods. To evaluate these protein candidates, an in silico validation was conducted using three protein-targeting prediction web programs (TargetP, ChloroP, and WoLF PSORT) or by searching for homology with known plastid proteins deposited in the NCBI non-redundant protein database, Swiss-Prot, and TAIR8. Consequently, 75 (14%) were found which were known or predicted to reside in cellular compartments other than plastids (listed in Supplementary Table S2 available at JXB online). These 75 proteins were considered as cross-contaminants, but the potential contribution of dual targeting must be kept in mind. The 75 potential cross-contaminants of plastids were then excluded from further analysis. A total of 418 proteins were tentatively considered to be plastid proteins based on: (i) the protein candidates were predicted to localize in plastids by at least two of the three web programs; or (ii) the protein candidates show significant homology (identity >70% or/and E-value <e−40) to plastid proteins annotated in the NCBI non-redundant protein database, Swiss-Prot, or TAIR8. As listed in Supplementary Table S1 at JXB online and in Fig. 4, the 418 putative citrus chromoplast proteins could be classified into 14 functional categories according to MapMan Bin based on functional annotations for their homologous proteins identified in various databases.
Fig. 4.
Functional classification of candidate citrus chromoplast proteins identified by GeLC-MS/MS. Proteins was classified into 14 functional categories according to MapMan Bin based on functional annotations for their homologous proteins.
Conservation and potential specialization of citrus chromoplast proteins
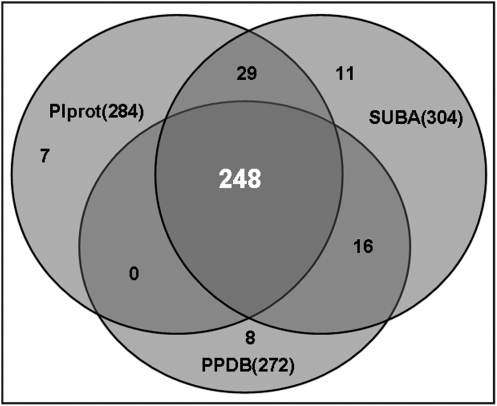
The 418 putative plastid proteins were compared with proteins deposited in three plastid protein databases (PlProt, SUBA, and PPDB). As shown in Fig. 5, homologues for 284, 272, and 304 citrus chromoplast proteins could be found in the PlProt, PPDB, and SUBA databanks, respectively. Among those citrus chromoplast proteins, homologues for the majority (248) could be found in all of the three databases; 45 in any two of the three databases and 26 only in one of the three databases. No apparent homologues were found for the remaining 99 putative citrus chromoplast proteins from these three databases. However, homologues of 10 of the 99 proteins could be found in plastid proteins from other plant species (Supplementay Table S1 at JXB online). Thus, the remaining 89 proteins that do not match any plastid proteins in the BLAST searches may represent novel chromoplast proteins, and they have been shaded in grey in Supplementary Table S1.
Fig. 5.
Venn diagram showing the presence of citrus pulp chromoplastic proteins in plastidial databases: SUBA, PPDB, and PlProt.
Major metabolic pathways in plastids envisaged by the citrus chromoplast proteome
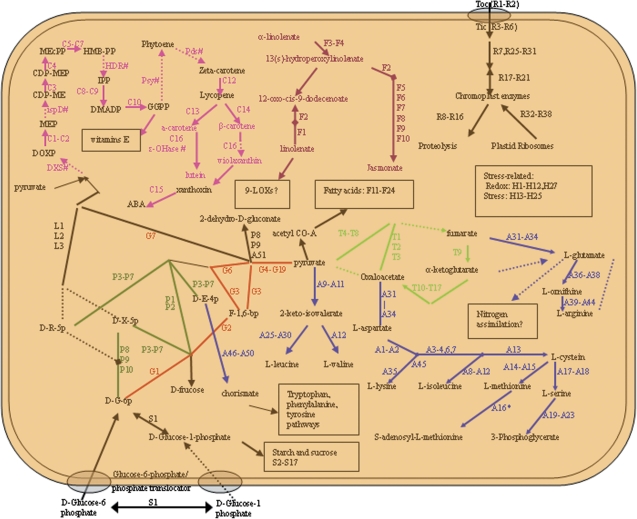
Despite the importance of plastids in plant growth and development, our current understanding on the metabolic functions of plastids is far from complete. Although shotgun (GeLC-MS/MS) proteomic analysis does not provide information about enzyme activity, the proteome profiles allow construction of a comprehensive view of the metabolic pathways in plastids (Kleffmann et al., 2004; von Zychlinski et al., 2005; Balmer et al., 2006; Daher et al., 2010). As depicted in Fig. 6, the major metabolic pathways in citrus chromoplast are carbohydrate metabolism, amino acid metabolism, protein regulation and transport, carotenoid metabolism, lipid metabolism, and oxidative stress related.
Fig. 6.
