Abstract
PYD1 (dihydropyrimidine dehydogenase) initiates the degradation of pyrimidine nucleobases and is located in plastids. In this study, a physiological analysis of PYD1 employing T-DNA knockout mutants and overexpressors was carried out. PYD1 knockout mutants were restricted in degradation of exogenously provided uracil and accumulated high uracil levels in plant organs throughout development, especially in dry seeds. Moreover, PYD1 knockout mutants showed delayed germination which was accompanied by low invertase activity and decreased monosaccharide levels. Abscisic acid (ABA) is an important regulator of seed germination, and ABA-responsive genes were deregulated in PYD1 knockout mutants. Together with an observed increased PYD1 expression in wild-type seedlings upon ABA treatment, an interference of PYD1 with ABA signalling can be assumed. Constitutive PYD1 overexpression mutants showed increased growth and higher seed number compared with wild-type and knockout mutant plants. During senescence PYD1 expression increased to allow uracil catabolism. From this it is concluded that early in development and during seed production PYD1 is needed to balance pyrimidine catabolism versus salvage.
Keywords: Arabidopsis, dihydropyrimidine dehydrogenase, Pyd1, pyrimidine, senescence, uracil
Introduction
Nucleotides are uniquely important since they represent building blocks of genetic information (DNA and RNA), represent major energy carriers, and pyrimidine nucleoside diphosphate sugars are activated intermediates in the synthesis of lipids, sucrose, and cell wall polysaccharides (Moffatt and Ashihara, 2002; Boldt and Zrenner, 2003; Kafer et al., 2004; Zrenner et al., 2006). Furthermore, nucleotides are core elements of cofactors such as NAD, FAD, S-adenosylmethionine, or coenzyme A (CoA), which serve in essential biochemical reactions, such as the synthesis of phospholipids and polysaccharides. By reviewing the recently published work on nucleotide metabolism, it becomes obvious that many facets of this important biochemical aspect of plant metabolism are still poorly understood. One possible reason for this is the complexity of a multitude of biochemical reactions that facilitate de novo synthesis, degradation, and interconversion (partial degradation and recycling) of nucleotides, nucleosides, and nucleobases. In contrast to nucleotides, nucleosides do not possess phosphate groups, and nucleobases lack the ribose moiety. The recycling of nucleosides and nucleobases is also known as salvage. In the salvage pathway, nucleobases and nucleosides are converted into nucleoside monophosphates by the action of phosphoribosyl-transferase and nucleoside kinase, respectively (Moffat and Ashihara, 2002; Zrenner et al., 2006; Islam et al., 2007; Mainguet et al., 2009). A great benefit of the salvage pathway is the lower energy cost compared with de novo synthesis. At3g53900 (UPP) has been described as being responsible for most of the uracil-phosphoribosyl-pyrophosphatase activity, and this enzyme is localized in the plastid (Mainguet et al., 2009). However, previous work contradicts these findings by claiming that At5g40870 (AtUK/UPRT1) localized to the cytosol exhibits activity for uridine kinase and uracil-phosphoribosyl-pyrophosphatase (Islam et al., 2007).
De novo synthesis leads to the formation of nucleoside monophosphates, in the case of pyrimidine synthesis to produce uridine-5'-monophosphate (UMP) which then can be converted to nucleotide di- and triphosphates (Moffatt and Ashihara, 2002; Stasolla et al., 2003). The recently identified uridine nucleosidase, NSH1, plays a pivotal role in the regulation of nucleoside, mainly uridine, degradation. NSH1 (formerly named URH1) from Arabidopsis was characterized by complementation of a yeast mutant and synthesized as a recombinant protein in Escherichia coli. The pure recombinant protein exhibits the highest hydrolase activity for uridine, followed by xanthosine and inosine; the corresponding Km values were 0.8, 1.4, and 0.7 mM, respectively (Jung et al., 2009, 2011). In soluble leaf extracts from Arabidopsis, higher affinities for uridine, inosine, and xanthosine hydrolysis were reported (Riegler et al., 2011). The reason for this discrepancy may be the presence of other non-homologous, as yet unknown proteins exhibiting nucleoside hydrolase activity, possibly residing in different subcellular compartments.
