Abstract
Label-free LC-MS/MS-based shot-gun proteomics was used to quantify the differential protein synthesis and metabolite profiling in order to assess metabolic changes during the development of citrus fruits. Our results suggested the occurrence of a metabolic change during citrus fruit maturation, where the organic acid and amino acid accumulation seen during the early stages of development shifted into sugar synthesis during the later stage of citrus fruit development. The expression of invertases remained unchanged, while an invertase inhibitor was up-regulated towards maturation. The increased expression of sucrose-phosphate synthase and sucrose-6-phosphate phosphatase and the rapid sugar accumulation suggest that sucrose is also being synthesized in citrus juice sac cells during the later stage of fruit development.
Keywords: Citrus, fruit development, juice sac cells, LC-MS/MS, proteomics
Introduction
Citrus is one of the most important and widely grown commodity fruit crops (Talon and Gmitter, 2008). Citrus has a non-climacteric fruit maturation behaviour and a unique anatomical fruit structure (Spiegel-Roy and Goldschmidt, 1996). The fruit contains two peel tissues, flavedo and albedo. The flavedo accumulates pigments and compounds which contribute to the fruit aroma, while the albedo comprises spongy cells rich in pectin. During the early stages of fruit development the albedo occupies most of the fruit volume and it becomes gradually thinner during fruit development as the juice cells in the pulp grow (Spiegel-Roy and Goldschmidt, 1996). Growth and development of the citrus fruit can be divided into three major stages (Bain, 1958; Katz et al., 2004). Stage I starts immediately after fruit set and is characterized by extensive cell division. During the transition to stage II, cell division ceases in all fruit tissues except the outermost flavedo layers and the tips of the juice sacs. During this stage, citrus fruit grows through cell expansion. Juice sac cell enlargement is mostly driven by the expansion of the vacuole, which occupies most of the cell volume. Stage III is the fruit maturation and ripening stage when fruit growth slows down and the pulp reaches its final size. Citrus fruit development is characterized by changes in primary and secondary metabolite content, with sugars and citric acid being the major components of the juice sac cells. Sucrose is translocated to the fruits from the leaves throughout fruit development, and constitutes about 50% of the total soluble sugars. The anatomy of the citrus fruit, where the juice sacs are disconnected from the vascular bundles present in the albedo, suggest apoplastic sucrose downloading (Lowell et al., 1989; Tomlinson et al., 1991; Koch, 2004). Sucrose can then be hydrolysed by cytosolic invertases or stored in the acidic vacuoles and hydrolysed by vacuolar acidic invertases (Echeverria, 1992; Echeverria and Burns, 1990). Accumulation of citric acid in the vacuole of the juice sac cells is correlated with vacuole acidification mediated by the proton pumping activity of the tonoplastic H+-ATPase. Citrate begins to accumulate during the second phase of fruit development. The accumulation continues for a few weeks, reaching a peak when the fruit volume is about 50% of its final value and then acid declines gradually as the fruit matures (Shimada et al., 2006). Citrate decline during the second half of fruit development is associated with the activity of CsCit1, a H+/citrate symporter (Shimada et al., 2006). It has been suggested that some of the citrate is targeted for amino acid biosynthesis generally induced during the second half of fruit development (Sadka et al., 2002). Indeed, there is an increase in some amino acid metabolizing genes, including those of the GABA shunt, and their corresponding enzymes during the citrate decline stage (Cercos et al., 2006; Katz, et al., 2007).
In the last few years, studies using transcriptome analysis and metabolite profiling demonstrated a tight regulation of fruit metabolism during fruit maturation (Carrari et al., 2006; Mounet et al., 2009; Zanor et al., 2009). However, comparison of mRNA expression levels, proteins amounts, and enzymatic activities have revealed low correlations between metabolome and transcriptome, indicating that transcriptome analysis was not sufficient to understand protein dynamics or biochemical regulation (Gygi et al., 1999; Gibon et al., 2006; Wienkoop et al., 2008). A more direct correlation is expected for proteins and metabolites (Wienkoop et al., 2008) and, therefore, quantitative mass spectrometric (MS) proteomics and metabolomics are becoming attractive approaches. Quantitative proteomics has been used for the quantification of complex biological samples (Bantscheff et al., 2007; America and Cordewener, 2008; Schulze and Usadel, 2010). Previously, LC-MS/MS was used to identify the proteome of various cellular fractions of the juice sac cell (Katz et al., 2007). More recently, a label-free differential quantitative mass spectrometry method was developed to follow protein changes in citrus juice sac cells. Two alternative methods, differential mass-spectrometry (dMS) and spectral counting (SC) were used to analyse the protein changes occurring during the earlier and late stages of fruit development (Katz et al., 2010). Along with the generation of a novel bioinformatics tool, iCitrus, the above method enabled the identification of approximately 1500 citrus proteins expressed in fruit juice sac cells and the quantification of changes in their expression during fruit development.
In this study, label-free LC-MS/MS-based shot-gun proteomic and metabolomic approaches were utilized to investigate citrus fruit development. These tools were used to identify and evaluate changes occurring in the metabolic pathways of juice sac cells which affect citrus fruit development and quality. Integration of proteomic and metabolomic analyses created a more comprehensive overview of changes in protein expression and metabolite composition of primary metabolism during citrus fruit development and maturation.
Materials and methods
Plant material and protein isolation
Orange Navel (Citrus sinensis cv. Washington) fruits at three different developmental stages, early stage II, stage II, and stage III (35, 55, and 80 mm in fruit diameter, respectively) (Katz et al., 2004) were obtained from the Lindcove Research Center, University of California, Exeter, CA. Juice sacs were collected from at least 20 fruits and pooled at each stage. Two independent biological repetitions from two consecutive years were used. Soluble and membrane-bound proteins were isolated as described by (Katz et al. 2007, 2010).
Mass spectrometry and data analysis
Digested peptides were separated by reverse-phase chromatography and the separated peptides were analysed in a Thermo-Scientific LTQ-FT Ultra mass-spectrometer (San Jose, CA) as described previously (Katz et al., 2010). Five technical replications of each pooled sample (older versus younger fruit) were run with blanks (washes) between each sample run. Tandem mass spectra were extracted with Xcalibur version 2.0.7. All MS/MS samples were analysed using SEQUEST (Protein Discoverer 1.1; Thermo-Scientific, San Jose, CA). SEQUEST was set up to search a FASTA file of the iCitrus Protein Database (Katz et al., 2010), assuming the digestion enzyme trypsin. SEQUEST parameters were as before (Katz et al., 2010). The filtering criteria consisted of Cross-correlation (xcorr) values larger than 1.5 for single-charged ions, 2.2 for double-charged ions, and 3.3 for triple-charged ions, for both half or fully tryptic peptides. This resulted in a false discovery rate of less than 5% using a decoy database search strategy.
For differential expression mass spectrometry (dMS), samples were analysed using a Thermo Scientific LTQ-FT mass-spectrometer and a Michrom-Paradigm HPLC. Peptides were separated according to Katz et al. (2010) and analysed using the label-free differential expression package SIEVE 1.3. (Thermo Scientific, San Jose Ca). Search results were filtered for a false discovery rate of 5% also employing a decoy search strategy utilizing a reverse database (Elias et al., 2005; Kall et al., 2008)
For spectral counting, all MS/MS samples were analysed using X! Tandem (www.thegpm.org; version TORNADO (2008.02.01.2)). X! Tandem was set up to search the 62,415 entries of iCitrus (Katz et al., 2010) assuming the digestion enzyme trypsin. Scaffold 2.06.00 (Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at greater than 95% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Unweighted spectral counts for the identified proteins obtained from the samples corresponding to two consecutive growth seasons were exported from Scaffold and analysed using QSpec (Choi et al., 2008) for significance analysis. Proteins were considered significantly different across sample conditions if QSpec reported a Bayes factor of >10. This corresponded to a false discovery rate (FDR) of approximately 5% (Katz et al., 2010).
Because of the use of different software packages and different FDR calculations, caution should be used when comparing the label-free data with the spectral counting.
Proteomics data set
The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash:
Cf3G8KatEeCbDv2kV1Gnw4njaSYARJgmtyzYl+5764Gsbb/M3LX+/oo1zcHnHK1Gs0ukuBM5Rk+Q1t5hpia109pVPXkAAAAAAAAoLg==
The hash may be used to prove exactly what files were published as part of this manuscript's data set, and the hash may also be used to check that the data have not changed since publication.
Extraction and derivatization of polar metabolites
Citrus juice sacs were immediately frozen after removal in liquid nitrogen and stored at –80 °C until further analysis. For extraction of polar metabolites, samples were lyophilized and ground, and the powder (10 mg) was mixed with 250 μl of 75% ice-cold methanol and two stainless steel balls (2.3 mm). Metabolites were extracted twice using a Retsch Mixer Mill (30 s at 30 cycles s−1) (Retsch Inc., Newtown, PA) and placed on dry ice for 15 min. A 250 μl aliquot of 25% methanol with an internal standard (12 μg ml−1 ribitol in water) was added to each sample and subjected to two more treatment cycles in the mixer mill. The mixture was vortexed, and then centrifuged at 13 000 rpm for 20 min. The liquid fraction was carefully transferred to a new vial and 200 μl of chloroform were added. The mixture was briefly vortexed and centrifuged at 2600 g for 20 min. The upper polar fraction was carefully aliquoted into 1.5 ml vials and dried in vacuo. Metabolites were methoximated with 80 μl of methoxylamine hydrochloride in pyridine (20 mg·ml−1) for 90 min at 45 °C. Metabolites were then trimethylsilylated with 80 μl of MSTFA+1%TMCS (N-methyl-N-trimethylsilyltrifluoroacetamide and trimethylchlorosilane, Pierce, Rockford, IL) for 30 min at 37 °C. After derivatization, a 1 μl aliquot was analysed by GC-MS (Roessner et al., 2000). Standard chemicals were derivatized with methoxyamine hydrochloride solution in pyridine and MSTFA as described above.