Schematic illustration of the main metabolic pathways based on the proteins identified in the citrus pulp chromoplasts. Metabolic pathway reconstruction was performed using the PlantCyc database (http://www.gramene.org/pathway/). Identified pathways were numbered according to column (A) in Supplementary Table S1 at JXB online, and are depicted by solid lines. Missing pathways are shown by broken lines, and missing proteins are indicated by abbreviations of the protein plus #. Different coloured lines represent different biochemical reactions: pink, isoprenoid metabolism; purple, pathway of lipoxygenase; red, glycolysis pathway; blue, amino acid metabolism; lawn green, pentose phosphate cycle; bright green, pyruvate cycle; black, other pathways. DOXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methylerythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2-C-methylerythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate; MEcPP, 2-C-methyl-D-erythritol 2,4-cyclopyrophosphate; HMB-PP, (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate; IPP, isopentenylpyrophosphate; DMAPP, dimethylallyl pyrophosphate; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; DXS, 1-deoxyxylulose-5-phosphate synthase; ispD, 4-diphosphocytidyl-2C-methyl-D-erythritol synthase; psy, phytoene synthase; Pds, phytoene desature; GGPP, geranylgeranyl diphosphate; ϵ-OHase, epsilon-carotene hydroxylase; ABA, abscisic acid; D-G-6P, D-glucose-6- phosphate; F-1, 6-bp, frucose-1, 6-bisphosphate; D-X-5P, D-xylulose-5 phosphate; D-E-4p, D-erythrose-4- phosphate; D-R-5p, D-ribose-5- phosphate.
Carbohydrate metabolism
Since the chromoplast is a kind of heterotrophic plastid, all proteins in this plastid are either imported from the cytoplasm or generated through oxidative processes to sustain anabolic reactions (Neuhaus and Emes, 2000). Cytosolic ATP import is also necessary for the synthesis of various metabolites such as amino acids and starch in heterotrophic plastids. Correspondingly, a plastid ATP/ADP transporter (Z1) and an ADP/ATP carrier protein (Z2) were identified in the present study. A glucose-6-phosphate/phosphate translocator (Z3, Fig. 6) was also detected, suggesting that glucose 6-phosphate could be imported into citrus chromoplasts. Some proteins involved in oxidative metabolism were identified. These include (i) a glucose-6-phosphate isomerase (G1) that catalyses the hydrolysis of fructose-6-phosphate into glucose-6-phospate to feed the oxidative pentose phosphate pathway (OPPP) and generate reducing power (NADPH) and metabolite precursors for essential anabolic pathways (Neuhaus and Emes, 2000); and (ii) 6-phosphogluconate dehydrogenase (P8–P9) and 6-phosphogluconolactonase (P10) that are involved in the OPPP by utilizing glucose 6-phosphate as a source of reducing power. Another pathway for NADP reduction in non-green plastids is glycolysis that generates ATP and pyruvate. Proteins of the glycolytic pathway were well represented, including 6-phosphofructokinase, fructose-bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, enolases, phosphoglyceromutase (G1–G18, S1), pyruvate dehydrogenase (G19–G27), etc. (Supplementary Table S1 at JXB online; Fig. 6). It is worth noting that two enzymes (transketolases and transaldolases) in the non-oxidative branch of the OPPP (P1–P10), which can feed anabolism- or glycolysis-related processes, were also detected. On the whole, the identification of these transporters in the present study reflects the energetic requirements of heterotrophic plastids.
Several proteins involved in the starch biosynthesis pathway were identified, including phosphoglucomutase (S1), glucose-6-phosphate isomerase (G1), and 1,4-α-glucan branching protein (S15). Likewise, enzymes in starch degradation, such as glucan phosphorylase (S13), 4-α-glucanotransferase (S9–S10),glucan water dikinase (S12), and hexokinase (S11), were also identified in this analysis, suggesting a rapid turnover of starch in citrus chromoplasts. In addition, a starch excess proteins 1 (S16) regulating starch accumulation was also identified. A mutant defective for starch excess 1 protein has been reported to cause starch accumulation in Arabidopsis (Yu et al., 2001). Together with earlier reports that both tomato and bell pepper chromoplast proteomes contain starch-degrading and starch excess proteins (Siddique et al., 2006; Barsan et al., 2010) and that starch grains exist in flower chromoplasts (Keresztes and Schroth, 1979), the results suggested that active starch metabolism/catabolism may occur in citrus chromoplasts to provide energy and primary substrates for various biological processes.
Similar to reported proteomes in non-photosynthetic plastids (von Zychlinski et al., 2005; Balmer et al., 2006; Siddique et al., 2006; Barsan et al., 2010), several photosynthesis-related proteins were also identified in orange chromoplasts, including Rubisco (L1–L3), PS1 reaction centre subunit III (L4), LHCP (L5–L6), cytochrome b6-f complex iron–sulphur subunit (L7), and thylakoid lumenal 29.8 kDa protein (L8). It is not surprising that photosynthetic proteins are retained in some non-photosynthetic plastids, such as tomato chromoplasts, as they are transited from chloroplasts (Alba et al., 2005) and the photosynthetic proteins may not be fully degraded. However, it seems impossible for this kind of transition to occur in citrus pulp chromoplasts, since they are fully separated from sunlight and do not perform photosynthesis during the whole ripening stages. Correspondingly, it was found that the citrus pulp plastids underwent white–light yellow–orange/red stages during the ripening process, while green plastids were never found (data not shown). In this context, together with the hypothesis that plastids originate from an intracellular bacterium, the photosynthetic and non-photosynthetic plastids may retain a conservative property during their transition or evolution. The maintenance of photosynthetic proteins in non-photosynthetic plastids may imply a basic role somehow rather than photosynthesis only.
