Abstract
The EID1-like protein 3 (EDL3) shows high similarity to EID1 (Empfindlicher im dunkelroten Licht 1), an F-box protein that functions as a negative regulator in the signalling cascade downstream of the phytochrome A photoreceptor in Arabidopsis thaliana. Analyses revealed a strong and rapid induction of EDL3 gene expression under osmotic stress, high salinity, and upon abscisic acid (ABA) application. Therefore, it was speculated that EDL3 is involved in the regulation of responses controlled by this plant hormone, which not only regulates many aspects of plant development but also integrates responses towards temperature, drought, osmotic, and salt stresses. Physiological data obtained with over-expresser lines and a conditional knock-down mutant demonstrated that EDL3 functions as a positive regulator in ABA-dependent signalling cascades that control seed germination, root growth, greening of etiolated seedlings, and transition to flowering. Results further demonstrate that EDL3 regulates anthocyanin accumulation under drought stress. The observed effects on physiological responses fit to tissue-specific expression patterns obtained with EDL3-promoter:GUS lines. Bimolecular Fluorescence Complementation assays and yeast two-hybrid analyses showed that EDL3 carries a functional F-box domain. Thus, the protein is presumed to act as a component of a ubiquitin ligase complex that specifically directs negatively acting factors in ABA signalling to degradation via the proteasome.
Keywords: ABA, anthocyanin, Arabidopsis, de-etiolation, EDL3, F-box protein, flowering, germination induction, root growth, water stress
Introduction
The plant hormone abscisic acid (ABA) plays an important role in numerous aspects of plant growth and development, including embryo maturation, seed dormancy, post-germinative growth, and leaf senescence. ABA also integrates and regulates plant response towards drought, cold, salt, and osmotic stresses. The plant hormone functions through a complex network of signalling pathways that starts from different receptors and includes protein kinases, phosphatases, RNA modifying proteins, and transcription factors (Finkelstein et al., 2002; Christmann et al., 2006; Pandey et al., 2009; Raghavendra et al., 2010; Umezawa et al., 2010; Hubbard et al., 2010). These transcription factors are most likely responsible for the regulation of transcriptional cascades that trigger the complex expression pattern of genes responding to ABA and osmotic, salt, or drought stresses (Shinozaki et al., 2003; Nakashima et al., 2006).
Signalling includes not only transcriptional and translational control, but also control at the protein level. Eukaryotes use ubiquitin as a polypeptide marker to target proteins for degradation through the 26S proteasome. Polyubiquitination is catalysed by E3 proteins that specifically interact with activated E2-ubiquitin conjugates and with target proteins. Higher eukaryotes have evolved with several classes of E3 proteins. Among these, the SCF protein complexes (for SKP1–CULLIN1–F-box protein) are essential to the post-translational regulation of many important factors involved in signal transduction (Craig and Tyers, 1999; Deshaies, 1999; Weissman, 2001; Lechner et al., 2006). CULLIN1 together with the RBX (RING box) proteins are essential for the interaction with E2-ubiquitin conjugates. CULLIN1 also interacts with SKP1 homologous proteins (for S-phase kinase associated protein 1), which link the so-called F-box domain proteins to the core complex. F-box proteins are compositionally complex and varied in their structural composition outside the F-box domain. They are present in high numbers within the genomes in all the eukaryotic organisms analysed and are thought to be responsible for the specific interaction between SCF E3 ligase complexes and target proteins (Craig and Tyers, 1999; Deshaies, 1999; Weissman, 2001; Lechner et al., 2006). Several F-box proteins have been characterized that play important roles during all phases of plant development and in diverse signalling cascades, including that of the plant hormones ethylene, gibberellic acid, jasmonate, and auxin (Lechner et al., 2006; Mockaitis and Estelle, 2008; McClellan and Chang, 2008; Fonseca et al., 2009; Schwechheimer and Willige, 2009).
EID1 is an F-box protein that functions as a negative regulator in the signalling cascade downstream of the phytochrome A photoreceptor (Buche et al., 2000; Dieterle et al., 2001; Zhou et al., 2002; Marrocco et al., 2006). The Arabidopsis genome contains three genes with similarities to EID1 that have been named EDL1 (At5g15440), EDL2 (At5g39360), and EDL3 (At3g63070) for EID1-like protein 1 to 3 (Marrocco et al., 2006). Aside from the putative F-box domain, the EDL proteins carry a conserved protein motif that might serve as a novel protein–protein interaction domain. Arabidopsis EDL3 shows a very high similarity to ZGT (zhong guang tiaokong=clock and light controlled) from tobacco, which is thought to function as a coupling agent between the central circadian oscillator and rhythmic expression of Chlorophyll-A/B-Binding Proteins (Xu and Johnson, 2001). Our results about the functional characterization of EDL3 are presented here. Interaction studies with Arabidopsis Skp1-like proteins (ASKs) and physiological experiments strongly indicate that ELD3 is a component of an SCF ubiquitin ligase complex that plays an important role in ABA signalling and in the regulation of floral transition in Arabidopsis.
Materials and methods
RNA isolation and RNA blot analysis
Seeds were sown on four layers of filter paper circles soaked with 4.5 ml distilled water and were incubated at 6 °C in darkness for 2 d. Seedlings were grown on moist filter paper under light/dark cycles consisting of 12 h white light and 12 h darkness (Sanyo MLR-351 growth chambers; Sanyo Electric, Osaka, Japan). After 4 d of growth at 23 °C, the upper layer of filter paper was transferred to new Petri dishes containing three layers of filter paper supplemented with 3.5 ml distilled water or solutions with 5 μM ABA (Sigma, Munich, Germany), 450 mM mannitol, or 150 mM NaCl. Samples were supplemented with 1 ml of the appropriate solutions that were equally dispersed on the filter paper. Seedlings were removed from the filter paper with a scalpel at the indicated time points, collected into two 1.5 ml tubes filled with seven glass beads of 1.7–2 mm (Roth, Karlsruhe, Germany), and immediately frozen in liquid nitrogen. If necessary, samples were stored at –70 °C. To grind the material, tubes were frozen in liquid nitrogen and shaken twice for 10 s each in a Silamat S5 shaker (Ivoclar Vivadent, Ellwangen, Germany). Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Ground material was extracted using Buffer RLT (RNeasy lysis buffer with guanidinium isothiocyanate) and samples were treated with DNaseI according to the manufacturer's specifications (Qiagen). RNA blot experiments were done as described by Zhou et al. (2002).
Quantitative Real-Time Polymerase Chain Reaction
Superscript III Reverse Transcriptase was used with a dT20-Oligomer and 500 ng of total RNA for cDNA synthesis according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany). Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) was carried out in 96-well format using a 7300 Real-Time PCR System and TaqMan probes (Applied Biosystems, USA). PCR reactions were performed using the ABsolute QPCR Rox Mix Kit following the manufacturer's instructions (ThermoScientific, Hamburg, Germany). TaqMan probes and primer pairs for each marker gene (see Supplementary Table S1 at JXB online) were designed using Primer Express Version 3.0 (Applied Biosystems). qGene software (http://www.gene-quantification.info) was used to calculate optimized standard curves and optimal CT-values from raw data for the different samples. CT-values from individual marker genes were further normalized to CT-values obtained for the constitutively expressed ACTIN1 transcripts, which served as an endogenous control for the efficiency of qRT-PCR. All the results presented were based on three biological replicates that were measured in triplicate. For semi-quantitative reverse-transcription PCR, cycle numbers were adapted to the amount of transcript present in the samples and agarose gels were stained with SYBR green (Invitrogen). The gels shown are representative of two experiments with independent RNA isolations.