GC/MS analysis
Samples were injected into a hot (230 °C) injector with a split ratio of 25:1. Compounds were separated on a non-polar Alltech AT-5ms column (25 m+5 m guard×0.25 mm ID×0.25 μm film thickness) using helium at 1 ml min−1 as the carrier gas. The oven programme was 70 °C for 5 min, 5 °C min−1 ramp to 310 °C, 1 min hold, and 2 min equilibration (Trace GC, Thermo Electron Corp.). The interface and ion source temperatures were 250 °C and 200 °C, respectively. Analytes were detected using a dual-stage single quadrupole mass selective detector (Trace DSQ, Thermo Electron Corp.). Mass spectra were recorded at 2 scans s−1 with a 50–600 m/z scanning range.
Metabolites were identified using spectral matching and retention indexes from custom in-lab libraries in AMDIS (automated mass spectral deconvolution and identification system, NIST, Gaithersburg, MD). Metabolite peak areas were integrated using the ICIS algorithm in Xcalibur v2.0. Statistical analysis of peak area and the calculation of targeted metabolites with external calibration curves was done using the SAS system v9.1 (SAS Institute, Cary, NC).
MAPMAN analysis
MapMan (http://mapman.gabipd.org/web/guest) BINs, currently used for Arabidopsis classification (Thimm et al., 2004; Usadel et al., 2005), were adopted for citrus using iCitrus (Katz et al., 2010). For visualization, the Arabidopsis homologues of citrus proteins were loaded into MapMan, which displays individual genes mapped on their pathway as false colour-coded rectangles. To facilitate comparison of the different colours, a legend explaining the changes is displayed by MapMan, which associates the colour representation with the log fold changes in protein expression.
RNA extraction
RNA was extracted from frozen juice sac tissues of Navel oranges, first by grinding 0.5 g of tissue in liquid nitrogen into a fine powder. The ground tissue was mixed with cold extraction buffer (TRIS/HCl pH 8 200 mM, EDTA 25 mM, NaCl 75 mM, SDS 1%, and β-mercaptoethanol 1 M). The same volume of phenol/chloroform/iodoacetamide (25/24/1, by vol.) was then added, mixed, and centrifuged at 10 000 g for 15 min. The supernatant was collected and an equal volume of pure ethanol was added, mixed by inversion and incubated at –20 °C for 15 min. This mixture was then centrifuged at 10 000 g for 10 min at 4 °C. The supernatant was collected and nucleic acids were precipitated by first adding 1/10 (v/v) of 3 M Na-acetate (pH 5.2) and 2 vols of 100% ethanol. After storing the samples at –20 °C for 20 min, they were then centrifuged at 12 000 g for 15 min. The pellet was retained and re-suspended in sterile water. RNA was selectively precipitated overnight at 4 °C by adding LiCl to a final concentration of 2 M, then the samples were centrifuged at 12 000 g for 15 min at 4 °C and then washed with 70% ethanol, after which samples were re-suspended in 50 μl of sterile water.
Quantitative PCR analysis
RNA was extracted from juice sac cells at early stage II, stage II, and stage III with three biological replicates. First-strand cDNA was synthesized from 1 μg of total RNA with the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). Primer3 software (ver. 0.4.0; http://frodo.wi.mit.edu/primer3/) was used for primer design. Quantitative PCR was performed on the StepOnePlus™ (Applied Biosystems, Foster City, CA, USA), using SYBR® Green. A total reaction volume of 15 μl was used. The reaction mix included 2 μl template, 0.3 μl of reverse primer, 0.3 μl of forward primer, 7.5 μl SYBR Green Master Mix, and 4.9 μl RNA-free water. A qPCR assay was performed using the following conditions: 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s. The 2–ΔΔCT method (Livak and Schmittgen, 2001) was used to normalize and calibrate transcript values relative to the endogenous citrus 18S ribosomal protein, whose expression did not change across citrus fruit developmental stages. Primer sequences are described in Supplementary Table S3 at JXB online.
Enzymatic assays
Protein extraction
Frozen juice sac samples were ground in liquid nitrogen with 1 mg of insoluble PVPP (polyvinyl polypyrrolidone) to remove polyphenols harmful to proteomics analysis. Total protein was extracted with 4 vols (w/v) of extraction buffer containing 50 mM HEPES-KOH pH 7.5, 10 mM MgCl2, 1 mM EDTA, 2 mM DTT, 1 mM PMSF, 0.1% (v/v) Triton X-100, and 10% glycerol. The extract was centrifuged at 4 °C and 12 000 g for 10 min. The supernatant was desalted with a Sephadex-G25 gel column pre-equilibrated with ice-cold extraction buffer containing no Triton X-100 nor PVPP. The protein fraction was collected in pre-chilled tubes and used for enzymatic assays. Protein concentration was determined according to Bradford (1976) using BSA as a standard.
Sucrose-phosphate synthase (SPS) assay
SPS activity was assayed by quantifying the fructosyl moiety of sucrose using the anthrone test (Baxter et al., 2003). Samples were incubated for 30 min at 25 °C in 200 μl of buffer containing 50 mM HEPES-KOH pH 7.5, 20 mM KCl, 4 mM MgCl2, 4 mM UDP-Glc, 2 mM Fru-6-P (in a 1:4 ratio with Glc-6-P), and 5 mM KH2PO4. The reaction was terminated by incubation at 95 °C for 5 min and samples were centrifuged at 4 °C and 12 000 g for 5 min. To neutralize unreacted hexose phosphate, 100 μl of the supernatant was mixed with 100 μl of 5 M KOH and incubated at 95 °C for 10 min. Samples were mixed with 4 vols of 0.14% (w/v) anthrone reagent in 14.6 M H2SO4 and incubated at 95 °C for 10 min. Absorbance was measured at 620 nm. Blank samples containing boiled protein and reaction buffer without hexose phosphates were prepared by the same procedure. The absolute amount of sucrose-6-P created by the reaction was calculated using a sucrose standard curve.
Sucrose-phosphate phosphatase assay (SPP)
SPP activity was assayed by quantifying the released orthophosphate from Suc-6-P (Lunn et al., 2000). Protein extract was mixed with 150 μl of buffer containing 25 mM HEPES-KOH pH 7.0, 8 mM MgCl2, and 1.25 mM Suc-6-P, and incubated for 30 min at 30 °C. The reaction was stopped by adding 30 μl of 2 M trichloroacetic acid. Orthophosphate in the sample was measured using an ascorbic acid–ammonium molybdate reagent (Harwood et al., 1969).
Statistical analysis
The JMP® 8.0 statistical package (SAS Institute, Cary, NC) was used for all statistical analyses. An agglomerative hierarchical procedure with an incremental sum of squares grouping strategy, known as Ward's method (Ward, 1963), was used for the purpose of classification the metabolites into similar expression groups.
Results
Changes in proteins associated with sugar metabolism and homeostasis during citrus fruit development
An extensive comparative proteomics study was conducted in order to identify protein changes occurring during citrus fruit growth and development. Samples were collected from three developmental stages; early stage II, stage II, and stage III (35, 55, and 80 mm fruit diameter, respectively). For proteomics analysis, two biological repetitions from two consecutive years were collected from at least 20 pooled fruits for each stage (Katz et al., 2010). For gene expression, enzyme activities, and metabolome analysis, three biological repetitions of three consecutive years were analysed. For better identification of differentially expressed proteins during fruit development and to decrease sample complexity, the juice sac cells were fractionated into soluble and membrane-bound proteins (Katz et al., 2010). Changes in protein expression were revealed by comparisons between spectra originated from fruit juice sac cells at different stages: (i) stage II versus early stage II and (ii) stage III versus stage II. The complete data of the differential proteins detected can be found in Supplementary Tables S1 and S2 at JXB online. The analysis revealed a significant metabolic change occurring during the transition from early stage II to stage II and from stage II to stage III (see Supplementary Fig. S1 at JXB online). Although these changes involved a wide range of processes, this study focuses on protein changes related to primary metabolism.
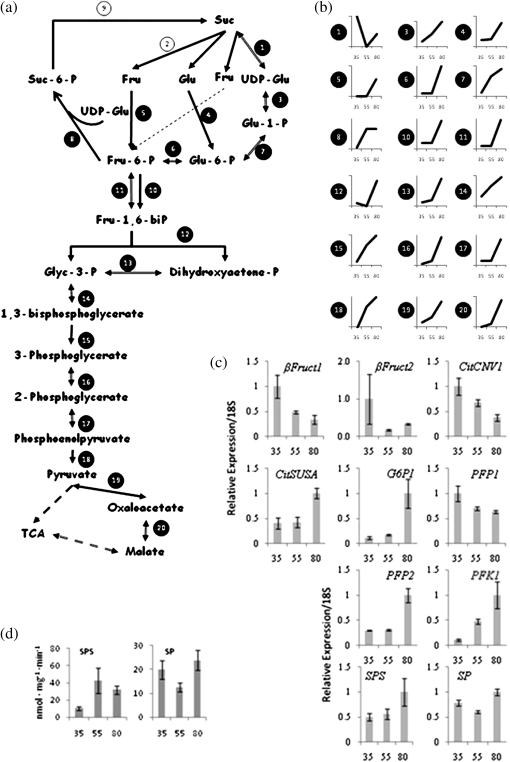
Processes involving sugar metabolism, the TCA cycle, amino acid metabolism, energy production, and cell wall-related metabolism changed significantly in citrus juice sac cells during fruit development. Citrus fruit accumulate large amounts of sugars, mainly sucrose, glucose, and fructose. Enzymes participating in sucrose metabolism were highly represented in the proteome analysis of citrus fruit juice sac cells. Most of the enzymes involved in sucrose degradation and glycolytic pathways were up-regulated during the transition from early stage II to stage II and were up-regulated toward maturation, emphasizing the regulatory role of glycolysis in sugar utilization to drive fruit growth during citrus fruit development (Table 1; Fig. 1). Hexokinase, fructokinase, glucose-6-phosphate isomerase, fructose-bisphosphate aldolase, ATP-dependent 6-phosphofructose-1-kinase, triosephosphate isomerase, and enolase protein expression did not change significantly during the early stages and were up-regulated during the transition from stage II to stage III. UDP-glucose pyrophosphorylase, phosphoglucomutase, glyceraldehyde-3-phosphate dehydrogenase, 2,3-biphosphoglycerate-independent phosphoglycerate mutase, phosphoglycerate kinase, phosphoenolpyruvate carboxylase, and phosphoenolpyruvate carboxykinase were up-regulated throughout fruit development. Two pyruvate kinases were identified: iCitrus ID 52671 that did not change during the transition from early stage II to stage II and iCitrus ID 28935 that was up-regulated at stage II compared witho early stage II. Both proteins were up-regulated during the transition from stage II to stage III. Sucrose synthase was found to be an interesting exception, since it was down-regulated during the transition from early stage II to stage II, and was up-regulated nearer to maturation (Fig. 1; Table 1; see Supplementary Fig. S1 at JXB online). Four citrus sucrose synthase isoforms derived from four different unigenes were identified and clustered into three groups according to their sequence homology. Group 1 consisted of isoforms with homology to unigenes related to CitSUSA (Komatsu et al., 2002), group 2 consisted of proteins derived from unigenes related to CitSUS1 (Komatsu et al., 2002), and group 3 comprised CitSUS4, shown in this study to be expressed in the fruit. The expression patterns of CitSUS1 and CitSUSA were in agreement with their corresponding transcripts and with previously characterized enzymatic activities (Komatsu et al., 2002; Katz et al., 2007). The CitSUS1 gene was shown to be expressed in the early stages of fruit development and its expression decreased towards maturation, while the CitSUSA gene was up-regulated towards maturation. In this study, it is shown that the amounts of both CitSUS1 isoforms decreased in the transition from early stage II to stage II while that of iCitrus ID 33038 increased during the transition from stage II to stage III (Table 1) similar to CitSUSA which was up-regulated towards maturation, in agreement with the gene expression profiles and enzyme activity (Komatsu et al., 2002). In addition, CitSUS4 was found to be significantly down-regulated between early stage II and stage II and was not detected in the later stage of fruit development (Table 1), thus indicating that its amounts remained constant.