The degradation of pyrimidine nucleobase occurs via the ‘reductive’ pathway in many organisms (Rawls, 2006; Piskur et al., 2007). Uracil or thymine degradation is initiated by dihydrouracil dehydrogenase [DHUDH; also called PYD1 (this name will be used herein), EC 1.3.1.2]. The resulting product of uracil reduction, dihydrouracil (DHU), can be further degraded by dihydropyrimidinase [DHPase; EC 3.5.2.2 (PYD2)] to β-ureidopropionate (N-carbamyl-β-alanine), finally leading to the release of β-alanine, CO2, and NH3 by β-ureidopropionase [β-UPase; EC 3.5.1.6 (PYD3), Zrenner et al. (2009)]. Each of the above-mentioned enzymes is encoded by a single gene (Zrenner et al., 2009). Chemically, pyrimidine catabolism represents a reversion to de novo synthesis (Zrenner et al., 2006). Green fluorescent protein (GFP) fusion studies revealed a cytosolic localization for NSH1 (Jung et al., 2009) and β-UPase, localization of PYD1 within the plastids, and of DHPase within the secretory system (Zrenner et al., 2009). The distribution of these enzymes within different subcellular compartments suggests a potentially complex regulation. β-Alanine, one of the products of pyrimidine catabolism, may become incorporated into pantothenate (vitamin B5), which is an essential precursor in CoA synthesis (Coxon, et al., 2005). However, there is experimental evidence that β-alanine from uracil does not contribute significantly to pantothenate biosynthesis in plants (for a discussion, see Zrenner et al., 2009).
Abscisic acid (ABA) is an important player in the early phase of seed germination. ABA inhibits endosperm rupture and seedling growth, but its contents decline sharply during stratification (Weitbrecht et al., 2011). After rehydration, activation of respiration and amino acid synthesis are very early metabolic events, followed by translation from stored RNAs (Weitbrecht et al., 2011). Nucleobase transporters AtUPS1 and AtUPS2 are localized in vascular tissues and may function as uracil transporters in Arabidopsis. High expression of AtUPS1 during the first days of germination may satisfy the initial nucleobase demand of seedlings (Schmidt, et al., 2004; Zrenner, et al., 2009). A major function of the pyrimidine catabolic pathway may be to regulate cellular levels of free pyrimidine nucleotides during growth and development, or in response to nitrogen limitation (Hewitt et al., 2005; Zrenner et al., 2009). The entire pathway recycles pyrimidine nitrogen to general nitrogen metabolism (Zrenner et al., 2009). Genome-wide expression analysis of primary and secondary metabolism in Arabidopsis liquid-cultured seedlings confirmed that reductions in nitrogen availability lead to increased PYD1 expression which can be rapidly reversed by the addition of nitrate to the growth medium (Scheible et al., 2004). In this study, a physiological analysis of the catabolic pathway enzyme PYD1 employing T-DNA knockout mutants and plants with increased activity of this enzyme was carried out. Evidence is provided that PYD1’s action is of major importance for proper seed germination and seed production. Moreover a function in recycling of pyrimidine nitrogen to general nitrogen metabolism during senescence is further substantiated.
Materials and methods
Plant material and growth conditions
Wild-type and transgenic Arabidopsis thaliana (L.) Heynh. plants (ecotype Columbia) were used throughout. Prior to germination, seeds were incubated for 24 h in the dark at 4 °C for imbibition, unless stated otherwise in standardized ED73 soil (Weigel and Glazebrook, 2002). Plant growth was carried out at 22 °C and 120 μmol quanta m−2 s−1 in a 10 h light/14 h dark regime in a growth chamber. As the light source, fluorecence tubes (Osram lumilux cool white and warm white alternating) were used. For growth experiments on sterile agar plates, surface-sterilized seeds were sown on Murashige and Skoog (MS) medium, supplemented with the indicated nucleoside analogues as described previously (Reiser et al., 2004). Growth of plants in liquid culture was performed as described in Scheible et al. (2004) with the modification that growth proceeded under the light regime given above.