Cloning of EDL3 constructs and plant transformation
A full-length EDL3 cDNA clone (RAFL08-11-G23; RIKEN BioResource Center, Japan; (Seki et al., 2002) was used as a template to create modified versions of the coding sequences with and without a stop codon in the pENTR1A Gateway vector (Invitrogen). Mutations were introduced by Polymerase Chain Reaction (PCR) using the primers listed in Supplementary Table S1 at JXB online. The modified fragment with stop codon was cloned behind a 35S promoter in a modified pPCVB T-DNA vector (Dieterle et al., 2001) to produce Arabidopsis over-expresser lines and into the pGBKT7 vector (Invitrogen) to perform yeast two-hybrid interaction assays. The modified fragment without stop codon was cloned into plasmids for Bimolecular Fluorescence Complementation assays (Stolpe et al., 2005). A 1141 bp fragment of the EDL3 promoter was amplified from genomic DNA by PCR (see Supplementary Table S1 at JXB online) and cloned into the AvrII/BamHI sites of pLITMUS28 (New England Biolabs, Ipswich, USA) for sequencing. The promoter fragment was transferred into the KpnI/EcoRV sites of the pENTR1A Gateway vector (Invitrogen) and, finally, introduced in front of a GUS:GFP reporter cassette in the pBGWFS7.0 plant transformation vector (Karimi et al., 2002). Arabidopsis transformation was done using the floral dip method (Clough and Bent, 1998). Plants exhibiting a 3:1 segregation ratio for the Basta selection marker were used to isolate homozygous lines.
Isolation of the T-DNA line
T-DNA insertion sites in the SALK_033705 T-DNA line (Alonso et al., 2003) were amplified by PCR using oligonucleotide combinations ELP3_Salk_5'/ROK2_RB1 for the right and ELD3_Salk_3'/LBb1 for the left border sequences (see Supplementary Table S1 at JXB online). Amplified fragments were cloned into pTOPO-TA vectors (Invitrogen) for sequencing. The mutant line was backcrossed twice with the Col wild type prior to phenotypic analysis.
Determination of GUS reporter gene activity
GUS assays were performed as described by Kretsch et al. (1995). Four-day-old seedlings were transferred to new filter papers supplemented with ABA, mannitol, NaCl, or water as described above (see RNA isolation and RNA blot analyses) and harvested after 6 h treatments. Tissue-specific GUS expression during Arabidopsis development was analysed in plants grown on soil under 16/8 h light/dark cycles. To analyse the effect of ABA and drought stress on reporter gene expression in rosette leaves, plants were grown on soil in a phytochamber under 12/12 h light/dark cycles for 3 weeks before they were either watered with 5 ml distilled water or 5 ml of 25 μM ABA at the middle of the light phase for 1 week. To apply drought stress, samples were kept dry until leaves started to wilt (∼1 week). After the removal of chlorophyll, the plant material was spread on water agar and photographed together with a size standard using a Zeiss Stemi SV6 binocular microscope supplemented with a Zeiss AxioCam MRc 5 digital camera (Zeiss, Oberkochen, Germany).
Determination of germination rates
To determine germination rates, seeds were sown on four layers of filter paper (80 mm diameter) soaked with 4.5 ml distilled water or distilled water supplemented with various concentrations of ABA and were kept at 6 °C in darkness for 2 d. Germination was induced with 8 h of red light and samples were transferred back to darkness at 25 °C for 4 d. The number of germinated and non-germinated seeds was counted under a magnifying glass. Seeds were regarded as being germinated when the radicle ruptured the testa and endosperm.
Determination of root length
For root growth measurements, seeds were sown on four layers of filter paper (80 mm diameter) soaked with 4.5 ml distilled water and were incubated at 6 °C in darkness for 2 d. Seedlings were grown on moist filter paper under continuous white light at 23 °C for 2 d, at which time the upper layer of filter paper was transferred to new Petri dishes containing three layers of filter paper supplemented with 3.5 ml of distilled water or solutions with various concentrations of ABA. Samples were then supplemented with 1 ml of water or ABA solutions that were equally dispersed on top of the seedlings. After two additional days, seedlings were spread on water agar and photographed together with a size standard using a Zeiss Stemi SV6 binocular microscope supplemented with a Zeiss AxioCam MRc 5 digital camera (Zeiss). Root lengths were measured using ImageJ software (rsb.info.nih.gov/ij/) as described by Kretsch (2010). Relative root length was calculated according to the water control.
Determination of chlorophyll content
Sowing and stratification were done on four layers of filter papers as described above. Germination was induced by 4 h of red light (39 μmol m−2 s−1). To analyse the influence of ABA on chlorophyll accumulation, seedlings were grown in darkness for 3 d. The upper layer of filter paper was then transferred to new Petri dishes containing water or various concentrations of ABA under green safe light and samples were kept in the dark for one additional day. Seedlings were exposed to continuous red light for 6 h to enable chlorophyll accumulation. Chlorophyll was extracted with dimethylformamide at 6 °C in darkness overnight. Chlorophyll was determined spectroscopically, and chlorophyll content was calculated with respect to fresh weight (Kretsch, 2010). Data represent the mean of at least six independent replicates.
Determination of anthocyanin accumulation
For experiments under conditions of water limitation, seeds were sown on a mixture of potting soil and vermiculite (2:1 v/v) in baskets of the ARASYSTEM (BETATECH, www.arasystem.com) and transferred to darkness at 6 °C for 3 d to complete stratification. Plants were grown at 23 °C in a 12/12 h light/dark regime in a phytochamber and were kept humid for 12 d before the cessation of watering. Baskets were removed from their plastic containment at day 14 to increase water evaporation. To reach water limitation under standardized conditions, samples were watered only once a day with 5 ml of water either at the beginning or end of the light phase. Control plants received one dose of water at dusk and dawn. The two youngest rosette leaves from each plant were harvested together at the onset of flowering, frozen in liquid nitrogen, and kept at –70 °C until anthocyanin extraction. Anthocyanin content was determined spectroscopically (Kretsch, 2010).
Determination of flowering times
To determine flowering times, seeds were sown on a mixture of potting soil and vermiculite (3:1 v/v) in baskets of the ARASYSTEM (BETATECH) and transferred to 6 °C for 3 d to complete stratification. Plants were grown at 23 °C in a phytochamber under different light/dark cycles. To analyse the influence of ABA on flowering induction, plants were grown under a 12/12 h light/dark cycle for 2 weeks before they were watered with either 5 ml of distilled water or 5 ml of distilled water supplemented with 25 μM ABA at the middle of the light phase for one week. After treatment, plants were kept humid until the onset of flowering. Flowering time is presented as the sum of rosette leaves and cauline leaves at the main stem.
Protein–protein interaction assays
Yeast two-hybrid interaction and BiFC assays were done as described elsewhere (Stolpe et al., 2005; Marrocco et al., 2006).
Statistical analysis
One way ANOVA was done by the Holm–Sidak method for samples drawn from normally distributed populations with the same standard deviations or by the Kruskal–Wallis test on ranks for non-normal populations using the SIGMASTAT 3.1 statistical tool (Sigma, Munich, Germany).
Results
EDL3 transcript accumulation induced by osmotic stress, high salinity, and upon ABA application
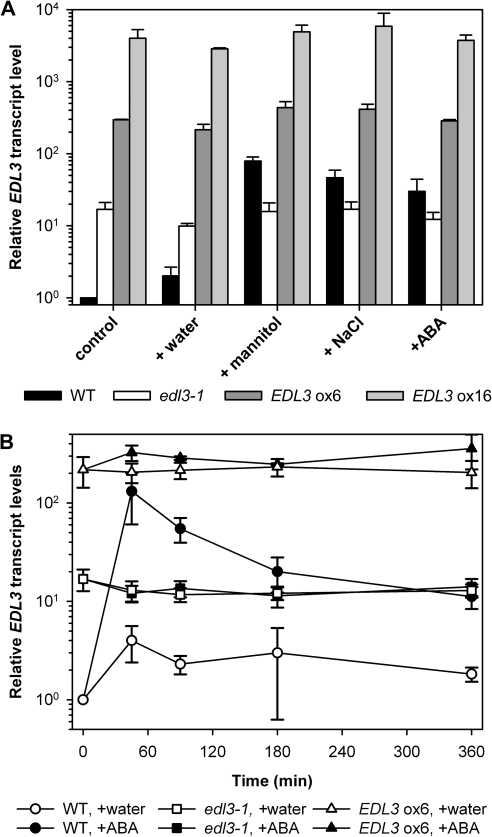
Analysis of publically available databases revealed a strong and rapid induction of EDL3 transcript accumulation under osmotic stress, high salinity, and upon ABA application (see Supplementary Fig. S1 at JXB online). These results were verified by qRT-PCR experiments using RNA samples from 4-d-old seedlings grown under continuous white light with either water or treated with 5 μM ABA, 450 mM mannitol, or 150 mM NaCl. These treatments induced a ∼30–80-fold increase in EDL3 transcript levels compared with the untreated water controls at 2 h (Fig. 1A). The kinetics of ABA-induced EDL3 transcript accumulation was analysed in greater detail using 4-d-old seedlings. EDL3 seems to sensitively respond to even weak stresses, as the transfer of seedlings to a new plate caused a ∼4-fold increase in transcript levels at 90 min (Fig. 1B). Wild-type (Col-0) plants exhibited a strong, rapid, and transient increase in EDL3 transcripts upon ABA treatment, with maximum levels (∼130-fold induction) reached after 45 min (Fig. 1B).
