Abstract
This study employs transcript profiling together with immunoblotting and co-immunopurification to assess the tissue-specific expression, protein:protein interactions, and post-translational modifications (PTMs) of plant- and bacterial-type phosphoenolpyruvate carboxylase (PEPC) isozymes (PTPC and BTPC, respectively) in the castor plant, Ricinus communis. Previous studies established that the Class-1 PEPC (PTPC homotetramer) of castor oil seeds (COS) is activated by phosphorylation at Ser-11 and inhibited by monoubiquitination at Lys-628 during endosperm development and germination, respectively. Elimination of photosynthate supply to developing COS by depodding caused the PTPC of the endosperm and cotyledon to be dephosphorylated, and then subsequently monoubiquitinated in vivo. PTPC monoubiquitination rather than phosphorylation is widespread throughout the castor plant and appears to be the predominant PTM of Class-1 PEPC that occurs in planta. The distinctive developmental patterns of PTPC phosphorylation versus monoubiquitination indicates that these two PTMs are mutually exclusive. By contrast, the BTPC: (i) is abundant in the inner integument, cotyledon, and endosperm of developing COS, but occurs at low levels in roots and cotyledons of germinated COS, (ii) shows a unique developmental pattern in leaves such that it is present in leaf buds and young expanding leaves, but undetectable in fully expanded leaves, and (iii) tightly interacts with co-expressed PTPC to form the novel and allosterically-desensitized Class-2 PEPC heteromeric complex. BTPC and thus Class-2 PEPC up-regulation appears to be a distinctive feature of rapidly growing and/or biosynthetically active tissues that require a large anaplerotic flux from phosphoenolpyruvate to replenish tricarboxylic acid cycle C-skeletons being withdrawn for anabolism.
Keywords: Enzyme phosphorylation, metabolic control, monoubiquitination, phosphoenolpyruvate carboxylase, post-translational modification, protein:protein interactions, tissue-specific gene expression
Introduction
Phosphoenolpyruvate carboxylase (PEPC, EC 4.1.1.31) is a tightly controlled cytosolic enzyme that catalyses the irreversible β-carboxylation of phosphoenolpyruvate (PEP) in the presence of HCO3− to yield oxaloacetate and Pi. The properties and control of PEPC have been extensively studied with regard to its critical function in catalysing the initial fixation of atmospheric CO2 in C4 and CAM photosynthesis (Chollet et al., 1996; Nimmo, 2003; Izui et al., 2004). However, PEPC also plays a wide variety of important roles in non-photosynthetic and C3 photosynthetic cells, particularly the anaplerotic replenishment of tricarboxylic acid (TCA) cycle intermediates withdrawn for biosynthesis or N-assimilation (O’Leary et al., 2011a). Non-photosynthetic PEPCs are generally less well described in terms of their genetic origin and post-translational controls.
Plant PEPCs belong to a small gene family encoding several plant-type PEPC (PTPC) isozymes, along with at least one distantly related bacterial-type PEPC (BTPC) isozyme (Sánchez and Cejudo, 2003; Mamedov et al., 2005; Gennidakis et al., 2007; O’Leary et al., 2011a). PTPC genes encode similar 100–110 kDa polypeptides that contain a conserved N-terminal seryl-phosphorylation site and a critical C-terminal tetrapeptide QNTG, and typically exist as homotetrameric Class-1 PEPCs (Izui et al., 2004). Owing to its location at a crucial branchpoint in primary metabolism, an impressive array of strategies has evolved to control plant PEPC activity post-translationally. Regulatory phosphorylation and monoubiquitination, as well as changes in intracellular pH and allosteric effector levels have all been described as mechanisms to regulate Class-1 PEPC activity in different plant tissues under various physiological conditions (Chollet et al., 1996; Nimmo, 2003; Izui et al., 2004; O’Leary et al., 2011a). The discovery of unusual Class-2 PEPC hetero-octameric complexes that arise from an interaction of a Class-1 PEPC with co-expressed BTPC subunits and are largely desensitized to allosteric effectors adds a further layer of complexity to the physiological functions and control of this important plant enzyme. A recent study identified a chloroplast stroma targeted Class-1 PEPC isozyme in rice, although the degree to which this phenomenon occurs in other plant species remains to be determined (Masumoto et al., 2010).
Plant BTPC genes encode 116–118 kDa polypeptides that exhibit low (∼40%) sequence identity with PTPCs and contain a prokaryotic-like (R/K)NTG C-terminal tetrapeptide (Sullivan et al., 2004; Sánchez et al., 2006; Gennidakis et al., 2007; Igawa et al., 2010). BTPC genes and transcripts have been well documented in vascular plants and green algae (Sullivan et al., 2004; Sánchez et al., 2006; Gennidakis et al., 2007; Igawa et al., 2010). However, insights into BTPC polypeptide occurrence and function have thus far been restricted to PEPC studies from unicellular green algae (Selenastrum minutum and Chlamydomonas reinhardtii), developing castor (Ricinus communis) oil seeds (COS), and developing lily (Lilium longiflorum) pollen (Rivoal et al., 1998, 2001; Blonde and Plaxton, 2003; Mamedov et al., 2005; Tripodi et al., 2005; Gennidakis et al., 2007; Moellering et al., 2007; Uhrig et al., 2008a; Igawa et al., 2010; O’Leary et al., 2009, 2011a, b). Native green algal and vascular plant BTPCs appear to only exist in physical association with PTPC subunits as part of a Class-2 PEPC heteromeric complex (Rivoal et al., 1998, 2001; Mamedov et al., 2005; Gennidakis et al., 2007; Igawa et al., 2010). Thus, two distinct classes of PEPC sharing the same 107 kDa PTPC subunit (p107; encoded by RcPpc3) were discovered in the triglyceride-rich endosperm of developing COS (Blonde and Plaxton, 2003; Gennidakis et al., 2007). The tight association of p107 with immunologically unrelated 118 kDa BTPC subunits (p118; encoded by RcPpc4) causes pronounced physical and kinetic differences between the 410 kDa Class-1 PEPC p107 homotetramer and 910 kDa Class-2 PEPC p118:p107 hetero-octamer (Blonde and Plaxton, 2003; Gennidakis et al., 2007; O’Leary et al., 2009). Both subunit types exhibit PEPC activity within the Class-2 PEPC complex. However, the PTPC subunits represent high PEP affinity and allosterically sensitive subunits, whereas the BTPC subunits exhibit a lower affinity for PEP, as well as remarkable insensitivity towards PTPC allosteric inhibitors such as L-malate and L-asparate (O’Leary et al., 2009, 2011b). Furthermore, COS BTPC also functions as a regulatory subunit by modulating the PEP binding, catalytic potential and allosteric effector sensitivity of associated PTPC subunits within the Class-2 PEPC complex (O’Leary et al., 2009, 2011b). Class-2 PEPC has been hypothesized to function as a ‘metabolic overflow’ mechanism that could maintain a significant flux from PEP to L-malate under physiological conditions that would largely inhibit the corresponding Class-1 PEPC (O’Leary et al., 2009). Although BTPCs lack the conserved N-terminal seryl phosphorylation site characteristic of PTPCs, BTPC from developing COS endosperm is in vivo phosphorylated at multiple sites, including Ser-425 (Uhrig et al., 2008a; O’Leary et al., 2011b). Immunoblotting with Ser-425 phosphorylation-site specific antibodies coupled with kinetic analyses of recombinant phosphomimetic COS BTPC mutants established that: (i) in vivo BTPC Ser-425 phosphorylation is dependent upon COS developmental and physiological status, (ii) Ser-425 phosphorylation attenuates the catalytic activity of BTPC subunits within the Class-2 PEPC complex, and (iii) the developmental pattern and kinetic influence of Ser-425 BTPC phosphorylation are highly distinct from the in vivo phosphorylation-activation of COS Class-1 PEPC’s PTPC subunits at Ser-11 (O’Leary et al., 2011b). Although no definitive physiological role has been attributed to COS Class-2 PEPC, it has been hypothesized to support a large flux of PEP to malate required for abundant fatty acid synthesis that dominates the leucoplast metabolism of developing COS.