In addition to wild-type plants, the following T-DNA insertion mutants were used: SAIL_363_E04 (designated PYD1-1; Sessions et al., 2002) and GK-251F09 (designated PYD1-2; Rosso et al., 2003) obtained from the NASC (National Arabidopsis Stock Center; Scholl et al., 2000)
Generation of mutants
To generate mutants expressing PYD1 under control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter, the complete coding region of PYD1 was initially amplified by PCR from Arabidopsis leaf cDNA using the primers mentioned in Supplementary Table S1 available at JXB online, and inserted into an EcoRV-cleaved vector (pBluescript, Stratagene, Heidelberg). The complete coding region was then excised from this vector with EcoRI/ClaI and inserted into pHannibal (Wesley et al., 2001) via the same restriction sites. The resulting pHannibal constructs were subcloned into the binary vector pART27 as detailed in Reiser et al. (2004). Subsequently, the plasmids were used for Agrobacterium tumefaciens transformation. Transformation of Arabidopsis was performed according to the floral-dip method (Clough and Bent, 1998).
Germination experiments
Endosperm rupture (germination) was monitored as described in Müller at al. (2006). Fifty seeds (9 months after harvest) were put on 0.1-fold strength MS agar plates and incubated under constant illumination (10 μmol quanta m−2 s−1) with white fluorescent tubes at 24 °C. Plates with seeds were stratified for 48 h at 4 °C in the dark when indicated. After the indicated time periods, endosperm rupture was monitored under a stereomicroscope (Leica MZ12, Leica, Germany).
Quantitative RT-PCR
Quantitative reverse transcription-PCR (RT-PCR) was performed as described in Leroch et al. (2005). Gene-specific primers used are listed in Supplementary Table S1. The gene At1g07930 encoding elongation factor 1α (EF1α) was used for quantitative normalization (Curie et al., 1991).
Extraction of total RNA and RNA gel-blot hybridization
Total RNA was isolated from frozen tissue samples by using the Purescript RNA extraction kit (Gentra Systems) according to the manufacturer's instructions. RNA samples (10 μg) were denatured and separated on a 1.2% agarose/2.5% formaldehyde gel. Ethidium bromide was included in the loading buffer medium to confirm equal sample loading. Isolated RNA was blotted onto a nylon membrane (Nytran-Plus, Schleicher & Schüll, Dassel, Germany), and PYD1, restricted from the corresponding pBluescript construct with EcoRI/ClaI, was 32P-labelled by random priming (Ready-To-Go DNA labeling beads, Amersham Pharmacia). Pre-hybridization and hybridization were performed at 65 °C in Church buffer. The final blots were visualized by a PhosphorImager (Packard, Dreieich, Germany).
Quantitation of [14C]pyrimidine degradation
For analysis of the [14C]nucleoside or nucleobase degradation, given as [14C]CO2 liberation, 7-day-old Arabidopsis seedlings grown in hydroponic culture were used (Scheible et al., 2004). Three seedlings were incubated in 2 ml reaction tubes with 800 μl of 5 μM of the indicated, 14C-labelled pyrimidines (1 GBq mmol−1) in growth media. On the inside at the top of this tube, a small 0.5 ml reaction vessel containing 100 μl of 1 M KOH was fixed with grease to allow [14C]CO2 absorption. The seedlings were allowed to float on the solution, and incubation was continued for 24 h in the dark. The reaction was stopped by adding 100 μl of 2 M HCl with a syringe through the closed lid. The hole in the lid was sealed with grease and, after subsequent incubation for 12 h, the released radioactively labelled [14C]CO2 was quantified in a scintillation counter.
Determination of metabolite contents and enzyme activities
Sugar quantification in fractionated samples was conducted by ion chromatography on an HPLC CarboPac PA1 column (Dionex) followed by amperometric quantitation as described (Zuther et al., 2004). Amino acids were quantified as described in Jung et al. (2009), and starch was quantified according to Reinhold et al. (2007). For the determination of protein and lipids in seeds, the method of Reiser et al. (2004) was applied. Uracil was determined using the same method as used to determine uridine (Jung et al., 2009). The relative contents of total carbon and total nitrogen were determined in dry powdered samples with an elemental analyser (Vario EL II, Elementaranalysensysteme GmbH, Hanau, Germany) according to the manufacturer's instructions.