Table 1.
Glycolysis and sugar metabolism related proteins identified by dMS and SC after search of the iCitrus database using X!Tandem with LC-MS/MS spectra
| Annotation | iCitrus ID | Blast Hit to TAIR | Stage II versus early stage II |
Stage III versus stage II |
|||||||||
| dMS | Ratio | Direction | SC | Fold change | dMS | Ratio | Direction | SC | Fold change | ||||
| No. peptides | Bayes factor | No. peptides | Bayes factor | ||||||||||
| Sucrose synthase | CitSUSA | 33122 | At4g02280 | – | – | – | – | – | 3 | 27.33 | – | – | – |
| CitSUS4 | 18627 | At5g20830 | 2 | 0.02 | – | – | – | – | – | – | – | – | |
| CitSUS1 | 25199 | At3g43190 | 2 | 0.03 | – | – | – | – | – | – | – | – | |
| 33038 | At1g73370 | 8 | 0.01 | –1 | 34022.5 | 37.08 | 3 | 6.86 | 1 | 57.57 | 6.96 | ||
| Hexokinase | 30768 | At2g19860 | 5 | 1.39 | 0 | 2.70 | 2.41 | 3 | 148.9 | 0 | 0.70 | 1.11 | |
| Fructokinase | 62294 | At5g51830 | – | – | 0 | 1.00 | 1.00 | – | – | 1 | 221.90 | 13.69 | |
| 29116 | At3g59480 | – | – | 0 | 4.91 | 6.50 | 3 | 3.93 | 0 | 0.67 | 1.21 | ||
| Sucrose phosphate synthase | 62092 | At1g04920 | – | – | 1 | 10.79 | 10.53 | – | – | 0 | 8.47 | 3.45 | |
| UDP-glucose pyrophosphorylase | 10418 | At3g03250 | 4 | 19.6 | – | – | – | 10 | 111.2 | 1 | 13129 | 32.79 | |
| 18602 | At5g17310 | 2 | 6.05 | – | – | – | 4 | 4.13 | – | – | – | ||
| Phosphoglucomutase | 60519 | At1g23190 | 7 | 37.1 | 0 | 6.65 | 3.87 | 6 | 5.68 | 0 | 6.53 | 2.32 | |
| Glucose-6-phosphate isomerase | 21361 | At5g42740 | – | – | 0 | 1.00 | 1.00 | – | – | 1 | 72.50 | 10.41 | |
| PPi-dependent 6-phosphofructose-1-kinase; | CitPFP1 | 28806 | At1g12000 | – | – | 0 | 1.00 | 1.00 | – | – | –1 | 54.30 | 6.23 |
| CitPFP2 | 61196 | At1g20950 | – | – | 0 | 1.25 | 3.23 | 3 | 9.55 | 0 | 1.29 | 1.62 | |
| ATP dependent 6-phosphofructose-1-pinase; PFK1 | 35527 | At4g26270 | – | – | 0 | 1.00 | 1.00 | – | – | 1 | 19.61 | 4.69 | |
| Fructose-bisphosphate aldolase | 22988 | At2g36460 | 9 | 0.61 | 0 | 0.63 | 1.41 | 16 | 45.05 | 0 | 0.30 | 1.31 | |
| 246 | At2g01140 | 5 | 0.67 | –1 | 257.71 | 21.93 | 9 | 5.80 | 0 | 0.71 | 1.16 | ||
| 21203 | At2g01140 | – | – | – | – | – | 4 | 3.16 | 0 | 0.97 | 1.34 | ||
| Triosephosphate isomerase | 43479 | At3g55440 | 5 | 0.77 | 0 | 1.47 | 1.69 | 5 | 3.20 | 0 | 0.77 | 1.32 | |
| 31271 | At3g55440 | 3 | 0.58 | 0 | 1.65 | 3.93 | 4 | 4.57 | 0 | 1.05 | 1.48 | ||
| Glyceraldehyde-3-phosphate | 37538 | At3g04120 | 4 | 16.4 | 0 | 0.44 | 1.75 | 5 | 15.33 | 0 | 0.75 | 1.65 | |
| dehydrogenase | 948 | At3g04120 | 5 | 10.1 | - | – | – | 5 | 7.24 | – | – | – | |
| 24071 | At1g13440 | 7 | 9.10 | 1 | 21.98 | 3.00 | 7 | 57.00 | 0 | 1.56 | 1.20 | ||
| Phosphoglycerate kinase | 58537 | At1g56190 | 7 | 42.0 | 1 | 6593.8 | 6.65 | 13 | 9.52 | 0 | 0.40 | 1.08 | |
| 4747 | At1g79550 | 2 | 36.0 | – | – | – | 5 | 5.60 | – | – | – | ||
| 2,3-biphosphoglycerate-independent | 40745 | At1g09780 | 2 | 3.10 | 0 | 1.00 | 1.00 | 8 | 6.46 | 1 | 16.61 | 3.44 | |
| Phosphoglycerate mutase | 28138 | At3g08590 | – | – | 0 | 0.98 | 1.03 | – | – | 0 | 0.71 | 1.18 | |
| Enolase | 61481 | At2g36530 | 11 | 0.90 | 0 | 1.72 | 1.25 | 19 | 6 | 0 | 0.53 | 1.08 | |
| 832 | At2g36530 | 4 | 0.99 | – | – | – | 6 | 3.10 | – | – | – | ||
| 15421 | At2g36530 | 4 | 0.73 | – | – | – | 4 | 4.54 | – | – | – | ||
| Pyruvate kinase | 52671 | At2g36580 | 2 | 1.42 | 0 | 0.62 | 1.17 | 3 | 2.15 | 0 | 0.60 | 1.13 | |
| 37060 | At2g36580 | – | – | – | – | – | 3 | 2.20 | – | – | – | ||
| 28935 | At5g08570 | 4 | 12.3 | 0 | 0.41 | 1.27 | 5 | 2.84 | 0 | 1.16 | 1.82 | ||
| Phosphoenolpyruvate carboxylase | 60990 | At1g53310 | 7 | 2.26 | 0 | 0.26 | 1.42 | 3 | 2.54 | 0 | 3.33 | 2.29 | |
| Phosphoenolpyruvate | 51484 | At5g65690 | 2 | 3.10 | 1 | 19.13 | 3.03 | 11 | 7.34 | 0 | 1.22 | 1.49 | |
| Carboxykinase invertase/pectin methylesterase inhibitor | 30454 | At3g17130 | 3 | 1.12 | 0 | 0.689 | 1.39 | 3 | 30.8 | 1 | 24.05 | 9.35 | |
Proteins identified by dMS were considered to be up-regulated when expression fold change >2, not changed when fold change >0.5 but <2, and down-regulated when fold change was <0.5. For SC, a Bayes factor of >10 was considered significantly different. The column ‘Direction’ under SC represents up-regulated=1, no change=0, down-regulated= –1.
Fig. 1.
Changes in glycolysis and sucrose metabolism during citrus fruit development. (a) Glycolysis and sucrose biosynthesis pathway. Enzymes that were found to be differentially expressed are numbered with a black background. Enzymes that remain unchanged are numbered with a white background. (1) Sucrose synthase. (2) Invertase. (3) UDP-Glu-pyrophosphorylase. (4) Hexokinase. (5) Fructokinase. (6) Glucose-6-P-isomerase. (7) Phosphoglucomutase. (8) Sucrose-phosphate-synthase. (9) Sucrose-phosphatase. (10) ATP-dependent phosphofructokinase. (11) PPi-dependent phosphofructokinase. (12) Fructose-bisphosphate aldolase. (13) Triose-phosphate-isomerase. (14) Glyceraldehyde-3-phosphate dehydrogenase. (15) Phosphoglycerate kinase. (16) Phosphoglycerate mutase. (17) Enolase. (18) Pyruvate kinase. (19) PEP carboxylase. (20) Malate dehydrogenase. (b) Protein expression changes of the above proteins during the transition from early stage II to stage II and from stage II to stage III. The figure shows enzyme changes as a function of development. The x-axis represents the three stages sampled: 35, early stage II; 55, stage II; 80, stage III. The y-axis represents the calculated fold change ratio where stage II was assigned the value 1 and the values in early stage II and stage III were calculated according to the fold change found by dMS. An average of the different isoforms was calculated in cases where different protein isoforms were found (see also Table 1). (c) Changes in patterns of gene expression of invertases (βFruct1, βFruct2, CitCNV1), sucrose synthase (CitSUSA), glucose-6-P-isomerase (G6PI), PPi-dependent phosphofructokinase 1 and 2 (PFP1 and PFP2), ATP-dependent phosphofructokinase (PFK1), sucrose-phosphate-synthase (SPS), and sucrose-phosphate phosphatase (SPP) during fruit development. The x-axis represents the three developmental stages as described in (b). (d) Sucrose-phosphate-synthase (SPS) and Sucrose-phosphate phosphatase (SPP) activities in juice sacs extract during fruit development.
