Abstract
Carotenoid cleavage dioxygenases (CCDs) are a class of enzymes involved in the biosynthesis of a broad diversity of secondary metabolites known as apocarotenoids. In plants, CCDs are part of a genetic family with members which cleave specific double bonds of carotenoid molecules. CCDs are involved in the production of diverse and important metabolites such as vitamin A and abscisic acid (ABA). Bixa orellana L. is the main source of the natural pigment annatto or bixin, an apocarotenoid accumulated in large quantities in its seeds. Bixin biosynthesis has been studied and the involvement of a CCD has been confirmed in vitro. However, the CCD genes involved in the biosynthesis of the wide variety of apocarotenoids found in this plant have not been well documented. In this study, a new CCD1 gene member (BoCCD1) was identified and its expression was charaterized in different plant tissues of B. orellana plantlets and adult plants. The BoCCD1 sequence showed high homology with plant CCD1s involved mainly in the cleavage of carotenoids in several sites to generate multiple apocarotenoid products. Here, the expression profiles of the BoCCD1 gene were analysed and discussed in relation to total carotenoids and other important apocarotenoids such as bixin.
Keywords: Annatto, apocarotenoids, Bixa orellana, bixin, carotenoid cleavage dioxygenase, CCD1, in situ RT-PCR, pigments
Introduction
Apocarotenoids are terpenoid compounds derived from the oxidative cleavage of carotenoids (Wahlberg and Eklund, 1998). They are generated when the double bonds in a carotenoid molecule are cleaved through a reaction involving molecular oxygen, with the formation of an aldehyde or ketone group in the cleavage site of each product. Carotenoids can be cleaved at any of their conjugated double bonds, resulting in a diverse set of apocarotenoids (Auldridge et al., 2006). These metabolites are widely distributed in plants, contributing to the flavour and/or aroma of flowers and fruits (i.e. β-cyclocitral, α- and β-damascenone, geranial, genaryl acetone, and β-ionone). In Bixa orellana an orange-red apocarotenoid known as bixin is accumulated in high concentrations, mainly on its seeds, and accounts for 80% of the total carotenoids present in the seeds (Rivera-Madrid et al., 2006). A bixin biosynthesis pathway has been documented in a heterologous system (Bouvier et al., 2003); the authors identified three B. orellana genes encoding three enzymes required for bixin synthesis from the linear C40 lycopene: lycopene cleavage dioxygenase (BoLCD); bixin aldehyde dehydrogenase (BoBADH); and norbixin methyltransferase (BoBMT). However, seed extracts contain a wide variety of apocarotenoids including both linear apocarotenoids [i.e. methyl (9Z)-apo-8′-lycopenoate] and cyclic apocarotenoids [i.e. methyl (all-E)-8′-apo-β-caroten-8′-oate] (Mercadante et al., 1996, 1997; Bittencourt et al., 2005). Nevertheless, apocarotenoid profiles in non-seed tissues and their accumulation patterns during different plant developmental stages remain to be clarified.
Although apocarotenoid formation can also occur via non-specific oxidation, biologically active forms with regulatory functions tend to be generated via site-specific cleavage. Several enzymes mediating the site-specific carotenoid cleavage are needed to generate biologically active apocarotenoids. On the basis of the substrates identified and their presumed mechanism of catalysis, these enzymes are referred to as carotenoid cleavage dioxygenases (CCDs) (Auldridge et al., 2006). The CCD family is ancient, with a large number of family members present in bacteria, animals, and plants. In Arabidopsis, the CCD enzyme family consists of nine members, which are further divided into two subfamilies: five 9-cis epoxycarotenoid dioxygenases (NCEDs) and four CCDs. The NCEDs are involved in the biosynthesis of the plant hormone abscisic acid (ABA) whereas CCDs are involved in the cleavage of carotenoids in several sites to generate multiple apocarotenoid products. CCDs are only distantly related to the NCEDs, and their substrate specificities and activities differ from those of the NCEDs (Ohmiya, 2009). Of the four CCD members in Arabidopsis, two CCD enzymes, now known as CCD7 and CCD8, produce a type of apocarotenoid that belongs to a novel class of plant hormones involved in shoot branching, strigolactones, (Gomez-Roldan et al., 2008; Goulet and Klee, 2010; Rameau, 2010). The function(s) of the remaining two, CCD4 and CCD1, appear to be related to specific carotenoids involved with colour, flavour, and aroma of fruits and flowers of many plant species (Ohmiya, 2009). In particular, plant CCD1s cleave numerous cyclic and linear all-trans-carotenoids (C5–C6, C7–C8, and C9–C10 double bonds) producing multiple apocarotenoid products (Vogel et al., 2008).
In this study, a cDNA coding for a new member of the CCD family (BoCCD1) in B. orellana was isolated. The putative BoCCD1 open reading frame (ORF) sequence differs from the CCD isolated in this plant (Bouvier et al., 2003) and shares a high homology with other plant CCD1s involved with the formation apocarotenoids. The mRNA expression of this new CCD1 member, BoCCD1, in B. orellana was documented during several plant development stages and during seed formation. The BoCCD1 expression pattern was correlated with carotenoid accumulation in this plant (mainly bixin). In situ hybridization methodology was used for the first time in this plant to investigate the expression of BoCCD1 and pigment accumulation in different tissues of B. orellana.
Materials and methods
Plant material
Leaves, floral buds, open flowers, immature fruits of 30 d post-anthesis, and immature and mature seeds (from mature and dehiscent fruit collected 60 d post-anthesis) were harvested from B. orellana plants cultivated at a commercial plantation in Chicxulub, Yucatán, México. Additionally, immature fruits from six different developmental stages were selected on the basis of the width and height of their valves (1.5×1.5 cm, 2×2.5 cm, 2.5×2.5 cm, 3.5×4 cm, 4.5×5 cm, and 5× 4.5 cm) and harvested to obtain immature seeds of six different stages of maturity. All tissues were obtained from a B. orellana variant with pink flowers and high pigment contents charaterized by Rivera-Madrid et al. (2006). In addition, 30-day-old B. orellana plantlets were obtained by germination of pre-treated seeds, as described by Narváez et al. (2001), and leaves, stalk, and roots were collected. Samples were immediately frozen by immersion in liquid N2 and stored at –80 °C until analysis. For in situ RT-PCR analysis and pigment localization, fresh tissues were cut into small pieces and immersed in 5 vols of AMBION® RNA Later solution (Applied Biosystems Inc., CA, USA). The fixed samples were kept at 4 °C prior to embedding and sectioning.