Amino acid metabolism
Sixty-four proteins (∼14% of the total) identified from citrus chromoplasts were predicted to be involved in amino acid synthesis (Supplementary Table S1 at JXB online). This result supports the notion that plastids are important sites for amino acid synthesis (Neuhaus and Emes, 2000). For example, the synthesis of methionine is known to be linked to the regulation of the aspartate pathway, and the assimilation and reduction of sulphur and its incorporation into cysteine (Hesse and Hoefgen, 2003). Consistent with this, a nearly entire set of proteins involved in the methionine pathway have been identified in the present analysis: threonine synthase (A8), aspartate semialdehyde dehydrogenase (A1–A2), and bi-functional aspartate kinase/homoserine dehydrogenases (A3–A7). Cysteine is used for the synthesis of methionine in a three-step reaction, with the first two catalysed by cystathionine γ-synthase (Cgs; A13), and cystathionine β-lyase [CBL; A14 (Martin et al., 2005; Wirtz and Droux, 2005)], respectively. The only enzyme not detected in the present study was methionine synthase (MetS) that catalyses the final step of methionine biosynthesis. Thus, the observations were well in line with previous studies showing that all enzymes required for methionine synthesis are located in plastids, except for MetS which seems to work in the cytosol (Hesse and Hoefgen, 2003; Ravanel et al., 2004). Proteins involved in threonine and serine synthesis were also detected. These included threonine synthase (A8), phosphoserine aminotransferase (A19), and serine hydroxymethyltransferase (A24). Additionally, five proteins were detected in the shikimate pathway (A46–A50) which also takes place in plastids (Herrmann and Weaver, 1999), starting with the condensation of erythrose 4-phosphate and phosphoenolpyruvate, and resulting in the formation of chorismate, the precursor of the aromatic amino acids phenylalanine, tyrosine, and tryptophan (Schmid and Amrhein, 1995). Further, the citrus chromoplast proteome appeared to contain many proteins responsible for the metabolism of branched chain amino acids. These included branched chain amino acid aminotransferase (A12), 3-isopropylmalate dehydratase (A28), 3-isopropylmalate dehydrogenase (A29-A30), 2-isopropylmalate synthase (A25–A27), and ketol-acid reductoisomerase (A10).
It is surprising that whereas proteins involved in amino acid metabolism seemed to have a better coverage in the proteome of citrus chromoplasts when compared with those identified in the proteomes of tomato chromoplasts (Supplementary Table S1 at JXB online), none of the proteins responsible for nitrogen assimilation was detected. Because MS/MS-based protein identification is recognized to be biased toward abundant proteins, it is assumed that the higher the coverage for the engaged enzymes, the higher activity the relevant metabolic pathway may have. In this respect, the present data implied that nitrogen assimilation is not a prevalent pathway, although there are high levels of amino acid metabolism proteins in the chromoplasts isolated from the pulp of ‘Hong Anliu’ oranges. If true, this may reflect specialization of biological processes in citrus chromoplasts.
Protein regulation and transport
Sixty proteins were associated with protein regulation or transport. Seven HSC70 proteins (R39–R45) and a 14-3-3 protein (R46) were identified, both of which are believed to be components of a guidance complex with protein precursors and facilitate targeting of cargo proteins to the outer envelope (Jarvis, 2008). Components of the TOC/TIC complexes (translocon at the outer/inner envelope membrane of chloroplasts) performing the import of nuclear-encoded precursor proteins into chloroplast (Jarvis, 2008) were also detected. These include TOC75 (R1–R2) and four TIC-associated proteins (R3–R6). TOC75 is the main pore of the TOC complex deeply embedded within the outer membrane, forming an aqueous protein-conducting channel (Hinnah et al., 2002; Vothknecht and Soll, 2005). In addition, IAP100 (R7), a protein involved in recruiting chaperonin for folding of newly imported proteins (Kessler and Blobel, 1996), was also detected.
Plastids undergo drastic changes morphologically and physiologically at different developmental stages and under different environmental conditions (see review by Egea et al., 2010). Keeping a balance between proteins biosynthesis and degradation is crucial for accomplishing these transitions and maintaining homeostasis (Sakamoto, 2006). Seven Cpn60 proteins (R25–R31) were characterized as chaperonin-associated machinery, and three elongation factors for protein biosynthesis (R32–R34) were identified. Plastid transition from one type to another entails a series of physiological processes such as chlorophyll degradation and turnover, and proteases appear to play essential roles in this process (Adam, 2000; Adam and Clarke, 2002). Therefore, it is not surprising that 20 proteins (R8–R20 and R47–R53) with a potential role in protein degradation were detected. For example, nine Clp proteases (R8–R16) were identified. These Clp proteases form a chaperone complex to drive import of proteins into the plastid, cleave the transit peptide, and recycle plastid components. The chaperone complex also facilitates the degradation of plastid proteins (Peltier et al., 2004; Pojidaeva et al., 2004; Sakamoto, 2006).