Fig. 1.
Osmotic stress, high salinity, and ABA induce EDL3 expression. EDL3 transcript levels were determined by quantitative RT-PCR. Results of experiments were first normalized according to the constitutively expressed ACTIN1 (AT2G37620) gene. Normalized expression values were then used to calculate fold induction with respect to untreated wild-type samples for which values were set to 1. Data represent the mean of three independent biological replicates ±SE. (A) Expression profiles of EDL3 in response to osmotic stress, high salinity, and ABA application. Seedlings were grown on moist filter paper under continuous white light for 4 d and then transferred to distilled water (control), 5 μM ABA, 450 mM mannitol, or 150 mM NaCl for 2 h before harvesting. (B) Temporal pattern of EDL3 transcript accumulation upon ABA treatment. After stratification seedlings were grown under a 12/12 h light/dark cycle for 4 d. Plants were transferred to new plates containing distilled water or 5 μM ABA at the onset of the light phase at the beginning of day 5. Samples were harvested at indicated time.
EDL3 promoter activity induced by osmotic stress, drought stress, high salinity, and ABA
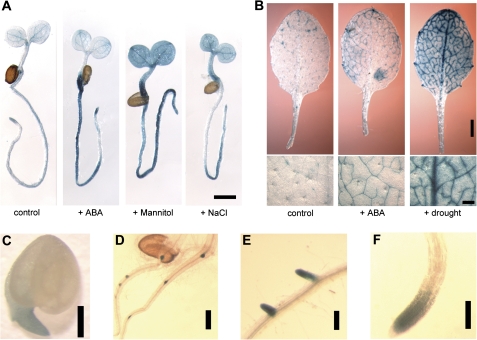
EDL3 transcript levels were up-regulated upon ABA treatment, under osmotic stress, or at high salinity (Fig. 1; see Supplementary Fig. S1 at JXB online). Comparable results were obtained with 4-d-old seedlings that had been transformed with a ProEDL3:GUS construct. In control plants, the reporter gene activity remained restricted to the tip of growing roots, the vascular bundle of primary roots, and the vascular tissues in cotyledons (Fig. 2A). ABA treatment mainly induced a strong increase in GUS staining in the radicle and the basal part of the hypocotyl, whereas cotyledons only exhibited a weak response outside of the vascular bundles. Osmotic and salt stresses strongly enhanced EDL3 promoter activity in nearly all seedling tissues (Fig. 2A).
Fig. 2.
Analyses of EDL3 promoter activity. (A) Histochemical analysis of ProEDL3:GUS activity in seedlings kept on water or exposed to ABA, osmotic stress or high salinity. Four-day-old seedlings were transferred to distilled water (control), 5 μM ABA, 450 mM mannitol, or 150 mM NaCl for 6 h before harvesting. Bar, 1 mm. (B) Detection of ProEDL3:GUS activity in rosette leaves of 4-week-old plants. Plants were grown on soil under 12/12 h light/dark cycles for 3 weeks and either watered with 5 ml distilled water (control) or 5 ml of 25 μM ABA (+ABA) in the middle of the light phase for 1 week. To apply drought stress, samples were kept dry until leaves started to wilt. Pictures give an overview of total leaf staining (upper row: bar, 1 mm) or show details of the leaf blade (lower row: bar, 0.2 mm). (C) ProEDL3:GUS activity in germinating seedlings. Seeds were harvested at the beginning of testa rupture. Bar, 0.2 mm. (D–F) Detection of ProEDL3-GUS activity during secondary root formation. Seedlings were grown under continuous white light on moist filter papers. Bars, 0.2 mm.
EDL3 promoter activity was also analysed in rosette leaves of adult plants. A patchy GUS staining pattern was detected in control plants, in a few cells associated with minor veins of the leaf (Fig. 2B). Treatment of adult plants with ABA enhanced reporter gene expression in rosette leaves and led to a strong, homogeneous GUS signal within minor veins. Reporter gene expression was further analysed in plants that were kept under dry conditions until rosette leaves started to wilt. Drought stress generated a strong increase in GUS staining around minor veins, at the major vein, and in cells of the leaf blade adjacent to the veins (Fig. 2B).
Transgenic ProELD3:GUS lines were also used to follow EDL3 expression patterns during Arabidopsis development. GUS activity was detectable in the root tip of germinating seeds (Fig. 2C). Strong EDL3 promoter activity was also prominent at the earliest stages of lateral root formation. At later stages, GUS staining was restricted to the meristem and elongation zone of growing roots, but was absent in the root tip (Fig. 2D–F). GUS staining was also seen in stipules at the base of leaf petioles, in young floral buds, in anthers at later stages of flower development, and in the abscission zone of floral leaves at the beginning of flower senescence (see Supplementary Fig. S2 at JXB online).
Characterization of T-DNA insertion and EDL3 over-expresser lines
The function of EDL3 in Arabidopsis plants was investigated through the isolation of homozygous T-DNA insertion and over-expresser lines. Among the six tested T-DNA lines with proposed insertions in the single EDL3 exon, only the SALK_033705 line could be verified and propagated to obtain homozygous plants. The respective line was named edl3-1. Sequencing of T-DNA border regions revealed that the integration event created a duplication of a small part of the gene and positioned the 35S promoter of the introduced DNA fragment about 900 bp upstream of the ATG codon normally coding for Met12 inside the protein (see Supplementary Fig. S3 at JXB online). Quantitative RT-PCR analyses demonstrated that the EDL3 transcript levels in edl3-1 plants were ∼14-times higher in the water control compared with the wild type, but remained unaltered upon application of ABA or treatment with high concentrations of mannitol and salt (Fig. 1A, B). These findings indicate that the accumulation of the artificial EDL3 transcript in the edl3-1 T-DNA line is under the control of the introduced 35S promoter. Furthermore, it is important to mention that the edl3-1 line behaves as a weak over-expresser under normal growth conditions, but as a conditional knock-down mutant under drought stress, high salinity, and ABA treatments that otherwise induced EDL3 transcripts by ∼10-fold in the wild type compared with the T-DNA line (Fig. 1A, B).
To create homozygous EDL3 over-expresser lines, the EDL3 coding region was cloned behind a 35S promoter (see Supplementary Fig. S3 at JXB online) and the construct was introduced into wild-type plants. Both homozygous over-expresser lines analysed exhibited a strong, constitutively enhanced EDL3 transcript accumulation independent of exogenously applied ABA, mannitol or salt (Fig. 1A, B). Transcript levels were increased ∼200-fold and ∼5000-fold compared with the untreated wild-type control in EDL3 ox6 and EDL3 ox16, respectively (Fig. 1A).