The reaction mediated by phosphofructokinase (PFK) is one of the key control points of glycolysis in plants (Mustroph et al., 2007). This reaction catalyses the interconversion of fructose-6-phosphate and fructose-1,6-bisphosphate. While most glycolytic enzymes are highly conserved between organisms, two types of phosphofructokinase isoforms exist in plants (Mustroph et al., 2007). In addition to the ATP-dependent phosphofructokinase (PFK), a pyrophosphate-fructose-6-phosphate-phosphotransferase (PFP) uses pyrophosphate as the phosphoryl donor. Phosphorylation of fructose-6-phosphate catalysed by PFK is virtually irreversible while PFP catalyses the reaction in both directions. In citrus juice cells, two PFPs and one PFK were found to be differentially expressed (Table 1). CitPFP1 and CitPFP2 remained unchanged during the transition from early stage II to stage II. CitPFP1 was down-regulated during the transition from stage II to stage III while CitPFP2 was up-regulated. Gene expression analysis of CitPFP1 and CitPFP2 showed similar expression patterns (Fig. 1c). In contrast to PFPs, only one PFK was identified (CitPFK1) and its expression did not change during the transition from early stage II to stage II but was up-regulated during the transition from stage II to stage III.
No changes in invertases, an important family of proteins responsible for sucrose degradation to glucose and fructose, were seen in our proteomics analysis. The activity of two citrus acid invertases were detected in juice sac cells (Kubo et al., 2001) with higher activities at the earlier stages of development. Since no differences in invertase proteins were detected in our comparisons, the expression of three citrus invertase genes, the vacuolar/acidic βFruct1 and βFruct2 and the neutral/alkaline invertase CitCINV1, was followed. The expression of these three genes peaked at early stage II and was down-regulated during the later stages of fruit development, suggesting a role of invertases during the early stages of fruit development (Fig. 1c). Interestingly, an invertase inhibitor protein (iCitrus ID 30454) was found between early stage II and stage II, and was up-regulated during the transition from stage II to stage III (detected in both dMS and SC). Invertase inhibitors are responsible for the decrease in invertase activity and function at the later stages of fruit development, highlighting the importance of sucrose synthase activity in sucrose degradation during these developmental stages.
Sucrose-phosphate synthase (SPS), an enzyme involved in sucrose synthesis, was up-regulated during the transition from early stage II to stage II and remained unchanged during the transition from stage II to stage III, while no changes were seen in sucrose-6-phosphate phosphatase (SPP), which mediates the formation of sucrose from sucrose-6-P. Some differences between SPS and SPP protein amounts and their levels of gene expression were noted. The expression of SPS was up-regulated only during the transition from stage II to stage III (Fig. 1c). Also, although no differences were seen in SPP protein amounts, the gene expression decreased slightly towards stage II and increased towards stage III (Fig. 1c). SPS and SPP activities correlated well with their protein expression patterns (Fig. 4d).
Fig. 4.
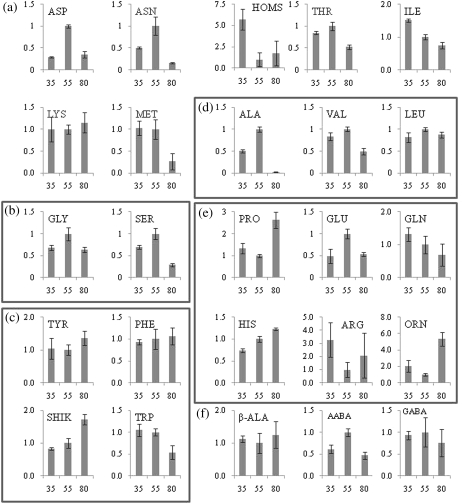
Amino acid and metabolite profiles of citrus juice sac cells during development. The relative content of primary metabolites was determined during three stages of citrus fruit development, early stage II, stage II, and stage III. Amino acids derived from (a) oxaloacetate, (b) 3-phosphoglycerate, (c) phosphoenolpyruvate, (d) pyruvate, and (e) 2-oxoglutarate are clustered in separate boxes. ASP, aspartate; ASN, asparagine; HOMS, homoserine, THR, threonine; ILE, isoleucine; LYS, lysine; MET, methionine; ALA, alanine; VAL, valine; LEU, leucine; GLY, glycine; SER, serine; PRO, proline; GLU, glutamate; GLN, glutamine; TYR, tyrosine; PHE, phenylalanine; HIS, histidine; ARG, arginine; ORN, ornithine; SHIK, shikimic acid; TRP, tryptophan; β-ALA, β-alanine; AABA, α-aminobutyrate; GABA, γ- aminobutyrate. The x-axis represents the three developmental stages as described in Figs 1 and 2. The y-axis represents the relative contents of the different metabolites with respect to the metabolite content at stage II (assigned the value of 1).
Changes in proteins involved in the TCA cycle during fruit development
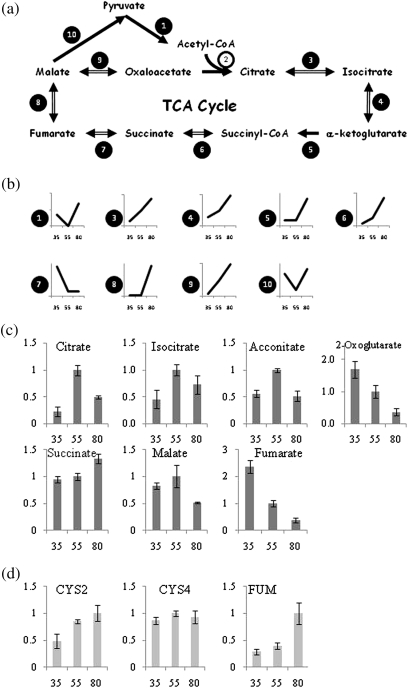
Citrus fruits accumulate large amount of organic acids, mainly citrate, in juice sac cells. In contrast to sugars, citrate is synthesized in the juice cells and not transported from other organs of the tree. Citrate is produced through the TCA cycle and accumulates in the vacuole during fruit development, reaching a maximum at late stage II and decreasing towards maturation (Shimada et al., 2006). Citrate is not only an intermediate metabolite in energy production in citrus juice cells, but also accumulates to high concentrations and is stored in the vacuole, contributing more than 90% of citrus fruit juice cells’ organic acids content (Canel et al., 1996; Spiegel-Roy and Goldschmidt, 1996). The mechanisms regulating citrate accumulation and degradation during pre- and post-harvest are unknown but play significant roles in determining the quality of many fruit species in general, and citrus fruit in particular. The pyruvate dehydrogenase enzyme complex links the TCA cycle to glycolysis. One of the pyruvate dehydrogenase complex (E1) proteins increased during the transition from early stage II to stage II while three others were down-regulated (Table 2). In addition, two other components of the pyruvate dehydrogenase complex, dihydrolipoamide S-acetyltransferase (E2) and dihydrolipoamide dehydrogenase (E3) were down-regulated during this transition. The pyruvate dehydrogenase complex (E1) was up-regulated during the transition from stage II to stage III (Fig. 2b; Table 2). Aconitase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, succinyl-CoA synthetase, fumarase, and malate dehydrogenase were up-regulated or remained unchanged during the transition from early stage II to stage II, and were up-regulated during the transition from stage II to stage III (Fig. 2b; Table 2). Three exceptions to these general trends in the TCA cycle protein expression were noted, pyruvate dehydrogenase, succinate dehydrogenase, and malic enzyme, which were down-regulated during the transition from early stage II to stage II and were up-regulated during the transition from stage II to stage III.
Table 2.