Assessment of carotenoid content
Total carotenoids were measured using spectrophotometric analysis, by extracting pigments from 10 mg of freeze-dried tissues ground with an electronic grinder, followed by adding 5 ml of ice-cold chloroform in semi-darkness in an ice water bath.
Each mixture was shaken vigorously for 3 min and was filtered through a nylon membrane (0.45 μm pore size). Subsequently, 50 μl of each extract was adjusted to 3 ml with chloroform and then measured with a DU 650 Beckman-Coulter spectrophotometer. Absorption spectra were obtained at a wavelength range of 404–480 nm and carotenoid content was calculated using the equation reported by Wellburn (1994).
For chromatographic [high-performance liquid chromatography (HPLC)] analysis of carotenoid and apocarotenoid compounds, 10 mg of freeze-dried tissues in chloroform were dried with nitrogen gas to remove the solvent and then dissolved in 1 ml of acetonitrile/methanol/isopropanol mixture (75:10:15 v/v/v). Samples were homogenized, centrifuged at 14 000 rpm for 10 min, and the supernatants were filtered through a PVDF membrane (0.22 μm pore size). A 20 μl aliquot of the sample was injected into a Hypersil ODS C-18 reverse phase column (250 mm×4.6 mm; 5 μm bead diameter) and analytes were separated by HPLC. The mobile phase consisted of solvent A [acetonitrile/methanol/isopropanol (75:10:15 v/v/v)] and solvent B [tetrahydrofuran (THF)], and chromatographic separation was performed at a flow rate of 1 ml min−1 as follows: step 1, 100% solvent A at injection, for 40 min; step 2, linear increase to 100% solvent B in 4 min; step 3, return to 100% solvent A in 5 min. Commercial standards from Sigma, lycopene, β-carotene, ABA, and bixin were used. All reagents used were HPLC grade and three replicates for each sample were carried out for both extraction and HPLC analysis.
Individual carotenoids and apocarotenoids were identified based on their column retention times, and their concentrations were calculated from specific peak area relative to a standard curve (Britton, 1995). Both HPLC and spectrophotometric data represented the mean of three replicates ±SE.
Bixin detection and distribution
For pigment distribution in plant tissues, fresh tissue samples were fixed with 4% neutral, buffered formalin and embedded in 5% melted agarose. Agarose blocks were mounted in wooden holders and sectioned transversely (30–40 μm slice thickness) with a microtome (Microm GmbH, mod. HM340E, Lab Tech). The agarose was eliminated from the tissue sections by rinsing with RNase-free water. The sections were subsequently stained using the periodic acid–Schiff (PAS) staining method. Fresh and stained sections were observed with a light transmission microscope for general morphology and pigment tissue localization. Photographs were taken using an Axiocam digital camera coupled to a microscope, and analysed by Axiovision-associated software.
Isolation of the BoCCD1 gene
Total RNA was obtained from immature seeds using the method previously reported by Rodríguez-Ávila et al. (2009), and the cDNA population was obtained by a reverse transcriptase reaction. BoCCD1 partial cDNA was amplified by PCR with degenerate primers (Fw1CCD, 5′-TGGTTYGAYGGNGAYGGNATG-3′; and Rv1CCD, 5′-GTDATNGCRAARTCRTGCATCAT-3′). This amplification was performed by conventional PCR in 50 μl (35 cycles of 94 °C for 1 min, 56 °C for 1.5 min, 72 °C for 1.5 min, and 45 s extension at 72 °C). For this first primer set, conserved sequences were identified by the alignment of several dioxygenases reported for plant CCD orthologous genes. To obtain a full BoCCD1 cDNA a 5′–3′ RACE (rapid amplification of cDNA ends) Kit (Invitrogen, San Diego, CA, USA) was used. To isolate the 3′ and 5′ends, gene-specific primers Fw2CCD, 5′-TGGTTTTATGCACGATCCAGTGCCG-3′; and Rv2CCD, 5′-GGGTTTGTCACCCTCGTTAAGTGCCA-3′ were designed using the partial BoCCD1 cDNA (540 bp). This reaction was performed according to the manufacturer's instructions (Invitrogen). PCR products of 3′ and 5′ RACE were purified using the Promega Wizard PCR Preps Purification System (Promega, Madison, WI, USA) and cloned using a TA cloning kit (Invitrogen). The cloned inserts were subsequently sequenced in an automated DNA sequencer (ABI PRISM™ 3700), and on an ABI 3730×1 automated sequencer (Applied Biosystems) by Macrogen Inc. (South Korea). Resulting ORF sequences were compared with the GenBank protein database using the Basic Local Alignment Sequence Tool (BLAST; National Center for Biotechnology Information), and domain homology was confirmed with the CLUSTALW2 tool (http://dot.imgen.bcm.tmc.edu:9331/multialign). Phylogenetic analysis was performed with the MEGA 4.1 program (Tamura et al. 2007) using the Neighbor–Joining (NJ) method. The reliability of the clusters was evaluated by bootstrapping with 1000 replicates.
Measurement of BoCCD1 expression level by reverse transcriptase-mediated PCR
Total RNA was obtained using the method previously reported by Rodríguez-Ávila et al. (2009), and the cDNA was synthesized using the SuperScript III First-Strand Synthesis System for the RT-PCR kit (Invitrogen) according to the manufacturer's instructions. After reverse transcription, the BoCCD1 transcript was amplified by PCR with 30 cycles and with specific primers (Fw:BoCCD1, 5′-ATGCAAGTCGAACCAACCAGGGGGATC-3′; Rv:BoCCD1, 5′-TGCCTGGTTCAGCAGATCCTTGTC-3′), designed from cloned BoCCD1 cDNA reported in this study (EF493214). A parallel reaction with 25 cycles and specific primers for the 18S rRNA gene (5'-CGGCTACCACATCCAAGGAA-3' and 5′-GCTGGAATTACCGCGGCT-3′, AF206868) was run as an expression control for each PCR. Replicates of each PCR were carried out to confirm the results. BoCCD1 expression relative to the 18S rRNA gene was assessed using a Bio-Image Analyzer GS25 (BioRad, Hercules, CA, USA). Preliminary PCR products were sequenced to verify their identity in all cases.