Import of precursor proteins into the plastid stroma and the removal and degradation of their transit peptides involve two types of metalloproteases termed stroma processing peptidases (SPPs) and pre-sequence protease (PreP) (Moberg et al., 2003; Zhong et al., 2003; Bhushan et al., 2005). Correspondingly, two zinc metallopeptidases (R17–R18) and PreP 2 (R47) implicated in this process (Krishna and Gloor, 2001; Richter et al., 2005) were identified in the present study. The oligopeptidases (R19–R20) cleave the target peptides into amino acids for recycling (Richter et al., 2005). Other proteases, such as FtsZ (R21–R23) and FtsH (R16) proteases, not only function to control plastid division, but are also involved in degrading the PSII reaction centre protein D1 (Sakamoto, 2006; Yoshioka et al., 2006). Because there is rare plastid division in mature citrus chromoplasts, these FtsZ proteins detected in citrus chromoplasts may play a major role in PSII degradation.
Carotenoid metabolism
Chromoplast development is accompanied by massive synthesis and accumulation of carotenoids, which is highly regulated by expression of the carotenoid biosynthetic genes during fruit ripening (Liu et al., 2007). It is not surprising that several enzymes involved in the 2-C-methyl-D-erythritol 4-phosphate pathway (MEP pathway) and some in the carotenoid biosynthesis pathway were identified. The MEP pathway provides most precursors for the synthesis of carotenoids, chlorophyll, quinones, and tocopherol. Except for 1-deoxyxylulose-5-phosphate synthase (DXS), 4-diphosphocytidyl-2C-methyl-D-erythritol synthase (ispD), and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), the majority of proteins in the MEP pathway are detected for the synthesis of isoprenoid precursors in plastids (Phillips et al., 2008; Cordoba et al., 2009). In this study, seven proteins involved in the MEP pathway were identified: DXR (C1–C2), ispE (C3), ispF (C4), HDS (C5–C7), ipI (C8–C9), GGPS (C10), and GGR (C11). The penultimate enzyme of the MEP pathway is hydroxymethylbutenyl 4-diphosphate synthase (HDS) (C5–C7), which catalyses the conversion of 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate into (E)-4-hydroxy-3-methylbut-2-enyl diphosphate. The next key regulatory enzyme is HDR, which catalyses the production of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Previous studies showed that knocking out HDS and HDR produced plants with albino leaves due to the impairment of the ability to synthesize chlorophylls and carotenoids via the MEP pathway (Page et al., 2004), suggesting that both enzymes are required for function of the MEP pathway. Surprisingly, as many as 32 peptides, occupying >50% of the total peptides in the MEP pathway, were identified as HDS in the present study (Supplementary Table S1 at JXB online). In contrast, no peptide was detected as HDR, implying that a special way of adjusting the balance of carotenoid metabolism exists in citrus chromoplasts and that HDS may be a key factor with a central role in carotenoid metabolism.
Almost all the enzymes dedicated to the biosynthesis of lycopene and two downstream proteins have been identified, including ZDS (C12), ϵ-LCY (C13), β-LCY (C14), β-OHase (C16), and NCED (C15). However, the rate-limiting enzyme phytoene synthase (PSY) was not detected, even though expression of PSY was reported to increase in the peel and juice sacs during the ripening of fruits (Ikoma et al., 2001; Kim et al., 2001; Liu et al., 2007). The cyclization of lycopene is a key branch point in the pathway of carotenoid biosynthesis in higher plants, and ϵ-LCY (C13) and β-LCY (C14) are two important lycopene cyclases (Cunningham et al., 1998). The expression of ϵ-LCY decreased rapidly in the juice sac of ‘Hong Anliu’ during fruit ripening, leading to lycopene accumulation in the juice sacs of citrus fruits (Liu et al., 2007). This hypothesis was further supported by the probably low expression levels of the ϵ-LCY or β-LCY proteins, because only one peptide sequence for each of these two enzymes was detected in the proteomic analysis with the mature citrus chromoplasts (Supplementary Table S1 at JXB online). Similarly, few peptides for ϵ-LCY or β-LCY were identified from protein samples prepared from mature pepper and tomato chromoplasts (Siddique et al., 2006; Barsan et al., 2010). Tocopherol cyclase (C17), an enzyme involved in vitamin E biosynthesis, was also detected, consistent with the hypothesis that there exists a close relationship between the biosynthesis of vitamin E and that of carotenoid plastids (DellaPenna and Pogson, 2006). It is well known that carotenoids were sequestrated by structural proteins such as PAP/fibrillin family proteins (Z14–Z17), and together they are major constituents of chromoplast plastoglobules (Deruere et al., 1994). It is not surprising that a large quantity of peptides of PAP was identified in the present analysis, suggesting that PAP indeed is one of the most abundant proteins in citrus chromoplasts.
Lipid metabolism
Lipoxygenases (LOXs) are ubiquitous enzymes in eukaryotes. Depending on the regiospecificity, they could be grouped into 9-LOX and 13-LOX which convert polyunsaturated fatty acids to 9-hydroperoxides and 13-hydroperoxides, respectively. Interestingly, four enzymes involved in the 13-LOX pathway [13-lipoxygenase (F1), LOX (F3), LOX2 (F4), and HPL1 (F2)] but none in the 9-LOX pathway were identified in citrus chromoplasts, indicating that citrus chromoplasts mainly use 13-LOX rather than 9-LOX in the turnover of unsaturated fatty acids. It is well documented that the 13-LOX pathway could ultimately lead to the biosynthesis of jasmonic acid (JA; Wasternack, 2007). Thus, as expected, several enzymes (F5–F10) involved in JA biosynthesis were detected, which had been predicted to be located in plastids (Ziegler et al., 2000; Wasternack, 2007). Of special interest is HPL1 (F2), which is thought to be responsible for the generation of flavours and tastes in plants, such as the odour of cucumber (Bate et al., 1998). Therefore, it is possible that citrus chromoplasts may play an important role in generating citrus-specific odour or flavour, just as has been proposed for a role of tomato chromoplasts in the synthesis of LOX-derived tomato volatiles (Barsan et al., 2010).