TCA cycle-related proteins identified by dMS and SC after search of the iCitrus database by X!Tandem using LC-MS/MS spectra
| Annotation | iCitrus ID | Blast hit to TAIR | Stage II versus early stage II |
Stage III versus stage II |
||||||||
| dMS | Ratio | Direction | SC | Fold change | dMS | Ratio | Direction | SC | Fold change | |||
| No. peptides | Bayes factor | No.peptides | Bayes factor | |||||||||
| Pyruvate dehydrogenase complex E1 | 22606 | At1g24180 | 4 | 9.90 | 0 | 1.87 | 2.14 | – | – | 0 | 0.69 | 1.08 |
| 1564 | At1g24180 | 2 | 0.01 | – | – | – | – | – | – | – | – | |
| 23699 | At5g50850 | 6 | 0.20 | –1 | 21.69 | 8.81 | 3 | 24.16 | 0 | 3.64 | 2.84 | |
| 17446 | At5g50850 | 2 | 0.22 | – | – | – | – | – | – | – | – | |
| Dihydrolipoamide S-acetyltransferase E2 | 42695 | At3g17240 | 2 | 0.11 | – | – | – | – | – | – | – | – |
| Dihydrolipoamide dehydrogenase E3 | 4911 | At1g48030 | 4 | 0.26 | – | – | – | – | – | – | – | – |
| 44669 | At1g48030 | 7 | 0.33 | –1 | 142.43 | 10.83 | 3 | 4.74 | 0 | 0.71 | 1.13 | |
| Aconitase | 43680 | At2g05710 | 2 | 25.29 | 0 | 1.00 | 1.00 | 2 | 13.64 | 1 | 143.42 | 12.24 |
| 45840 | At2g05710 | 4 | 49.72 | 0 | 0.40 | 1.08 | 4 | 4.77 | 0 | 0.58 | 1.29 | |
| 39802 | At2g05710 | 3 | 24.94 | 0 | 0.66 | 1.88 | 4 | 33.48 | 1 | 30.46 | 5.52 | |
| Isocitrate dehydrogenase (NADP+) | 30767 | At1g54340 | 2 | 2.15 | 0 | 4.329 | 7.19 | 7 | 8.92 | 0 | 0.744 | 1.03 |
| 2385 | At1g65930 | – | – | – | – | – | 2 | 8.40 | – | – | – | |
| 3923 | At1g65930 | – | – | – | – | – | 3 | 4.43 | – | – | – | |
| 24612 | At3g09810 | 2 | 1.31 | 0 | 1.16 | 2.03 | 2 | 0.80 | 0 | 1.57 | 1.95 | |
| 149 | At4g35260 | 3 | 0.53 | 0 | 4.43 | 2.77 | – | – | 0 | 0.80 | 1.28 | |
| 2-oxoglutarate dehydrogenase E1 | 53498 | At3g55410 | – | – | 0 | 1.76 | 3.59 | 2 | 335.26 | 1 | 22.20 | 4.96 |
| 1249 | At3g55410 | – | – | 0 | 1.52 | 2.86 | 2 | 39.54 | 0 | 0.70 | 1.22 | |
| 38819 | At3g55410 | – | – | 0 | 0.98 | 1.03 | 2 | 136.04 | 1 | 22.11 | 4.73 | |
| succinyl-CoA ligase | 26895 | At2g20420 | 3 | 5.29 | 1 | 104.00 | 17.39 | 4 | 298.60 | 0 | 0.65 | 1.05 |
| 5556 | At2g20420 | – | – | – | – | – | 3 | 327 | – | – | – | |
| 22272 | At5g08300 | 2 | 0.95 | 0 | 0.62 | 1.23 | 2 | 3.41 | 0 | 1.19 | 1.53 | |
| Succinate dehydrogenase | 61503 | At2g18450 | 4 | 0.20 | 0 | 0.54 | 1.27 | x | x | 0 | 0.78 | 1.20 |
| 55144 | At5g40650 | – | – | –1 | 14.30 | 6.42 | – | – | 0 | 9.83 | 3.68 | |
| 1184 | At5g66760 | 2 | 0.24 | – | – | – | – | – | – | – | – | |
| Fumarase | 23918 | At2g47510 | – | – | 0 | 1.57 | 3.97 | 4 | 23.59 | 1 | 26.92 | 5.03 |
| 920 | At2g47510 | – | – | – | – | – | 2 | 6.64 | – | – | – | |
| Malate dehydrogenase | 2641 | At1g04410 | 2 | 11.92 | – | – | – | 6 | 4.65 | – | – | – |
| 2305 | At3g15020 | 3 | 0.49 | 0 | 1.63 | 2.90 | 2 | 2.55 | 0 | 0.87 | 1.64 | |
| 33986 | At5g43330 | 3 | 6.84 | 1 | 12.60 | 3.68 | 9 | 9.00 | 1 | 41.23 | 4.25 | |
| NADP-malic enzyme | 37162 | At1g79750 | 7 | 0.48 | 0 | 0.70 | 1.08 | 11 | 2.57 | 0 | 0.23 | 1.35 |
| 1735 | At1g79750 | 4 | 0.45 | – | – | – | 6 | 2.57 | – | – | – | |
| 35283 | At5g25880 | 2 | 0.42 | – | – | – | 2 | 4.86 | – | – | – | |
| 4879 | At5g25880 | 2 | 0.42 | – | – | – | – | – | – | – | – | |
| 36024 | At2g13560 | 2 | 0.10 | 0 | 2.47 | 5.78 | – | – | 0 | 1.00 | 1.00 | |
| 30233 | At2g13560 | 2 | 0.26 | –1 | 24.60 | 11.91 | – | – | 0 | 1.65 | 2.07 | |
Proteins identified by dMS were considered to be up-regulated when expression fold change >2, not changed when fold change >0.5 but <2, and down-regulated when fold change was <0.5. For SC, a Bayes factor of >10 was considered significant difference. The column ‘Direction’ under SC represents up-regulated=1, no change=0, own-regulated= –1.
Fig. 2.
Changes in the TCA cycle during citrus fruit development. (a) TCA cycle. Enzymes that were found to be differentially expressed are numbered with a black background. Enzymes that remained unchanged are numbered with a white background. (1) Pyruvate dehydrogenase. (2) Citrate synthase. (3) Aconitase. (4) Isocitrate dehydrogenase. (5) α-Ketoglutarate dehydrogenase/2-oxoglutarate dehydrogenase. (6) Succinyl-CoA synthetase. (7) Succinate dehydrogenase. (8) Fumerase. (9) Malate dehydrogenase. (10) Malic enzyme. (b) Protein expression changes of the above-mentioned proteins during the transition from early stage II to stage II and from stage II to stage III. The figure shows enzyme changes as a function of development. The x-axis represents the three stages sampled: 35, early stage II; 55, stage II; 80, stage III. The y-axis represents the calculated fold change ratio where stage II was assigned the value 1 and the values in early stage II and stage III were calculated according to the fold change found by dMS. An average of the different isoforms was calculated in cases where different protein isoforms were found (see also Table 1). (c) Relative content of TCA cycle intermediate accumulation was determined during citrus fruit development. Metabolite concentrations were normalized according to the concentration of each metabolite at stage II. (d) Quantitative PCR analysis for citrate synthase genes CYS2, CYS4, and fumarase FUM. The x-axis represents the three developmental stages as described in (b).
Changes in organic acids, sugars, and sugar alcohols during fruit development
After determining protein changes in juice cells during fruit development, our focus turned to changes in core metabolite accumulation. The amounts of organic acids changed dramatically during fruit development. Organic acids of the TCA cycle, citrate, isocitrate, aconitate, and malate, were highest at stage II and decreased towards stage III, while 2-oxoglutarate and fumarate gradually decreased during fruit development and succinate accumulated mainly during stage III (Fig. 2c).
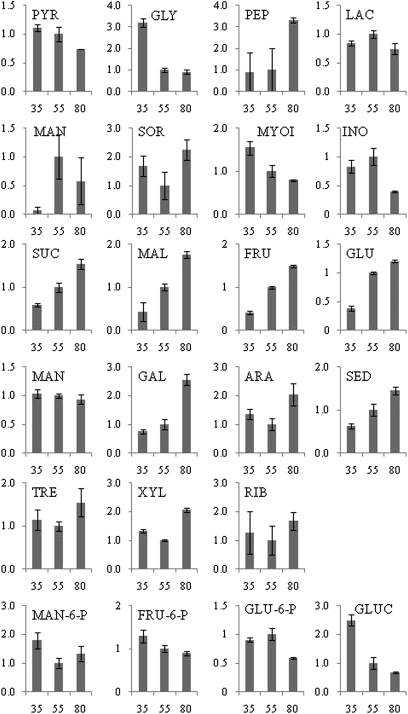
Sucrose, glucose, fructose, maltose, and sedoheptulose accumulated in an essentially linear manner during fruit development (Fig. 3). Galactose and trehalose rapidly accumulated in the juice sac cells towards maturation. Mannose gradually decreased during fruit development. Sugar alcohols displayed different accumulation patterns; inositol reached a maximum at stage II and decreased towards stage III, sorbitol decreased towards stage II and accumulated again towards maturation, while myo-inositol reached a maximum at the early stages of development and decreased gradually towards fruit maturation. Mannitol displayed considerable variation during development, increasing at stage II and decreasing towards maturation. Sugar phosphates and gluconate were higher at the earlier stages of fruit development and decreased during fruit maturation.
Fig. 3.
Metabolic profiles of citrus juice sac cells during development. The relative content of glycolysis intermediates, sugars, sugar-phosphates, and sugar alcohols was determined during three stages of citrus fruit development, early stage II, stage II, and stage III. PYR, pyruvate; GLY, glycerate; PEP, phosphoenolpyruvate; LAC, lactate; MAN, mannitol; SOR, sorbitol; MYOI, myo-inositol; INO, inositol; SUC, sucrose; MAL, maltose; FRU, fructose; GLU, glucose; MAN, mannose; GAL, galactose; ARA, arabinose; SED, sedoheptulose; TRE, trehalose; XYL, xylose; RIB, ribose; MAN-6-P, mannose-6-phosphate; FRU-6-P, fructose-6-phosphate; GLU-6-P, glucose-6-phosphate; GLUC, gluconate. Metabolite concentrations were normalized according to the concentration of each metabolite at stage II. The x-axis represents the three developmental stages as described in Figs 1 and 2. The y-axis represents the relative contents of the different metabolites with respect to the metabolite content at Stage II (assigned the value of 1).
Changes in amino acid accumulation and proteins involved amino acid metabolism during fruit development
Plants assimilate inorganic nitrogen into four major amino acids, glutamate, glutamine, aspartate, and asparagine. These amino acids are usually transported from source to sink tissues and are used as a nitrogen source for metabolism and growth. The carbon skeletons for amino acids are derived from 3-phosphoglycerate, phosphoenolpyruvate or pyruvate generated during glycolysis or from 2-oxoglutarate and oxaloacetate generated in the citric acid cycle. The amounts of amino acids were highly variable during fruit development (Fig. 4). A gradual decline was noted for isoleucine and glutamine while a gradual increase was noted for shikimic acid, histidine, and tyrosine. Aspartate, asparagine, threonine, alanine, valine, leucine, glycine, serine, glutamate, and α-aminobutyrate peaked at stage II and decreased towards maturity. Homoserine, proline, arginine, and ornithine decreased towards late stage II and increased again towards maturity. Methionine and tryptophan showed a decrease only towards maturation and phenylalanine, β-alanine, lysine, and GABA (γ-aminobutyric acid) did not change during fruit development (Fig. 4).
