Abstract
Lipid-derived molecules produced by acylhydrolases play important roles in the regulation of diverse cellular functions in plants. In Arabidopsis, the DAD1-like phospholipase A1 family consists of 12 members, all of which possess a lipase 3 domain. In this study, the biochemical and cellular functions of AtDLAH, an Arabidopsis thaliana DAD1-like acylhydrolase, were examined. Bacterially expressed AtDLAH contained phospholipase A1 activity for catalysing the hydrolysis of phospholipids at the sn-1 position. However, AtDLAH displayed an even stronger preference for 1-lysophosphatidylcholine, 1-monodiacylglycerol, and phosphatidic acid, suggesting that AtDLAH is a sn-1-specific acylhydrolase. The AtDLAH gene was highly expressed in young seedlings, and its encoded protein was exclusively localized to the mitochondria. AtDLAH-overexpressing transgenic seeds (35S:AtDLAH) were markedly tolerant to accelerated-ageing treatment and thus had higher germination percentages than wild-type seeds. In contrast, the atdlah loss-of-function knockout mutant seeds were hypersusceptible to accelerated-ageing conditions. The 35S:AtDLAH seeds, as opposed to the atdlah seeds, exhibited a dark red staining pattern following tetrazolium treatment under both normal and accelerated-ageing conditions, suggesting that AtDLAH expression is positively correlated with seed viability. The enhanced viability of 35S:AtDLAH seeds was accompanied by more densely populated epidermal cells, lower levels of accumulated lipid hydroperoxides, and higher levels of polar lipids as compared with wild-type and atdlah mutant seeds. These results suggest that AtDLAH, a mitochondrial-localized sn-1-specific acylhydrolase, plays an important role in Arabidopsis seed viability.
Keywords: DAD1-like acylhydrolase, lipid peroxidation, mitochondrial targeting, seed viability
Introduction
Seed viability is important for reproduction and propagation in higher plant species. Seeds also play a significant role in food sources for animals and humans. Therefore, seed quality and longevity are critical factors for ecology, agriculture, and the economy (Chrispeels and Sadava, 2003; Walters et al., 2005; Li and Pritchard, 2009). Seed quality is characterized by the seed's capability to germinate and maintain its storage contents (Coolbear, 1995). However, seeds are continuously exposed to harsh environments, such as high humidity, extreme temperatures, strong sun rays, and pathogen infections, during their development, harvest, and storage. Thus, most seeds suffer gradual deterioration, including decreased germination percentages, storability, and stress tolerance (Nakayama et al., 1981; McDonald, 1999). A number of mechanisms of seed deterioration have been suggested, including disruption of nucleic acids, proteins, storage lipids, and membranes (Osborne, 1980; ReuZeau et al., 1992; Bewley and Black, 1994; Sun and Leopold, 1995; Thapliyal and Connor, 1997; Pukacka, 1998). The disruption of membrane phospholipids and storage lipids by peroxidation is regarded as the primary reason for seed deterioration, because these lipids can be easily damaged by oxidative stress caused by unfavourable environmental conditions (Smirnoff, 1993). However, the molecular and biochemical events underlying lipid peroxidation-mediated seed deterioration have not been well characterized (Clerkx et al., 2004).
Several lines of evidence have suggested that seed deterioration is associated with lipid peroxidation caused by peroxyl radicals. Sattler et al. (2004) reported that tocopherol-deficient mutants had significantly reduced seed longevity and elevated levels of lipid hydroperoxides (LOOHs) during germination. Therefore, protection of membrane lipids and oils by tocopherols (lipid-soluble antioxidants) against various oxidative stresses is crucial for seed germination. Regeneration of ascorbate may play an important role in protecting storage reserves that serve as essential energy sources for seed germination (Eastmond, 2007). Arabidopsis mutants with defects in the peroxisomal membrane monodehydroascorbate reductase isoform, a protein that generates reduced ascorbate, exhibited elevated levels of H2O2, lipid peroxidation, and protein oxidation, resulting in impaired seedling establishment. This finding suggests that detoxifying H2O2 and preventing peroxisomal release of H2O2 are critical for protecting membrane lipids and storage oils. Phospholipase Dα1 (PLDα1), a membrane lipid-hydrolysing phospholipase, plays a role in Arabidopsis seed deterioration and ageing (Devaiah et al., 2007). Therefore, knockout mutants of PLDα1 exhibited an increased tolerance to accelerated and natural ageing. PLDα1-deficient seeds lost fewer unsaturated fatty acids and accumulated fewer lipid peroxides than wild-type seeds after storage or exposure to adverse conditions. This result supports the theory that production of phosphatidic acid (PA) from phospholipids by PLD is an initial step of membrane degradation and seed deterioration, and that PA-derived lipid peroxyl radicals will subsequently attack phospholipids, resulting in a chain reaction of membrane lipid peroxidation (Thompson, 1988; Samama and Pearce, 1993).
In higher plants, acylhydrolases play important roles in the regulation of diverse cellular metabolic functions, including seed germination, cell elongation, anther dehiscence, jasmonate-mediated defence signalling, and leaf senescence (Ishiguro et al., 2001; He and Gan, 2002; Lee et al., 2003; Eastmond, 2006; Hyun et al., 2008). Based on their lipolytic specificities, acylhydrolases are classified into different types of lipases, such as galactolipases, triacylglycerol (TAG) lipases, and phospholipases (Brady et al., 1990; Beisson et al., 2003). The DEFECTIVE IN ANTHER DEHISCIENCE 1 (DAD1) (At2g44810) was originally identified as an Arabidopsis PLA1 that catalysed the initial step for jasmonic acid production in chloroplasts (Ishiguro et al., 2001). The DAD1-like acylhydrolase family consists of 12 members, all of which contain sn-1-specific acylhydrolase activity, and is further divided into three subgroups based on predicted subcellular localizations (Beisson et al., 2003; Ryu, 2004; Seo et al., 2008, 2009; Kim et al., 2011). Seven proteins, including DAD1, belong to class I, which is typified by a putative N-terminal chloroplast-targeting signal, while four cytosolic proteins belong to class II. The sole class III protein, At1g30370, was predicted to localize to the mitochondria with unknown function. The class II enzyme At2g42690 was suggested to be involved in the PR-1-mediated defensive response to ultraviolet-B (UV-B) irradiation (Lo et al., 2004). The chloroplast-targeted DONGLE (DGL) (At1g05800) participates in wound-induced jasmonate formation and is functionally redundant with DAD1 in Arabidopsis (Ishiguro et al., 2001; Hyun et al., 2008; Ellinger et al., 2010). In this study, the biochemical and cellular properties of the class III DAD1-like acylhydrolase isoform AtDLAH encoded by At1g30370 was analysed. Transgenic Arabidopsis seeds that overexpress AtDLAH exhibited strongly enhanced resistance to lipid peroxidation and ageing treatments compared with wild-type and atdlah knockout mutant plants, suggesting that AtDLAH plays a significant role in Arabidopsis seed viability and longevity.
Materials and methods
Plant materials
Wild-type Arabidopsis thaliana (ecotype Columbia-0) and the T-DNA insertion AtDLAH (At1g30370) loss-of-function mutant line (WiscDsLox489-492N9) were obtained from the Ohio State University Arabidopsis Biological Resources Center (ABRC, Columbus, OH, USA). The atdlah mutant was confirmed by genotyping PCR using the T-DNA left-border primer and gene-specific primers (Supplementary Table S1 available at JXB online). Full-length AtDLAH cDNA was cloned into the binary vector pBI121 (ABRC stock number CD3-388), and the resulting plasmid was transformed into Arabidopsis as previously described (Seo et al., 2008). 35S:AtDLAH transgenic lines were selected due to their resistance to kanamycin (30 μg ml−1). Expression levels of the AtDLAH gene in leaves and seeds of transgenic and mutant plants were examined by reverse transcription-PCR (RT-PCR) using gene-specific primers (Supplementary Table S1).
RNA extraction and cDNA synthesis
Total RNA was isolated from developing seeds (0, 12, and 21 d after pollination) and germinating seeds (0, 1, 2, 3, and 4 d after imbibition) as previously described (Ruuska and Ohlrogge, 2001). RNA samples were extracted using an RNAiso RNA purification kit according to the manufacturer's protocol (Takara, Shiga, Japan) and then treated with DNase I for 30 min. First-strand cDNA synthesis was performed as previously described (Kim et al., 2010). RT-PCR was conducted using gene-specific primer sets (Supplementary Table S1) with the following conditions: 25 cycles were conducted, each consisting of 45 s at 95 °C, 1 min at 60 °C, and 90 s at 72 °C in an automatic thermal cycler (Applied Biosystems, Carlsbad, CA, USA).
Construction of the MBP–AtDLAH recombinant protein
AtDLAH cDNA lacking the N-terminal transit peptide sequence was amplified by PCR using gene-specific primers (Supplementary Table S1). The products were introduced into the pMal-c2X plasmid (New England BioLabs, Hertfordshire, UK). The fusion protein was expressed in the Escherichia coli BL21 (DE3) strain and purified by affinity chromatography using amylose resin (New England BioLabs) as previously described (Seo et al., 2009).