Thanks to the pioneering work of Harry Beevers and co-workers in the early 1960s (Benedict and Beevers, 1961; Canvin and Beevers, 1961), the COS has been widely developed as an important model system for studies of oilseed development and germination, carbon metabolism, and triacylglyceride storage and mobilization. The recent sequencing and annotation of the castor genome constitute an important foundation for further identification of the regulatory and metabolic networks underpinning castor-oil biosynthesis (Chan et al., 2010). However, biochemical analyses of the expressed native proteins must continue as detailed information about the control of enzymes and metabolism cannot be solely deduced from annotated gene sequences and the application of related genomic tools. In the current study, mRNA profiling was integrated with immunoblotting and co-immunopurification studies using monospecific antibodies against the COS PTPC and BTPC, as well as immunoblotting with phosphorylation site-specific antibodies to examine the expression, post-translational modifications (PTMs), and protein:protein interactions of PTPC and BTPC isozymes in various tissues of the castor plant. The tissue-specific distribution of monoubiquitinated PTPC was also assessed and the results indicate that monoubiquitination rather than phosphorylation is the dominant PTPC PTM that occurs in planta.
Materials and methods
Plant material
Castor (cv. Baker 296) plants were cultivated in a greenhouse at 24 °C and 70% humidity under natural light supplemented with 16 h of artificial light. Female flowers at approximately 5 d post-anthesis (corresponding to stage I or proembryo developing COS (Greenwood and Bewley, 1982)) were harvested, and their inner integument and pericarp (Fig. 1) rapidly dissected. Male flowers were harvested at maturity. Pods containing stage VII (full cotyledon) developing COS (Fig. 1) (Greenwood and Bewley, 1982) were harvested at midday, and cotyledon and endosperm were dissected. For depodding treatments, stems containing intact pods of stage VII developing COS were excised and placed in water in the dark for 72 h at 24 °C. For germination, castor seeds were surface-sterilized for 5 min in 1% (v/v) sodium hypochlorite and imbibed overnight with running water. Endosperm, cotyledon, roots, and hypocotyls of germinated seedlings were harvested following incubation of the imbibed seeds in moist vermiculite in darkness for 5 d at 30 °C and 80% humidity. Leaf buds and expanding and fully mature leaves (Fig. 1) were harvested at midday. All harvested tissues were snap frozen in liquid N2 and stored at –80 °C until used. Arabidopsis thaliana (cv. Columbia) seeds were germinated on sterile agar plates containing half-strength Murashige–Skoog medium and 1% (w/v) sucrose and the seedlings were harvested after 7 d.
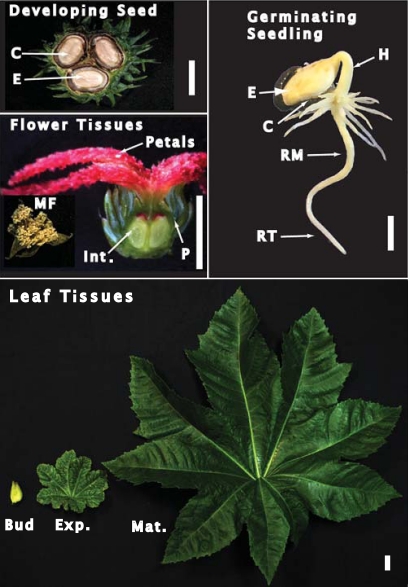
Fig. 1.
Images of the castor tissues analysed in this study. Developing seed tissues were harvested from stage VII (full cotyledon) COS, germinating seeding tissues were harvested at 5 d post-imbibition, male flowers were harvested at maturity, whereas female flowers were harvested at 5 d post-anthesis (corresponding to the proembryo or stage I COS). Cross-sections of a stage VII developing COS and the base of the female flower are depicted. The white scale bar is equivalent to 1 cm. Abbreviations are as follows: E, endosperm; C, cotyledon; MF, male flower; Int., integument; P, pericarp; H, hypocotyl; RM, root middle; RT, root tip; Bud, leaf bud; Exp., expanding leaf; Mat., mature leaf.
Extraction of total RNA and RT-PCR
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) with an additional on-column DNase digestion step to eliminate genomic DNA. cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s protocol. Castor actin and PEPC isoforms were amplified with gene-specific primer pairs as follows: RcPpc1 (forward 5′-CATCACGATCTCCACCCATCCA-3′, reverse 5′-GATTCAAGCTGCATTCCGCACA-3′); RcPpc3 (forward 5′-TCCCCGTAAACTTGATGAGC-3′, reverse 5′-TATAGGAAACCGATCCAAAAGCAAA-3′); RcPpc4 (forward 5′-TGCTGGAGAAGCAGCTGGCATTGG-3′, reverse 5′-ACCTCCTGCTTGCAAACTGTGTCA-3′); and RcActin (forward 5′-TTGCAGACCGTATGAGCAAG-3′, reverse 5′-TATAGGTCATACTCGCCCTTGGAAA-3′). Typical cycle parameters were 95 °C for 5 min followed by 28–32 cycles of 95 °C for 30 s, 50 °C for 30 s, and 68 °C for 1 min, followed by 68 °C for 5 min. All amplifications were limited to sub-saturating levels for each primer pair with actin as a reference. PCR products were resolved on 1.5% (w/v) agarose gels and visualized with ethidium bromide. All RT-PCR experiments were replicated a minimum of three times with representative results shown in the figures.
Preparation of clarified extracts, PEPC activity assays, and protein concentration determination
Quick-frozen tissues were extracted as previously described (Gennidakis et al., 2007) in 50 mM HEPES-KOH (pH 7.5) containing 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 1 mM , 1 mM Na2MoO4, 0.1% (v/v) Triton X-100, 20% (v/v) glycerol, 10 mM MgCl2, 5 mM thiourea, 1% (w/v) poly(vinylpolypyrrolidone) (Sigma-Aldrich, catalogue no. P-6755), 2 mM phenylmethylsulphonyl fluoride, 2 mM 2,2′-dipyridyl disulphide, 5 μl ml−1 ProteCEASE-100 (G-Biosciences), and 50 nM microcystin-LR. The leaf homogenization buffer also contained 4% (w/v) polyvinylpyrrolidone (BDH Chemicals, catalogue no. B29579) and an additional 1% (w/v) poly(vinylpolypyrrolidone). Homogenates were centrifuged at 4 °C and 15 000 g for 15 min, and the resulting clarified extracts rapidly prepared for non-denaturing and SDS-PAGE, and assayed for PEPC activity and total protein. To increase PEPC concentration prior to non-denaturing PAGE, clarified extracts were prepared as described above from 5 g each of the inner integument and germinating roots and subjected to 15–50% (saturation) ammonium sulphate fractionation. Following centrifugation, the resulting pellets were dissolved in 1 ml of 100 mM KH2PO4 (pH 7.0) containing 1 mM EDTA, 2 mM MgCl2, 5 mM malate, 15% (v/v) glycerol, 25 mM NaF, and 5 μl ml−1 ProteCEASE-100.