Two aspartate aminotransferases were up-regulated during the transition from early stage II to stage II and from stage II to stage III and one was up-regulated during the transition from stage II to stage III (Table 3). In addition, two glutamine synthetases (iCitrus IDs 25117 and 678), catalysing the reaction synthesis of glutamine from glutamate, were up-regulated during the transition from early stage II to stage II, while two other isoforms (iCitrus IDs 2123 and 41697) remained stable. During the transition from stage II to stage III , iCitrus 41697 was up-regulated, iCitrus 2123 and iCitrus 678 remained unchanged, and iCitrus 25117 was down-regulated (Table 3). Another player in the amino acid core biosynthetic pathway is glutamate dehydrogenase (GDH) catalysing the reversible conversion of 2-oxoglutarate to glutamate. Two GDH proteins were identified; GDH3 was down-regulated during the transition from early stage II to stage II and was up-regulated during the transition from stage II to stage III and GDH2 was up-regulated during the transition from stage II to stage III (Table 3). The GABA shunt is suggested to be essential for plant growth (Bouché et al., 2003) and to be pivotal in regulating citric acid degradation and fruit acidity during the later stage of citrus fruit development (Cercos et al., 2006). Although significant changes in GABA were not detected, the three components of the GABA shunt [i.e. glutamate decarboxylase (GAD), GABA transaminase, and succinic semialdehyde dehydrogenase (SSADH)] displayed changes during fruit development. These enzymes, catalysing the decarboxylation of glutamate to GABA and CO2, were up-regulated during the transition from early stage II to stage II and from stage II to stage III (Table 3). The level of GABA transaminase, catalysing the conversion of GABA to succinate semialdehyde, increased towards stage III. SSADH, mediating the oxidation of succinate semialdehyde to succinate, accumulated during the transition from early stage II to stage II and remained unchanged during the transition from stage II to stage III (Table 3).
Table 3.
Amino acid metabolism related proteins identified by dMS and SC after search of the iCitrus database using X!Tandem with LC-MS/MS uninterpreted spectra
| Annotation | iCitrus ID | Blast hit to TAIR | Stage II versus early stage II |
Stage III versus stage II |
||||||||
| dMS | Ratio | Direction | SC | Fold change | dMS | Ratio | Direction | SC | Fold change | |||
| No. peptides | Bayes factor | No. peptides | Bayes factor | |||||||||
| Acetyl-CoA C-acyltransferase | 55738 | at1g04710 | – | – | 0 | 1.25 | 3.23 | – | – | 1 | 11.30 | 7.63 |
| BCAT-2; branched-chain-amino-acid transaminase | 15175 | at1g10070 | – | – | 0 | 0.98 | 1.03 | – | – | 0 | 3.69 | 2.90 |
| Aminomethyltransferase | 40906 | at1g11860 | – | – | 0 | 0.98 | 1.03 | 2 | 13.50 | 0 | 1.51 | 1.81 |
| Glycine dehydrogenase | 56681 | at2g26080 | 3 | 0.20 | –1 | 452.08 | 20.99 | – | – | 0 | 0.99 | 1.24 |
| ALAAT1 (alanine aminotransferase) | 35404 | at1g17290 | – | – | 0 | 1.00 | 1.00 | – | – | 1 | 587.23 | 15.56 |
| PGDH (3-phosphoglycerate dehydrogenase) | 33085 | at1g17745 | 7 | 23.49 | 0 | 0.32 | 1.19 | 4 | 2.04 | 0 | 5.82 | 2.29 |
| PGDH (3-phosphoglycerate dehydrogenase) | 4033 | at4g34200 | 3 | 0.73 | – | – | – | 2 | 2.26 | – | – | – |
| AGT2 (alanine:glyoxylate aminotransferase 2) | 12839 | at4g39660 | – | – | – | – | – | 3 | 18.30 | – | – | – |
| AGT2 (alanine:glyoxylate aminotransferase 2) | 27165 | at4g39660 | – | – | 0 | 0.98 | 1.03 | 4 | 16.42 | 0 | 1.69 | 1.93 |
| ASP4 (aspartate aminotransferase 4) | 40919 | at1g62800 | 2 | 9.08 | – | – | – | 3 | 41.05 | – | – | – |
| ASP4 (aspartate aminotransferase 4) | 25228 | at1g62800 | 2 | 9.08 | 1 | 64.43 | 7.15 | 4 | 38.73 | 1 | 14.81 | 3.34 |
| ASP3 (aspartate aminotransferase 3) | 14798 | at5g11520 | – | – | – | – | – | 2 | 7.30 | – | – | – |
| GAD2 (glutamate decarboxylase 2) | 45590 | at1g65960 | 2 | 1924.40 | 0 | 1.00 | 1.00 | 4 | 2.15 | 0 | 1.75 | 2.1 |
| GAD2 (glutamate decarboxylase 2) | 60356 | at1g65960 | 2 | 239.09 | 1 | 696.64 | 26.44 | 5 | 2.06 | 0 | 0.49 | 1.06 |
| GAD1 (glutamate decarboxylase 1) | 58418 | at3g17760 | – | – | – | – | – | 2 | 1.47 | – | – | – |
| GDH3(glutamate dehydrogenase 3) | 45569 | at3g03910 | – | – | -1 | 41.11 | 11.21 | 2 | 6.64 | 0 | 2.31 | 2.23 |
| GDH2 (glutamate dehydrogenase 2) | 37770 | at5g07440 | – | – | – | – | – | 2 | 6.64 | – | – | – |
| GLN1;3 (glutamine synthetase) | 2123 | at3g17820 | – | – | 0 | 1.00 | 1.00 | – | – | 0 | 7.38 | 6.85 |
| GLN1;1-GSR1(glutamine synthetase) | 25117 | at5g37600 | 5 | 161.07 | 1 | 1197.34 | 4.24 | 6 | 0.67 | –1 | 16.77 | 1.88 |
| GLN1;1-GSR1(glutamine synthetase) | 41697 | at5g37600 | – | – | 0 | 1.00 | 1.00 | – | – | 1 | 92.46 | 12.23 |
| GLN1;1-GSR1(glutamine synthetase) | 678 | at5g37600 | 2 | 180.37 | – | – | – | – | – | – | – | – |
| ?-Aminobutyrate transaminase | 22281 | at3g22200 | – | – | – | – | – | 3 | 13.87 | – | – | – |
| P5CS1 (delta1-pyrroline-5-carboxylate synthase 1) | 37246 | at2g39800 | 3 | 0.32 | –1 | 24.08 | 7.09 | – | – | 0 | 1.07 | 1.48 |
| ALDH12A1, 1-pyrroline-5-carboxylate dehydrogenase | 27988 | at5g62530 | 2 | 0.17 | 0 | 7.46 | 8.52 | – | – | 0 | 1.00 | 1.00 |
| MS2 (methionine synthase 2) | 41927 | at3g03780 | 5 | 0.14 | – | – | – | 4 | 1.16 | – | – | – |
| MS2 (methionine synthase 2) | 43263 | at3g03780 | 11 | 0.16 | –1 | 13256.47 | 31.34 | 11 | 5.38 | 0 | 0.51 | 1.60 |
| MS1 (methionine synthase 1) | 24003 | at5g17920 | – | – | – | – | – | 3 | 16.69 | – | – | – |
| SAHH2 (S-adenosyl-L-homocysteine hydrolase 2) | 11482 | at3g23810 | – | – | – | – | – | 2 | 1.73 | – | – | – |
| SAHH2 (S-adenosyl-L-homocysteine hydrolase 2) | 49170 | at3g23810 | 9 | 0.17 | –1 | 130.58 | 4.57 | 14 | 170.98 | 0 | 0.86 | 1.41 |
| SAHH2 (S-adenosyl-L-homocysteine hydrolase 2) | 51358 | at3g23810 | 7 | 0.17 | 0 | 0.19 | 1.13 | 14 | 170.98 | 0 | 0.86 | 1.41 |
| SAHH1 (S-adenosyl-L-homocysteine hydrolase 1) | 1927 | at4g13940 | 7 | 0.16 | – | – | – | 9 | 182.30 | – | – | – |
| Ketol-acid reductoisomerase | 22397 | at3g58610 | 2 | 0.26 | –1 | 23.24 | 10.72 | – | – | –1 | 88.90 | 12.39 |
| O-acetylserine(thiol)lyase | 11065 | at4g14880 | 2 | 14.31 | 1 | 33.25 | 12.26 | – | – | 0 | 9.65 | 3.28 |
| Acetyl-CoA C-acyltransferase | 33619 | at5g47720 | – | – | 0 | 1.00 | 1.00 | – | – | 0 | 1.05 | 2.23 |
| Succinate semialdehyde dehydrogenase | 56374 | at1g79440 | 2 | 2.3 | 0 | 1.27 | 1.74 | – | – | 0 | 0.75 | 1.07 |
Proteins identified by dMS were considered to be up-regulated when expression fold change >2, not changed when fold change >0.5 but <2, and down-regulated when fold change was <0.5. For SC, a Bayes factor of >10 was considered a significant difference. The column ‘Direction’ under SC represents up-regulated=1, no change=0, down-regulated= –1.
Pyruvate supplies the carbon skeleton for alanine, leucine, and valine. Alanine aminotransferase and alanine:glyoxylate aminotransferase, both mediating alanine synthesis, remained unchanged at the early stages and were up-regulated during the late stage of development (Table 3). In the aspartate-derived amino acid biosynthesis pathway changes were identified in S-adenosyl-L-homocysteine hydrolases, methionine synthase, and ketol-acid reductiosomerase. S-adenosyl-L-homocysteine hydrolase and methionine synthase, active in the SAM cycle, were both down-regulated during the transition from early stage II to stage II and were up-regulated during the transition from stage II to stage III (Table 3).