In vitro lipase assay
The in vitro assay for measuring lipase activity was performed as previously described (Seo et al., 2009). To examine phospholipase A1 activity using radiolabelled phosphatidylcholine (PC) with asymmetric fatty acids, a reaction mixture containing 15 pmol 1-palmitoyl-2-[14C]palmitoyl-PC (2.22 GBq mmol−1, GE Healthcare, Uppsala, Sweden) was incubated with 20 μg of recombinant fusion proteins for 30 min at 30 °C in a final volume of 200 μl of 0.2% Triton X-100, 100 mM NaCl, and 50 mM sodium phosphate buffer (pH 6.8). Reaction products were separated by thin-layer chromatography (TLC) (Silica Gel 60; Merck, Whitehouse Station, NJ, USA) and developed with chloroform/methanol/CH3COOH/water (85:15:12.5:3.5, v/v/v/v). The colorimetric assay for substrate specificity was conducted with a mixture containing various lipid substrates, including PC, phosphatidylethanolamine (PE), phosphatidic acid (PA), monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), triolein (TAG), 1,2-diacylglycerol, 1,3-diacylglycerol, 1-monoacylglycerol, 2-monoacylglycerol, and 1-lysophosphatidylcholine (1-LPC). The recombinant fusion protein was incubated with the mixtures for 30 min at 30 °C, and then released free fatty acids were measured using NEFA-HR colorimetric kits (Wako Pure Chemicals, Osaka, Japan) according to the manu-facturer's protocol.
Protoplast transient assay
A full-length AtDLAH cDNA clone and a synthetic nuclear localization signal (NLS; Woo et al., 2010) were ligated into a soluble-modified green fluorescent protein (GFP) plasmid (psmGFP) (Cho et al., 2008) to construct 35S:AtDLAH-GFP and 35S:NLS-GFP, respectively. The GFP fusion constructs were transformed into protoplasts prepared from wild-type and mt-yk CS16264 Arabidopsis rosette leaves by polyethylene glycol (PEG) treatment (Seo et al., 2008). The mt-yk CS16264 plant was used as a mitochondria-localized marker (Nelson et al., 2007). After 16 h of incubation, the expression of 35S:AtDLAH-GFP and 35S:NLS-GFP was monitored with a cooled CCD camera and a BX51 fluorescence microscope (Olympus, Tokyo, Japan) as previously described (Son et al., 2009).
Purification of chloroplasts and mitochondria
Chloroplasts and mitochondria were isolated from light-grown 2-week-old leaves from wild-type and 35S:AtDLAH-HA T4 transgenic plants as previously described (Tanaka et al., 2004) with some modifications. The collected leaves were homogenized in an isolation solution containing 50 mM HEPES-KOH (pH 7.4), 0.33 M sorbitol, 1 mM MnCl2, 2 mM EDTA, and 0.2% bovine serum albumin (BSA). The homogenate was filtered through four layers of nylon mesh.
To fractionate chloroplasts, extracts were centrifuged at 350 g for 5 min at 4 °C and the resulting supernatant was layered on to an uncontinuous gradient consisting of 30% and 60% (v/v) Percoll in isolation solution. The gradients were centrifuged at 8000 g for 15 min at 4 °C. The intact chloroplasts distributed around the 30/60% Percoll interface were isolated and diluted with the isolation solution. After samples were centrifuged at 4000 g for 10 min at 4 °C to remove Percoll, pellets were re-suspended in isolation solution.
To separate mitochondria, the filtered extracts were centrifuged at 3000 g for 5 min at 4 °C and supernatants were re-centrifuged at 22 000 g for 15 min at 4 °C. Pellets were resuspended with isolation solution and centrifuged at 3000 g for 15 min at 4 °C. To obtain mitochondrial pellets, supernatants were centrifuged at 22 000 g for 15 min at 4 °C and pellets were resuspended with isolation solution. The quality of the purified chloroplasts and mitochondria was assessed by an immunoblot analysis using a specific antibody for the mitochondrial protein VDAC1 (voltage-dependent anion-selective channel protein 1) (Clausen et al., 2004). The signals were detected with an ECL Western Detection kit (Millipore, Billerica, MA, USA).
Measurement of mitochondrial lipase enzyme activities in wild-type, 35S:AtDLAH transgenic, and atdlah mutant plants
Total proteins were isolated from purified mitochondrial fractions from wild-type, 35S:AtDLAH transgenic, and atdlah knock-out mutant seedlings as previously described by Seo et al. (2008). Cell extracts were used for colorimetric assays (40 μg total protein) as previously described by Seo et al. (2008).
Accelerated-ageing treatment and germination tests
For normal growth conditions, freshly harvested seeds were surface-sterilized and imbibed at 4 °C for 5 d. These seeds were plated on 0.5× MS medium (Duchefa, Haarlem, The Netherlands) and 0.7% phytoagar (Duchefa), pH 5.7, and then incubated in a growth chamber. For the accelerated-ageing treatment, seeds were incubated for 48 h at 43 °C and 100% relative humidity in a closed bottle before the cold imbibition (Sattler et al., 2004). In order to validate comparisons, all seeds are grown under the same conditions and harvested at the same time. Each day for 5 d, germination was scored by radicle emergence from the seeds.
Scanning electron microscopy
Dry seeds were coated with platinum–palladium in a sputter-coater as previously described by Ryu et al. (2009). The surface structure was subjected to high-resolution scanning electron microscopy (model S-800, FESEM, Hitachi, Tokyo, Japan) at an accelerating voltage of 3 kV under high vacuum conditions (Penfield et al., 2001; Atia et al., 2009).
FOX assay
To determine the level of LOOHs, total lipids were extracted and assayed as previously described (Griffiths et al., 2000; Zhu et al., 2009) with minor modifications. Briefly, the lipid extracts were incubated with FOX solution [90% methanol (v/v), 25 mM H2SO4, 4 mM butylated hydroxytoluene (BHT), 250 μM ferrous ammonium sulphate hexahydrate, and 100 μM xylenol orange] for 30 min at 25 °C. Absorbancies were immediately measured at 560 nm. Because the reactivity of 18:2-derived hydroperoxides with the FOX reagent was reported to be nearly identical to that of hydrogen peroxide (DeLong et al., 2002), serial concentrations (0, 0.05, 0.10, 0.20, 0.25, 0.30, and 0.50 mM) of hydrogen peroxide were used to make a standard curve, and the levels of LOOHs in the seeds were calculated using the standard curve.
TLC analysis of total polar lipid contents
Total lipids were extracted from 30 mg of dry seeds as previously described (Welti et al., 2002; Devaiah et al., 2006). To inhibit phospholipase activities, seeds were homogenized in 1 ml of isopropanol with 0.01% BHT at 75 °C. For lipid extraction, the ground samples were extracted several times with chloroform. The lipid extracts were separated on TLC plates and stained with iodine vapour. Stained bands were quantified using Multi Gauge v.3.1 (Fuji Film, Tokyo, Japan).
Results
Characterization of AtDLAH
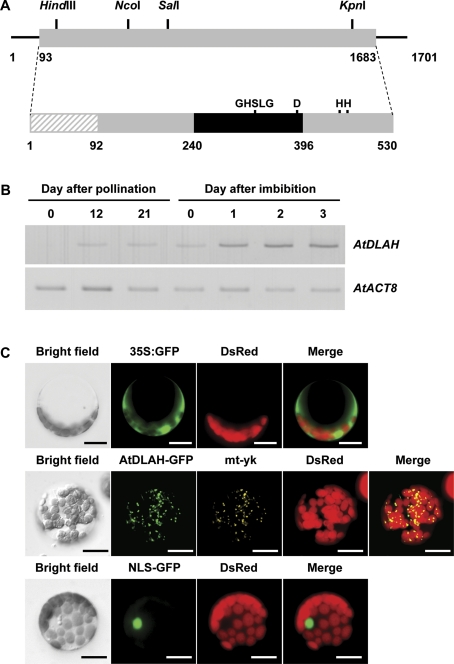
At1g30370 is classified as the only member of the class III PLA1 family because it contains a putative N-terminal transit peptide for localizing to the mitochondria (Ryu, 2004). The At1g30370 gene is comprised of a single 1,590 bp exon, and encodes a 530 amino acid protein containing a lipase 3 domain (Fig. 1A). The predicted molecular mass of the At1g30370 protein was determined to be 60.8 kDa and the calculated pI was 9.9. Sequences common to lipase active sites, such as the GXSXG motif and a catalytic triad (serine, aspartate, and histidine residues), were conserved in the lipase 3 domain, indicating that At1g30370 has typical lipase features. Therefore, At1g30370 was termed AtDLAH (Arabidopsis thaliana DAD1-like acylhydrolase). A database search revealed that AtDLAH was most closely related to a poplar protein (Populus trichocarpa, XP_002314049.1) and a castor bean triacylglycerol lipase (Ricinus communis, XP_002531054.1) with 68% and 63% identities, respectively (Supplementary Fig. S1 at JXB online). AtDLAH also shared relatively high sequence identity with a grape protein (Vitis vinifera, XP_002272780.1; 59% identity) and a rape chloroplast lipase (Brassica napus, ACJ76846.1; 45% identity). The cellular functions of these putative plant lipases are currently unknown. Protein sorting signal prediction programs, such as PSORT (http://psort.ims.u-tokyo.ac.jp/form.html) and TargetP (http://www.cbs.dtu.dk/services/TargetP), predicted that AtDLAH has a 92 amino acid N-terminal transit peptide for targeting to the mitochondria (TargetP score: 0.513). These results strongly suggest that AtDLAH is a mitochondria-localized DAD1-like acylhydrolase.