Assessment of BoCCD1 mRNA localization by in situ reverse transcriptase-mediated PCR
Fresh samples fixed as described above were rinsed three times for 10 min in a mixture of ethanol:acetic acid:water (63:5:32 v/v/v) and once more with 1× phosphate-buffered saline (PBS). Samples were then embedded in 5% melted agarose (in 1× PBS). Sample blocks were mounted in wooden holders and sliced in a microtome (Microm GmbH Type HM 340E). Sections (30–40 μm thickness) were rinsed with RNase-free water and incubated for 1 h with DNase (1 U ml−1 at 37 °C; Invitrogen). RT-PCRs were performed with a ‘One Step RT/PCR reaction kit’ (Invitrogen) using 0.1 μM of specific BoCCD1 primers and 15 μM digoxigenin (DIG)-dUTP. DIG-labelled PCR products were detected with alkaline phosphatase-coupled anti-DIG antibodies and alkaline phosphatase reaction with NBT/BCIP (Roche, USA). As negative controls, DNase-treated sections were incubated with RNase solution immediately before reverse transcription. In addition, in situ RT-PCR analysis was performed with β-actin-specific primers as positive controls. Digital images of the samples were captured with a Leica DFC320 digital camera adapted to the Leica MZFL III stereoscopic microscope and were processed using Leica IM 50 software.
Results
Pigment content in organs of B. orellana plants
Pigment quantification for different plantlet and adult tissues of B. orellana showed a large amount of variation in the accumulation of different carotenoids. Bixin was the most abundant apocarotenoid observed in both immature and mature seeds (Table 1). The concentrations of bixin seem to change according to the developmental stage of the seeds, continuously increasing during their development until the seed reaches its maximum size (Table 1). Although bixin has been widely reported in the mature seeds of B. orellana (Rivera-Madrid et al., 2006), the presence of this pigment in different tissues of B. orellana has not been documented before. In this study, HPLC data indicated that bixin was present in all of the tissues analysed, with the highest amounts in immature and mature seeds. The lowest quantities of bixin were registered in the plantlet root and green fruit of adult plants (Table 1). Other carotenes, such as β-carotene and lycopene, and the apocarotene ABA were also measured in order to understand their relative roles in bixin synthesis (Table 1). As expected, young and adult plant leaves accumulated important amounts of β-carotene (5–15 mg DW g−1), since it is involved in the photosynthetic apparatus. However, β-carotene was not detected in other organs (Table 1). Lycopene, the precursor of several carotenoids, was not detected in samples analysed in this study, probably suggesting a rapid turnover of this compound into the carotenoid biosynthetic pathways. Finally, ABA was abundant during seed development, with values ranging from 0.4 mg DW g−1 to 1.22 mg DW g−1, and in green tissues such as leaves and green fruits (Table 1). Vegetative organs and flowers accumulated high amounts of carotenoids in their tissues (Table 1).
Table 1.
Accumulation of carotenes and apocarotenoids during different development stages in B. orellana
| Lycopene | β-Carotene | Bixin (mg DW g−1) | ABA | Total carotenoids | |
| Adult tissues | |||||
| Leaves | ND | 15.36±1.22 a | 2.59±0.39 i | 0.49±0.19 d,e | 33.50±3.70 c |
| Flower bud | ND | ND | 2.44±0.18 i | 1.24±0.23 b | 28.36±1.03 d |
| Flower | ND | ND | 3.77±0.46 g,h | ND | 42.28±3.02 b |
| Green fruit | ND | ND | 0.34±0.02 j | 1.89±0.15 a | 4.90±0.38 i |
| Immature seed | ND | ND | 8.61±0.72 f | 0.25±0.08 e,f | 10.21±0.34 g,h |
| Mature seed | ND | ND | 52.41±0.82 a | 0.35±0.06 e | 65.18±1.86 a |
| Plantlet tissues | |||||
| Leaves | ND | 12.64±2.62 b | 2.95±0.31 h,i | 0.80±0.06 c | 26.58±0.55 d,e |
| Stalk | ND | 5.7±1.89 c | 3.24±0.20 h,i | 0.17±0.01 f | 45.21±1.57 b |
| Roots | ND | ND | 0.42±0.09 j | 0.12±0.02 f | 0.67±0.01 j |
| Seed development | |||||
| Stage 1 (0.1×0.1 mm) | ND | ND | 4.69±0.44 g | 0.41±0.03 e | 7.68±0.43 h,i |
| Stage 2 (0.1×0.15 mm) | ND | ND | 10.62±0.18 e | 0.46±0.02 d,e | 13.33±2.02 f,g |
| Stage 3 (0.15×0.2 mm) | ND | ND | 12.27±0.18 d | 1.05±0.11 b,c | 16.16±0.15 f |
| Stage 4 (0.2×0.3 mm) | ND | ND | 17.05±0.17 c | 1.22±0.05 b | 23.90±0.40 e |
| Stage 5 (0.3×0.4 mm) | ND | ND | 18.37±0.40 b | 0.86±0.09 c | 25.47±0.15 d,e |
| Stage 6 (0.4×0.5 mm) | ND | ND | 18.61±0.59 b | 0.98±0.04 b,c | 27.57±0.24 d,e |
*Values are presented as the means ±SD of three replicates. Different letters indicate significant differences determined by the statistical test ANOVA with P <0.001. ND, not detected.