Correlation in metabolite changes during fruit development
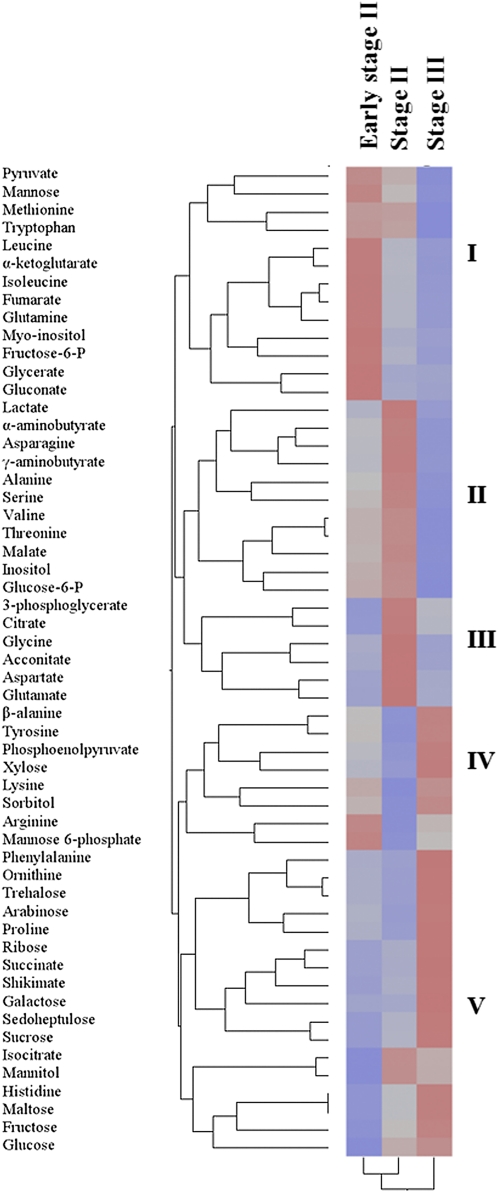
The use of two-way hierarchical clustering of metabolite amounts allowed the grouping of metabolites according to their accumulation trends (Fig. 5). Metabolites were separated into five different clusters: (i) pyruvate, mannose, methionine, tryptophan, leucine, 2-oxoglutarate, isoleucine, fumarate, glutamine, myo-inositol, fructose-6-P, glycerate, and gluconate were clustered together (Fig. 5) and their amounts declined during development and maturation (Figs. 2, 3, 4, 6). In two clusters: (ii) lactate, α-aminobutyrate, asparagine, γ-aminobutyrate, alanine, serine, valine, threonine, malate, inositol, and glucose-6-P; and (iii) citrate, 3-phosphoglycerate, glycine, aconitate, aspartate, and glutamate amounts increased from early stage II to stage II followed by a decline in their amounts in stage III. Two additional clusters of metabolites (iv) including β-alanine, tyrosine, phosphoenolpyruvate, xylose, lysine, sorbitol, arginine, and mannose-6-phosphate and (v) phenylalanine, ornithine, trehalose, arabinose, proline, ribose, succinate, shikimate, galactose, sedoheptulose, sucrose, isocitrate, mannitol, histidine, maltose, fructose, and glucose increased during both stage II and stage III. Interestingly, almost all sugars (except for mannose) displayed the same trend of accumulation towards fruit maturation.
Fig. 5.
Two-way hierarchical clustering of metabolites levels. Juice cells of citrus at early stage II, stage II, and stage III were collected. The red colour represents higher values and green represents lower relative values when compared with the mean metabolite value across all samples.
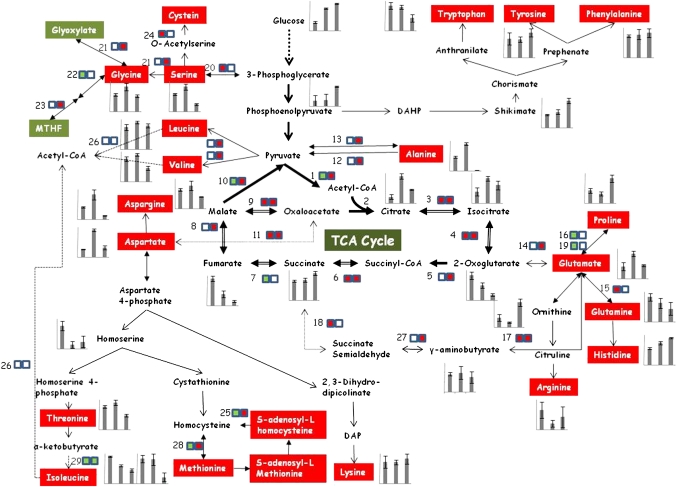
Fig. 6.
Illustration of metabolism flow during citrus fruit development. Graphs represent the accumulation of organic and amino acids during early stage II, stage II, and stage III. Numbered boxes represent the protein expression trend; the left side represents the comparison of stage II versus early stage II and the right side represents the comparison of stage III versus stage II. Green represents down-regulation, white represents no change, and red represents up-regulation. (1) Pyruvate dehydrogenase. (2) Citrate synthase. (3) Aconitase. (4) Isocitrate dehydrogenase. (5) α-Ketoglutarate dehydrogenase/2-oxoglutarate dehydrogenase. (6) Succinyl-CoA synthetase. (7) Succinate dehydrogenase. (8) Fumarase. (9) Malate dehydrogenase. (10) Malic enzyme. (11) Aspartate aminotransferase. (12) Alanine:glyoxylate aminotransferase. (13) Alanine aminotransferase. (14) Glutamate dehydrogenase. (15) Glutamine synthetase. (16) 1-Pyrroline-5-carboxylate dehydrogenase. (17) Glutamate decarboxylase. (18) Succinate-semialdehyde dehydrogenase. (19) Delta1-pyrroline-5-carboxylate synthase 1. (20) PGDH (3-phosphoglycerate dehydrogenase). (21) Alanine:glyoxylate aminotransferase. (22) Glycine decarboxylase P-protein 2. (23) Aminomethyltransferase. (24) Cysteine synthase. (25) S-adenosyl-L-homocysteine hydrolase. (26) Branched-chain amino acid transaminase. (27) γ-Aminobutyrate transaminase. (28) Methionine synthase. (29) Ketol-acid reductoisomerase.
Discussion
Metabolic shift during citrus fruit development
In this study, differential quantitative proteomics and metabolite profiling were used to assess developmental changes of citrus fruits. Most of the organic acids and many of the amino acids branching out from glycolysis and the TCA cycle peaked at stage II and declined during stage III of development. On the other hand, most of the sugars increased during stage III. No correlation was found between citrate accumulation and the expression of enzymes participating in citrate biosynthesis and degradation. Interestingly, citrate synthase protein amounts remained constant while aconitase, mediating the first step of citrate catabolism, isomerizing citrate to isocitrate, was up-regulated during fruit development.
The TCA cycle maintains a cyclic flux in order to generate reducing NADH and FADH2 facilitating ATP synthesis by oxidative phosphorylation. Beyond the maintenance of a cyclic flux, the TCA cycle also functions to provide carbon skeletons for biosynthetic pathways as well as to metabolize organic acids generated from other pathways (Sweetlove et al., 2010). The reduced expression of some of the proteins involved in the TCA cycle during the transition from early stage II to stage II such as pyruvate dehydrogenase, succinate dehydrogenase, and malic enzyme suggest a non-cyclic flux controlled by the influx of citrate and malate from the vacuole into the cytosol for its use in the TCA cycle. The changes in metabolite amounts throughout fruit development, with little correlation with protein expression levels, would also suggest that a large proportion of metabolism regulation occurs at the post-translational level (Gibon et al., 2004; Usadel et al., 2005; Carrari et al., 2006; Kummel et al., 2006). The increase in glutamate dehydrogenase, glutamate decarboxylase, and γ-aminobutyrate transaminase protein expression and the patterns of GABA and succinate accumulation indicates that the GABA shunt is active.
In citrus, sucrose is transported into the juice cells through the apoplast and accumulates mainly in the vacuole (Koch, 1984; Koch and Avigne, 1990). Sucrose is then degraded to glucose and fructose by invertases or to fructose and UDP-glucose by sucrose synthase (Lowell et al., 1989). While invertases did not appear to change at the protein level, the expression of three known invertase genes decreased during fruit development (Fig. 1c). Early studies have shown that acidic invertase activities in both grapefruit and Satsuma mandarin were initially high and decreased to very low levels at fruit maturation (Lowell et al., 1989). The role of invertases in plant development is well established and the cleavage of sucrose is of key importance in the generation of hexoses needed for metabolism and signalling (Vargas and Salerno, 2010). In addition to transcriptional control, invertase activity can be regulated post-translationally by the action of invertase inhibitors (Jin et al., 2009). Interestingly, an invertase inhibitor was up-regulated towards maturation, suggesting its potential role in the previously described decrease in invertase activity in citrus fruits (Lowell et al., 1989). As invertase inhibitor proteins have also been implicated in the regulation of sugar metabolism in grape and peach fruits (da Silva et al., 2005; Ziliotto et al., 2008), their role in the control of fruit quality warrants further investigation. Sucrose synthase proteins decreased during the transition from early stage II to stage II and increased during the transition from stage II to stage III. This pattern correlated well with sucrose synthase enzymatic activity reported by Komatsu et al. (2002). Sucrose synthase can play important roles in sink strength and it was suggested that CitSUS1 may have a role in supplying UDP-glucose and fructose for cell wall synthesis during the cell division stage while CitSUSA and SUS1 may supply substrates for sucrose synthesis during maturation (Komatsu et al., 2002). Two isozymes of sucrose synthase, each active during different stages of development (immature and mature), were also reported in pear (Suzuki et al., 1996). Our results support the notion of a balance between the action of sucrose synthases and the regulation of invertase activities.
The accumulation of arabinose, galactose, xylose, and ribose, important for cell wall synthesis was correlated with proteins associated with cell wall metabolism such as cellulose synthase, pectin methylesterases, β-chitinase, PR4, β-1,3-glucanase, polygalacturonase inhibiting protein, and UDP-glucose 6-dhydrogenase.