Fig. 1.
Structure, expression, and subcellular localization of Arabidopsis AtDLAH. (A) Schematic representation of AtDLAH (At1g30370) cDNA and its deduced protein. Solid lines depict the 5'- and 3'-untranslated regions. The coding region (grey box) with restriction enzyme sites, the lipase 3 domain (black box), and the putative N-terminal transit peptide (hatched box) are represented. The lipase consensus sequence (GHSLG) and the catalytic triad (serine, aspartate, and two candidate histidine residues) are indicated. (B) AtDLAH expression in developing and early germinating seeds was analysed by RT-PCR. AtACT8 was used as a loading control. (C) Subcellular localization of AtDLAH in Arabidopsis protoplasts. The 35S:AtDLAH-GFP fusion gene was introduced into protoplasts using a PEG-mediated method. The 35S:GFP and 35S:NLS-GFP constructs were used as controls for cytosolic and nuclear proteins, respectively. The mt-yk plant was used as a mitochondria-localized marker. Scale bars=10 μm. (This figure is available in colour at JXB online.)
Expression and subcellular localization of AtDLAH
Using semi-quantitative RT-PCR with gene-specific primers, the temporal expression patterns of AtDLAH in developing and early germinating Arabidopsis seeds were examined. Figure 1B shows that AtDLAH transcript was slightly detected during seed development stages, while its level was elevated in germinating seeds after imbibition. Since the mRNA for AtDLAH was gradually expressed in germinating seeds, AtDLAH probably plays a role in early Arabidopsis seedling development.
To determine if AtDLAH was localized to the mitochondria as predicted, a protoplast transient assay using AtDLAH-fused GFP as a fluorescent marker was performed. Figure 1C shows that control GFP was uniformly distributed throughout the cytosolic fractions of protoplasts, and GFP fused to the synthetic NLS sequence (NLS–GFP) was exclusively localized to the nuclei. In contrast, the AtDLAH–GFP fusion protein displayed a spotted pattern. Because similar spotted signals were previously detected for mitochondria-localized proteins (Kabeya and Sato, 2005; Sheahan et al., 2005), there is a possibility that the AtDLAH–GFP fusion protein is also localized to the mitochondria. To test this possibility, the AtDLAH–GFP fusion protein was expressed in protoplasts of organelle-specific marker plants (mt-yk CS16264), which contain a mitochondrial protein fused with yellow fluorescence protein (YFP) (Nelson et al., 2007). The GFP signals were overlaid with the yellow fluorescence signal of the mitochondrial marker. These results indicate that the AtDLAH–GFP fusion protein is primarily localized to the mitochondria of Arabidopsis leaf protoplasts.
In vitro enzyme activity and substrate specificity of AtDLAH
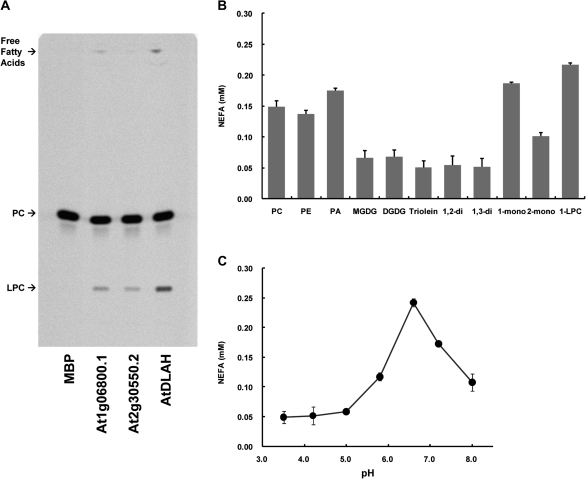
Ishiguro et al. (2001) previously reported that DAD1 exhibits PLA1 activity. Because AtDLAH has a highly conserved lipase 3 domain similar to that in DAD1, AtDLAH is also considered to be a member of the PLA1 family (Ryu, 2004). To test whether the AtDLAH protein exhibits PLA1 activity, AtDLAH was expressed without its transit peptide in E. coli as a fusion with maltose-binding protein (MBP). The cleavage site of the transit peptide for the protein was determined using the TargetP program (probability score: 0.513) and analysis of previous reports (Ishiguro et al., 2001; Padham et al., 2007; Hyun et al., 2008). The purified MBP–AtDLAH protein produced 1-LPC after incubation with 1-palmitoyl-2-[14C]linoleoyl-PC as a substrate (Fig. 2A). These results indicate that AtDLAH can catalyse the hydrolysis of PC at the sn-1 position in vitro.
Fig. 2.
AtDLAH enzyme assays. (A) AtDLAH catalyses the hydrolysis of PC at the sn-1 position. PLA1 activity was measured by the production of radiolabelled lysophosphatidylcholine (LPC) after incubation with 1-palmitoyl-2-[14C]palmitoyl-PC. The resultant 14C-labelled LPC was detected by TLC. MBP was used as a negative control, and At1g06800.1 and At2g30550.2, which have PLA1 activity (Seo et al., 2009), were used as positive controls. (B) Lipolytic enzyme assays to determine substrate specificity. Lipolytic activities were determined using an NEFA-HR kit with PC, PE, PA, MGDG, DGDG, triolein, 1,2-diacylglycerol, 1,3-diacylglycerol, 1-monodiacylglycerol, 2-monodiacylglycerol, and 1-LPC as substrates. Results are expressed as the means ±SD from four independent experiments. (C) Optimal pH for AtDLAH activity. Lipase activity of AtDLAH was determined by quantifying the release of free fatty acids from 1-LPC in phosphate buffers with different pHs at 30 °C for 30 min. Results are expressed as the means ±SD from four independent experiments.
Although AtDLAH has PLA1 activity in vitro, it could possess activities for other lipid substrates as reported previously (Padham et al., 2007; Hyun et al., 2008; Seo et al., 2009). To examine this possibility, various lipid substrates, including PC, PE, PA, MGDG, DGDG, triolein, 1,2-diacylglycerol, 1,3-diacylglycerol, 1-monodiacylglycerol, 2-monodiacylglycerol, and 1-LPC, were used for in vitro enzyme assays. Under the experimental conditions used in this study, MBP–AtDLAH displayed a strong preference for 1-LPC, 1-monodiacylglycerol, and PA, and, to a lesser extent, MBP–AtDLAH possessed phopholipase activity toward PC and PE (Fig. 2B). Therefore, AtDLAH contains lipase activity toward a broad range of lipid substrates with preferential specificities for lipids with an acyl chain on their sn-1 position. Additionally, AtDLAH functions optimally at a pH of 6.6 with 1-LPC as a substrate (Fig. 2C).
Generation and characterization of AtDLAH-overexpressing transgenic and atdlah loss-of-function mutant plants
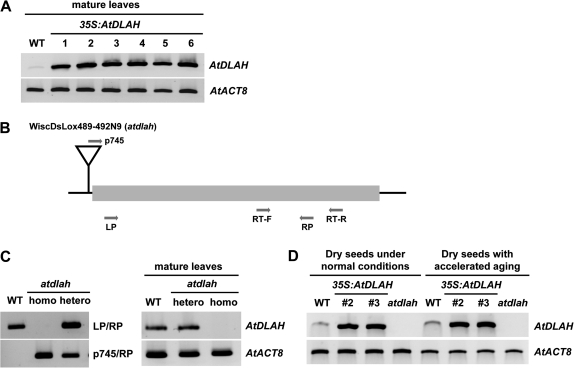
To address the cellular function of AtDLAH, overexpression and reverse-genetic approaches were used. Transgenic Arabidopsis plants (35S:AtDLAH) that ectopically expressed AtDLAH under the control of the Cauliflower mosaic virus (CaMV) 35S promoter were developed. Overexpression of AtDLAH in independent T4 transgenic lines was confirmed by RT-PCR (Fig. 3A). The loss-of-function T-DNA knockout mutant (WiscDsLox489_492N9; atdlah) for AtDLAH was also identified by genotyping PCR and RT-PCR (Fig. 3B, C). In normal and accelerated-ageing-treated seeds, transcript levels of AtDLAH in wild-type, 35S:AtDLAH, and mutant plants were highly similar to those in leaves (Fig. 3D). These plants were subsequently used for phenotypic analysis.
Fig. 3.