Oxidative stress related
Reactive oxygen species (ROS) burst is an intrinsic feature of plant cells/organelles during senescence and fruit ripening. To ensure normal biological processes in plastids, two classes of antioxidative systems (non-enzymatic and enzymatic) are recruited to remove overproduced ROS to maintain a steady-state level of ROS in the organelle. For the non-enzymatic system, cell metabolites such as glutathione, ascorbate (AsA; hydrophilic), carotenoids, and α-tocopherol may function as ROS scavengers to protect thylakoid membranes and photosynthetic structures in chloroplasts from oxidative damage (Havaux et al., 2005; Kruk et al., 2005; Penuelas and Munne-Bosch, 2005). Carotenoids, especially lycopene, had been suggested to play an important role in redox homeostasis in citrus fruits (Pan et al., 2009). The enzymatic antioxidative system is believed to be more effective (Foyer and Noctor, 2009). Several key components of the Asc–glutathione (GSH) cycle and other important antioxidants such as stromal- and thylakoid-bound ascorbate peroxidases (sAPXs and tAPXs; H1–H3), glutathione peroxidases (GPXs; H7–H8), dehydroascorbate reductase (H10), peroxiredoxins (PRXs; H9), and superoxide dismutases (SODs; H4–H6) were identified in the present study. In the chloroplasts of higher plants, the water–water cycle is carried out by Cu/Zn-SOD and tAPX (H2–H3) isoenzyme. Additionally, the AsA–GSH cycle, which is catalysed by sAPX (H1) and the AsA-regenerating systems including dehydroascorbate reductase (H10) also occurs in the stroma as a second active oxygen species (AOS)-scavenging system (Yabuta et al., 2002). Like chloroplasts, APXs encountered in the chromoplasts may be indicative of the presence of the AsA-dependent water–water cycle which reduces H2O2 using AsA as reductant (Asada, 2006). Among the SODs, Zn-SOD and Fe-SOD are targeted to plastids and their overexpression has been shown to enhance plant tolerance to different environmental stresses (Lee et al., 2007). The identification of 15 proteins (Z7–Z8, H1–H12, and H27) involved in oxidative stress response in the present study is indicative of high levels of redox activity in citrus chromoplasts and suggestive of an important role for ROS metabolism in citrus fruit ripening. In support of the presence of a functional antioxidant system in chromoplasts, it has been demonstrated that high coverage of redox proteins was present in tomato chromoplasts (Barsan et al., 2010); and the activity of SOD and enzymes in the Asc–GSH cycle was up-regulated during ripening of pepper fruit (Marti et al., 2009).
The citrus chromoplast proteome as compared with the tomato chromoplast proteome
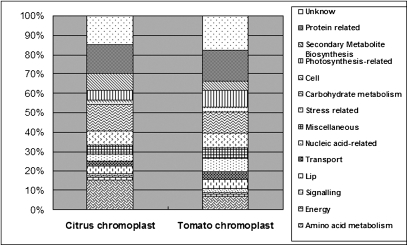
Citrus has an obvious non-climacteric fruit ripening behaviour, which is different from that of the model fruit tomato which is a typical climacteric fruit. Here, the chromoplast proteome of citrus was compared with that of tomato (Barsan et al., 2010) to explore similarities and differences between these two fruit types. Functional analysis revealed that almost 70% of the proteins in each proteome were grouped into the same five functional categories: amino acid metabolism, carbohydrate metabolism, stress related, protein related, and secondary metabolism (Fig. 7), suggesting that the general function of citrus chromoplasts resembled that of tomato chromoplasts. However, according to Fig. 7 and proteins listed in Supplementary Table S1 at JXB online, it was noticed that the citrus chromoplast is characterized by more extensive carotenoid synthesis, extensive amino acid synthesis without nitrogen assimilation, and evidence for lipid metabolism concerning JA synthesis. These differences may reflect functional characteristics of citrus chromoplasts in comparison with tomato chromoplasts.
Fig. 7.
Comparison of functional classification of the chromoplast proteins identified in citrus and tomato. The tomato chromoplast proteins were taken from Supplementary Table S1 of the study by Barsan et al. (2010) Journal of Experimental Botany 61(9), 2413–2431. Proteins were functionally classified according to MapMan Bin based on functional annotations for their homologous proteins.