Maximum PEPC activity was assayed at 25 °C by following NADH oxidation at 340 nm using a Molecular Devices Spectramax Kinetics Microplate reader and the following optimized assay conditions: 50 mM HEPES-KOH (pH 8.0) containing 5 mM PEP, 5 mM KHCO3, 5 mM MgCl2, 2 mM DTT, 0.15 mM NADH, 10% (v/v) glycerol, and 5 units ml−1 desalted porcine muscle malate dehydrogenase. One unit of activity is defined as the amount of PEPC resulting in the production of 1 μmol oxaloacetate min−1. All assays were linear with respect to time and concentration of enzyme assayed. Activity values are reported as the means ±SE of duplicate assays performed with a minimum of three biological replicates. Protein concentrations were determined by the Coomassie Blue G-250 dye-binding method using bovine γ-globulin as the protein standard (Blonde and Plaxton, 2003).
Partial PEPC purification from expanding castor leaves
Expanding castor leaves (50 g) (Fig. 1) were ground to a powder under liquid N2 and homogenized (1:2 w/v) as described above. The extract was centrifuged for 15 min at 17 500 g and the supernatant filtered through two layers of Miracloth. (NH4)2SO4 (20% saturation) was added and the sample was centrifuged for 15 min at 30 000 g. The supernatant was loaded at 2 ml min−1 on to a column (1.6×5 cm) of Butyl-Sepharose Fast Flow (GE Biosciences) equilibrated with 50 mM HEPES-KOH (pH 8.0), 5 mM MgCl2, 20 mM NaF, 2 mM 2,2′-dipyridyl disulphide, and 20% (saturation) (NH4)2SO4. After the A280 approached baseline, PEPC was eluted with the same buffer containing 5% (saturation) (NH4)2SO4. Pooled peak fractions were concentrated to 1 ml with an Amicon Ultra-15 centrifugal filter unit (100 kDa cut-off), frozen in liquid N2, and stored at –80 °C.
Electrophoresis and immunoblotting
Non-denaturing and SDS-PAGE using a Bio-Rad Protean III mini-gel apparatus, in-gel PEPC-activity staining, and immunoblotting were as described previously (Blonde and Plaxton, 2003). Antigenic polypeptides were visualized using an alkaline phosphatase-conjugated secondary antibody and chromogenic detection (Blonde and Plaxton, 2003). All gel and immunoblot results were replicated a minimum of three times with representative results shown in the figures. Preparation of COS PEPC antibodies were all described elsewhere: anti-RcPPC3-IgG (anti-PTPC) and its corresponding anti-(phospho-Ser-11 specific)-IgG (anti-pSer11) (Tripodi et al., 2005), anti-RcPPC4-IgG (anti-BTPC) (O’Leary et al., 2009) and its corresponding anti-(phospho-Ser-425 specific)-IgG (anti-pSer425) (O’Leary et al., 2011b). The anti-pSer11 and anti-pSer425 immunoblots were probed with antibodies raised against synthetic phosphopeptides that matched the in vivo phosphorylation sites of COS PTPC and BTPC, respectively. The corresponding dephosphopeptide was used to block any non-specific antibodies raised against that non-phosphorylated sequence. Anti-ubiquitin-IgG (anti-ubiquitin) was purchased from Millipore Canada (catalogue no. 05-944).
PEPC deubiquitination, dephosphorylation, and co-immunopurification
Deubiquitination was performed as previously described (Uhrig et al., 2008b). Desalted clarified extracts containing 50 μg of protein were incubated in the presence and absence of 5 μM of recombinant human ubiquitin specific protease 2 catalytic domain (USP2c) (Progenra Inc.) for 1 h at 37 °C. Phosphatase treatment of clarified extracts was performed as previously described (O’Leary et al., 2009) in 50 μl reactions containing 300 μg of protein and 400 units of λ-phosphatase (New England Biolabs). Clarified extracts from the endosperm and cotyledon of stage VII developing COS, and the inner integument of stage I developing COS were also subjected to immunoaffinity chromatography using anti-PTPC and anti-BTPC co-immunopurification (co-IP) columns as previously described (Uhrig et al., 2008a; O’Leary et al., 2009).
Results
Semi-quantitative RT-PCR analysis of castor PTPC and BTPC transcripts
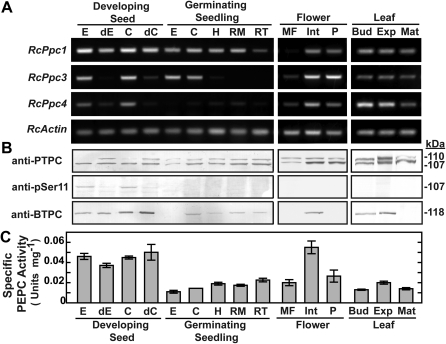
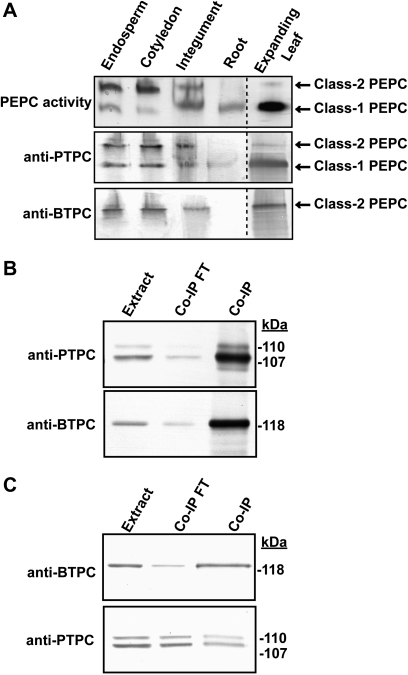
Interrogation of the castor genome database identified three PEPC genes: RcPpc1 and RcPpc3 encode PTPCs, whereas RcPpc4 encodes a BTPC. Their full-length cDNAs were previously isolated and sequenced from a developing COS endosperm cDNA expression library (Gennidakis et al., 2007). RcPpc1 and RcPpc3 are predicted to encode 110.6 kDa PTPC polypeptides sharing high (>90%) amino acid sequence identity, whereas RcPpc4 encodes a 118.5 kDa BTPC polypeptide exhibiting low (41%) identity with either PTPC (see Supplementary Fig. S1 at JXB online). The expression of this PEPC gene family in several castor tissues (Fig. 1) was assessed using semi-quantitative RT-PCR with gene-specific primers (Fig. 2A). Although RcPpc1 and RcPpc3 transcripts were widely distributed, the expression of RcPpc3 appeared to be more variable. In general, the expression patterns of RcPpc3 and the BTPC-encoding RcPpc4 were well correlated, with both being highly expressed in the endosperm and cotyledon of developing COS, and expanding leaves. Both transcripts also showed a pronounced down-regulation in endosperm and cotyledon following the depodding of developing COS. A major difference between the expression of these two genes was the absence of RcPpc4 transcripts in germinating COS endosperm and the absence of RcPpc3 transcripts in the hypocotyls and roots of germinating COS (Fig. 2A). In order to assess the degree to which RcPpc1, RcPpc3, and RcPpc4 transcripts correlate with PTPC or BTPC polypeptide levels, clarified protein extracts from the various tissues were immunoblotted using anti-PTPC and anti-BTPC. Notably, the down-regulation of RcPpc3 and RcPpc4 transcripts caused by COS depodding was not paralleled by down-regulation of immunoreactive BTPC or PTPC polypeptides (Fig. 2B). Maximal PEPC activity occurred in clarified extracts from endosperm and cotyledon of stage VII (full cotyledon) developing COS, as well as the inner integument of stage I (proembryo) female flowers (Fig. 2C).
Fig. 2.