Molecular characterization of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants. (A) RT-PCR analysis of wild-type and six independent AtDLAH-overexpressing T4 transgenic plants (lines #1, #2, #3, #4, #5, and #6) in leaves. The AtACT8 gene was used as a loading control. (B) Schematic representation of the atdlah mutant. The T-DNA insertion is shown as an inverted triangle. The shaded bar indicates the coding region, and gene-specific (LP and RP) and T-DNA-specific (p745) primers used for genotyping and RT-PCR are indicated with arrows. (C) PCR analysis of the atdlah mutant in leaves. Genomic PCR analysis of the atdlah loss-of-function mutant using gene-specific and T-DNA-specific primers (left panel). RT-PCR analysis of the atdlah mutant (right panel). The AtACT8 gene was used as a loading control. DNA sequences of primers used in this study are shown in Supplementary Table S1 at JXB online. (D) RT-PCR analysis of wild-type, 35S:AtDLAH (transgenic lines #2 and #3), and atdlah mutant seeds without or with accelerated-ageing treatment. The AtACT8 gene was used as a loading control.
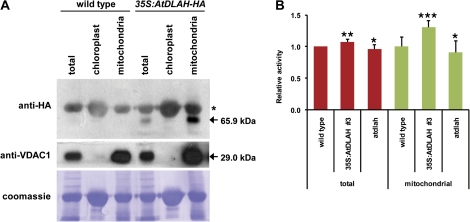
To ensure further the ectopic expression of AtDLAH and its mitochondrial localization at the protein level, chloroplast and mitochondrial fractions were isolated from light-grown 2-week-old leaves of wild-type and 35S:AtDLAH-HA T4 transgenic leaves. Protein extracts were prepared from each fraction and subsequently analysed by protein gel blotting using an anti-haemagglutinin (HA) antibody. As shown in Fig. 4A, the 65.9 kDa band specific to AtDLAH-HA was predominantly present in the mitochondrial fraction, confirming its mitochondrial localization. VDAC1, a mitochondria-specific marker protein, was exclusively detected in the corresponding fraction.
Fig. 4.
Mitochondrial localization of AtDLAH-HA and mitochondrial lipase activities in wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants. (A) Cellular fractionation analysis of the AtDLAH-HA fusion protein. Extracts of total, chloroplast, and mitochondrial fractions were prepared from wild-type and 35S:AtDLAH-HA T4 transgenic plants. Total proteins from each fraction were analysed with anti-HA and anti-VDAC1 antibodies (control for mitochondrial proteins). Loaded proteins were visualized by Coomassie staining. Arrows indicate AtDLAH-HA (65.9 kDa) and VDAC1 (29.0 kDa) proteins. An asterisk indicates non-specific binding to a 70 kDa protein by the anti-HA antibody. (B) Mitochondrial lipase enzyme assays. Total and mitochondrial proteins were prepared from wild-type, AtDLAH-overexpressing transgenic (line #3), and atdlah mutant plants and incubated with 1-palmitoyl-2-[14C]palmitoyl-PC as the substrate at 30 °C for 30 min. Lipase activities were determined by quantifying the release of 14C-labelled lyso-PC as described in Fig. 2A. Results are expressed as the means ±SD from three independent experiments. The data were analysed by Student's t-test. The statistical significance was determined at ***P <0.01, **P <0.05, and *P <0.1, respectively. (This figure is available in colour at JXB online.)
AtDLAH lipase activity was measured using mitochondrial protein extracts from wild-type, 35S:AtDLAH-HA, and atdlah leaves. The results in Fig. 4B demonstrate that the level of mitochondrial lipase activity in 35S:AtDLAH-HA (lines #2 and #3) was 1.3 times greater than that of the wild-type leaves. These results indicate that ectopic expression of AtDLAH caused a small but specific increase in mitochondrial lipase activity. However, the predicted decrease in mitochondrial lipase activity in the atdlah mutant as compared with that of the wild-type leaves was not identified.
AtDLAH-overexpressing transgenic seeds had higher germination percentages after ageing treatment as compared with wild-type and atdlah mutant seeds
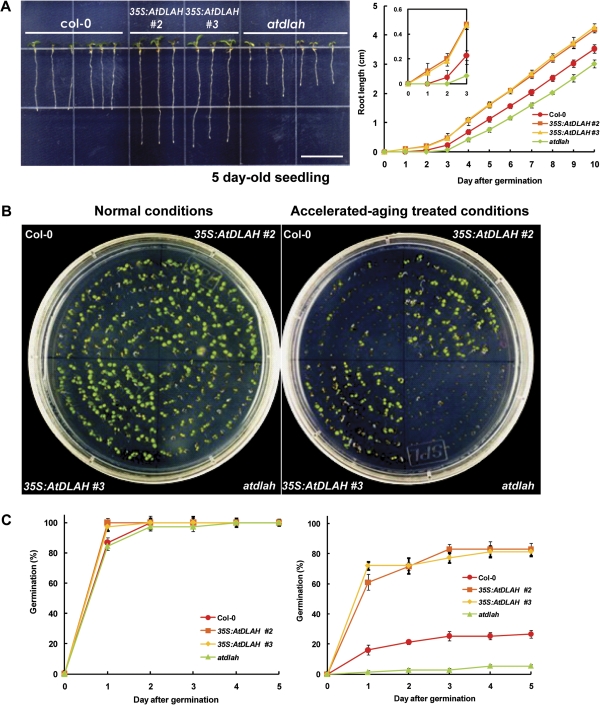
The morphological comparison of light-grown wild-type, 35S:AtDLAH (lines #2 and #3), and atdlah mutant seedlings at an early stage of development is presented in Fig. 5A. The AtDLAH-overexpressing transgenic plants displayed significantly longer roots than control seedlings 5 d after germination, while the atdlah mutant plants exhibited shorter roots than did the wild-type plants (Fig. 5A). To investigate whether the longer roots in 35S:AtDLAH seedlings were due to enhanced cell elongation and/or cell division, expression patterns of cell elongation- and division-associated genes were monitored. The expression levels of several cell elongation/division marker genes, including AtCYCD3, AtCDC2b, AtEXP5, and AtPCNA (Seo et al., 2008), were indistinguishable between the three types of seedlings (Supplementary Fig. S2 at JXB online), suggesting that the longer roots in the 35S:AtDLAH seedlings may not be a consequence of enhanced cell elongation or cell cycle progression.
Fig. 5.
Phenotypes of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants at early stages of development. (A) Morphologies of 5-day-old wild-type, AtDLAH-overexpressing transgenic (lines #2 and #3), and atdlah mutant seedlings (left panel). Root length was monitored for 10 d after germination (right panel). Results are expressed as means ±SD from three independent experiments. The inset shows the root length for 0–3 d after germination. (B) Germination of wild-type, AtDLAH-overexpressing, and atdlah mutant seeds under normal germination conditions (left panel) or following accelerated-ageing conditions (43 °C for 48 h at 100% relative humidity) (right panel). (C) Germination percentages of wild-type, AtDLAH-overexpressing (lines #2 and #3), and atdlah mutant seeds were monitored under normal (left panel) and accelerated-ageing (right panel) conditions for 5 d after germination. Germination was defined as radicle emergence. Results are expressed as means ±SD from three independent experiments. (This figure is available in colour at JXB online.)
The germination percentages of wild-type, 35S:AtDLAH transgenic, and atdlah mutant seeds were examined. Under normal germination conditions, 35S:AtDLAH transgenic seedlings (lines #2 and #3) displayed higher (∼15%) germination percentages 1 d after germination than the wild-type and mutant seedlings (left panels in Fig. 5B, C). Thereafter, the germination percentages were very similar among the three different types of plants, though the germination percentage of the mutant plant was slightly lower (∼3%). The higher germination percentage of the transgenic seeds probably resulted in the slightly larger cotyledons in the AtDLAH-overexpressing plants 5 d after germination (Fig. 5A, B). Therefore, the longer roots of the 35S:AtDLAH seedlings were also probably not due to increased cell elongation/division, but rather due to quicker germination. The slightly quicker germination in overexpressors and later germination in the mutant were also supported by the finding that there were apparent differences in root length even in the earlier germination stage (1–3 d after imbibition) (the inset graph of the right panel in Fig. 5A). In contrast, the root elongation rates remained constant in wild-type, 35S:AtDLAH, and atdlah seedlings for 10 d after germination (right panel in Fig. 5A), further supporting the role of AtDLAH in germination.
The germination of wild-type, 35S:AtDLAH, and atdlah mutant seeds was further analysed following treatment with accelerated-ageing conditions. Seeds were incubated with high temperature (43 °C) and high humidity (100% relative humidity) for 2 d, conditions known to accelerate seed ageing (Tesnier et al., 2002; Devaiah et al., 2007; Oge et al., 2008; Rajjou et al., 2008). Aged seeds were imbibed at 4 °C for 5 d to break dormancy, and then placed at 22 °C to germinate while root radicle emergence was monitored (Sattler et al., 2004). In response to the accelerated-ageing treatment, the germination percentage of wild-type seeds was reduced to the level of 16–27% at 1–5 d after germination (right panels in Fig. 5B, C). In contrast, aged 35S:AtDLAH seeds (lines #2 and #3) displayed markedly higher germination percentages (61–83%) than aged wild-type seeds, whereas aged mutant seeds rarely germinated (1–5%).