Pigment histological analysis
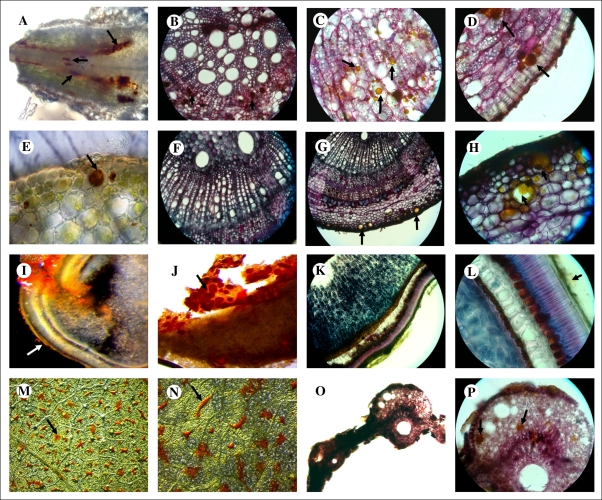
Both fresh and PAS/Alcian blue-stained preparations of B. orellana tissues showed the presence of bixin in specific cells. In the root apex (Fig. 1A), bixin was detected in pericycle cells, which are located between the vascular pith and endodermis. In addition, in plantlet roots, bixin was mainly accumulated in the cortex (Fig. 1B–D). Oily droplets of pigments, larger than starch grains accumulated in cells of root tissues, were observed in the cytosol (Fig. 1C, D). Higher accumulation of bixin was observed in plant stalks, where it was only accumulated in collenchyma cells in the stem cortex (Fig. 1E–H). In immature seeds, bixin was detected in aril cells, observed as oily inclusions contained in a thin cell layer covering the seed surface (Fig. 1I–L). Pigment inclusions were distributed throughout the aril layer (Fig. 1J), over a palisade-like cell layer covering the endosperm tissue (Fig. 1K). It was observed that in mature aril cells, bixin occupied the whole cell volume (Fig. 1J), suggesting that mature aril cells are used for pigment storage and lose their biosynthetic functions. Based on this, mature seeds were not viable for further molecular studies. Finally, stereoscopic observations of the leaf abaxial surface showed specific bixin accumulation in cell patches uniformly distributed across each leaf (Fig. 1M, N). Histological tissue samples of the plantlet leaves showed the accumulation of bixin droplets in the spongy mesophyll cells (Fig. 1O, P).
Fig. 1.
Histological analysis of different bixin-accumulating Bixa orellana plant organs. (A–D) Plantlet root (A, longitudinal section; B–D, transversal section). (E–H) Stalk (E, longitudinal section; F and G, transversal sections). (I–L) Immature seed (I, longitudinal section; J, aril cells; K and L, transversal sections). (M–P) Leaves (M and N, blade section; O and P, transversal section). A, E, I, J, M, and N are unstained tissue sections. Arrows indicate bixin accumulation. Scale bars in G, M, N, O, P=400 μm; A, B, F, I, K=200 μm; C, D, E, H, L=100 μm.
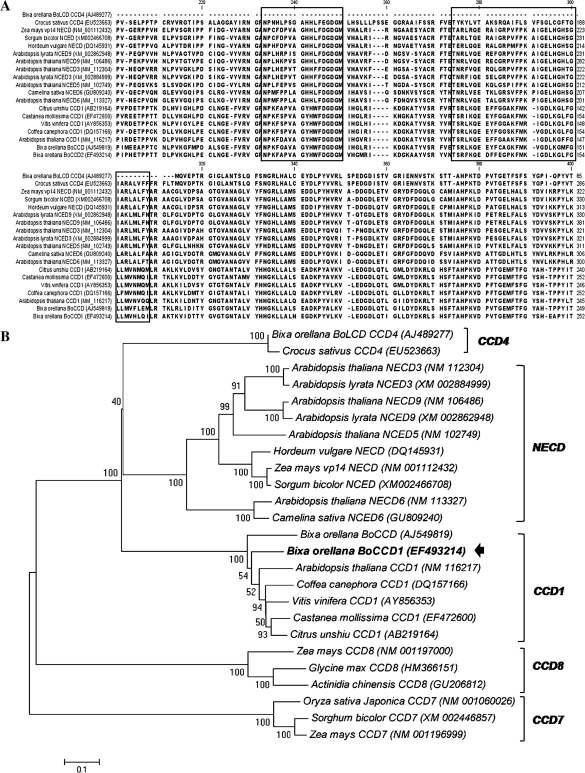
Bioinformatic analysis of the cloned BoCCD1 gene
The BoCCD1 cDNA sequence showed high identity with CCD protein sequences available in GenBank (Fig. 2). A phylogenetic tree was constructed using the NJ method (Fig. 2), showing a short distance of 0.01 with reference to other plant CCD1 genes. The amino acid sequence also reflected the presence of a conserved domain (RPE65), which is characteristic of the enzymes involved in the biosynthesis of apocarotenoids (Kloer and Schulz, 2006). A graphic scheme showing the conformation and the sites of the sequence isolated coding for the RPE65 domain described is presented in Fig. 2. This bioinformatics analysis shows at least three CCD members isolated in this plant (BoCCD1, BoCCD, and BoLCD). In addition, a high stringency Southern blot analysis was conducted to detect the gene copy number likely to be encoding a CCD. In all the experiments at least five fragments sharing homology with BoCCD1 were detected, suggesting the presence of a multigene family of CCDs in this plant (data not shown). It is important to mention that the BoCCD1 isolated in this study exhibits identity with another CCD (BoCCD) previously isolated in B. orellana (AJ549819).
Fig. 2.
(A) Translated sequence alignment of the CCD probe isolated with other plant dioxygenases. Boxes indicate the location of the RPE65 domain. (B) Phenogram obtained from the Neighbor–Joining phylogenetic analysis of the B. orellana CCD isolated. The percentage of parsimonious trees in which the associated taxa clustered together (bootstrap value >1000 repetitions) is shown next to the branches. The arrow indicates the gene under study. Brackets indicate the main carotenoid cleavage dioxygenase families. Accession numbers are in parentheses.