Tissue-specific distribution of castor PTPC and BTPC transcripts and polypeptides and PEPC specific activity. (A) Semi-quantitative RT-PCR analysis of RcPpc1, RcPpc3, and RcPpc4 gene expression in castor. PEPC transcript levels were measured in various tissues using primers specific for the three castor PEPC genes. Starting template levels were adjusted to normalize RcActin band intensity, which served as a control for equal template loading. Primers pairs for RcPpc1, RcPpc3, RcPpc4, and RcActin yielded fragments of the expected size. (B) Clarified extracts were subjected to SDS-PAGE and immunoblotting. For anti-PTPC, anti-pSer11, and anti-BTPC immunoblots, lanes with developing COS extracts contained 5, 25 or 10 μg of protein, respectively, whereas all other lanes contained 10, 50 or 40 μg of protein, respectively. (C) Specific PEPC activities represent the means ±SE of duplicate determinations on a minimum of three biological replicates. Abbreviations are as described in the legend to Fig. 1 in addition to the following: dC and dE, cotyledon and endosperm, respectively, from stage VII developing COS that had been depodded for 72 h.
Tissue-specific distribution and post-translational modifications of castor PTPC polypeptides
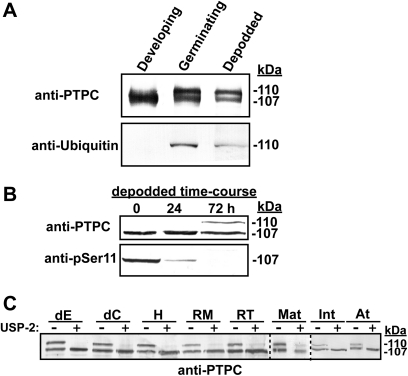
An immunoreactive 107 kDa PTPC polypeptide (p107) was detected on immunoblots of each castor extract that was surveyed, although its relative abundance and PTMs varied (Fig. 2B). Parallel immunoblots probed with anti-pSer11 corroborated previous reports (Tripodi et al., 2005; Murmu and Plaxton, 2007) that in vivo p107 phosphorylation at Ser-11 occurs in developing COS endosperm. Although p107 phosphorylation was also apparent in the cotyledon of developing COS, this PTM was not evident in any other tissue (Fig. 2B). Rather, the majority of surveyed tissues also contained a comparable amount of an immunoreactive 110 kDa PTPC polypeptide (p110). Notably, p110 was absent only in the developing COS endosperm and cotyledon (where p107 phosphorylation at Ser-11 occurred), but appeared following 72 h of depodding treatment of these tissues (Figs 2B, 3B). That p110 arises from the in vivo monoubiquitination of p107 was suggested by the fact that COS endosperm germination is accompanied by monoubiquitination of 50% of p107 (RcPPC3) subunits at its conserved Lys-628 residue (see Supplementary Fig. S1 at JXB online) leading to the formation of a p110:p107 heterotetrameric Class-1 PEPC (Uhrig et al., 2008b). Anti-ubiquitin immunoblots of immunopurified PTPC from depodded developing COS endosperm confirmed that p110, but not p107, was monoubiquitinated (Fig. 3A). Our previous co-IP proteomics study of the COS PEPC interactome demonstrated that the depodding-induced p110 is encoded by RcPpc3, the same PTPC gene that encodes the phosphorylated p107 of non-depodded developing COS endosperm (Uhrig et al., 2008a). A COS depodding time-course revealed that the in vivo dephosphorylation of p107 at Ser-11 preceded the appearance of monoubiquitinated p110 subunits in the endosperm (Fig. 3B). The detection of an anti-PTPC immunoreactive p110:p107 doublet in the majority of non-COS tissues that were surveyed (Fig. 2B) indicated that their p110 PTPC subunit may also be monoubiquitinated. This was verified by immunoblotting clarified extracts that had been preincubated with USP2c, which effectively deubiquitinates a large number of substrates in vitro (Lin et al., 2001). Uhrig et al. (2008b) used USP2c to cleave ubiquitin from the native Class-1 PEPC purified from 3-d-old germinating COS endosperm, resulting in the in vitro conversion of the p110:p107 heterotetramer into a p107 homotetramer. Similarly, the immunoreactive p110 band disappeared from anti-PTPC immunoblots of each castor tissue extract that had been preincubated with 5 μM USP2c at 37 °C for 1 h (Fig. 3C). This experiment was extended to include PEPC from Arabidopsis seedlings where an anti-PTPC immunoreactive p110:p107 doublet was also observed. Preincubation of a desalted Arabidopsis extract with USP2c also resulted in the disappearance of the immunoreactive p110 (Fig. 3C), confirming the existence of monoubiquitinated PTPC in Arabidopsis.
Fig. 3.
Monoubiquitination is the most prevalent PTM of castor PTPC. (A) Immunopurified PTPC from developing and 72 h depodded COS endosperm (stage VII), and 6 d germinating endosperm were immunoblotted with anti-PTPC (0.5 μg protein per lane) and anti-ubiquitin (5 μg protein per lane). (B) Immunopurified PTPC from an endosperm depodding time-course (stage VII COS) was immunoblotted with anti-PTPC (0.5 μg protein per lane) and anti-pSer11 (5 μg protein per lane). (C) Clarified, desalted protein extracts from various castor tissues, as well as 7-d-old Arabidopsis seedlings were incubated ±USP2c for 1 h, followed by SDS-PAGE and immunoblotting with anti-PTPC (10 μg protein per lane). Abbreviations are as described in the legends to Figs 1 and 2, in addition to the following: At, 7-d-old Arabidopsis seedlings.
Tissue-specific distribution and post-translational modification of castor BTPC polypeptides
Immunodetection of the p118-RcPPC4 using anti-BTPC has thus far been restricted to the endosperm and cotyledon of developing COS (Gennidakis et al., 2007; O’Leary et al., 2011b). However, the presence of RcPpc4 transcripts in several other castor tissues (Fig. 2A) was correlated with the appearance of immunoreactive p118-BTPC on anti-BTPC immunoblots (Fig. 2B). Maximal levels of immunoreactive p118 occurred in the endosperm and cotyledon of developing COS, followed by the inner integument of female flowers. The p118-BTPC was also present during leaf development, but was absent from fully mature leaves (Fig. 2B). A comparatively low level of p118-BTPC was detected on immunoblots of root and cotyledon extracts from 5-d-old germinating seedlings, but not on immunoblots of extracts prepared from germinating endosperm, male flower (pollen), or female flower pericarp.
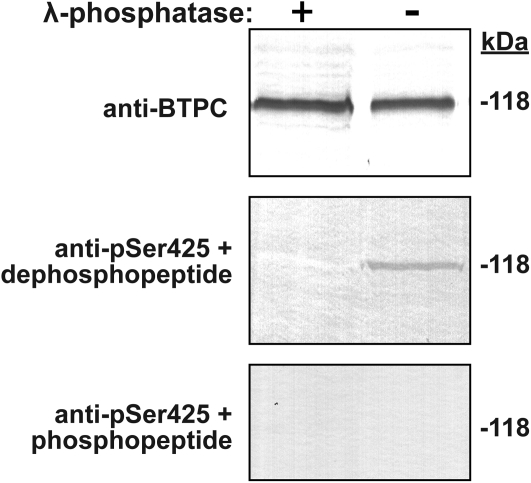
Immunoblots probed with anti-pSer425 recently confirmed that BTPC from developing COS endosperm and cotyledons is subject to in vivo proline-directed phosphorylation at Ser-425 (O’Leary et al., 2011b). It is interesting that the p118-BTPC of the inner integument of the stage I (proembryo) female flowers also appears to be phosphorylated at Ser-425 since it cross-reacted with the anti-pSer425 (Fig. 4). This cross-reaction was eliminated when an integument immunoblot was probed with anti-pSer425 in the presence of 10 μg ml−1 of the corresponding blocking phosphopeptide or when the integument extract was preincubated with exogenous λ-phosphatase (Fig. 4). By contrast, parallel immunoblotting with anti-pSer425 was unable to detect p118 phosphorylation at Ser-425 in other BTPC-containing tissues; for example, expanding leaves and leaf buds, as well as the roots, hypocotyls, and cotyledons of germinated COS (see Supplementary Fig. S2 at JXB online).