Abscisic acid (ABA) is a plant hormone that inhibits seed germination (Leung and Giraudat, 1998). To test whether the higher germination percentages of 35S:AtDLAH seeds under normal and accelerated-ageing treatments were due to reduced sensitivity to ABA, seed germination percentages of wild-type, 35S:AtDLAH, and atdlah plants in the presence of various ABA concentrations (0.1, 0.5, or 1 μM) were examined. All three types of plants displayed highly similar sensitivities to the various concentrations of exogenously applied ABA (Supplementary Fig. S3 at JXB online). Thus, the greater germination percentage of the 35S:AtDLAH seeds was probably due to increased seed viability, not decreased sensitivity to ABA. Taken together, the results in Fig. 5 suggest that AtDLAH activity is positively associated with germination ability, and this effect was more evident after accelerated-ageing treatments.
AtDLAH activity was positively correlated with seed viability and longevity
Seed coats play an important role in seed viability and longevity since they provide the primary line of defence against unfavourable environmental conditions (Mohamed-Yasseen et al., 1994). Development of the epidermal layer in the Arabidopsis seed coat is a complex process, involving cell growth, biosynthesis and secretion of pectinaceous mucilage, and production of a secondary cell wall (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). Because of the water-holding capacity of the mucilage layer in Arabidopsis seeds, it is difficult to monitor seed viability (Debeaujon and Koornneef, 2000). Therefore, the tetrazolium uptake assay was employed to assess seed viability. Upon entry of the tetrazolium solution into seeds, the aleurone layer of live embryos stain red, whereas seeds with low viability stain a whitish colour (Rossetto et al., 2004; Oge et al., 2008). The 35S:AtDLAH (lines #2 and #3) seeds had a dark-red staining pattern after tetrazolium treatment under both normal and accelerated-ageing conditions (Supplementary Fig. S4A at JXB online). However, the atdlah mutant seeds clearly had less of a red-stained pattern as compared with the AtDLAH overexpressors. As expected, the wild-type seeds had an intermediate degree of tetrazolium staining (Supplementary Fig. S4A). Therefore, the level of AtDLAH expression was positively correlated with seed viability.
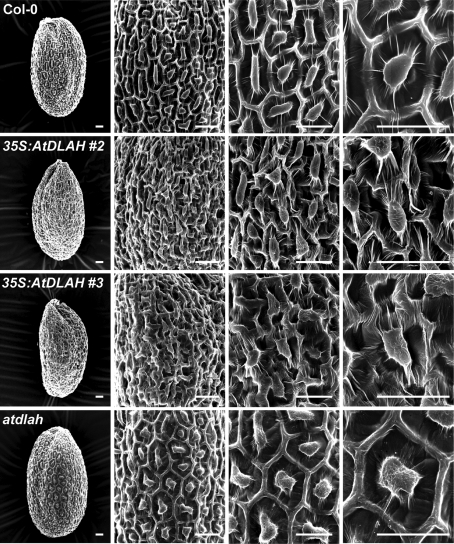
The structural aspects of the seed coat of wild-type, 35S:AtDLAH transgenic, and atdlah mutant seeds was examined using a scanning electron microscope. In general, the epidermal layers of Arabidopsis seed coats have hexagonal morphologies, a volcano-shaped structure known as the columella, and mucilage. The epidermal cells of wild-type and mutant seed coats displayed these typical properties (Fig. 6). However, the AtDLAH-overexpressing transgenic seed coats exhibited unusual epidermal cells that were more populous and had amorphous shapes (Fig. 6). To determine if this structural abnormality affects the extrusion of pectinaceous mucilage, wild-type, 35S:AtDLAH, and atdlah mutant seeds were subjected to ruthenium red staining that selectively stains pectinaceous mucilage layers (Arsovski et al., 2009). The mucilage layers extruded from seed coats were equally stained with ruthenium red, regardless of the plant type (Supplementary Fig. S4B at JXB online), indicating that the unusual morphology of the cell layers of the 35S:AtDLAH seed coat did not alter the mucilage layers. The overexpression of AtDLAH resulted in dense and amorphous epidermal cells in seed coats, which correlated with the enhanced seed longevity of 35S:AtDLAH transgenic plants.
Fig. 6.
Structural aspects of the seed coat of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants. Scanning electron microscopic examination was performed with wild-type, AtDLAH-overexpressing, and atdlah mutant seeds. Scale bars=150 μm.
Analysis of lipid peroxidation and total polar lipid contents after ageing treatments
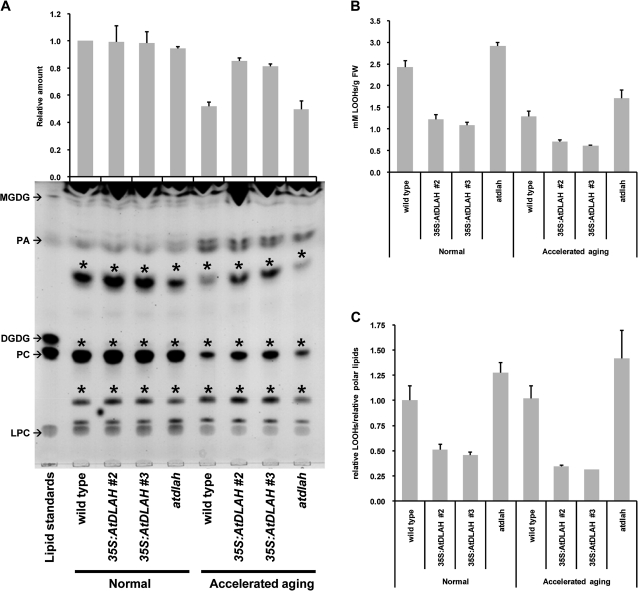
To examine further why 35S:AtDLAH seeds have a higher germination percentage, qualitative and quantitative changes in total seed lipid content were evaluated. Total lipids from wild-type, 35S:AtDLAH, and atdlah mutant seeds with or without accelerated-ageing treatment were extracted, separated on TLC plates, and visualized with iodine vapour. Under normal growth conditions, the total polar lipid content was highly similar among these seeds. More detailed inspection, however, indicates that there were detectable differences in intensities of 2–3 polar lipid bands on TLC plates. These lipid bands are indicated by an asterisk in Fig. 7A. Furthermore, after accelerated-ageing treatments, the quantities of polar lipids in wild-type and atdlah mutant seeds were reduced by ∼50% as compared with seeds under normal conditions (Fig. 7A), a finding consistent with previous results (Pearce and Abdel-Samad, 1980; Pukacka and Kuiper, 1988; Ouzouline et al., 2009). However, the 35S:AtDLAH seeds (line #2 and #3) lost only 15–19% of their polar lipids during accelerated-ageing treatments relative to the seeds under normal conditions, indicating that the decrease in polar lipids in 35S:AtDLAH seeds was not as drastic as those in wild-type and mutant seeds. On the other hand, the levels of neutral storage lipids, including TAGs and diacylglycerols, were not significantly changed in response to accelerated-ageing treatments (Supplementary Fig. S5 at JXB online). These results suggest that enough polar lipids remain in 35S:AtDLAH seeds to germinate following accelerated-ageing treatment, as overexpression of AtDLAH resulted in a smaller reduction of polar lipids.
Fig. 7.
Lipid peroxidation and lipid content in wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant seeds. (A) Total lipid content in wild-type, AtDLAH-overexpressing (lines #2 and #3), and atdlah mutant seeds under normal and accelerated-ageing conditions. Total polar lipids were separated by TLC and visualized with iodine vapour. The lipid standards from top to bottom are MGDG, PA, DGDG, PC, and LPC. The amounts of individual polar lipids (represented above the plate) were quantified from the TLC plates using Multi Gauge v.3.1 (Fuji Film, Tokyo, Japan). The lipid bands which showed the detectable differences in intensity on the TLC plate among wild-type, 35S:AtDLAH, and ataldh are indicated by asterisks (*). Results are expressed as means ±SD from three independent experiments. (B) Absolute levels of lipid hydroperoxide in wild-type, AtDLAH-overexpressing (lines #2 and #3), and atdlah mutant seeds under normal and accelerated-ageing conditions. The levels of lipid hydroperoxides (LOOHs) were measured by FOX assay. The different amounts of H2O2 were used for generating a standard curve and the amount of LOOH in seeds was calculated as a H2O2 level. LPC (40 μg) was used as a control for lipid extraction from each seed sample. Results are expressed as means ±SD from three independent experiments. (C) Relative levels of LOOHs per polar lipid in wild-type, AtDLAH-overexpressing (lines #2 and #3), and atdlah mutant seeds under normal and accelerated-ageing conditions. Each ratio was calculated by dividing the amount of LOOH by the relative amount of polar lipids. Results are expressed as means ±SD from three independent experiments.