EDL3 involvement in the ABA-dependent regulation of germination induction
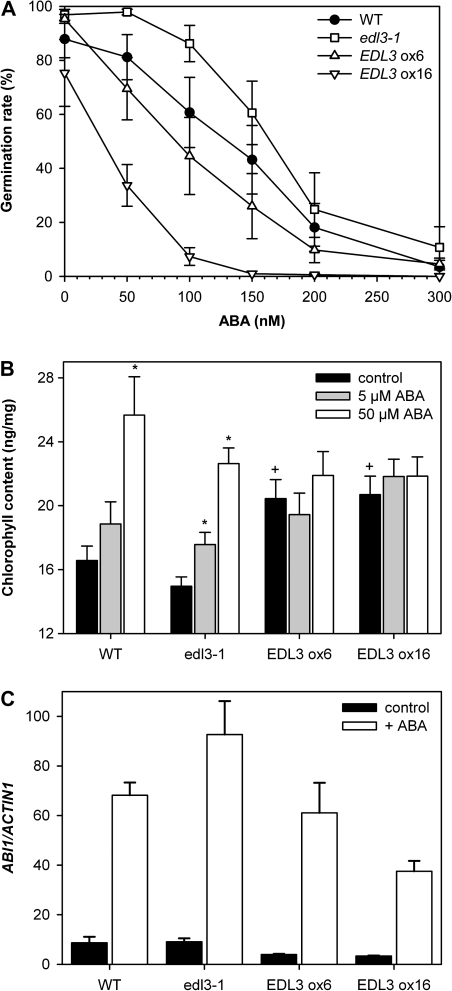
Because EDL3 transcript accumulation was strongly up-regulated by ABA, the involvement of EDL3 in downstream ABA signalling cascades was investigated through the analysis of the induction of seed germination. Germination of wild- type seeds was reduced at concentrations above 50 nM ABA and nearly completely blocked at 300 nM ABA (Fig. 3A). Germination rates of edl3-1 seeds were always greater than those of the wild type for all ABA concentrations tested, indicative of a reduced ABA sensitivity. By contrast, ABA sensitivity was clearly enhanced in both over-expresser lines. Consistent with the observed differences in transcript accumulation, EDL3 ox16 seeds exhibited a stronger inhibition of germination compared to EDL3 ox6.
Fig. 3.
Germination, greening of etiolated seedlings, and ABI1 transcript accumulation upon ABA application. (A) Seeds of wild-type, edl3-1, and EDL3 over-expressing lines were sown on four layers of moist filter papers containing variable concentrations of ABA and incubated at 6 °C for 2 d in darkness for stratification. Samples were irradiated with continuous red light for 8 h for germination induction and were then transferred back to darkness for 4 d. The number of germinated seeds was counted. The results are expressed as percentage of the total amount of seeds plated. Each data point represents the mean of 5–7 independent experiments ±SE. (B) After germination induction, seedlings were grown on moist filter papers in darkness for 3 d. The upper filter paper was then transferred to new layers of filter papers supplemented with the indicated concentrations of ABA under green safe light. After one additional day in darkness, seedlings were exposed to continuous red light for 6 h. Data presented as mean ±SE of 8 independent biological replicates. (*) Significant difference compared with the water control of the respective genotype; +, significant difference to wild-type control (one way ANOVA; P ≤0.05). (C) To determine ABA-induced ABI1 transcript accumulation, seedlings were grown for 4 d under continuous white light before transfer to new layers of filter papers supplemented with distilled water or 100 μM ABA for 3 h. Results of experiments were normalized according to the constitutively expressed ACTIN1 (AT2G37620) gene. Data represent the mean of three independent biological replicates ±SE.
EDL3 involvement in ABA-dependent chlorophyll accumulation during seedling de-etiolation
In order to test for the involvement of ABA and EDL3 in greening, 3-d-old, etiolated seedlings were pre-treated with various concentrations of ABA for 24 h in darkness before transfer to continuous red light for 6 h. Unexpectedly, ABA pre-treatment stimulated chlorophyll accumulation during red-light-induced greening of dark-grown seedlings in a dose-dependent manner (Fig. 3B). Compared with the wild type, only slightly reduced chlorophyll levels were found in edl3-1 in the water control and upon ABA treatments at 6 h of irradiation. Both EDL3 over-expresser lines exhibited significantly enhanced chlorophyll contents in the water control, with levels similar to wild-type seedlings pre-treated with 50 μM ABA. The application of ABA did not further enhance chlorophyll accumulation in the over-expresser lines, indicating that increased levels of EDL3 caused a constitutive ABA response (Fig. 3B).
ABA-dependent ABI1 transcript accumulation
ABI1 plays a central role as negative regulator in ABA signalling together with additional Type 2C protein phosphatases (Umezawa et al., 2010). To analyse the involvement of EDL3 in ABI1 transcript accumulation, seedlings were grown under continuous white light on filter papers supplemented with distilled water for 4 d and then transferred to new dishes containing with either distilled water or 100 μM ABA for 3 h. ABI1 transcript levels were slightly enhanced in the wild type and edl3-1 compared with the EDL3 over-expressers in the controls (Fig. 3C). ABA treatment caused a strong increase in ABI1 transcript accumulation in wild-type seedlings. Compared with the wild type, edl3-1 accumulated higher ABI1 transcript levels upon ABA-treatment, whereas ABI1 gene expression was reduced in the strong EDL3 ox16 over-expresser line. EDL3 ox6 seedlings did not exhibit a clear reduction in ABA-induced ABI1 transcript accumulation compared with the wild type (Fig. 3C).
EDL3 involvement in ABA-dependent inhibition of root growth
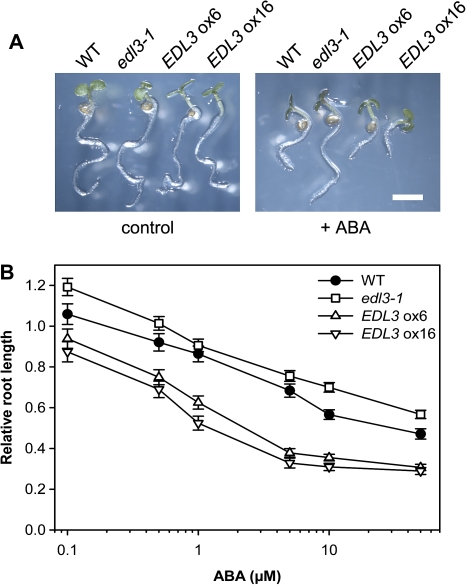
Inhibition of root growth is another well-established physiological response that is influenced by ABA (Nambara et al., 2000; Finkelstein et al., 2002; Lopez-Molina et al., 2002). To determine root growth, seedlings were grown on filter papers supplemented with distilled water for 2 d and then transferred to new dishes containing variable concentrations of ABA for two additional days. Increasing concentrations of ABA caused a reduction in root growth in wild-type seedlings (Fig. 4A, B). Whereas ABA sensitivity is slightly reduced in the T-DNA line, both EDL3 over-expresser lines exhibited an increased sensitivity compared with the wild type. Again, the EDL3 ox16 over-expresser line was slightly more sensitive to ABA than EDL3 ox6 (Fig. 4A, B).
Fig. 4.
Root growth of wild-type, edl3-1 T-DNA insertion, and EDL3 over-expressing lines in the presence of variable concentrations of exogenous ABA. Seedlings were grown on moist filter papers under continuous white light for 2 d to complete germination. The upper filter paper was then transferred to new layers of filter papers supplemented with distilled water or with distilled water containing the indicated concentrations of ABA. Root length was detected after two additional days of incubation. (A) Pictures of control plants or plants treated with 5 μM ABA. (B) Root length is expressed as percentage of the root length of the water control. All data represent the mean of at least 30 seedlings analysed in two independent experiments ±SE.