Fig. 4.
BTPC of COS integument is phosphorylated at Ser-425. Clarified integument extracts were incubated in the presence (+) and absence (–) of λ-phosphatase prior to SDS-PAGE and immunoblotting with anti-BTPC or anti-pSer425 with 10 μg ml−1 of the corresponding dephospho- or phospho-blocking peptide (50 μg protein per lane).
Non-denaturing PAGE and co-immunopurification analysis of BTPC:PTPC interactions
The p118-BTPC and p107-PTPC from developing COS endosperm, as well as lily pollen, tightly interact to form a unique 910 kDa hetero-octameric Class-2 PEPC (Blonde and Plaxton, 2003; Gennidakis et al., 2007; Igawa et al., 2010). It was therefore of interest to determine whether this interaction occurs in other castor tissues that contain immunologically detectable BTPC and PTPC polypeptides. Clarified extracts of the developing cotyledon, plus ammonium sulphate-concentrated PEPC samples from the inner integument and germinating roots were subjected to non-denaturing PAGE followed by in-gel PEPC activity staining and parallel immunoblotting with the anti-PTPC and anti-BTPC (Fig. 5A). The developing cotyledon and integument samples yielded similar PEPC-activity banding patterns as the developing endosperm indicating that they also contain Class-1 and Class-2 PEPCs. The corresponding immunoblots displayed the expected cross-reaction, with PTPC polypeptides present in both Class-1 and Class-2 PEPCs, and BTPC subunits restricted to the Class-2 PEPC. The germinating root sample only displayed a Class-1 PEPC activity staining and anti-PTPC immunoreactive band following non-denaturing PAGE (Fig. 5A), although it is possible that a small amount of Class-2 PEPC was present but below the detection limit achieved here. The inner integument and root Class-1 PEPC bands migrated slightly slower during non-denaturing PAGE relative to the Class-1 PEPC from developing endosperm or cotyledon (Fig. 5A). This difference in molecular mass is probably attributable to the monoubiquitination of the PTPC subunits that occurs in the integument and root, but not in the developing endosperm or cotyledon (Fig. 2A). Non-denaturing PAGE of a clarified expanding leaf extract only revealed the presence of a Class-1 PEPC (results not shown). However, upon enrichment of the PEPC from this tissue by ammonium sulphate fractionation and hydrophobic interaction chromatography, a Class-2 PEPC activity staining band was observed (Fig. 5A). The corresponding non-denaturing PAGE anti-(PTPC and BTPC) immunoblots confirmed the presence of Class-2 PEPC in the expanding leaves. This appears to be the first evidence for the existence of a Class-2 PEPC complex in a photosynthetic tissue of vascular plants.
Fig. 5.
Non-denaturing PAGE and co-immunopurification of castor PEPC isozymes. (A) Non-denaturing PAGE followed by in-gel PEPC activity staining and immunoblotting was performed on clarified extracts from stage VII (full cotyledon) developing COS endosperm and cotyledon (5 mU per lane), alongside ammonium sulphate-concentrated extracts from the inner integument of stage I (proembryo) COS (10 mU per lane) and 5 d germinating roots (3 mU per lane). Non-denaturing PAGE was also performed on PEPC from expanding castor leaves which had been partially purified by ammonium sulphate fractionation and Butyl-Sepharose hydrophobic chromatography (15 mU PEPC for activity stain and anti-BTPC, 5 mU PEPC activity for anti-PTPC). (B, C) A clarified extract originating from 10 g of the inner integument was subjected to immunopurification using 2 ml anti-PTPC (B) or anti-BTPC (C) immunoaffinity columns. After washing non-absorbed proteins with Pi-buffered saline, the bound proteins were eluted using 100 mM glycine-HCl (pH 2.8), neutralized, and concentrated. The initial extract (Extract; 5 μg per lane), concentrated flow through fractions (Co-IP FT; 5 μg per lane), and concentrated eluates (Co-IP; 5 μg for anti-PTPC, 1 μg for anti-BTPC) were subjected to SDS-PAGE followed by immunoblotting with anti-PTPC and anti-BTPC.
The tight interaction of BTPC with PTPC in the integument tissue was corroborated by the co-IP of both p107-PTPC and p118-BTPC during chromatography of integument extracts on either anti-PTPC or anti-BTPC immunoaffinity columns (Fig. 5B, C). As the anti-BTPC column only bound about 15% of the total integument PEPC activity that was loaded on to the column, the proportion of Class-2 PEPC in the integument appears to be low compared with developing cotyledon or endosperm, in which Class-2 PEPC accounts for at least 50% of the total PEPC activity (Fig. 5A) (Blonde and Plaxton, 2003; O’Leary et al., 2009).
Discussion
To the best of our knowledge, the present study represents the first examination of the tissue-specific expression and PTMs of BTPC and PTPC isozymes in any plant. The physiology and underlying metabolism of the sampled castor tissues were highly diverse, and the multilevel analysis sought to correlate the occurrence and PTMs of castor PTPC and BTPC polypeptides to a specific metabolic status. Interrogation of the castor genome revealed three PEPC genes: RcPpc1 and RcPpc3 encode closely related PTPCs, whereas RcPpc4 encodes the distantly related BTPC isozyme (see Supplementary Fig. S1 at JXB online). Semi-quantitative RT-PCR analysis of transcriptional expression of these genes revealed two obvious patterns. First, RcPpc3 and RcPpc4 expression were variable between tissues compared with the more constant level of RcPpc1 expression (Fig. 2A). Secondly, the expression patterns of RcPpc3 and RcPpc4 were quite similar (with the exception of germinating endosperm where only RcPpc1 and RcPpc3 were expressed, and germinating roots where only RcPpc1 and RcPpc4 were expressed). This pattern of co-expression may be significant as RcPPC3 and RcPPC4 tightly interact in endosperm of developing COS to form the novel hetero-octameric Class-2 PEPC complex (Gennidakis et al., 2007; Uhrig et al., 2008a). Several additional lines of evidence support the hypothesis that native vascular plant and green algal BTPCs only exist in vivo in physical association with co-expressed PTPC subunits as a Class-2 PEPC enzyme complex that exhibits altered physical, kinetic, and regulatory properties (Rivoal et al., 1998, 2001; Mamedov et al., 2005; O’Leary et al., 2009; Igawa et al., 2010). Supporting this hypothesis, it was shown here that apart from the developing endosperm, BTPC also interacts with PTPC subunits in the cotyledon of developing COS, the inner integument of female flowers, and expanding leaves (Fig. 5). Mass spectrophotometric analyses of tryptic peptides as well as N-terminal microsequencing have consistently demonstrated that RcPPC4 and/or RcPPC3 are the subunits of native PEPC isoforms purified from developing or germinating COS endosperm, whereas no RcPPC1-PTPC polypeptides were ever detected in COS (Blonde and Plaxton, 2003; Gennidakis et al., 2007; Uhrig et al., 2008b). By contrast, it appears that the Class-1 PEPC expressed in roots is an example of a functional RcPPC1 enzyme (Fig. 5A), as suggested by the presence of RcPpc1 transcripts and the absence of RcPpc3 expression in this tissue (Fig. 2A).