The role of phosphoenolpyruvate (PEP) carboxylase in the metabolism of malate and citrate is intriguing. Pyruvate is generally the major product of glycolysis arising from PEP via the activity of pyruvate kinase. However, plant cells can convert PEP to malate via oxaloacetate in reactions catalysed by PEP carboxylase (PEPC) and malate dehydrogenase (MDH). The resulting malate may be utilized as a respiratory substrate in the TCA cycle, or be converted to pyruvate via the activity of malic enzyme. As shown in potato tubers with reduced NAD-dependent malic enzyme activity, the conversion of malate to pyruvate can influence glycolytic flux (Jenner et al., 2001; Sweetman et al., 2009). Studies in tomato and grape fruits suggested the occurrence of gluconeogenesis in fruits, particularly during the ripening stage when sugars are accumulating rapidly. A correlation between citrate, malate, and oxaloacetate loss and the activities of PEPC and PEPCK (PEP carboxykinase) and gluconeogenesis in fruits was demonstrated (Sweetman et al., 2009). Both of these proteins were up-regulated during citrus fruit development.
Our results suggest that organic acid and amino acid accumulation shifted toward sugar synthesis during the later stage of citrus fruit development. The notion of a metabolic shift during fruit maturation is supported by work on grape, strawberry, and tomato fruit maturation (Carrari et al., 2006; Deluc et al., 2007; Fait et al., 2008). Gene expression analysis of maturing grapes showed that a decrease in expression of transcripts associated with organic acid accumulation was accompanied by the increased expression of genes associated with the TCA cycle and genes encoding enzymes mediating sugar accumulation (Deluc et al., 2007). The observed increase in SPS and SPP activity in the later stages of fruit development, concomitant with the rapid accumulation of sucrose, suggest that this sugar is also being synthesized in citrus juice sac cells during fruit development and ripening.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Protein expression data using spectral counting (dMS).
Supplementary Table S2. Protein expression data using spectral counting (SC).
Supplementary Table S3. List of primers used for qPCR analysis as described in the Materials and methods.
Supplementary Fig. S1. Visualization of metabolism overview using MapMan: (a) stage II versus early stage II, (b) stage III versus stage II.
Acknowledgments
This work was supported by a research grant No. US-4010-07C from BARD, the United States-Israel Binational Agricultural Research and Development Fund, and by the Will W Lester Endowment, University of California.
References
- America AHP, Cordewener JHG. Comparative LC-MS: a landscape of peaks and valleys. Proteomics. 2008;8:731–749. doi: 10.1002/pmic.200700694. [DOI] [PubMed] [Google Scholar]
- Bain JM. Morphological, anatomical, and physiological changes in the developing fruit of the Valencia orange, Citrus sinensis (L.) Osbeck. Australian Journal of Botany. 1958;6:1–23. [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical Biochemistry. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Foyer CH, Turner J, Rolfe SA, Quick WP. Elevated sucrose-phosphate synthase activity in transgenic tobacco sustains photosynthesis in older leaves and alters development. Journal of Experimental Botany. 2003;54:1813–1820. doi: 10.1093/jxb/erg196. [DOI] [PubMed] [Google Scholar]
- Bouché N, Fait A, Bouchez D, Møller SG, Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proceedings of the National Academy of Sciences, USA. 2003;100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein–dye binding. Anaytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Canel C, Bailey-Serres JN, Roose ML. Molecular characterization of the mitochondrial citrate synthase gene of an acidless pummelo (Citrus maxima) Plant Molecular Biology. 1996;31:143–147. doi: 10.1007/BF00020613. [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiology. 2006;142:1380–1396. doi: 10.1104/pp.106.088534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercos M, Soler G, Iglesias DJ, Gadea J, Forment J, Talon M. Global analysis of gene expression during development and ripening of citrus fruit flesh. a proposed mechanism for citric acid utilization. Plant Molecular Biology. 2006;62:513–527. doi: 10.1007/s11103-006-9037-7. [DOI] [PubMed] [Google Scholar]
- Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Molecular and Cellular Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva FG, Iandolino A, Al-Kayal F, et al. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiology. 2005;139:574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Grimplet J, Wheatley M, Tillett R, Quilici D, Osborne C, Schooley D, Schlauch K, Cushman J, Cramer G. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics. 2007;8:429. doi: 10.1186/1471-2164-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria E. Activities of sucrose metabolising enzymes during sucrose accumulation in developing acid limes. Plant Science. 1992;85:125–129. [Google Scholar]
- Echeverria E, Burns JK. Sucrose breakdown in relation to fruit growth of acid lime (Citrus aurantifolia) Journal of Experimental Botnay. 1990;41:705–708. [Google Scholar]
- Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nature Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiology. 2008;148:730–750. doi: 10.1104/pp.108.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. The Plant Cell. 2004;16:3304–3325. doi: 10.1105/tpc.104.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing O, Kamlage B, Hoehne M, Trethewey R, Stitt M. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biology. 2006;7:R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Molecular and Cellular Biology. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood JE, van Steenderen RA, Kühn AL. A rapid method for orthophosphate analysis at high concentrations in water. Water Research. 1969;3:417–423. [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA. NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiology. 2001;126:1139–1149. doi: 10.1104/pp.126.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL. Post-translational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. The Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. Journal of Proteome Research. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- Katz E, Fon M, Eigenheer RA, Phinney BS, Sadka A, Blumwald E. A label-free differential quantitative mass spectrometry method for the characterization and identification of protein changes during citrus fruit development. Proteome Science. 2010;8:68. doi: 10.1186/1477-5956-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Fon M, Lee Y, Phinney B, Sadka A, Blumwald E. The citrus fruit proteome: insights into citrus fruit metabolism. Planta. 2007;226:989–1005. doi: 10.1007/s00425-007-0545-8. [DOI] [PubMed] [Google Scholar]
- Katz E, Lagunes PM, Riov J, Weiss D, Goldschmidt EE. Molecular and physiological evidence suggests the existence of a system II-like pathway of ethylene production in non-climacteric citrus fruit. Planta. 2004;219:243–252. doi: 10.1007/s00425-004-1228-3. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Koch K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Koch KE. The path of photosynthate translocation into citrus fruit. Plant, Cell and Environment. 1984;7:647–653. [Google Scholar]
- Koch KE, Avigne WT. Post-phloem, non-vascular transfer in citrus: kinetics, metabolism, and sugar gradients. Plant Physiology. 1990;93:1405–1416. doi: 10.1104/pp.93.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T. Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. Journal of Experimental Botany. 2002;53:61–71. [PubMed] [Google Scholar]
- Kubo T, Hohjo I, Hiratsuka S. Sucrose accumulation and its related enzyme activities in the juice sacs of satsuma mandarin fruit from trees with different crop loads. Scientia Horticulturae. 2001;91:215–225. [Google Scholar]
- Kummel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Molecular Systems Biology. 2006;2:0034. doi: 10.1038/msb4100074. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowell CA, Tomlinson PT, Koch KE. Sucrose-metabolizing enzymes in transport tissues and adjacent sink structures in developing citrus fruit. Plant Physiology. 1989;90:1394–1402. doi: 10.1104/pp.90.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Ashton AR, Hatch MD, Heldt HW. Purification, molecular cloning and sequence analysis of sucrose-6F -phosphate phosphohydrolase from plants. Proceedings of the National Academy of Sciences, USA. 2000;97:12914–12919. doi: 10.1073/pnas.230430197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, et al. Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiology. 2009;149:1505–1528. doi: 10.1104/pp.108.133967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Sonnewald U, Biemelt S. Characterisation of the ATP-dependent phosphofructokinase gene family from. Arabidopsis thaliana. FEBS Letters. 2007;581:2401–2410. doi: 10.1016/j.febslet.2007.04.060. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. The Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Sadka A, Dahan E, Or E, Cohen L. NADP(+) isocitrate dehydrogenase gene expression and isozyme activity during citrus fruit development. Plant Science. 2000;158:173–181. doi: 10.1016/s0168-9452(00)00328-9. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annual Review of Plant Biology. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- Shimada T, Nakano R, Shulaev V, Sadka A, Blumwald E. Vacuolar citrate/H+ symporter of citrus juice cells. Planta. 2006;224:472–480. doi: 10.1007/s00425-006-0223-2. [DOI] [PubMed] [Google Scholar]
- Spiegel-Roy P, Goldschmidt EE. Biology of citrus. Cambridge University Press; 1996. [Google Scholar]
- Suzuki A, Kanayama Y, Yamaki S. Occurrence of two sucrose synthase isozymes during maturation of Japanese pear fruit. Journal of the American Society for Horticultural Science. 1996;121:943–947. [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. Not just a circle: flux modes in the plant TCA cycle. Trends in Plant Science. 2010;15:462–470. doi: 10.1016/j.tplants.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70:1329–1344. doi: 10.1016/j.phytochem.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Talon M, Gmitter FG. Citrus genomics. International Journal of Plant Genomics doi:10.1155/2008/528361. 2008 doi: 10.1155/2008/528361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tomlinson PT, Duke ER, Nolte KD, Koch KE. Sucrose synthase and invertase in isolated vascular bundles. Plant Physiology. 1991;97:1249–1252. doi: 10.1104/pp.97.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiology. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas WA, Salerno GL. The Cinderella story of sucrose hydrolysis: alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Science. 2010;178:1–8. [Google Scholar]
- Ward JH. Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
- Wienkoop S, Morgenthal K, Wolschin F, Scholz M, Selbig J, Weckwerth W. Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Molecular Cell Proteomics. 2008;7:1725–1736. doi: 10.1074/mcp.M700273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor MI, Rambla J-L, Chaib J, Steppa A, Medina A, Granell A, Fernie AR, Causse M. Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. Journal of Experimental Botany. 2009;60:2139–2154. doi: 10.1093/jxb/erp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziliotto F, Begheldo M, Rasori A, Bonghi C, Tonutti P. Transcriptome profiling of ripening nectarine (Prunus persica L. Batsch) fruit treated with 1-MCP. Journal of Experimental Botany. 2008;59:2781–2791. doi: 10.1093/jxb/ern136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.