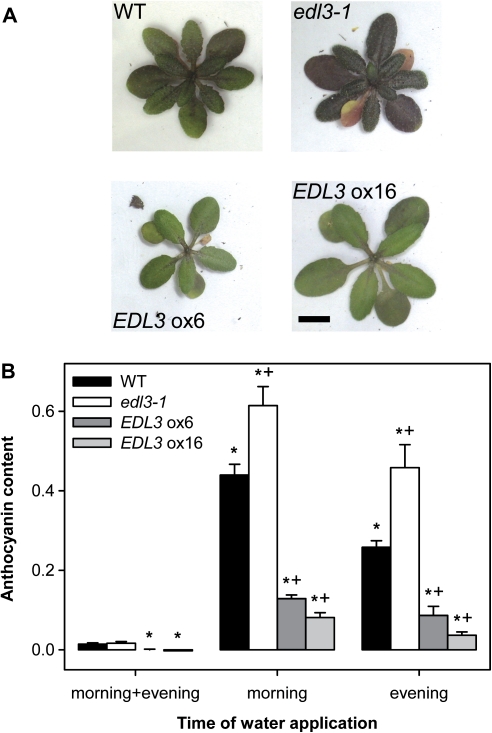
Anthocyanin accumulation under drought stress conditions in wild-type, edl3-1 T-DNA insertion, and EDL3 over-expresser lines
Drought stress caused the accumulation of anthocyanin in rosette leaves of Arabidopsis wild-type plants (Fig. 5A). Compared with the wild type, edl3-1 accumulated much higher amounts of anthocyanin, whereas EDL3 over-expresser lines did not exhibit strong accumulation of the purple pigment under conditions of water limitation. In order to quantify anthocyanin accumulation under standardized water-stress conditions, plants received a minimum volume of water either at the beginning or before the end of the daily 12 h light phase. Control plants were watered twice a day, at dusk and dawn. Limitation of water supply led to a significant increase of anthocyanin accumulation in the two youngest rosette leaves in the wild type (Fig. 5B). Stronger effects were obtained with plants that were watered only in the morning and, thus, were exposed to an increased water deficit through the night, when humidity is expected to be high. The edl3-1 knock-down line did not exhibit significant differences in anthocyanin accumulation in control treatment, but accumulated much higher levels of the pigment under drought-stress conditions. Both over-expresser lines accumulated reduced levels of anthocyanin in well-watered control plants. Exposure to drought stress induced only modest accumulation of the pigment compared with both edl3-1 and the wild type. In good agreement with the differences obtained in EDL3 transcript levels, the ox6 line exhibited a weaker reduction compared with ox16 (Fig. 5B).
Fig. 5.
Anthocyanin accumulation under drought-stress conditions in wild-type, edl3-1 T-DNA insertion, and EDL3 over-expresser lines. Plants were watered twice a day with 5 ml (morning+evening) or once a day immediately after the start (morning) or before the end (evening) of the 12 h light-phase. (A) Phenotype of adult plants under drought-stress conditions. Plants were watered once a day in the morning. Pictures were taken immediately before budding of major stems. (B) Anthocyanin content in rosette leaves under drought-stress conditions. Anthocyanin was extracted from the two youngest leaves at the start of flowering. Data presented as mean ±SE. (*) Significant difference compared with the unstressed control of the respective genotype; +, significant difference to the wild type within each treatment group treated (one way ANOVA; P ≤0.05)
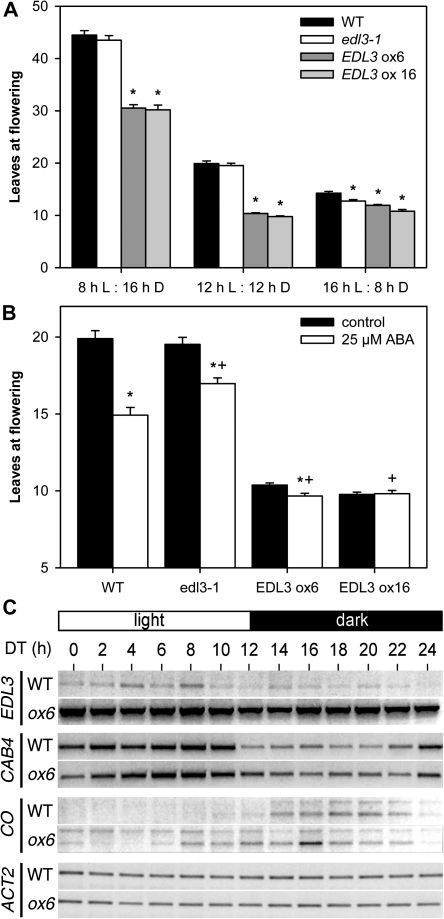
Transition to flowering in wild-type, edl3-1 T-DNA insertion, and EDL3 over-expresser lines
Over-expresser lines exhibited an early flowering phenotype during the propagation of T1 plants. Therefore, flower induction of wild type, edl3-1, and homozygous over-expresser lines was examined in greater detail under different day-lengths. As expected for a conditional long-day plant, Arabidopsis flowered much earlier when the dark phases were shortened (Fig. 6A). The edl3-1 mutant did not show a significant difference in leaf numbers at flowering under either an 8/16 h or a 12/12 h light/dark cycle. Time to flowering seemed to be shortened in the T-DNA line under long day conditions (16/8 h light/dark cycle), but the difference was subtle and did not extend to more than one leaf. Both over-expresser lines exhibited an early flowering phenotype under all tested day-lengths, even though enhanced EDL3 levels did not completely abolish photoperiodic responses (Fig. 6A).
Fig. 6.
EDL3 affects flowering and the diurnal expression of CONSTANS. (A) Flowering time under different day length. Flowering time is presented as the sum of rosette leaves and cauline leaves at the main stem. A minimum of 23 plants from both wild type and edl3-1 were analysed under short-day and long-day conditions, whereas 15 plants were counted for the two EDL3 over-expresser lines. A minimum of 39 plants from each genotype was counted under 12/12 h light/dark cycles. All experiments were repeated at least twice. An asterisk (*) indicates significant difference to the respective wild-type control for each day-length (Kruskal–Wallis One Way Analysis of Variance on Ranks; P ≤0.001). Bars, SE; L, white light; D, darkness. (B) Flowering time upon ABA treatment. Plants were grown under 12/12 h light/dark cycles. Two-week-old plants were either treated with distilled water or distilled water supplemented with ABA at the middle of the light phase for 1 week. A minimum of 39 plants from each genotype were counted. Experiments were done in triplicate. Bars, SE; *, significant difference compared to the untreated control of the respective genotype; +, significant difference to wild-type treated with 25 μM ABA (Mann–Whitney Rank Sum Test; P ≤0.001). (C) Diurnal transcript accumulation patterns of different genes in the wild type and EDL3 over-expressing lines. Seedlings were grown under 12/12 h light/dark cycles for 2 weeks. On day 14, samples were taken every 2 h. Transcript levels were determined by semi-quantitative reverse-transcription PCR using identical cDNA samples. ACTIN1 was used as a constitutive control. The gels shown are representative of two experiments with independent RNA isolations. ACT1, ACTIN1; CAB4, CHLOROPHYLL-A/B-BINDING PROTEIN 4; CO, CONSTANS; DT, day-time; ox6, EDL3 ox6 over-expresser line; WT, wild type.
Because variations in EDL3 levels can alter ABA signalling, leaf numbers at flowering were examined after hormone application. Plants were grown under a 12/12 h light/dark cycle for 2 weeks before either an ABA solution or water was applied every day at noon for one additional week. Following treatment, plants were kept humid, and the number of leaves at flowering was determined. Wild-type plants flowered earlier with ABA treatment (Fig. 6B). The ABA effect is significantly reduced in the edl3-1 line, even though the plants still responded to hormone treatment. As mentioned above, both tested over-expressers exhibited an early flowering phenotype without ABA treatment (Fig. 6B). No further reduction in leaf numbers at flowering was observed for the strong over-expresser EDL3 ox16 upon ABA application, whereas the weaker over-expresser line EDL3 ox6 exhibited a slightly earlier flowering.
EDL3 involvement in the diurnal regulation of CONSTANS transcript accumulation
EDL3 exhibits high sequence similarity to tobacco ZGT, which is thought to function as a coupling agent between the central circadian oscillator and rhythmic expression of CAB genes (Xu and Johnson, 2001). To analyse the influence of EDL3 on diurnal transcript accumulation patterns, EDL3, CAB4, and CONSTANS (CO) transcripts were followed by semi-quantitative RT-PCR in the wild type and the EDL3 ox6 over-expresser line together with a constitutive ACTIN1 (ACT1) control. CO transcripts were analysed because its protein product functions as a central component in the photoperiodic pathway of flowering induction and becomes strongly regulated by the circadian clock (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002).