The precise physiological role(s) of castor Class-2 PEPC complexes remains to be firmly established. However, their unusual kinetic/regulatory properties and developmental profiles have led to the hypothesis that the Class-2 PEPCs may have evolved to provide a ‘metabolic overflow’ mechanism that could maintain a significant flux of PEP to malate under physiological situations where the corresponding Class-1 PEPC isoform would be largely suppressed by feedback inhibitors such as L-malate and L-asparate (O’Leary et al., 2009). Further development of this hypothesis has been hampered because studies of Class-2 PEPC complexes and/or BTPC polypeptides in vascular plants have thus far been restricted to developing COS endosperm (Blonde and Plaxton, 2003; Tripodi et al., 2005; Uhrig et al., 2008a; O’Leary et al., 2009, 2011b), and, more recently, developing lily pollen (Igawa et al., 2010). Results of the current study revealed that the p118-BTPC is relatively abundant in the cotyledon and inner integument of developing COS, but also occurs at a low level in most of the other tissues that were analysed (Fig. 2B). Insights into the role of BTPC were also gained by its distinct developmental pattern in castor leaves where the expression of p118-BTPC polypeptides was restricted to early development (Fig 2B). It is notable that the only tissue in which RcPpc4 transcripts were undetectable is germinating COS endosperm (Fig. 2A; Gennidakis et al., 2007), a tissue whose carbon metabolism is dominated by the conversion of storage triacylglycerides into sucrose needed to fuel early seedling growth (Canvin and Beevers, 1961). In lily and Arabidopsis, BTPC expression is initiated during pollen development after the final mitosis, coinciding with the appearance of storage organelles (Igawa et al., 2010). It is tempting to speculate that increased BTPC and thus Class-2 PEPC expression in vascular plants is a characteristic feature of rapidly growing and/or highly biosynthetically active tissues requiring a large anaplerotic flux of PEP to malate and other TCA cycle intermediates being withdrawn for anabolism. Class-2 PEPCs might also function in the support of osmoregulation during rapid cellular expansion, consistent with the transcriptional up-regulation of the BTPC-encoding gene AtPpc4 during osmotic stress of Arabidopsis seedlings (Sánchez et al., 2006). In either case, the purpose of the BTPC subunits would be to facilitate the accumulation of a pool of organic acids which can either generate osmotic potential and/or act as a source of carbon skeletons and reducing power to support anabolism.
The phosphorylation of castor BTPC at Ser-425 occurs adjacent to a Pro residue within its intrinsically disordered region (see Supplementary Fig. S1 at JXB online) and causes regulatory inhibition of BTPC subunits within the Class-2 PEPC complex (O’Leary et al., 2011b). In vivo BTPC phosphorylation at this site has been previously documented in developing endosperm and cotyledons by mass spectrometry and/or immunoblotting using the anti-pSer425 (Uhrig et al., 2008a; O’Leary et al., 2011b). The stoichiometry of Ser-425 phosphorylation increases throughout COS endosperm development and in response to depodding, whereas the level of Ser-425 phosphorylation in the cotyledon appears to remain relatively constant (O’Leary et al., 2011b). BTPC from the inner integument of female castor flowers is also phosphorylated at this site in vivo (Fig. 4). It will be of interest to characterize the BTPC protein kinase and related signal transduction pathways that lead to in vivo phosphorylation of RcPPC4 at Ser-425 in various tissues of the developing COS.
The activation of Class-1 PEPCs due to the phosphorylation of their PTPC subunits at a conserved N-terminal seryl residue is well understood (Chollet et al., 1996; Nimmo, 2003; Izui et al., 2004). This regulatory phosphorylation is tightly linked to C4 and CAM photosynthesis, but has also been documented for a number of non-photosynthetic Class-1 PEPC isozymes (Duff and Chollet, 1995; Zhang et al., 1995; Osuna et al., 1996; Law and Plaxton, 1997; Nimmo, 2003, Tripodi et al., 2005, Gregory et al., 2009, O’Leary et al., 2011a). Our understanding of the post-translational control of plant PEPC was further complicated by the discovery that COS germination is accompanied by monoubiquitination of 50% of the endosperm’s p107-RcPPC3 subunits to form the p110:p107 heterotetrameric Class-1 PEPC, and that this PTM results in an increased Km(PEP) and enhanced sensitivity to allosteric effectors (Uhrig et al., 2008b). A subsequent study revealed that PTPC monoubiquitination also occurs in developing lily pollen, and that the monoubiquitinated lily PTPC appears to form a functional Class-2 PEPC complex with co-expressed BTPC polypeptides (Igawa et al., 2010). The current study demonstrated that all sampled castor tissues, other than developing endosperm or cotyledon, yielded an immunoreactive p110:p107 PTPC doublet that co-migrated with the monoubiquitinated Class-1 PEPC from germinated COS endosperm (Fig. 2B). Incubation of desalted extracts with USP2c followed by immunoblotting with anti-PTPC provided definitive proof that the immunoreactive p110 observed on immunoblots of the various extracts is a monoubiquitinated form of the corresponding p107 (Fig. 3C). As with lily pollen (Igawa et al., 2010), our examination of PEPC from expanding COS leaves and the inner integument of stage I developing COS suggested that monoubiquitinated PTPC subunits also tightly interact with co-expressed BTPC polypeptides. Conversely, phosphorylation of the p107-PTPC (RcPPC3) subunits at Ser-11 appears to be restricted to the endosperm and cotyledon of developing COS (Fig. 2B) (Tripodi et al., 2005). Furthermore, depodding of the developing COS caused the p107-PTPC subunits of the endosperm and cotyledon to become dephosphorylated (Tripodi et al., 2005), concomitant with the disappearance of PTPC-protein kinase activity (Murmu and Plaxton, 2007), and then subsequently monoubiquitinated (Figs 3A, B). Because depodding abolishes photosynthate delivery to COS, it is conceivable that the carbohydrate metabolism of the depodded developing endosperm is rearranged to mimic that of germinating COS.