The loss of polar lipids during ageing is mainly due to lipid peroxidation, and this oxidative stress is a major contributor to seed deterioration (Bailly et al., 1996, 1998; Devaiah et al., 2006, 2007; Ouzouline et al., 2009). To determine the level of lipid peroxidation in wild-type, 35S:AtDLAH, and atdlah mutant seeds, the quantity of LOOHs in seeds was measured using ferrous oxidation–xylenol orange (FOX) assays (DeLong et al., 2002; Zhu et al., 2009). Total lipids were extracted from seeds and incubated with FOX solution, and LOOH levels were measured by spectrophotometry. Because the reactivity of 18:2-derived hydroperoxides with the FOX reagent was reported to be nearly identical to that of hydrogen peroxide (DeLong et al., 2002), different concentrations of H2O2 were used for generating a standard curve, and the levels of LOOHs were calculated as H2O2 levels. The mean LOOH level in wild-type seeds was 2.42±0.15 mM g−1 FW (Fig. 7B). The mean concentrations of LOOHs in 35S:AtDLAH (lines #2 and #3) and atdlah mutant seeds were 1.11±0.068–1.23±0.11 mM g−1 FW and 3.08±0.084 mM g−1 FW, respectively. Therefore, the 35S:AtDLAH seeds accumulated a significantly lower level of LOOHs than wild-type seeds, whereas the mutant seeds contained the highest level of lipid hydroperoxidation (Fig. 7B). After accelerated-ageing treatments, this lipid hydroperoxidation trend was maintained, even though the absolute amounts of LOOHs in all seeds were reduced to nearly half of their starting levels (Fig. 7B). In addition, the ratio of LOOH to polar lipids in 35S:AtDLAH seeds (lines #2 and #3) was further reduced by one-third of that in the wild-type seeds in response to accelerated-ageing treatment, whereas the relative level was increased 1.4 times in atdlah mutant seeds (Fig. 7C). These results indicate that propagation of lipid peroxidation during seed ageing was repressed by overexpression of AtDLAH and accelerated by ablation of the gene. Thus, overexpression of AtDLAH decreased the reduction of polar lipids and reduced lipid peroxidation, which probably led to the increased seed viability.
Discussion
In this study, AtDLAH, an Arabidopsis DAD1-like acylhydrolase homologue encoded by the At1g30370 gene, was characterized. AtDLAH has a well-conserved GHSLG lipase consensus sequence and a catalytic triad similar to fungal and animal lipases (Fig. 1A) (Brady et al., 1990; Winkler et al., 1990). Similar to DAD1, AtDLAH also has a highly conserved lipase 3 domain, and therefore, it belongs to the Arabidopsis PLA1 family (Ryu, 2004). Several recent reports suggested that DAD1-like PLA1 family members contain lipase activities in addition to their PLA1 activity (Padham et al., 2007; Hyun et al., 2008; Seo et al., 2009). Consistently, bacterially expressed AtDLAH had PLA1 activity for catalysing the hydrolysis of phospholipids at the sn-1 position (Fig. 2A); however, it displayed an even stronger preference toward 1-LPC, 1-monodiacylglycerol, and PA (Fig. 2B). These results suggest that AtDLAH has lipase activity toward a broad range of lipid substrates with a preferential acylhydrolase activity on the sn-1 position of its lipid substrates.
In silico analyses indicated that AtDLAH is a mitochondria-localized class III PLA1 (Ishiguro et al., 2001; Ryu, 2004). AtDLAH was predominantly found in the mitochondria in both the protoplast transient assay (Fig. 1C) and the cellular fractionation study (Fig. 4A). In addition, 35S:AtDLAH transgenic lines had significantly higher lipase activities in their mitochondrial fractions relative to those of the wild-type and atdlah mutant plants (Fig. 4B). Thus, AtDLAH is most probably a mitochondria-localized DAD1-like acylhydrolase. A number of mitochondrial lipases with critical cellular functions, such as preventing oxidant-induced lipid peroxidation and rescuing cells from death, have been identified in various species, including humans, animals, and yeast (Claycomb and Kilsheimer, 1971; Schousboe, 1976; Demant, 1978; Andersen et al., 2009). In contrast, only a few plant lipases, such as peanut lipase and potato PLA, have been identified as mitochondria-targeted lipases (Jacks et al., 1967; Hasson and Laties, 1976). Research elucidating functional roles of these putative plant mitochondrial lipases is scarce.
These results prompted this investigation of the cellular functions of AtDLAH. This initial phenotypic analysis demonstrated that AtDLAH-overexpressing young seedlings contained significantly longer roots than wild-type and atdlah seedlings (Fig. 5A). This apparent phenotype was reminiscent of the transgenic Arabidopsis plants that constitutively expressed a hot pepper phospholipase 1 (CaPLA1). The root length of 35S:CaPLA1 seedlings was longer than that of wild-type plants, and was caused by promotion of the cell cycle and enhanced fatty acid metabolism (Seo et al., 2008). However, the longer-root phenotype of 35S:AtDLAH was unlikely to be due to increased cell elongation or cell cycle progression (Supplementary Fig. S2 at JXB online), but was probably due to enhanced seed germination under normal conditions (Fig. 5B, C). More importantly, 35S:AtDLAH seeds were markedly tolerant to accelerated-ageing treatments, while most atdlah seeds failed to germinate under the accelerated-ageing conditions. Because 35S:AtDLAH and atdlah seeds displayed very similar sensitivities to ABA in terms of germination percentages (Supplementary Fig. S3 at JXB online), AtDLAH probably plays a positive role in protecting and/or maintaining seed contents that are important for germination. This view is further supported by the finding that AtDLAH-overexpressing seeds were more effectively stained by tetrazolium (Supplementary Fig. S4A) and contained more densely populated epidermal cells with amorphous shapes (Fig. 6) in comparison with the wild-type and atdlah mutant seeds.
Seed deterioration during storage is accompanied by a progressive loss of membrane lipids (Stewart and Bewley, 1980; Samama and Pearce, 1993; Al-Maskri et al., 2003; Sattler et al., 2006; Devaiah et al., 2007). This type of polar lipid degradation is mainly due to lipid peroxidation caused by natural ageing and environmental factors, such as temperature, humidity, and oxygen (Smirnoff, 1993; Bailly et al., 1996, 1998; Rajjou and Debeaujon, 2008; Ouzouline et al., 2009; Mene-Saffrane et al., 2010). A previous study using accelerated-ageing-treated, tocopherol-deficient Arabidopsis mutants (vte1 and vte2 plants), indicated that tocopherols can prevent membrane lipid peroxidation during seed storage, germination, and early seedling development, thus preserving seed viability (Sattler et al., 2004). The 35S:AtDLAH seeds harboured greater amounts of polar lipids following accelerated-ageing treatments than wild-type and atdlah knockout mutant seeds (Fig. 7A). Furthermore, the lipid peroxidation level of 35S:AtDLAH seeds was almost 2-fold lower than that of wild-type and mutant seeds under normal conditions, and this difference was increased to 3-fold following accelerated-ageing treatments (Fig. 7B). Therefore, cellular levels of AtDLAH are inversely correlated to the peroxidation of polar lipids in mature seed embryos and, in turn, the reduction of lipid hydroperoxide content enhances ageing tolerance and seed viability.
For germination, Arabidopsis seeds obtain most of their metabolic energy from sucrose in the cotyledons of mature embryos, rather than from storage lipids (Baud et al., 2002; Dekkers et al., 2004; Cernac et al., 2006; Andre and Benning, 2007). On the other hand, storage lipid mobilization is essential for subsequent seedling establishment, including elongation of the hypocotyl and root, greening of the cotyledons, and transition from a heterotroph to a photoautotroph (Eastmond, 2006). Therefore, the increased ageing tolerance of 35S:AtDLAH seeds may have been caused by limiting the peroxidation and degradation of the polar lipids in structural embryonic membranes, not by reduction of oxidative damage in neutral storage lipids. The results of the current study suggest that there was no difference in the quantity of neutral lipids among wild-type, 35S:AtDLAH, and atdlah seeds following accelerated-ageing treatments (Supplementary Fig. S5 at JXB online). Additionally, root elongation rates were constant in these seedlings 1–10 d after germination (Fig. 5A), suggesting that the differences in seed viabilities were not a result of the mobilization ability of the storage lipids. In addition, although the total amounts of the polar lipids seemed to be indistinguishable among wild-type, overexpressors, and mutant seeds, the intensities of 2–3 polar lipid bands on TLC plates were apparently different among these lines under normal conditions (Fig. 7A). These results suggest that the differences in some kinds of polar lipid contents resulting from the changes in expression of AtDLAH are possibly attributed to germination under normal conditions (Fig. 5A). These differences were further exacerbated by accelerated-ageing treatment (Fig. 7) sufficient to produce large differences in germination ability (Fig. 5B, C).