It is worth noting that a total of 197 peptides were identified as plastid small heat shock proteins (psHSPs; H13–H15, H21–H25), among which were 103 peptides corresponding to the plastid small HSP21 (H14). The total peptides of psHSPs account for as much as 9.3% of all the detected peptides in citrus chromoplasts (Supplementary Table S1 at JXB online). In contrast, although there are 62 peptides corresponding to psHSPs in the tomato chromoplast, only one peptide of psHSP21 was detected in the tomato chromoplast (Barsan et al., 2010). Though HSPs have been suggested to contribute to fruit ripening, it seems that only psHSP21 was functionally evidenced to play a dual role in protecting PSII from oxidative stress and promoting colour changes during fruit maturation (Neta-Sharir et al., 2005). This kind of plastid-derived signal, such as psHSP21, was suggested to be involved in plastid conversion perhaps by a feedback to ethylene which plays a vital role in chromoplast differentiation and fruit ripening (Lopez-Juez, 2007). However, the chromoplast differentiation or ripening of non-climacteric citrus fruits must be ethylene independent. Therefore, the role of psHSPs might be different between citrus and tomato. Regarding the larger portion of psHSP peptides in citrus chromoplasts as compared with tomato, the psHSPs, especially psHSP21, may play a distinct and more important role in citrus ripening.
Conclusion
In this study, a new protocol was developed for the purification of citrus chromoplasts by a combination of three consecutive density gradient centrifugations. Subsequent proteomic and in silico homology analyses identified 418 proteins as candidate proteins located in citrus chromoplasts. The majority of these proteins show homology to confirmed or putative plastid proteins reported in other plant species, while 89 proteins may be novel plastid residents. The analyses of the likely biochemical functions of these proteins indicate that proteins participating in carbohydrate metabolism, amino acid metabolism, protein regulation and transport, carotenoid metabolism, and lipid metabolism dominate the citrus chromoplast proteome. In conclusion, this study revealed the functional complexity of citrus chromoplasts at the proteome level as an important first step towards understanding of the fruit ripening mechanisms of sweet orange at the subcellular level.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Characteristics of the 418 citrus pulp chromoplast plastid protein candidates identified by GeLC-MS/MS.
Table S2. List of the 75 contaminants according to BLAST search homology.
Supplementary Material
Acknowledgments
This work was supported by the National Basic research programme of China (973 programme; No. 2011CB100601), and the National Natural Science Foundation of China (No. 30830078, 30921002). We would like to thank Professor S. Xiao (Center for Biosystems Research, University of Maryland Biotechnology Institute, USA) for his critical review of the manuscript. We also thank Shanghai Applied Protein Technology Co. Ltd for technology support and helpful advice.
References
- Adam Z. Chloroplast proteases: possible regulators of gene expression? Biochimie. 2000;82:647–654. doi: 10.1016/s0300-9084(00)00612-x. [DOI] [PubMed] [Google Scholar]
- Adam Z, Clarke AK. Cutting edge of chloroplast proteolysis. Trends in Plant Science. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates III, JR, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiology. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Siddique A, Gruissem W. Proteome analysis of tobacco bright yellow-2 (BY-2) cell culture plastids as a model for undifferentiated heterotrophic plastids. Journal of Proteome Research. 2004;3:1128–1137. doi: 10.1021/pr0499186. [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, DuPont FM, Buchanan BB, Hurkman WJ. Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. Journal of Experimental Botany. 2006;57:1591–1602. doi: 10.1093/jxb/erj156. [DOI] [PubMed] [Google Scholar]
- Barsan C, Sanchez-Bel P, Rombaldi C, Egea I, Rossignol M, Kuntz M, Zouine M, Latche A, Bouzayen M, Pech JC. Characteristics of the tomato chromoplast revealed by proteomic analysis. Journal of Experimental Botany. 2010;61:2413–2431. doi: 10.1093/jxb/erq070. [DOI] [PubMed] [Google Scholar]
- Bate NJ, Sivasankar S, Moxon C, Riley JMC, Thompson JE, Rothstein SJ. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiology. 1998;117:1393–1400. doi: 10.1104/pp.117.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biology. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Stahl A, Nilsson S, et al. Catalysis, subcellular localization, expression and evolution of the targeting peptides degrading protease, AtPreP2. Plant and Cell Physiology. 2005;46:985–996. doi: 10.1093/pcp/pci107. [DOI] [PubMed] [Google Scholar]
- Chen CX, Zhou P, Choi YA, Huang S, Gmitter FG. Mining and characterizing microsatellites from citrus ESTs. Theoretical and Applied Genetics. 2006;112:1248–1257. doi: 10.1007/s00122-006-0226-1. [DOI] [PubMed] [Google Scholar]
- Cordoba E, Salmi M, Leon P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. Journal of Experimental Botany. 2009;60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Daher Z, Recorbet G, Valot B, Robert F, Balliau T, Potin S, Schoefs B, Dumas-Gaudot E. Proteomic analysis of Medicago truncatula root plastids. Proteomics. 2010;10:2123–2137. doi: 10.1002/pmic.200900345. [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. Vitamin synthesis in plants: tocopherols and carotenoids. Annual Review of Plant Biology. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- Deruere J, Romer S, Dharlingue A, Backhaus RA, Kuntz M, Camara B. Fibril assembly and carotenoid overaccumulation in chromoplasts—a model for supramolecular lipoprotein structures. The Plant Cell. 1994;6:119–133. doi: 10.1105/tpc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian WP, Purgatto E, Latche A, Chervin C, Bouzayen M, Pech JC. Chromoplast differentiation: current status and perspectives. Plant and Cell Physiology. 2010;51:1601–1611. doi: 10.1093/pcp/pcq136. [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. Proteomics of the chloroplast envelope membranes from. Arabidopsis thaliana. Molecular and Cellular Proteomics. 2003;2:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxidants and Redox Signaling. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Goldschmidt EE. Regulatory aspects of chloro-chromoplast interconvensions in senescing Citrus fruit peel. Israel Journal of Botany Basic and Applied Plant Sciences. 1988;47:123–130. [Google Scholar]