Invertase activity was quantified spectrophotometically in a coupled enzyme assay at pH 7.4 after extraction in HEPES/KOH pH 7.4 (50 mM), MgCl2 (5 mM), EDTA (1 mM), and phenylmethylsulphonyl fluoride (PMSF; 0.1 mM).
Statistical analysis
Statistical analysis was carried out using one-way analysis of variance (ANOVA) with Dunnett's post-test compared with the corresponding wild type. The analysis software was embedded in the GraphPad Prism software 5 (GraphPad software Inc.). P-values are indicated by asterisks (*P <0.05, **P <0.01, ***P <0.005).
Results and Discussion
Analysis of PYD1 expression and biochemical function using mutants
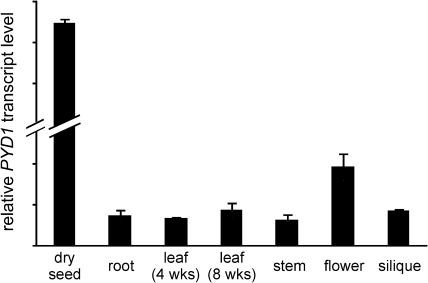
PYD1 (At3g17810) was recently shown to catalyse the conversion of uracil to dihydrouracil in Arabidopsis. This was achieved by the use of corresponding T-DNA insertion mutants which showed lowered catabolism of exogenously supplied uracil (Zrenner et al., 2009). In the analysis presented here, the results of Zrenner et al. (2009) are supported and a comprehensive physiological analysis highlighting an important function of PYD1 during early and late Arabidopsis development is presented. As a starting point, the expression of PYD1 was monitored by quantitative RT-PCR in Arabidopsis tissues from soil-grown plants. Whereas PYD1 expression was quite similar in roots, 4- and 8-week-old leaves, stems, and siliques, a 2-fold higher expression was seen in flowers (Fig. 1). It was striking to see a very marked expression in dry seeds which was ∼28 times higher compared with roots or leaves (Fig. 1). Microarray data support this expression profile (Zimmermann et al., 2004). Further examples of the accumulation of translatable RNAs in dry seeds are LEA (late embryo abundant) mRNA, and mRNAs for seed storage proteins and for heat shock factors. In addition, transcripts from genes encoding enzymes of metabolism such as an aquaporin, isocitrate lyase, and malate synthase have been found (Kimura and Nambara, 2010). For the further analysis of PYD1 function, T-DNA insertion mutants were analysed and, in addition constitutive CaMV-35S overexpression lines (35S:PYD1) were generated.
Fig. 1.
Relative expression of PYD1 in Arabidopsis tissues. PYD1 expression was monitored by quantitative RT-PCR using EF1α as the reference gene.
Two knockout lines were analysed, GK-251F09 (designated PYD1-1) and SAIL_363_E04 (designated PYD1-3; Supplementary Fig. S1A at JXB online). The former line has been described as a PYD1 knockout previously (Zrenner et al., 2009). First, the presence of a T-DNA insertion in both mutants was checked by PCRs with PYD1-specific primers (PYD1 KO fwd/rev) and T-DNA-specific primers (SAIL_LB for PYD1-3 and GK_LB for PYD1-1) (Supplementary Fig. S1B, C). Secondly, the absence of an amplification product with gene-specific primers on genomic DNA in homozygous plants was checked (Supplementary Fig. S1B) and, furthermore, the lack of a gene-specific PCR product in cDNA preparations from both PYD1-1 and PYD1-3 mutants was confirmed using primers PYD1_fw and PYD1_rev (Supplementary Fig. S1D). As a control, cDNA prepared from wild-type plants gave rise to a PCR band of the expected size of 1281 bp (Supplementary Fig. S1D).