In contrast to tobacco ZGT, EDL3 did not exhibit a clear diurnal rhythm in transcript accumulation (Fig. 6C). Furthermore, EDL3 over-expression did not result in an altered transcript accumulation pattern of CAB4 (Fig. 6C) or in a loss of dampening of CAB transcript accumulation after transfer of entrained plants to continuous darkness (see Supplementary Fig. S4 at JXB online). Interestingly, a clear difference in diurnal expression patterns was observed for both splicing variants of the CONSTANS (CO) gene between the wild type and the EDL3 over-expresser. In good accordance to published results, CO transcript accumulation started at daytime (DT) 8 h to 10 h after the onset of light, reached a maximum between DT 16 h and 18 h, and fell below the detection level at DT 24 h in wild-type plants (Fig. 6C). In the EDL3 over-expresser line, CO transcripts were detectable at nearly all time points. Re-accumulation of CO transcripts started as early as DT 6 h and reached high levels between DT 8 h to DT 22 h. Thus, EDL3 over-expression clearly leads to the accumulation of high CO transcript levels during the light phase of the 12/12 h light/dark cycle.
Interactions between EDL3 and ASK proteins
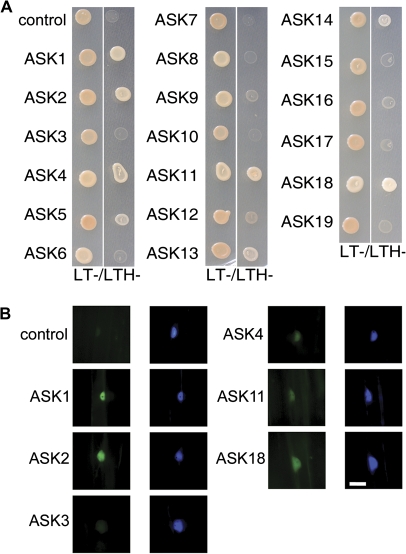
Sequence comparisons between EID1 and EDL proteins from different plant species revealed that the protein family exhibits the highest degree of similarity at the ELP and F-box domains of EID1 (see Supplementary Fig. S5 at JXB online). Nevertheless, publically available protein analysis tools did not predict an F-box domain in EDL3, even though the carboxy terminal half of the putative EDL3 F-box domain yielded several hits with the Pfam domain signature. To check whether EDL3 is an F-box protein, interactions between EDL3 and ASKs were analysed with our established yeast and in planta interaction assays (Stolpe et al., 2005; Marrocco et al., 2006).
EDL3 was cloned behind a GAL4 binding domain and the resulting vector was introduced into haploid yeast cells. Yeast growth was analysed on LTH- selection media after mating with strains expressing 19 different ASK proteins. Rapid yeast growth was observed with ASK1, ASK2, ASK4, ASK11, and ASK18, and slow growth was seen with ASK5, ASK13, and ASK14. No other combinations allowed yeast colony formation on selective media, similar to empty vector controls (Fig. 7A).
Fig. 7.
EDL3 interacts with a specific subset of ASK proteins. (A) Yeast two-hybrid interaction assays. After mating, yeast was grown at 28 °C for 3 d on non selective (LT-) and selective media (LTH-). Empty activation domain vector was included as a control. (B) In planta Bimolecular Fluorescence Complementation assays of the interaction between EDL3 and different ASKs. Three-day-old, dark-grown mustard seedlings were co-transfected by particle bombardment with EDL3-YC and one of 19 different ASK-YN fusion proteins. ASK1-YN was co-transfected with empty YC vector as a control. Images were recorded using YFP- and CFP-specific filter sets. The CFP-specific filter set shows the signal from the CPRF2-CFP transfection control, which is mainly localized in the nucleus of transfected cells. Bar, 40 μm.
For in planta Bimolecular Fluorescence Complementation (BiFC) assays, EDL3-YC and different ASK-YN fusion proteins were co-transfected into cells of etiolated mustard seedlings together with a CPRF2-CFP transfection control. Strong YFP signals became visible with ASK1 and ASK2. Weak fluorescence was detected with ASK4, ASK11, and ASK18, whereas all other ASK constructs did not give a signal exceeding the YN background control, as seen in the data for ASK3 (Fig. 7B; data not shown). All YFP signals were restricted to the nucleus in a manner similar to the CPRF2-CFP transfection control. This finding is in good agreement with the strict nuclear localization observed for EDL3-YFP fusion proteins in transfected mustard cells (see Supplementary Fig. S6 at JXB online).
Discussion
EDL3 is involved in the regulation of ABA responses
Several lines of evidence clearly indicate that EDL3 plays an important role as a positively acting factor in ABA signalling in Arabidopsis. First, the EDL3 gene is strongly and rapidly induced upon ABA treatments and by osmotic, salt, and drought stress (Figs 1, 2; see Supplementary Fig. S1 at JXB online). The observed EDL3 gene expression is consistent with the proposed function of ABA as an integrator downstream of different exogenous stressors (Zhu, 2002; Shinozaki et al., 2003; Umezawa et al., 2010). Second, results with YFP fusion proteins and BiFC in transfected mustard seedlings exhibited a strict nuclear localization of EDL3, which makes it very likely that the protein functions in the regulation of gene expression (Fig. 7; see Supplementary Fig. S6 at JXB online). Third, physiological experiments with over-expresser and T-DNA lines demonstrated that changes in EDL3 transcript levels led to measurable alterations in ABA sensitivity for seed germination, root growth, greening of etiolated seedlings, ABI1 transcript accumulation and induction of flowering (Figs 3, 4, 6). Differences observed between the wild type and the edl3-1 mutant were sometimes small compared with results obtained with the two over-expresser lines. This observation is in accordance with transcript accumulation data, which demonstrates that edl3-1 only becomes a knock-down mutant under conditions that induce high EDL3 transcript levels in wild-type plants (Fig. 1). Finally, tissue-specific expression of promoter–GUS constructs activity is in good agreement with its proposed functions in ABA responses (Fig. 2). Promoter activity was high in root tips and at leaf vascular bundles that are thought to be involved in the regulation of germination, root growth, and floral transition (Finch-Savage and Leubner-Metzger, 2006; Christmann et al., 2006; Corbesier et al., 2007; Kobayashi and Weigel, 2007; Finkelstein et al., 2008; Turck et al., 2008).
Inhibition of germination, root growth, and flowering are well known physiological responses that are influenced by ABA (Wijanyanti et al., 1997; Nambara et al., 2000; Finkelstein et al., 2002; Lopez-Molina et al., 2002; Koiwa et al., 2002; Bezerra et al., 2004; Wilmowicz et al., 2008; Pandey et al., 2009). Several studies also demonstrate that chlorophyll accumulation, ABA signalling, and plastid development are interlaced. Beside its enzymatic activity in chlorophyll synthesis, the Mg-chelatase H subunit is shown to act in plastid-to-nucleus signal transduction and as an ABA receptor (Mochizuki et al., 2001; Shen et al., 2006). ABI4 (ABA INSENSITIVE 4) has been proposed to function downstream of GENOMES UNCOUPLED (GUN) factors in retrograde signalling pathways to regulate expression of nuclear genes encoding plastid proteins (Koussevitzky et al., 2007). Results obtained with abi3 further indicate that ABA plays an important role in plastid development and greening in dark-grown Arabidopsis seedlings (Rohde et al., 2000). Unexpectedly, hormone pre-treatment or EDL3 over-expression stimulated chlorophyll accumulation during de-etiolation of dark-grown seedlings (Fig. 3B), whereas most published studies showed reduced greening of seedlings upon ABA application (Finkelstein et al., 2002; Penfield et al., 2006; Guo et al., 2009; Park et al., 2009). The discrepancies might be caused by different experimental approaches: In most greening assays, seeds were directly sown on ABA-containing media and seedlings were grown under continuous light for several days. Therefore, this experimental design might mainly detect sensitivity towards the inhibitory effect of ABA on germination and photomorphogenic seedling development. By contrast, our experimental approach tested the influence of the plant hormone during skotomorphogenesis and at early phases of plastid development. EDL3 over-expresser lines resemble the greening after extended darkness 1 (ged1) mutant, which accumulates higher chlorophyll levels after de-etiolation and exhibits increased ABA-sensitivity for inhibition of germination (Choy et al., 2008).