- Hadjeb N, Gounaris I, Price CA. Chromoplast-specific proteins in capsicum-annuum. Plant Physiology. 1988;88:42–45. doi: 10.1104/pp.88.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dormann P. Vitamin E protects against photoinhibition and photooxidative stress in. Arabidopsis thaliana. The Plant Cell. 2005;17:3451–3469. doi: 10.1105/tpc.105.037036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis subcellular database. Nucleic Acids Research. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM, Weaver LM. The shikimate pathway. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R. Molecular aspects of methionine biosynthesis. Trends in Plant Science. 2003;8:259–262. doi: 10.1016/S1360-1385(03)00107-9. [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Wagner R, Sveshnikova N, Harrer R, Soll J. The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophysical Journal. 2002;83:899–911. doi: 10.1016/S0006-3495(02)75216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikoma Y, Komatsu A, Kita M, Ogawa K, Omura M, Yano M, Moriguchi T. Expression of a phytoene synthase gene and characteristic carotenoid accumulation during citrus fruit development. Physiologia Plantarum. 2001;111:232–238. [Google Scholar]
- Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytologist. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- Katz E, Fon M, Eigenheer RA, Phinney BS, Fass JN, Lin DW, Sadka A, Blumwald E. A label-free differential quantitative mass spectrometry method for the characterization and identification of protein changes during citrus fruit development. Proteome Science. 2010;8:68. doi: 10.1186/1477-5956-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Fon M, Lee YJ, Phinney BS, Sadka A, Blumwald E. The citrus fruit proteome: insights into citrus fruit metabolism. Planta. 2007;226:989–1005. doi: 10.1007/s00425-007-0545-8. [DOI] [PubMed] [Google Scholar]
- Keresztes A, Schroth A. Light and electron-microscopic investigation of invitro starch synthesis in chromoplasts. Cytobios. 1979;26:185–191. [PubMed] [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries in the chloroplast. Proceedings of the National Academy of Sciences, USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Ko KC, Kim CS, Chung WI. Isolation and expression patterns of a cDNA encoding phytoene synthase in Citrus. Journal of Plant Physiology. 2001;158:795–800. [Google Scholar]
- Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S. plprot: a comprehensive proteome database for different plastid types. Plant and Cell Physiology. 2006;47:432–436. doi: 10.1093/pcp/pcj005. [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Current Biology. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Krishna P, Gloor G. The Hsp90 family of proteins in. Arabidopsis thaliana. Cell Stress and Chaperones. 2001;6:238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk J, Hollander-Czytko H, Oettmeier W, Trebst A. Tocopherol as singlet oxygen scavenger in photosystem II. Journal of Plant Physiology. 2005;162:749–757. doi: 10.1016/j.jplph.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ahsan N, Lee KW, Kim DH, Lee DG, Kwak SS, Kwon SY, Kim TH, Lee BH. Simultaneous overexpression of both CuZn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. Journal of Plant Physiology. 2007;164:1626–1638. doi: 10.1016/j.jplph.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu YZ, Zhao XL, Deng XX, Guo LL, Gu JQ. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck) Journal of Experimental Botany. 2007;58:4161–4171. doi: 10.1093/jxb/erm273. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. International Journal of Developmental Biology. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E. Plastid biogenesis, between light and shadows. Journal of Experimental Botany. 2007;58:11–26. doi: 10.1093/jxb/erl196. [DOI] [PubMed] [Google Scholar]
- Marti MC, Camejo D, Olmos E, Sandalio LM, Fernandez-Garcia N, Jimenez A, Sevilla F. Characterization and changes in the antioxidant system of chloroplasts and chromoplasts isolated from green and mature pepper fruits. Plant Biology. 2009;11:613–624. doi: 10.1111/j.1438-8677.2008.00149.x. [DOI] [PubMed] [Google Scholar]
- Martin MN, Tarczynski MC, Shen B, Leustek T. The role of 5'-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynthesis Research. 2005;86:309–323. doi: 10.1007/s11120-005-9006-z. [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Current Opinion in Plant Biology. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Moberg P, Stahl A, Bhushan S, Wright SJ, Eriksson A, Bruce BD, Glaser E. Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. The Plant Journal. 2003;36:616–628. doi: 10.1046/j.1365-313x.2003.01904.x. [DOI] [PubMed] [Google Scholar]