Expression of the BoCCD1 gene in several plant organs and developmental stages
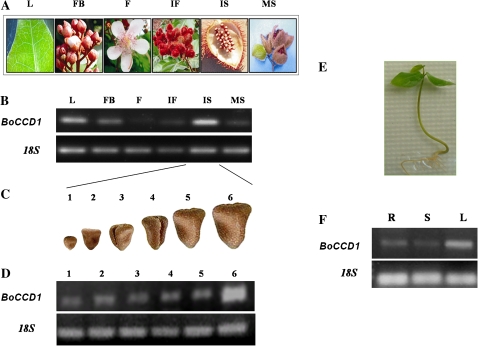
BoCCD1 mRNA expression was evaluated in vegetative organs (young and mature leaves, stems, and roots) and in reproductive tissues (flower bud, flower, immature fruit, and seeds in different developmental stages) by RT-PCR analysis (Fig. 3). Adult and plantlet leaves, flower buds, and immature seeds exhibited a high expression of the BoCCD1 gene and, to a lesser degree, in flower, immature fruit, and mature seeds (Fig. 3B, F). A more detailed analysis was carried out in order to obtain information concerning how the BoCCD1 expression profile could be related to bixin accumulation in immature seeds. BoCCD1 expression increased throughout seed maturation, reaching maximum levels at stage 6 of the immature seeds (Fig. 3D). After seed maturation and desiccation, BoCCD1 expression was weak (Fig. 3B). Plantlet root and stalk showed a lower expression of this gene, while in the leaves BoCCD1 mRNA was up-regulated (Fig. 3F).
Fig. 3.
BoCCD1 expression of different organs of B. orellana plants. (A) Images of the different organs used for semi-quantitative RT-PCR analysis. L, leaf; FB, floral bud; F, flower; IF, immature fruit; IS, immature seed; MS, mature seed. (B) BoCCD1 RT-PCR expression profile obtained for the tissues shown in (A). (C) Six different developmental stages of the immature seeds. (D) Expression profile of BoCCD1 obtained during seed development. (E) Photograph of a 30-day-old plantlet. (F) BoCCD1 expression profile obtained for B. orellana plantlets. BoCCD1 and 18S rRNA gene RT-PCR analyses were conducted with 30 and 25 cycles, respectively. R, root; S, stalk; L, leaf.
BoCCD1 gene localization
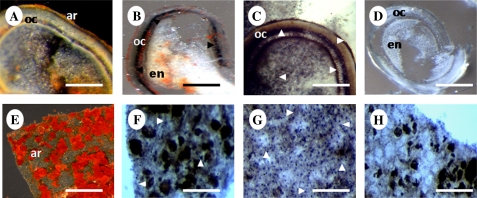
An in situ RT-PCR assay was carried out in the tissues where apocarotenoid production was observed, with the aim of identifying the location of the BoCCD1 mRNA. First the carotenoid content and distribution between different tissues and cellular types were assessed. Seeds and several other tissues with elevated amounts of apocarotenoids were chosen to analyse the presence of this gene in detail in several specific cells. In seeds, the BoCCD1 probe was detected in the layer of oily cells located immediately below the aril (Fig. 4B) and in a high number of cells located in the aril layer adjacent to bixin deposits (Fig. 4F); however, it was not detected in the endosperm of the seeds (Fig. 4B). This suggests that the BoCCD1 gene could be involved with apocarotenoid production, specifically bixin, since its expression in these tissues coincided with the highest pigment contents detected (Table 1).
Fig. 4.
BoCCD1 gene location in seed preparations (A–D) and in the dissected aril layer (E–H) from the immature seeds of B. orellana. Seed and dissected fresh aril preparations are shown in A and E. In situ RT-PCR tissue preparations are shown in B–D and F–H, with the following treatments: (B) and (F) BoCCD1 amplification; (C) and (G) actin amplification; (D) and (H) digestion with RNase prior to BoCCD1 amplification. Arrowheads point to RT-PCR-positive cells. A, B, and E are unstained tissue sections Tissue abbreviations: ar, aril; oc, oily cells; en, endosperm. Scale bars=400 μm.
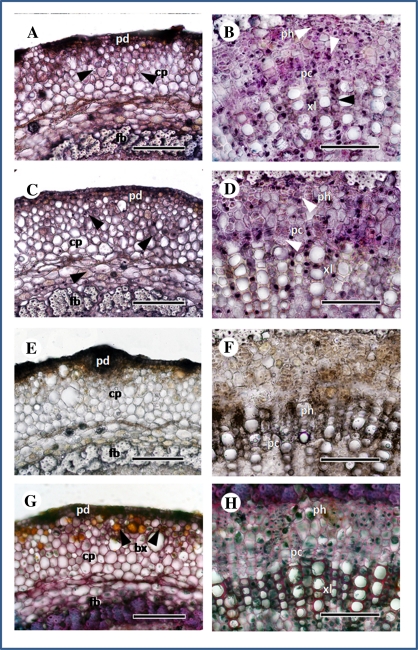
Due to the fact that during the RT-PCR assay the BoCCD1 transcripts were observed in the stalk of plantlets, the tissue where pigment contents, specifically the apocarotenoid bixin, were highest (Table 1), an in situ RT-PCR assay was carried out on stalk transversal sections. The results obtained from the analysis derived from the PAS/Alcian blue preparations showed bixin to be accumulated specifically in the cells that make up the peridermis (Fig. 5G). Similarly, the BoCCD1 signal was clearly detected in cells corresponding to the xylem and surrounding the tubes of the phloem and in tissues where pigment accumulation was detected, as well as in the cells of the parenchyma and peridermis in the leaves (Fig. 5A, B).
Fig. 5.
BoCCD1 gene location in stalk preparations. (A and B) BoCCD1; (C and D) actin; (E and F) RNase+BoCCD1; (G and H) PAS staining. All panels show transversal sections. Scale bars=100 μm. pc, pericycle; ph, phloem; xl, xylem; cp, cells of parenchyma; pd, peridermis; fb, fibres; pt, pith; ph, phloem; bx, bixin inclusions. Arrowheads: RT-PCR-positive cells.