EDL3 regulates drought stress-induced anthocyanin accumulation
The over-expresser and knock-down lines also exhibited significant differences in drought-stress-induced anthocyanin accumulation (Fig. 5). The pigment is thought to function as a sunscreen that protects the photosynthetic apparatus against photodamage (Steyn et al., 2002). Increased levels of the pigment might help to reduce impairment of the photosynthetic apparatus under drought-stress conditions and at high light intensities, two stressors that often occur simultaneously in natural environments. The observed increase in ABA sensitivity in over-expresser lines might help to protect against drought stress and, thus, would reduce stress-induced anthocyanin accumulation. By contrast, reduced sensitivity in edl3-1 would cause an increase in anthocyanin levels similar to Arabidopsis plants grown under conditions of limited water supply.
The observed alterations in EDL3-dependent anthocyanin accumulation fit the results obtained with mutants in the Elongator histone acetyl-transferase complex and ABI3 (ABA-INSENSITIVE 3), an important transcriptional regulator involved in ABA signalling. Mutants in the Elongator histone acetyl-transferase complex exhibited reduced root growth, ABA hypersensitivity, and an increased accumulation of anthocyanin similar to EDL3 over-expresser lines (Zhou et al., 2009). ABI3 synergistically controls anthocyanin accumulation together with its homologues FUSCA3 and LEC1 (LEAFY COTYLEDON 1) during seed maturation, when they function as negative regulators (Parcy et al., 1997).
EDL3 might regulate transition to flowering through modulation of CO expression
EDL3 shows high similarity to the tobacco ZGT protein. Tobacco ZGT was proposed to function as a coupling factor between the central oscillator and its dependent genes because of its clear circadian expression and because over-expression of the gene caused a loss of dampening for circadian CAB expression after entrainment and transfer to continuous darkness (Xu and Johnson, 2001). In contrast to findings in tobacco, the Arabidopsis EDL3 gene exhibited no clear oscillation of transcript levels even under diurnal conditions, and EDL3 over-expression did not lead to a loss of dampening of CAB transcripts after transfer to darkness (see Supplementary Fig. S4 at JXB online). Therefore, despite the observed sequence similarity, the factors seem to fulfil different functions in the divergent plant species.
Nevertheless, a function of EDL3 in clock-regulated gene expression cannot be fully excluded, because EDL3 over-expression severely affected the diurnal expression pattern of CO, a central component of the photoperiodic pathway in Arabidopsis. In Arabidopsis, a conditional long-day plant, CO protein accumulation takes place during the light phase and becomes stabilized during subsequent darkness, which leads to flower induction (Laubinger et al., 2006; Imaizumi and Kay, 2006; Corbesier et al., 2007; Kobayashi and Weigel, 2007; Turck et al., 2008). Because CO transcript accumulation in the wild type starts during the late afternoon, coincidence of CO protein accumulation and light is only obtained under long-day conditions. According to our data, EDL3 over-expression results in increased CO transcript levels much earlier in the day and, thus, would lead to a coincidence of CO accumulation and light at shorter day-lengths. Thus, the change in diurnal CO expression might explain the observed early flowering phenotype of EDL3 over-expresser lines. Comparable results were obtained with mutants in repressors of CO that cause enhanced expression throughout the day (Fowler et al., 1999; Suarez-Lopez et al., 2001; Imaizumi et al., 2005; Mizoguchi et al., 2005; Chen and Ni, 2006). By contrast, flowering is delayed in mutants depressing CO expression (Fowler et al., 1999; Mizoguchi et al., 2005).
The observed strong EDL3 promoter activity along leaf veins is in agreement with its proposed function in flowering induction and the observed effect on CO expression. Several studies demonstrate that CO has to be expressed along vascular tissues in leaves, where it controls the expression of the FT (flowering locus T), which is thought to function as a signalling mediator between leaves and the apical meristem (Ayre and Turgeon, 2004; Kobayashi and Weigel, 2007; Corbesier et al., 2007; Turck et al., 2008).
Data about ABA and its effect on flower induction are scarce and sometimes ambiguous (Bezerra et al., 2004; Pandey et al., 2009; Koiwa et al., 2002). Data with the obligatory short-day plant Pharbitis nil showed that the hormone can either stimulate or inhibit flowering to different degrees, depending on the time of hormone application during and around an inductive dark treatment (Wijanyanti et al., 1997; Wilmowicz et al., 2008). Fluctuations and ambiguities in ABA-induced flowering can easily be explained if the plant hormone functions by a modulation of CO expression. Because CO expression fluctuates in a clock-dependent manner (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002) and because the CO protein accumulation requires light (Imaizumi and Kay, 2006; Kobayashi and Weigel, 2007; Corbesier et al., 2007; Turck et al., 2008), responsiveness toward ABA treatments should also vary during daytime and the duration of light phases. EDL3 might be involved in this regulatory process, because it becomes rapidly induced by ABA and because increased levels of the regulatory factor seem to enhance CO gene expression. The model would also explain the delay of ABA-induced flowering in edl3-1 compared with the wild type, because the mutant accumulates reduced levels of EDL3 which leads to reduced levels of CO only upon treatment with the plant stress hormone.
EDL3 most probably functions as a component of an SCF ubiquitin ligase complex
F-box proteins are involved in the regulation of a multitude of biological processes in higher plants. During plant development, F-box proteins are shown to participate in the regulation of floral development, shoot branching, leaf senescence, root branching, and pollen self-incompatibility (Lechner et al., 2006). Furthermore, F-box proteins function in signalling cascades that regulate light responses, synchronization of the circadian clock, and responses towards different plant hormones, including auxin, jasmonate, ethylene, gibberellins, and ABA (Mockaitis and Estelle, 2008; Zhang et al., 2008; McClellan and Chang, 2008; Fonseca et al., 2009; Schwechheimer and Willige, 2009).
Because F-box domains exhibit a high variability in their amino acid composition, the presence of the respective domain can only be unequivocally proven by interaction assays with ASKs (Craig and Tyers, 1999; Deshaies, 1999). EDL3 interacts with a specific subset of ASK proteins in yeast and in planta. The interaction with ASK proteins strongly indicates that EDL3 functions as a component of an SCF ubiquitin ligase complex. EDL3 over-expression leads to reduced sensitivity for all tested ABA responses, whereas the conditional edl3-1 knock-down line very often shows an antagonistic effect compared with the over-expresser lines. These results indicate that EDL3 functions as a positive regulator of ABA signalling. Because EDL3 most probably functions as a positive regulator in ABA signalling and because it carries a functional F-box domain, it is worthwhile speculating that the corresponding SCFEDL3 ubiquitin ligase complex targets negatively acting regulators in the ABA signalling pathway to degradation in the proteasome.
The data indicate that EDL3 negatively regulates ABI1 transcript accumulation. Together with other plant type 2C protein phosphatases (PP2Cs) ABI1 functions as negative regulator in ABA signalling by opposing phosphorylation of positively acting SnRK2 kinases (sugar non-fermenting (SNF1)-related protein kinases). PP2Cs play a central role in ABA signalling, because their activities are regulated by direct interaction with PYR/PYL/RCAR (pyrabactin resistence 1/PYR-like/regulatory component of ABA receptor 1) ABA receptors (Hubbard et al., 2010; Raghavendra et al., 2010; Umezawa et al., 2010). According to these findings, it might be speculated that altered ABA responses in edl3-1 and the over-expresser lines are caused by altered expression of negatively acting PP2Cs leading to increased ABA sensitivity in the conditional knock-down mutant or reduced hormone sensitivity upon over-expression of EDL3.