An immunoreactive PTPC doublet reminiscent of monoubiquitination has been frequently observed on PEPC immunoblots of extracts from a wide variety of plant tissues, including hydrilla (Hydrilla verticillata) leaves (Rao et al., 2006), broad bean (Vicia faba) stomata (Denecke et al., 1993), cucumber (Cucumis sativus) roots (De Nisi and Zocchi, 2000), germinating seeds of wheat (Triticum aestivum), barley (Hordeum vulgare), and sorghum (Sorghum bicolor) (González et al., 1998, Osuna et al., 1999, Nhiri et al., 2000), ripening banana (Musa cavendishii) fruit (Law and Plaxton, 1995), and Arabidopsis roots , stems, siliques, flowers, and rosettes (Sánchez et al., 2006). Here, the p110 PTPC subunits extracted from Arabidopsis seedlings were shown to be monoubiquitinated because the immunoreactive p110 also disappeared following incubation of the extract with USP2c (Fig. 3C). High throughput proteomic screens have identified numerous ubiquitinated metabolic enzymes in Arabidopsis, including PEPC; however, it was not determined whether the various targets were poly- or monoubiquitinated (Igawa et al., 2009; Saracco et al., 2009). This discrimination is crucial for future studies of the plant ubiquitome since polyubiquitination targets many proteins for their proteolytic elimination by the 26S proteasome, whereas monoubiquitination is a non-destructive and reversible PTM. Monoubiquitination typically influences protein:protein interactions and subcellular localization to control diverse processes in eukaryotic cells including gene expression, DNA repair, endocytosis, and signal transduction pathways (Schnell and Hicke, 2003; Mukhopadhyay and Riezman, 2007). PTPC provides what appears to be the first well-documented example in nature of in vivo regulatory monoubiquitination of a metabolic enzyme. The collective findings of the current and recent (Uhrig et al., 2008b; Igawa et al., 2010) studies leads to the surprising conclusion that monoubiquitination is the customary PTM for most PTPCs. Moreover, the distinctive developmental patterns of PTPC phosphorylation versus monoubiquitination in the castor plant indicates that these two PTMs may be mutually exclusive. By contrast, it has been well documented that certain E3-ubiquitin ligase substrates must first be phosphorylated before an E3 ligase can ubiquitinate them (Schnell and Hicke, 2003). It will be of considerable interest to establish further the interplay between in vivo phosphorylation versus monoubiquitination of PTPCs. In vivo PTPC phosphorylation appears to be related to physiological conditions that necessitate a high flux through a Class-1 PEPC that can be readily controlled, as occurs during photosynthate partitioning to storage end-products in developing COS. In other species, PTPC phosphorylation-activation has been mainly described in tissues in which a high and tightly controlled flux of PEP to oxaloacetate has an obvious metabolic role; for example, as occurs during atmospheric CO2 assimilation in C4 and CAM leaves, rapid N-assimilation following -resupply to N-limited cells, provision of malate as a respiratory substrate for N2-fixing bacteroids of legume root nodules, or following nutritional Pi starvation (Zhang et al., 1995; Chollet et al., 1996; Nimmo, 2003; Sullivan et al., 2004; Fukayama et al., 2006; Chen et al., 2007; Gregory et al., 2009). By contrast, PTPC monoubiquitination at Lys-628 in germinating COS is inhibitory in nature as it causes a significant increase in this Class-1 PEPC's Km(PEP) and sensitivity to allosteric inhibitors (Uhrig et al., 2008b). It seems logical that phosphorylation and monoubiquitination would elicit opposing kinetic effects on Class-1 PEPCs since they appear to occur in a reciprocal fashion. However, as monoubiquitination has been generally implicated in mediating protein:protein interactions and influencing protein subcellular localization in eukaryotic cells (Schnell and Hicke, 2003), future research needs to characterize specific ubiquitin-binding domain proteins that might interact with the ubiquitin ‘docking site’ of monoubiquitinated PTPCs, as well as the possible influence of this PTM on the subcellular location of PTPCs. There is also an obvious need to document the signalling pathways and specific E3-ubiquitin ligase that lead to tissue-specific PTPC monoubiquitination in vascular plants, as well as how this PTM is co-ordinated with PTPC protein kinase-mediated phosphorylation. Could PTPC phosphorylation recruit a deubiquitinating enzyme that deubiquitinates and/or suppresses PTPC monoubiquitination?
In summary, vascular plant PEPCs are represented by a small family of isozymes that are becoming attractive targets for engineering desired modifications of C metabolism in various tissues. However, several attempts at metabolic engineering of PEPC have failed owing to a lack of consideration or knowledge of the post-translational controls of this important anaplerotic enzyme (Miyao and Fukayama, 2003). As documented for castor PEPC, these controls involve a complex suite of novel mechanisms that work hand-in-hand with the well-established regulation of PTPCs by allosteric effectors and reversible phosphorylation and include: (i) the tight interaction between unrelated PTPC and BTPC subunits leading to the formation of unique and allosterically desensitized Class-2 PEPC enzyme complexes, (ii) regulatory phosphorylation of Class-1 PEPC’s PTPC subunits at Ser-11 and Class-2 PEPC’s BTPC subunits at Ser-425, and (iii) regulatory monoubiquitination of Class-1 PEPC’s PTPC subunits at Lys-628. An understanding of the occurrence, function, and interaction between these post-translational controls in normal developmental and stressed conditions will be required to describe fully the roles and metabolic control of plant PEPC. A major challenge will be to link the various post-translational controls of Class-1 and Class-2 PEPCs with the in vivo regulation of PEP partitioning to specific anabolic pathways in vivo.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Sequence alignment of cDNA-deduced primary structures of castor PTPC (RcPPC1 and RcPPC3) and BTPC (RcPPC4) isozymes.
Supplementary Fig. S2. Clarified extracts from several BTPC-containing castor tissues were subjected to SDS-PAGE and immunoblotting with anti-pSer425 in the presence of 10 μg ml−1 of the corresponding dephospho-peptide.
Acknowledgments
This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Queen’s Research Chairs program (to WCP), NSERC post-graduate scholarships (to BO and ETF), and a Queen’s-Province of Ontario Post-doctoral Fellowship (to JP).
Glossary
Abbreviations
- co-IP
co-immunopurification
- COS
castor (Ricinus communis) oil seed
- p107
p110, and p118, 107, 110, and 118 kDa polypeptides, respectively
- PEP
phosphoenolpyruvate
- PEPC
PEP carboxylase
- PTM
post-translational modification
- PTPC and BTPC
plant- and bacterial-type PEP carboxylase, respectively
- RT
reverse transcription
- TCA
tricarboxylic acid
- USP2c
ubiquitin specific protease 2 catalytic domain
References
- Benedict CR, Beevers H. Formation of sucrose from malate in germinating castor beans.1. Conversion of malate to phosphoenol-pyruvate. Plant Physiology. 1961;36:540–544. doi: 10.1104/pp.36.5.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonde J, Plaxton WC. Structural and kinetic properties of high and low molecular mass phosphoenolpyruvate carboxylase isoforms from the endosperm of developing castor oilseeds. Journal of Biological Chemistry. 2003;278:11867–11873. doi: 10.1074/jbc.M211269200. [DOI] [PubMed] [Google Scholar]
- Canvin DT, Beevers H. Sucrose synthesis from acetate in germinating castor bean: kinetics and pathway. Journal of Biological Chemistry. 1961;236:988–995. [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, et al. Draft genome sequence of the oilseed species Ricinus communis. Nature Biotechnology. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Jenkins GI, Nimmo HG. pH and carbon supply control the expression of phosphoenolpyruvate carboxylase kinase genes in Arabidopsis thaliana. Plant, Cell and Environment. 2007;31:1844–1850. doi: 10.1111/j.1365-3040.2008.01885.x. [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O’Leary M. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- De Nisi P, Zocchi G. Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. Journal of Experimental Botany. 2000;51:1903–1909. doi: 10.1093/jexbot/51.352.1903. [DOI] [PubMed] [Google Scholar]
- Denecke M, Schulz M, Fischer C, Schnabl H. Partial purification and characterization of stomatal phosphoenolpyruvate carboxylase from Vicia faba. Physiologia Plantarum. 1993;87:96–102. [Google Scholar]