- Muccilli V, Licciardello C, Fontanini D, Russo MP, Cunsola V, Saletti R, Recupero GR, Foti S. Proteome analysis of Citrus sinensis L. (Osbeck) flesh at ripening time. Journal of Proteomics. 2009;73:134–152. doi: 10.1016/j.jprot.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Nair R, Rost B. Sequence conserved for subcellular localization. Protein Science. 2002;11:2836–2847. doi: 10.1110/ps.0207402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neta-Sharir I, Isaacson T, Lurie S, Weiss D. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. The Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- Page JE, Hause G, Raschke M, Gao WY, Schmidt J, Zenk MH, Kutchan TM. Functional analysis of the final steps of the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiology. 2004;134:1401–1413. doi: 10.1104/pp.103.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZY, Liu Q, Yun Z, Guan R, Zeng WF, Xu Q, Deng XX. Comparative proteomics of a lycopene-accumulating mutant reveals the important role of oxidative stress on carotenogenesis in sweet orange (Citrus sinensis L. Osbeck) Proteomics. 2009;9:5455–5470. doi: 10.1002/pmic.200900092. [DOI] [PubMed] [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Molecular and Cellular Proteomics. 2006;5:114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. The Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy J, van Wijk KJ. Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. Journal of Biological Chemistry. 2004;279:4768–4781. doi: 10.1074/jbc.M309212200. [DOI] [PubMed] [Google Scholar]
- Penuelas J, Munne-Bosch S. Isoprenoids: an evolutionary pool for photoprotection. Trends in Plant Science. 2005;10:166–169. doi: 10.1016/j.tplants.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Phillips MA, Leon P, Boronat A, Rodriguez-Concepcion M. The plastidial MEP pathway: unified nomenclature and resources. Trends in Plant Science. 2008;13:619–623. doi: 10.1016/j.tplants.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Pojidaeva E, Zinchenko V, Shestakov SV, Sokolenko A. Involvement of the SppA1 peptidase in acclimation to saturating light intensities in Synechocystis sp strain PCC 6803. Journal of Bacteriology. 2004;186:3991–3999. doi: 10.1128/JB.186.12.3991-3999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rebeille F, Douce R. Methionine metabolism in plants—chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. Journal of Biological Chemistry. 2004;279:22548–22557. doi: 10.1074/jbc.M313250200. [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma CL, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. The Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–16. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Richter S, Zhong R, Lamppa G. Function of the stromal processing peptidase in the chloroplast import pathway. Physiologia Plantarum. 2005;123:362–368. [Google Scholar]
- Sakamoto W. Protein degradation machineries in plastids. Annual Review of Plant Biology. 2006;57:599–621. doi: 10.1146/annurev.arplant.57.032905.105401. [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. Journal of Biological Chemistry. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- Schmid J, Amrhein N. Molecular organization of the shikimate pathway in higher plants. Phytochemistry. 1995;39:737–749. [Google Scholar]
- Siddique MA, Grossmann J, Gruissem W, Baginsky S. Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant and Cell Physiology. 2006;47:1663–1673. doi: 10.1093/pcp/pcl033. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Research. 2008;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zychlinski A, Kleffmann T, Krishnamurthy N, Sjolander K, Baginsky S, Gruissem W. Proteome analysis of the rice etioplast: metabolic and regulatory networks and novel protein functions. Molecular and Cellular Proteomics. 2005;4:1072–1084. doi: 10.1074/mcp.M500018-MCP200. [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Soll H. Chloroplast membrane transport: interplay of prokaryotic and eukaryotic traits. Gene. 2005;354:99–109. doi: 10.1016/j.gene.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M. Synthesis of the sulfur amino acids: cysteine and methionine. Photosynthesis Research. 2005;86:345–362. doi: 10.1007/s11120-005-8810-9. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Motoki T, Yoshimura K, Takeda T, Ishikawa T, Shigeoka S. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. The Plant Journal. 2002;32:915–925. doi: 10.1046/j.1365-313x.2002.01476.x. [DOI] [PubMed] [Google Scholar]
- Yoshioka M, Uchida S, Mori H, Komayama K, Ohira S, Morita N, Nakanishi T, Yamamoto Y. Quality control of photosystem II—cleavage of reaction center D1 protein in spinach thylakoids by FtsH protease under moderate heat stress. Journal of Biological Chemistry. 2006;281:21660–21669. doi: 10.1074/jbc.M602896200. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiology. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Hausler RE, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. The Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Z, Li WY, Pan ZY, Xu J, Cheng YJ, Deng XX. Comparative proteomics analysis of differentially accumulated proteins in juice sacs of ponkan (Citrus reticulata) fruit during postharvest cold storage. Postharvest Biology and Technology. 2010;56:189–201. [Google Scholar]
- Zhong R, Wan JX, Jin RG, Lamppa G. A pea antisense gene for the chloroplast stromal processing peptidase yields seedling lethals in Arabidopsis: survivors show defective GFP import. in vivo. The Plant Journal. 2003;34:802–812. doi: 10.1046/j.1365-313x.2003.01772.x. [DOI] [PubMed] [Google Scholar]
- Ziegler J, Stenzel I, Hause B, Maucher H, Hamberg M, Grimm R, Ganal M, Wasternack C. Molecular cloning of allene oxide cyclase—the enzyme establishing the stereochemistry of octadecanoids and jasmonates. Journal of Biological Chemistry. 2000;275:19132–19138. doi: 10.1074/jbc.M002133200. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Friso G, Kim J, Rudella A, Rodriguez VR, Asakura Y, Sun Q, van Wijk KJ. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Molecular and Cellular Proteomics. 2009;8:1789–1810. doi: 10.1074/mcp.M900104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.