F-box proteins and different E3 ubiquitin ligases control several aspects of ABA signalling. The F-box protein DOR (drought tolerance repressor) functions as a negative regulator for ABA-induced stomata closure and drought-stress responses in Arabidopsis (Zhang et al., 2008). DOR is mainly expressed in guard cells and becomes suppressed by ABA treatments. Thus, with respect to many aspects of ABA responses, DOR seems to execute opposite functions compared with EDL3, which functions as a positive regulator of ABA signalling. The ABI3-interacting protein 2 (AIP2) contains a RING motif and targets ABI3 for degradation in the proteasome (Zhang et al., 2005). Several data indicate that ABI5 abundance seems to be regulated by DWA1 and DWA2 (DDB1 binding WD hypersensitive to ABA1 and 2; Lee et al., 2010) and the RING-type E3 protein KEEP ON GOING (Liu and Stone, 2010). Results further indicate ABI5 stability is regulated by the ABI5 binding protein AFP (Lopez-Molina et al., 2003). Thus, EDL3 seems to be only one additional component in a complex network of ubiquitin-dependent protein degradation that regulates ABA signalling.
Conclusions
The EDL3 protein shows a high similarity to EID1, an F-box protein that functions as a negative regulator in phytochrome A-dependent light signalling in Arabidopsis. In order to unravel the functional relevance of EDL3, knock-down and over-expression lines were isolated. Physiological analyses of these lines demonstrated that EDL3 functions as a positive regulator in ABA-dependent signalling cascades regulating germination induction, root growth, greening of etiolated seedlings, and flowering. Ectopic over-expression of EDL3 leads to early flowering under short days. This effect is most probably mediated by an extended diurnal expression peak of CONSTANS, an important component of the photoperiodic pathway in Arabidopsis. The observed physiological responses are in good accordance with the observed EDL3 promoter activity, which is high in root tips and at leaf vascular bundles and becomes strongly induced by ABA, osmotic stress, drought stress and high salinity. Because EDL3 expression is rapidly and strongly increased by ABA and water stress, it is worthwhile to speculate that EDL3-dependent modulation of ABA sensitivity is at least partially regulated on the transcriptional level. EDL3 carries a functional F-box domain and is thus presumed to act as a component of a ubiquitin ligase complex that specifically targets negatively acting regulatory proteins in ABA signalling to degradation in the proteasome.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Microarray data of EDL3 transcript accumulation under osmotic stress, high salinity, and in the presence of exogenous ABA.
Supplementary Fig. S2. Analysis of EDL3 promoter activity during Arabidopsis development.
Supplementary Fig. S3. Schematic overview of the EDL3 wild-type gene, the construct used to create over-expresser lines, and the T-DNA integration site in the edl3-1 line.
Supplementary Fig. S4. Circadian rhythm of CAB transcript accumulation after transfer to continuous darkness.
Supplementary Fig. S5. Domain structure of EDL3 proteins.
Supplementary Fig. S6. Subcellular localization of EDL3-YFP in cells of transfected mustard seedlings.
Supplementary Table S1. Oligonucleotides used for this study.
Acknowledgments
We thank the Salk Institute and the Nottingham Stock Centre for providing T-DNA insertion mutants of Arabidopsis, the RIKEN Institute for the RAFL01-11-G23 cDNA clone, Martina Krenz for her helpful technical assistance, and Anita K Snyder for her helpful comments on the manuscript. This work was supported by the Arabidopsis Functional Genomic Network programme of the Deutsche Forschungsgemeinschaft (KR 2020/1-3 and 1-4) to SP, PK, and TK, the DFG grant ‘Analysis of phytochrome A-dependent light signalling in Arabidopsis thaliana’ (KR2020/2-3) to CK and TK, a grant of the European Union (HPRN-CT-2002-00333) to KMS and TK, and the financial support of the Freiburg Institute for Advanced Studies (FRIAS), School of Life Sciences to MI and TK.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Ayre BG, Turgeon R. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiology. 2004;135:2271–2278. doi: 10.1104/pp.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. The Plant Journal. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- Buche C, Poppe C, Schafer E, Kretsch T. eid 1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. The Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ni M. RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. The Plant Journal. 2006;46:823–833. doi: 10.1111/j.1365-313X.2006.02740.x. [DOI] [PubMed] [Google Scholar]
- Choy MK, Sullivan JA, Theobald JC, Davies WJ, Gray JC. An Arabidopsis mutant able to green after extended dark periods shows decreased transcripts of seed protein genes and altered sensitivity to abscisic acid. Journal of Experimental Botany. 2008;59:3869–3884. doi: 10.1093/jxb/ern227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. Integration of abscisic acid signalling into plant responses. Plant Biology. 2006;8:314–325. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Craig KL, Tyers M. The F-box: a new motif for ubiquitin-dependent proteolysis in cell cycle regulation and signal transduction. Progress in Biophysics and Molecular Biology. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annual Review of Cell and Developmental Biology. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schafer E, Funk M, Kretsch T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes and Development. 2001;15:939–944. doi: 10.1101/gad.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE., Leubner-Metzger G. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Review of Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14, Supplement:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Current Opinion in Plant Biology. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO Journal. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG. RACK1 is a negative regulator of ABA responses in Arabidopsis. Journal of Experimental Botany. 2009;60:3819–3833. doi: 10.1093/jxb/erp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes and Development. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes and Development. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proceedings of the National Academy of Sciences, USA. 2002;99:10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Kretsch T. Phenotypic characterization of photomorphogenic responses during plant development. Methods in Molecular Biology. 2010;655:189–202. doi: 10.1007/978-1-60761-765-5_13. [DOI] [PubMed] [Google Scholar]
- Kretsch T, Emmler K, Schafer E. Spatial and temporal pattern of light-regulated gene expression during tobacco seedling development: the photosystem II-related genes Lhcb (Cab) and PsbP (Oee2) The Plant Journal. 1995;7:715–729. [Google Scholar]
- Laubinger S, Marchal V, Le GJ, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Current Opinion in Plant Biology. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. The Plant Cell. 2010;22:1716–1732. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. The Plant Cell. 2010;22:2630–2641. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes and Development. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Marrocco K, Zhou Y, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T. Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. The Plant Journal. 2006;45:423–438. doi: 10.1111/j.1365-313X.2005.02635.x. [DOI] [PubMed] [Google Scholar]
- McClellan CA, Chang C. The role of protein turnover in ethylene biosynthesis and response. Plant Science. 2008;175:24–31. doi: 10.1016/j.plantsci.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. The Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of the National Academy of Sciences, USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Molecular Biology. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Developmental Biology. 2000;220:412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. The Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends in Plant Science. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Rohde A, De RR, Beeckman T, Engler G, Van MM, Boerjan W. ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. The Plant Cell. 2000;12:35–52. doi: 10.1105/tpc.12.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Willige BC. Shedding light on gibberellic acid signalling. Current Opinion in Plant Biology. 2009;12:57–62. doi: 10.1016/j.pbi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJ, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytologist. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Stolpe T, Susslin C, Marrocco K, Nick P, Kretsch T, Kircher S. In planta analysis of protein–protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma. 2005;226:137–146. doi: 10.1007/s00709-005-0122-6. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant and Cell Physiology. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nature Reviews Molecular Cell Biology. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wijanyanti L, Fujioka S, Kobayashi M, Sakurai A. Involvement of abscisic acid and indole-3-acetic acid in the flowering of Pharbitis nil. Journal of Plant Growth Regulation. 1997;16:115–119. [Google Scholar]
- Wilmowicz E, Kesy J, Kopcewicz J. Ethylene and ABA interactions in the regulation of flower induction in Pharbitis nil. Journal of Plant Physiology. 2008;165:1917–1928. doi: 10.1016/j.jplph.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Xu Y, Johnson CH. A clock- and light-regulated gene that links the circadian oscillator to LHCB gene expression. The Plant Cell. 2001;13:1411–1425. doi: 10.1105/tpc.13.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes and Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y. F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiology. 2008;148:2121–2133. doi: 10.1104/pp.108.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hua D, Chen Z, Zhou Z, Gong Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. The Plant Journal. 2009;60:79–90. doi: 10.1111/j.1365-313X.2009.03931.x. [DOI] [PubMed] [Google Scholar]
- Zhou YC, Dieterle M, Buche C, Kretsch T. The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome A-specific light signaling. Plant Physiology. 2002;128:1098–1108. doi: 10.1104/pp.010811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.