Discussion
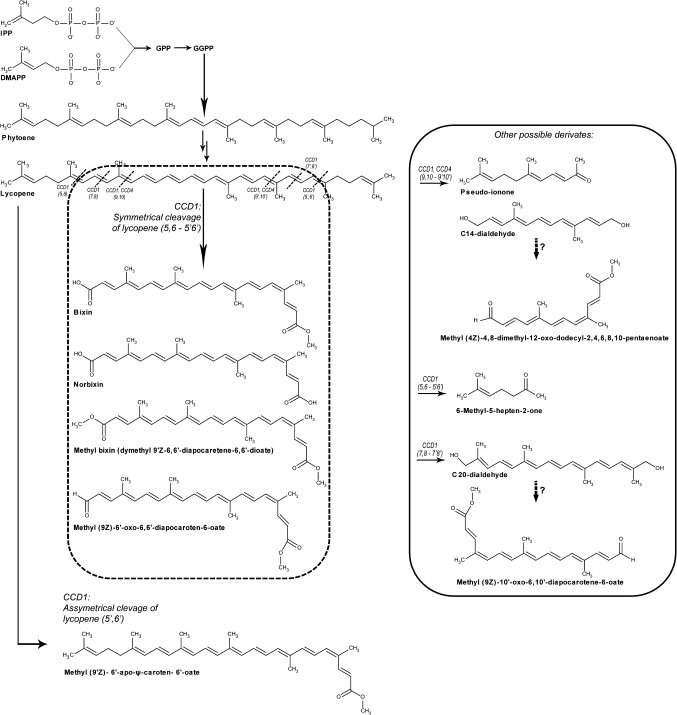
A plethora of apocarotenoid compounds have been reported in the seeds of B. orellana, including methyl (9′Z)-apo-6′-lycopenoate, methyl (9Z)-6′-oxo-6,6′-diapocarotenoate, and methyl (all-E)-8′-apo-β-caroten-8′-oate, with bixin being the most abundant (Mercadante et al., 1997; Bittencourt et al., 2005) (Fig. 6). So far, only one study has investigated an expression response of carotene-related genes during development in B. orellana (Rodríguez-Avila et al., 2011). In that study, expression responses of dxs, psy, pds, β-lcy, and ϵ-lcy genes were associated with pigment overaccumulation in two different annatto cultivars. However, in vivo apocarotenoid-related gene analysis and apocarotenoid production during plant development have not been conducted in this plant.
Fig. 6.
Hypothetical biosynthesis pathway of linear apocarotenoids reported for Bixa orellana. The possible cleavage sites and CCDs involved were determined based on the substrates and cleavage sites of type 1 CCDs (such as the one isolated in this study) and type 4 (BoLCD; Bouvier et al., 2003), the regioselectivity of which has been well charaterized in this and in other plants. The question marks relate to possible precursors and routes of biosynthesis inferred on the basis of the chemical structure observed. The small dotted lines indicate the possible cleavage sites of the CCDs, whose hypothetical function is indicated.
The results obtained in the present study showed that bixin production is not restricted to seed tissues since this pigment was detected in all the tissues analysed, particularly in the cells surrounding the vascular tissues and in the epidermal cells. In mature seeds, bixin is located exclusively in the aril occupying the whole cell volume, suggesting that mature aril cells are used for pigment storage and lose their biosynthetic functions. Functional changes in the aril cells from meristem cells to storage cells (large central vacuole and peripheral nucleus) during the last steps of aril development have been previously described by Rodríguez-Riaño et al. (2006). A constant increment in bixin production was observed during the development of immature seeds.
On the other hand, ABA was detected in all the adult plant tissues analysed, and accumulated in greater amounts in floral buds and immature fruits in accordance with its role in development regulation and fruit maturation (Chernys and Zeevaart, 2000; Rodrigo et al., 2006). Other pigments related to carotene precursors of bixin such as lycopene (Bouvier et al., 2003) were undetected in all tissues analysed, suggesting a rapid turnover of this molecule during pigment biosynthesis. Finally, β-carotene was detected only in green tissues, in accordance with its role in photosynthesis (Grotewold, 2006).
CCDs are enzymes involved in the production of apocarotenoids such as bixin. Two CCD genes have previously been isolated for B. orellana; however, only one of these (BoLCD) has been proved to be responsible for bixin production and its role has been shown in a heterologous system (Bouvier et al., 2003). Here, a new CCD gene, BoCCD1, was isolated (EF493214) and its sequence showed high homology with other plant CCD1s. Furthermore, the highly conserved RPE65 domain, which is responsible for the catalytic activity of the CCDs, was found to be encoded in this sequence (Kloer and Schulz, 2006) (Fig. 2). Phylogenetic analysis conducted with the sequences of the previously isolated CCD genes for B. orellana (AJ489277 and AJ549819), and for other plants, showed that BoCCD1 shares the same ancestor gene as another CCD isolated for B. orellana (the CCD gene, AJ549819). However, BoLCD (AJ489277), the gene involved in bixin biosynthesis (Bouvier et al., 2003), is located in a distant clade (CCD4 clade), suggesting a different gene ancestor.
BoCCD1 has the same phylogenetic origin as the CCD1 genes isolated from Coffea canephora (CcCCD1) or Petunia hybrida (PhCCD1), these enzymes catalysing the symmetrical 9,10 (9',10') cleavage of multiple linear and cyclized carotenoids (Simkin et al., 2004, 2008). This cleavage results in the formation of a diverse variety of C13 cyclohexone apocarotenoids and a C14 dialdehyde, corresponding to the central portion of the original carotenoid (Simkin et al., 2004). In addition, CCD1 cleavage activity on 5,6 (5',6') double bonds in vitro has also been reported (Vogel et al., 2008).