Recently, polar lipid profiling and lipid peroxidation analyses with a PLDα1 knockout mutant indicated that the activity of PLDα1 may promote membrane lipid degradation and reduce seed viability (Devaiah et al., 2006, 2007). After storage and accelerated ageing, the pldα1 mutant seeds exhibited higher germination percentages, decreased reduction in oil content, and significantly lower lipid hydroperoxide levels than did wild-type seeds that were similar to the 35S:AtDLAH seeds. Therefore, it is possible that PLDα1 and AtDLAH have conflicting roles in peroxidation and degradation of polar lipids. One possible mechanism is that PLDα1 and AtDLAH compete for the same phospholipid substrates. This hypothesis is partially supported by the finding that PLDα1 is found not only in the plasma membrane fractions but also in the mitochondria (Fan et al., 1999) where AtDLAH is predominantly localized (Figs 1C, 4).
PA is a strong candidate for a signalling molecule for generating reactive oxygen species (ROS) and inducing lipid peroxidation (Sang et al., 2001; Park et al., 2004; Devaiah et al., 2007). In Arabidopsis leaves, PA activates NADPH oxidase activity to produce superoxide, which is immediately converted to H2O2. These PA-induced ROS, in turn, promote the death of leaf cells (Sang et al., 2001; Park et al., 2004). There is circumstantial evidence that PA produced by PLDα1 enhances production of lipid peroxidation in Arabidopsis seeds, thereby decreasing seed quality during natural and accelerated ageing processes (Devaiah et al., 2007). Under the in vitro lipase enzyme assay conditions used in this study, PA was one of the preferred substrates for AtDLAH (Fig. 2B). Therefore, PA produced by natural and accelerated ageing conditions could be degraded by AtDLAH in transgenic 35S:AtDLAH seeds, resulting in increased seed longevity.
Alternatively, PLDα1 and AtDLAH may work independently. The overall level of PA detected by TLC was similar in wild-type, 35S:AtDLAH, and atdlah seeds (Fig. 7A). Thus, more detailed quantifications of PA and other signalling lipids are required to elucidate the roles of PLDα1 and AtDLAH further. Current efforts are focused on analysing total and mitochondrial lipid profiles in wild-type, 35S:AtDLAH, and atdlah seeds using an electrospray ionization tandem mass spectrometer (ESI-MS/MS), rather than TLC, to elucidate the effect of AtDLAH expression on both qualitative and quantitative traits of seed lipids. In addition, analysis of phenotypes and cellular functions of AtDLAH with fine-regulated ageing treatments under lower relative humidity conditions, which may be more similar to natural ageing conditions (Oge et al., 2008), is also being conducted. These results will clarify the mode of action of mitochondrial DAD1-like acylhydolase in seed viability and longevity.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence analysis of Arabidopsis AtDLAH.
Figure S2. Expression levels of cell cycle- and cell elongation-related genes in wild-type, 35S:AtDLAH, and atdlah mutant seedlings.
Figure S3. Germination analysis of wild-type, 35S:AtDLAH, and atdlah mutant seeds in response to ABA.
Figure S4. Vital staining with tetrazolium and mucilage release of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant seeds.
Figure S5. Neutral lipid content of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant seeds.
Table S1. Primer sequences used for cloning AtDLAH cDNA, construction of the MBP fusion protein, the GFP fusion protein, and transgenic plants, genotyping PCR, and RT-PCR.
Supplementary Material
Acknowledgments
This work was supported by grants from the Technology Development Program for Agriculture and Forestry (Project no. 309017-5 funded by the Ministry for Agriculture, Forestry and Fisheries, Republic of Korea), the National Research Foundation (Project no. 2009-0078317 funded by the Ministry of Education, Science, and Technology, Republic of Korea), and the National Center for GM Crops (Project no. PJ008152 of the Next Generation BioGreen 21 Program funded by the Rural Development Administration, Republic of Korea) to WTK.
Glossary
Abbreviations
- DGDG
digalactosyldiacylglycerol
- LOOH
lipid hydroperoxide
- 1-LPC
1-lysophosphatidylcholine
- MGDG
monogalactosyldiacylglycerol
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- TAG
triacylglycerol
References
- Al-Maskri AY, Khan MM, Khan IA, Al-Habsi K. Effect of accelerated aging on viability, vigor (RGR), lipid peroxidation and leakage in carrot (Daucus carota L.) seeds. International Journal of Agriculture and Biology. 2003;5:580–584. [Google Scholar]
- Andersen AD, Poulsen KA, Lambert IH, Pedersen SF. HL-1 mouse cardiomyocyte injury and death after simulated ischemia and reperfusion: roles of pH, Ca2+-independent phospholipase A(2), and Na+/H+ exchange. American Journal of Physiology-Cell Physiology. 2009;296:C1227–C1242. doi: 10.1152/ajpcell.00370.2008. [DOI] [PubMed] [Google Scholar]
- Andre C, Benning C. Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiology. 2007;145:1670–1680. doi: 10.1104/pp.107.108340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atia A, Debez A, Barhoumi Z, Abdelly C, Smaoui A. Histochemical localization of essential oils and bioactive substances in the seed coat of the halophyte Crithmum maritimum L. (Apiaceae) Journal of Plant Biology. 2009;52:448–452. [Google Scholar]
- Arsovski AA, Villota MM, Rowland O, Subramaniam R, Western TL. MUM ENHANCERS are important for seed coat mucilage production and mucilage secretory cell differentiation in Arabidopsis thaliana. Journal of Experimental Botany. 2009;60:2601–2612. doi: 10.1093/jxb/erp102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D. Changes in malondialdehyde content and in superoxide dismutase, catalase and glutathione reductase activities in sunflower seeds as related to deterioration during accelerated aging. Physiologia Plantarum. 1996;97:104–110. [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D. Free radical scavenging as affected by accelerated ageing and subsequent priming in sunflower seeds. Physiologia Plantarum. 1998;104:646–652. [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiology and Biochemistry. 2002;40:151–160. [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inze D. Histological study of seed coat development in Arabidopsis thaliana. Journal of Plant Research. 2000;113:139–148. [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiology. 2003;132:681–697. doi: 10.1104/pp.103.022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: physiology of development and germination. New York: Plenum Press; 1994. [Google Scholar]
- Brady L, Brzozowski AM, Derewenda ZS, Dodson E, Dodson G, Tolley S, Turkenburg JP, Christiansen L, Huge-Jensen B, Norskov L. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C. WRI1 is required for seed germination and seedling establishment. Plant Physiology. 2006;141:745–757. doi: 10.1104/pp.106.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. The Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels MJ, Sadava DE. Plants, genes, and crop biotechnology. Boston: Jones and Bartlett Publishers, Inc.; 2003. [Google Scholar]
- Clausen C, Ilkavets I, Thomson R, Philippar K, Vojta A, Mohlmann T, Neuhaus E, Fulgosi H, Soll J. Intracellular localization of VDAC proteins in plants. Planta. 2004;220:30–37. doi: 10.1007/s00425-004-1325-3. [DOI] [PubMed] [Google Scholar]
- Claycomb WC, Kilsheimer GS. Purification and properties of a lipase from rat liver mitochondria. Journal of Biological Chemistry. 1971;246:7139–7143. [PubMed] [Google Scholar]
- Clerkx EJM, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M. Genetic differences in seed longevity of various Arabidopsis mutants. Physiologia Plantarum. 2004;121:448–461. [Google Scholar]
- Coolbear P. Mechanism of seed deteroration. In: Basra AS, editor. Seed quality: basic mechanisms and agricultural implications. New York: Food Product Press; 1995. pp. 223–277. [Google Scholar]
- Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiology. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC. Glucose delays seed germination in Arabidopsis thaliana. Planta. 2004;218:579–588. doi: 10.1007/s00425-003-1154-9. [DOI] [PubMed] [Google Scholar]
- DeLong JM, Prange RK, Hodges DM, Forney CF, Bishop MC, Quilliam M. Using a modified ferrous oxidation–xylenol orange (FOX) assay for detection of lipid hydroperoxides in plant tissue. Journal of Agricultural and Food Chemistry. 2002;50:248–254. doi: 10.1021/jf0106695. [DOI] [PubMed] [Google Scholar]
- Demant EJ. Fatty acid inhibition of triacylglycerol lipase in mitochondrial fractions from baker's yeast. FEBS Letters. 1978;85:109–113. doi: 10.1016/0014-5793(78)81259-9. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Pan XQ, Hong YY, Roth M, Welti R, Wang XM. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. The Plant Journal. 2007;50:950–957. doi: 10.1111/j.1365-313X.2007.03103.x. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry. 2006;67:1907–1924. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. The Plant Cell. 2006;18:665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. The Plant Cell. 2007;19:1376–1387. doi: 10.1105/tpc.106.043992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, Stingl N, Kubigsteltig, Bals T, Juenger M, Pollmann S, Berger S, Schuenemann D, Mueller MJ. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: redundant lipases contribute to jasmonate formation. Plant Physiology. 2010;153:114–127. doi: 10.1104/pp.110.155093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zheng S, Cui D, Wang X. Subcellular distribution and tissue expression of phospholipase Dα, DβΔ, and Dγ in Arabidopsis. Plant Physiology. 1999;119:1371–1378. doi: 10.1104/pp.119.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Leverentz M, Silkowski H, Gill N, Sanchez-Serrano JJ. Lipid hydroperoxide levels in plant tissues. Journal of Experimental Botany. 2000;51:1363–1370. [PubMed] [Google Scholar]