- Duff SMG, Chollet R. In vivo regulation of wheat-leaf phosphoenolpyruvate carboxylase by reversible phosphorylation. Plant Physiology. 1995;107:775–782. doi: 10.1104/pp.107.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Tamai T, Taniguchi Y, Sullivan S, Miyao M, Nimmo HG. Characterization and functional analysis of phosphoenolpyruvate carboxylase kinase genes in rice. The Plant Journal. 2006;47:258–268. doi: 10.1111/j.1365-313X.2006.02779.x. [DOI] [PubMed] [Google Scholar]
- Gennidakis S, Rao S, Greenham K, Uhrig RG, O’Leary B, Snedden WS, Lu C, Plaxton WC. Bacterial- and plant-type phosphoenol-pyruvate carboxylase polypeptides interact in the hetero-oligomeric Class-2 PEPC complex of developing castor oil seeds. The Plant Journal. 2007;52:839–849. doi: 10.1111/j.1365-313X.2007.03274.x. [DOI] [PubMed] [Google Scholar]
- González M, Osuna L, Echevarría C, Vidal J, Cejudo FJ. Expression and localization of phosphoenolpyruvate carboxylase in developing and germinating wheat grains. Plant Physiology. 1998;116:1249–1258. doi: 10.1104/pp.116.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JS, Bewley JD. Seed development in Ricinus communis (castor bean). I. Descriptive morphology. Canadian Journal of Botany. 1982;60:1751–1760. [Google Scholar]
- Gregory AL, Hurley BA, Tran HT, Valentine AJ, She Y, Knowles VL, Plaxton WC. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochemical Journal. 2009;420:57–65. doi: 10.1042/BJ20082397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Takahashi H, Sawasaki T, Endo Y, Seki M, Shinozaki K, Fukao Y, Yanagawa Y. Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. Journal of Experimental Botany. 2009;60:3067–3073. doi: 10.1093/jxb/erp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Tanaka I, Fukao Y, Yanagawa Y. Characterization of bacterial-type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biology. 2010;10:200. doi: 10.1186/1471-2229-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y. Phosphoenolpyruvate carboxylase: A new era of structural biology. Annual Review of Plant Biology. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- Law RD, Plaxton WC. Purification and characterization of a novel phosphoenolpyruvate carboxylase from banana fruit. Biochemical Journal. 1995;307:807–816. doi: 10.1042/bj3070807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RD, Plaxton WC. Regulatory phosphorylation of banana fruit phosphoenolpyruvate carboxylase by a copurifying phosphoenolpyruvate carboxylase-kinase. European Journal of Biochemistry. 1997;247:642–651. doi: 10.1111/j.1432-1033.1997.00642.x. [DOI] [PubMed] [Google Scholar]
- Lin H, Yin L, Reid J, Wilkinson KD, Wing SS. Divergent N-terminal sequences of a deubiquitinating enzyme modulate substrate specificity. Journal of Biological Chemistry. 2001;276:20357–20363. doi: 10.1074/jbc.M008761200. [DOI] [PubMed] [Google Scholar]
- Mamedov TG, Moellering ER, Chollet R. Identification and expression analysis of two inorganic C- and N-responsive genes encoding novel and distinct molecular forms of eukaryotic phosphoenolpyruvate carboxylase in the green microalga Chlamydomonas reinhardtii. The Plant Journal. 2005;42:832–843. doi: 10.1111/j.1365-313X.2005.02416.x. [DOI] [PubMed] [Google Scholar]
- Masumoto C, Miyazawa S, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proceedings of the National Academy of Sciences, USA. 2010;107:5226–5231. doi: 10.1073/pnas.0913127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao M, Fukayama H. Metabolic consequences of overproduction of phosphoenolpyruvate carboxylase in C3 plants. Archives of Biochemistry and Biophysics. 2003;414:197–203. doi: 10.1016/s0003-9861(03)00117-6. [DOI] [PubMed] [Google Scholar]
- Moellering ER, Ouyang Y, Mamedov TG, Chollet R. The two divergent PEP-carboxylase catalytic subunits in the green microalga Chlamydomonas reinhardtii respond reversibly to inorganic-N supply and co-exist in the high-molecular-mass, hetero-oligomeric class-2 PEPC complex. FEBS Letters. 2007;581:4871–4876. doi: 10.1016/j.febslet.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Murmu J, Plaxton WC. Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta. 2007;226:1299–1310. doi: 10.1007/s00425-007-0551-x. [DOI] [PubMed] [Google Scholar]
- Nhiri M, Bakrim N, Bakrim N, El Hachimi-Messouak Z, Echevarria C, Vidal J. Posttranslational regulation of phosphoenolpyruvate carboxylase during germination of Sorghum seeds: influence of NaCl and -malate. Plant Science. 2000;151:29–37. [Google Scholar]
- Nimmo H. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Archives of Biochemistry and Biophysics. 2003;414:189–196. doi: 10.1016/s0003-9861(03)00115-2. [DOI] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC. The remarkable diversity of plant phosphoenolpyruvate carboxylase (PEPC): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochemical Journal. 2011a;436:15–34. doi: 10.1042/BJ20110078. [DOI] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Kim J, Plaxton WC. Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel Class-2 PEPC complex of vascular plants. Journal of Biological Chemistry. 2009;284:24797–24805. doi: 10.1074/jbc.M109.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Plaxton WC. Phosphorylation of a bacterial-type phosphoenolpyruvate carboxylase at serine-425 provides a further tier of enzyme control in developing castor oil seeds. Biochemical Journal. 2011b;433:65–74. doi: 10.1042/BJ20101361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna L, Gonzalez MC, Cejudo FJ, Vidal J, Echevarria C. In vivo and in vitro phosphorylation of the phosphoenolpyruvate carboxylase from wheat seeds during germination. Plant Physiology. 1996;111:551–558. doi: 10.1104/pp.111.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna L, Pierre J, González M, Alvarez R, Cejudo FJ, Echevarría C, Vidal J. Evidence for a slow-turnover form of the Ca2+-independent phosphoenolpyruvate carboxylase kinase in the aleurone-endosperm tissue of germinating barley seeds. Plant Physiology. 1999;119:511–520. doi: 10.1104/pp.119.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Reiskind J, Bowes G. Light regulation of the photosynthetic phosphoenolpyruvate carboxylase (PEPC) in Hydrilla verticillata. Plant and Cell Physiology. 2006;47:1206–1216. doi: 10.1093/pcp/pcj091. [DOI] [PubMed] [Google Scholar]
- Rivoal J, Plaxton WC, Turpin D. Purification and characterization of high- and low-molecular-mass isoforms of phosphoenolpyruvate carboxylase from Chlamydomonas reinhardtii. Biochemical Journal. 1998;331:201–209. doi: 10.1042/bj3310201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoal J, Trzos S, Gage D, Plaxton WC, Turpin D. Two unrelated phosphoenolpyruvate carboxylase polypeptides physically interact in the high molecular mass isoforms of this enzyme in the unicellular green alga Selenastrum minutum. Journal of Biological Chemistry. 2001;276:12588–12601. doi: 10.1074/jbc.M010150200. [DOI] [PubMed] [Google Scholar]
- Sánchez R, Cejudo FJ. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiology. 2003;132:949–957. doi: 10.1104/pp.102.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Flores A, Cejudo FJ. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta. 2006;223:901–909. doi: 10.1007/s00425-005-0144-5. [DOI] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. The Plant Journal. 2009;59:344–358. doi: 10.1111/j.1365-313X.2009.03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. Journal of Biological Chemistry. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- Sullivan S, Jenkins GI, Nimmo HG. Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiology. 2004;135:2078–2087. doi: 10.1104/pp.104.042762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi K, Turner W, Gennidakis S, Plaxton WC. In vivo regulatory phosphorylation of novel phosphoenolpyruvate carboxylase isoforms in endosperm of developing castor oil seeds. Plant Physiology. 2005;139:969–978. doi: 10.1104/pp.105.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, O’Leary B, Spang HE, MacDonald JA, She Y, Plaxton WC. Coimmunopurification of phosphorylated bacterial- and plant-type phosphoenolpyruvate carboxylases with the plastidial pyruvate dehydrogenase complex from developing castor oil seeds. Plant Physiology. 2008a;146:1346–1357. doi: 10.1104/pp.107.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, She Y, Leach CA, Plaxton WW. Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. Journal of Biological Chemistry. 2008b;283:29650–29657. doi: 10.1074/jbc.M806102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Li B, Chollet R. In vivo regulatory phosphorylation of soybean nodule phosphoenolpyruvate carboxylase. Plant Physiology. 1995;108:1561–1568. doi: 10.1104/pp.108.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.