In accordance with the above, molecular studies were performed to analyse the role of this new CCD1 isolated from B. orellana in carotenoid production. In this sense, the primary expression profiles were obtained for different B. orellana tissues which have been analysed and characterized in previous studies (Rivera-Madrid et al., 2006). BoCCD1 mRNA was present in all of the plantlet tissues analysed (leaves, stalk, and roots), being up-regulated again in the leaves of the plantlets analysed. It was possible to relate BoCCD1 expression to total carotenoid accumulation. Expression analyses carried out on the CCD1 genes of several plant species have shown that individual genes have a tissue-specific expression profile, which is controlled in accordance with the production of apocarotenoids such as those involved in development or stress response (Prescott and John, 1996; Simkin et al., 2004, 2008). Here, BoCCD1 expression was high in immature seeds, although the peak in total carotenoids and bixin was observed in mature seeds. These results could mean that BoCCD1 expression is induced when pigment production begins and is down-regulated when its biosynthesis is accomplished. Consequently, a study was designed to observe the role of the BoCCD1 gene during the accumulation of total carotenoids in seeds. Immature seeds corresponding to six different developmental stages were selected to analyse the expression profile of BoCCD1 by RT-PCR. Interestingly, BoCCD1 mRNA showed a coordinated increase in its expression with the accumulation of carotenoids, particularly bixin, observed in the different developmental stages of the seeds. Similarly, the expression of CCD1s in several plant species is controlled in accordance with the production of important apocarotenoids such as β-ionone, a fragrance volatile in petunia flowers, or C13-norisoprenoids, considered flavour compounds in grape berries (Simkin et al., 2004; Mathieu et al., 2005).
The results obtained from the histological analysis revealed that the pigment granules (mainly bixin) accumulated in the surface of the seeds, settling in a layer of oily cells which cover the aril. In addition, the in situ RT-PCR assay showed that the BoCCD1 transcripts were also located in this oily layer, suggesting a relationship between this gene and the accumulation of apocarotenoids. A new in situ RT-PCR assay was carried out specifically on the aril layer of the seed. The strong signal of the BoCCD1 gene obtained in this seed tissue indicated that BoCCD1 mRNA location is concordant with the production of total carotenoids and specific apocarotenoids such as bixin. The importance of this gene in carotenoid turnover during seed maturation has been demonstrated in Arabidopsis by the construction of a CCD1 mutant, which shows a normal appearance but higher levels of mature seed carotenoids (Auldridge et al., 2006).
In the in situ RT-PCR assay carried out on the stalk, BoCCD1 expression was located in the cells surrounding the xylem and phloem tissues. Results derived from the histological analysis showed that in the stalk, pigments (mainly bixin) are accumulated specifically in the cells that make up the peridermis. Both results suggest that pigment apocarotenoids are probably synthesized in the vascular tissues and later exported to the epidermal cells; accordingly, the translocation of bixin from the stalk and leaves into seeds could also be possible. Some important apocarotenoids, such as the plant hormone ABA, act in tissues different from where they were synthesized (Tan et al., 2003). Thus, the expression of the BoCCD1 gene is probably related to the high apocarotenoid diversity registered in different tissues of this plant and may be related to the accumulation of some specific apocarotenoids present in seeds not analysed here [i.e. methyl (all-E)-8′-apo-β-caroten-8′-oate] (Mercadante et al., 1996, 1997; Bittencourt et al., 2005).
The cleavage sites of CCD1 suggest its possible involvement in the synthesis of several apocarotenoids of B. orellana (Walter and Strack, 2011) (Fig. 6). It has been discovered that in rice, maize, and tomato plants, CCD1s also act on the C5–C6/C5′–C6′ double bonds of acyclic carotenoids, resulting in the volatile C8 ketone 6-methyl-5-hepten-2-one. By means of in vivo and in vitro assays, they have shown that rice CCD1 converts lycopene into three different volatiles: 6-methyl-5-hepten-2-one, pseudoionone, and geranial (C10). Furthermore, comparison of the relative amounts of the C19 and C17 aldehydes accumulated in the incubations with the natural substrates lycopene and 3-OH-c-carotene indicated that the preference of OsCCD1 for the C5'–C6' and C7'–C8' double bonds depends on the nature of the substrate (Ilg et al., 2009). In the present study, the carotenoid lycopene was absent in all tissues analysed, which strongly suggests the rapid metabolism of these molecules to produce their derivatives.
Finally, based on the current knowledge of the cleavage sites and substrates of this type of enzyme (Rubio et al., 2008; Ilg et al., 2009; Walter and Strack, 2011), it is considered that this BoCCD1 enzyme is possible related to the cleavage of lycopene at the double bonds 5–6/5′–6′ leading to the formation of the second precursor of bixin (Fig. 6). However, biochemical analyses remain to be carried out on the protein product of this gene in order to clarify this. The above idea is in agreement with the presence of transcripts of this gene (in almost all tissues) in relation to the accumulation of bixin, mainly in seed maturation developmental stages. Although a previous CCD4 (BoLCD) has been reported during bixin formation (Bouvier et al., 2003), the other CCD members, particularly CCD1, could be involved in the synergic production of bixin and other important apocarotenoids derived from its putative CCD1 activity. As previously reported in Crocus sativus, four CCDs were isolated, two CCD1s and two CCD4s, and all of them were able to cleave β-carotene at the 9, 10 (9′, 10′) positions to yield β-ionone. It seems to be common in secondary metabolism for gene families with the same enzymatic activity to be present (Rubio et al., 2008). The data of Rubio et al. (2008) strongly suggest that each class of enzymes could be responsible for the metabolism of carotenoids at specific and different subcellular sites, during normal development and in response to conditions of stress. Thus in B. orellana, taking into account that BoLCD (Bouvier et al., 2003) seems to be a putative CCD4 protein located in plastids, and BoCCD1 seems to be cytosolic, both could, in a coordinated or alternating manner, produce several apocarotenoids in B. orellana including bixin, as in the case of C. sativus for the production of other apocarotenoids (Rubio et al., 2008). Although the biosynthetic pathway of bixin was revealed several years ago (Bouvier et al., 2003), the predicted 5–6/5'–6' cleavage specificity of the putative BoLCD has not yet been clarified (Sergeant et al., 2009; Walter and Strack, 2011). However, for these hypotheses, additional research needs to be conducted in planta to obtain further direct evidence that BoCCD1 is involved in bixin biosynthesis and apocarotenoid diversification in B. orellana.
Acknowledgments
This work was supported by the International Foundation for Science (IFS) F/2932-3 and by the Consejo Nacional de Ciencia y Tecnología (CONACYT) 46541, 98508; and UC Mexus-CONACYT. NLRA was supported by a CONACYT PhD grant (no. 196432). We would like to thank Dr Yumi Nakazawa for her technical assistance provided in the RACE analysis. Thanks also to the two anonymous reviewers for improving the manuscript.