- Hasson EP, Laties GG. Purification and characterization of an a type phospholipase from potato and its effect on potato mitochondria. Plant Physiology. 1976;57:148–152. doi: 10.1104/pp.57.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Gan SS. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. The Plant Cell. 2002;14:805–815. doi: 10.1105/tpc.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Choi S, Hwang HJ, et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Developmental Cell. 2008;14:183–192. doi: 10.1016/j.devcel.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. The Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks TJ, Yatsu LY, Altschul AM. Isolation and characterization of peanut spherosomes. Plant Physiology. 1967;42:585–597. doi: 10.1104/pp.42.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Sato N. Unique translation initiation at the second AUG codon determines mitochondrial localization of the phage-type RNA polymerases in the moss Physcomitrella patens. Plant Physiology. 2005;138:369–382. doi: 10.1104/pp.105.059501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Seo YS, Lee H, Kim WT. Constitutive expression of CaSRP1, a hot pepper small rubber particle protein homolog, resulted in fast growth and improved drought tolerance in transgenic Arabidopsis plants. Planta. 2010;232:71–83. doi: 10.1007/s00425-010-1149-2. [DOI] [PubMed] [Google Scholar]
- Kim EY, Seo YS, Kim WT. AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. Journal of Plant Physiology. 2011;168:1705–1709. doi: 10.1016/j.jplph.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Lee HY, Bahn SC, Kang YM, Lee KH, Kim HJ, Noh EK, Palta JP, Shin JS, Ryu SB. Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. The Plant Cell. 2003;15:1990–2002. doi: 10.1105/tpc.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annual Review of Plant Physiology. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Li D, Pritchard H. The science and economics of ex situ plant conservation. Trends in Plant Science. 2009;14:614–621. doi: 10.1016/j.tplants.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Lo M, Taylor C, Wang L, Nowack L, Wang TW, Thompson J. Characterization of an ultraviolet B-induced lipase in Arabidopsis. Plant Physiology. 2004;135:947–958. doi: 10.1104/pp.103.036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MB. Seed deterioration: physiology, repair and assessment. Seed Science and Technology. 1999;27:177–237. [Google Scholar]
- Mene-Saffrane L, Jones AD, Dellapenna D. Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2010;107:17815–17820. doi: 10.1073/pnas.1006971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed-Yasseen Y, Barringer SA, Splittstoesser WE, Costanza S. The role of seed coats in seed viability. Botanical Review. 1994;60:426–439. [Google Scholar]
- Nakayama Y, Sayo K, Kito M. Decomposition of phospholipids in soybean during storage. Cereal Chemistry. 1981;58:260–264. [Google Scholar]
- Nelson B, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Oge L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. The Plant Cell. 2008;20:3022–3037. doi: 10.1105/tpc.108.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne DJ. Senescence in plants. Boca Raton, FL: CRC Press; 1980. [Google Scholar]
- Ouzouline M, Tahani N, Demandre C, El Amrani A, Benhassaine-Kesri G, Caid HS. Effects of accelerated aging upon the lipid composition of seeds from two soft wheat varieties from Morocco. Grasas Y Aceites. 2009;60:367–374. [Google Scholar]
- Padham AK, Hopkins MT, Wang TW, McNamara LM, Lo M, Richardson LG, Smith MD, Taylor CA, Thompson JE. Characterization of a plastid triacylglycerol lipase from Arabidopsis. Plant Physiology. 2007;143:1372–1384. doi: 10.1104/pp.106.090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Gu Y, Lee Y, Yang ZB, Lee Y. Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiology. 2004;134:129–136. doi: 10.1104/pp.103.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RS, Abdel-Samad IM. Change in fatty acid content of polar lipids during ageing of seeds of peanut (Arachis hypogea L.) Journal of Experimental Botany. 1980;31:1283–1980. [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. The Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukacka S. Changes in membrane fatty acid composition during desiccation of seeds of silver maple. Seed Science and Technology. 1998;26:535–540. [Google Scholar]
- Pukacka S, Kuiper PJC. Phospholipid composition and fatty acid peroxidation during ageing of Acer platanoides seeds. Physiologia Plantarum. 1988;72:89–93. [Google Scholar]
- Rajjou L, Debeaujon I. Seed longevity: survival and maintenance of high germination ability of dry seeds. Comptes Rendus Biologies. 2008;331:796–805. doi: 10.1016/j.crvi.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghaz M, Job C, Job D. Proteome-wide characterization of seed aging in Arabidopsis. A comparison between artificial and natural aging protocols. Plant Physiology. 2008;148:620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ReuZeau C, Goffner D, Cavalie G. Relations between protein composition and germination capacity of sunflowers seeds. Seed Science Research. 1992;2:223–230. [Google Scholar]
- Rossetto M, Gross CL, Jones R, Hunter J. The impact of clonality on an endangered tree (Elaeocarpus williamsianus) in a fragmented rainforest. Biological Conservation. 2004;117:33–39. [Google Scholar]
- Ruuska SA, Ohlrogge JB. Protocol for small-scale RNA isolation and transcriptional profiling of developing Arabidopsis seeds. Biotechniques. 2001;31:752–758. doi: 10.2144/01314bm08. [DOI] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokinin signaling pathway, in Arabidopsis. Molecules and Cells. 2009;28:375–382. doi: 10.1007/s10059-009-0132-x. [DOI] [PubMed] [Google Scholar]
- Ryu SB. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends in Plant Science. 2004;9:229–235. doi: 10.1016/j.tplants.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Samama AM, Pearce RS. Ageing of cucumber and onion seeds—phospholipase-D, lipoxygenase activity and changes in phospholipid content. Journal of Experimental Botany. 1993;44:1253–1265. [Google Scholar]
- Sang Y, Cui D, Wang X. Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiology. 2001;126:1449–1458. doi: 10.1104/pp.126.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity, and for preventing lipid peroxidation during germination. The Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mene-Saffrane L, Farmer EE, Krischke M, Mueller MJ, DellaPennaa D. Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. The Plant Cell. 2006;18:3706–3720. doi: 10.1105/tpc.106.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe I. Properties of triacylglycerol lipase in a mitochondrial fraction from baker's yeast (Saccharomyces cerevisiae) Biochimica et Biophysica Acta. 1976;450:165–174. doi: 10.1016/0005-2760(76)90088-6. [DOI] [PubMed] [Google Scholar]
- Seo YS, Kim EY, Kim JH, Kim WT. Enzymatic characterization of class I DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Letters. 2009;583:2301–2307. doi: 10.1016/j.febslet.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Seo YS, Kim EY, Mang HG, Kim WT. Heterologous expression, and biochemical and cellular characterization of CaPLA1 encoding a hot pepper phospholipase A1 homolog. The Plant Journal. 2008;53:895–908. doi: 10.1111/j.1365-313X.2007.03380.x. [DOI] [PubMed] [Google Scholar]
- Sheahan MB, McCurdy DW, Rose RJ. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. The Plant Journal. 2005;44:744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Son O, Cho SK, Kim EY, Kim WT. Characterization of three Arabidopsis homologs of human RING membrane anchor E3 ubiquitin ligase. Plant Cell Reports. 2009;28:561–569. doi: 10.1007/s00299-009-0680-8. [DOI] [PubMed] [Google Scholar]
- Stewart RRC, Bewley JD. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiology. 1980;65:245–248. doi: 10.1104/pp.65.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WO, Leopold AC. The Millard reaction and oxidative stress during aging of soybean seeds. Physiologia Plantarum. 1995;94:94–104. [Google Scholar]
- Tanaka N, Fujita M, Handa H, et al. Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Molecular Genetics and Genomics. 2004;271:566–576. doi: 10.1007/s00438-004-1002-z. [DOI] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, Van Pijlen JG, Van der Geest AHM, Bino RJ, Groot SPC. A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Science and Technology. 2002;30:149–165. [Google Scholar]
- Thapliyal RC, Connor KF. Effects of accelerated ageing on viability, leachate exudation, and fatty acid content of Dalbergia sissoo Roxb seeds. Seed Science and Technology. 1997;25:311–319. [Google Scholar]
- Thompson JE. Senescence and aging in plants. New York: Academic Press; 1988. 52–84. [Google Scholar]
- Walters C, Wheeler L, Grotenhuis J. Longevity of seeds stored in a genebank: species characteristics. Seed Science Research. 2005;15:1–20. [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. Journal of Biological Chemistry. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiology. 2000;122:345–355. doi: 10.1104/pp.122.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM. Arabidopsis seed coat development: morphological differentiation of the outer integument. The Plant Journal. 2000;22:483–493. doi: 10.1046/j.1365-313x.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Winkler FK, D'Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Lee U, Song IJ, Lee HY, Nam HG, Lim PO. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. Journal of Experimental Botany. 2010;61:3947–3957. doi: 10.1093/jxb/erq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhao D-X, Miao Q, Xue T-T, Li X-Z, Zheng C-C. Arabidopsis thaliana metallothionein, AtMT2a, mediates ROS balance during oxidative stress. Journal of Plant Biology. 2009;52:582–592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.