References
- Auldridge ME, McCarty DR, Klee HJ. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Current Opinion in Plant Biology. 2006;9:315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bittencourt C, Felicissimo MP, Pireaux JJ, Houssiau L. ToF-SIMS characterization of thermal modifications of bixin from Bixa orellana fruit. Journal of Agricultural and Food Chemistry. 2005;53:16–19. doi: 10.1021/jf0505271. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Dogbo O, Camara B. Biosynthesis of the food and cosmetic plant pigment bixin (annato) Science. 2003;300:2089–2091. doi: 10.1126/science.1085162. [DOI] [PubMed] [Google Scholar]
- Britton G. UV/visible spectroscopy. In: Britton S, Liaaen-Jensen, Pfander H, editors. Carotenoids. Vol. 1B. Basel: Birkhäuser; 1995. pp. 13–62. [Google Scholar]
- Chernys JT, Zeevaart JAD. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiology. 2000;124:343–353. doi: 10.1104/pp.124.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Goulet C, Klee HJ. Climbing the branches of the strigolactones pathway one discovery at a time. Plant Physiology. 2010;154:493–496. doi: 10.1104/pp.110.161026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;7:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Ilg A, Beyer P, Al-Babili S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS Journal. 2009;276:736–747. doi: 10.1111/j.1742-4658.2008.06820.x. [DOI] [PubMed] [Google Scholar]
- Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cellular and Molecular Life Sciences. 2006;63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S, Terrier N, Procureur J, Bigey F, Günata Z. A carotenoid cleavage dioxygenase from Vitis vinifera L.: functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. Journal of Experimental Botany. 2005;56:2721–2731. doi: 10.1093/jxb/eri265. [DOI] [PubMed] [Google Scholar]
- Mercadante AZ, Steck A, Pfander H. Isolation and structure elucidation of minor carotenoids from annatto (Bixa orellana L.) seeds. Phytochemistry. 1997;46:1379–1383. [Google Scholar]
- Mercadante AZ, Steck D, Rodriguez-Amaya, Pfander H, Britton G. Isolation of methyl 9'Z-apo-6'-lycopenoate from Bixa orellana. Phytochemistry. 1996;41:1201–1203. [Google Scholar]
- Narváez JA, Flores-Perez P, Herrera-Valencia V, Castillo F, Ku-Cauich R, Canto-Canche B, Buzzy N, Rivera-Madrid R. Development of molecular techniques for studying the metabolism of carotenoids in Bixa orellana L. HortScience. 2001;36:982–986. [Google Scholar]
- Ohmiya A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnology. 2009;26:351–358. [Google Scholar]
- Prescott AG, John P. Dioxygenases: molecular structure and role in plant metabolism. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:245–271. doi: 10.1146/annurev.arplant.47.1.245. [DOI] [PubMed] [Google Scholar]
- Rameau C. Strigolactones, a novel class of plant hormone controlling shoot branching. Comptes Rendus Biologies. 2010;333:344–349. doi: 10.1016/j.crvi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Rivera-Madrid R, Escobedo-Medrano RM, Balam-Galera E, Vera-Ku M, Huges H. Preliminary studies toward genetic improvement of annatto (Bixa orellana L.) Scientia Horticulturae. 2006;109:165–172. [Google Scholar]
- Rodrigo MJ, Alquezar B, Zacarías L. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck) Journal of Experimental Botany. 2006;57:633–643. doi: 10.1093/jxb/erj048. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ávila NL, Narváez-Zapata JA, Aguilar-Espinosa ML, Rivera-Madrid R. Full-length genes enrichment by using an optimized RNA isolation protocol in Bixa orellana recalcitrant tissues. Molecular Biotechnology. 2009;42:89–94. doi: 10.1007/s12033-008-9138-4. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ávila NL, Narváez-Zapata JA, Aguilar-Espinosa M, Rivera-Madrid R. Regulation of pigment-related genes during flower and fruit development of Bixa orellana. Plant Molecular Biology Reporter. 2011;29:43–50. [Google Scholar]
- Rodríguez-Riaño T, Valtueña FJ, Ortega-Olivencia A. Megasporogenesis, megagametogenesis and ontogeny of the aril in Cytisus striatus and C. multiflorus (Leguminosae: Papilionoideae) Annals of Botany. 2006;98:777–791. doi: 10.1093/aob/mcl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Rambla JL, Santaella M, Gomez MD, Orzaez D, Granell A, Gomez-Gomez L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in β-ionone release. Journal of Biological Chemistry. 2008;283:24816–24825. doi: 10.1074/jbc.M804000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant MJ, Li JJ, Fox C, Brookbank N, Rea D, Bugg TD, Thompson AJ. Selective inhibition of carotenoid cleavage dioxygenases: phenotypic effects on shoot branching. Journal of Biological Chemistry. 2009;284:5257–5264. doi: 10.1074/jbc.M805453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Moreau H, Kuntz M, Pagny G, Lin C, Tanksley S, McCarthy J. An investigation of carotenoid biosynthesis in Coffea canephora and Coffea Arabica. Journal of Plant Physiology. 2008;165:1087–1106. doi: 10.1016/j.jplph.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Underwood BA, Auldridge M, Loucas HM, Shibuya K, Schmelz E, Clark DG, Klee HJ. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of beta-ionone, a fragance volatile of petunia flowers. Plant Physiology. 2004;136:3504–3514. doi: 10.1104/pp.104.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. The Plant Journal. 2003;35:44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Tan BC, McCarty DR, Klee HJ. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. Journal of Biological Chemistry. 2008;283:11364–11373. doi: 10.1074/jbc.M710106200. [DOI] [PubMed] [Google Scholar]
- Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Natural Products Reporter. 2011;28:651–844. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- Wahlberg I, Eklund AM. Degraded carotenoids. In: Britton G, Liaaen-Jensen S, Pfander H, editors. Carotenoids. Vol. 3. Basel: Birkhaüser; 1998. pp. 195–216. [Google Scholar]
- Wellburn AR. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology. 1994;144:307–313. [Google Scholar]