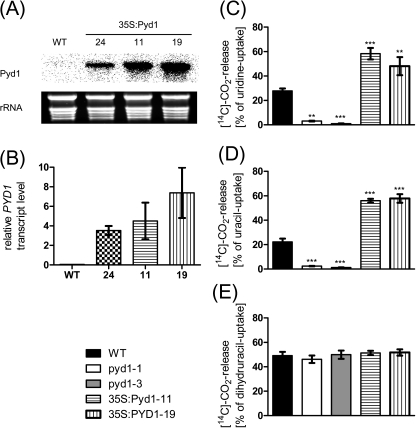
Three independent, constitutive overexpression lines (35S:PYD1) were also generated, as described in the Materials and methods. All three lines accumulated significantly increased transcript levels as compared with the wild type (Fig. 2A). Specifically, plants from lines #24, #11, and #19 showed 74-, 95-, and 156-fold increased transcript levels, respectively (Fig. 2B). As no direct enzyme activity assay is available for PYD1 (for discussion, see Zrenner et al., 2009), changes in activity of all mutants were monitored by the release of radiocarbon ([14C]CO2) after feeding [14C]uridine, [14C]uracil, or [14C]dihydrouracil to 10-day-old liquid-grown seedlings (Fig. 2C–E). The CO2 release by wild-type plants from uridine, uracil, and dihydrouracil accounted for 28, 22, and 49% of the imported metabolite, respectively. In contrast, PYD1-1 and PYD1-3 knockout lines released only 3% and 1% of imported uridine, and 2% and 1% of imported uracil (Fig. 2C–E). In contrast, the CO2 release by overexpressors from uridine was 2-fold and from uracil 2.5 fold higher compared with the wild-type situation (Fig. 2C, D). No change was observed in the degradation of dihydrouracil in a comparison of all mutants (Fig. 2E). In Zrenner et al. (2009) the effect of PYD1 inactivation in corresponding knockout mutants was less drastic. Whereas in that work wild-type seedlings liberated 48% of incorporated uracil as CO2, the mutants still liberated 29%. The higher uracil catabolism observed may be due to the higher uracil concentration of 2 mM that was applied (Zrenner et al., 2009) compared with 5 μM used here. A higher external concentration will affect the internal concentration and drive more uracil towards catabolism, even in the PYD1 knockout, indicating the presence of one or more further enzymes exhibiting DHPDH activity. The lower concentration of 5 μM uracil was used as this fits to the observed affinities of the known plasma membrane uracil transporters UPS1 and UPS2 of ∼6 μM (Schmidt et al., 2004). Nevertheless it is not known whether other transport proteins capable of uracil import into cells exist. Nevertheless, these results indicate that PYD1 exerts marked control over uracil degradation. This is further illustrated by the marked susceptibility of both PYD1:T-DNA insertion lines towards 100 μM toxic flurouracil (Supplementary Fig. S2A at JXB online). In contrast, PYD1 overexpressors tolerate higher concentrations (200 μM and 500 μM) compared with wild-type seedlings (Supplementary Fig. S2B). This indicates that PYD1 activity is limiting for the detoxification of flurouracil under the experimental conditions used. The observed increased degradation of uridine in overexpressor lines (Fig. 2C) further indicates that the upstream reaction catalysed by NSH1 (formerly known as URH1; Jung et al., 2009, 2011) is accelerated. Also the reciprocal effect (acceleration of pyrimidine catabolism by overexpression of NSH1 (Jung. et al., 2009) was observed. This may be due to changed uracil concentrations or point to a concerted regulation of both NSH1 and PYD1, activity. In addition to uracil, degradation of the only other pyrimidine nucleobase occurring in plants, thymine, is also markedly impaired in PYD1 knockouts but increased in PYD1 overexpressor lines (Supplementary Fig. S3).
Fig. 2.
PYD1 transcript accumulation in overexpression mutants (35S:PYD1) and pyrimidine degradation in PYD1 knockout and overexpression lines. (A) Expression analysis of PYD1 overexpression mutants by northern blot hybridization and (B) quantitative RT-PCR. Values represent means ±SE of three biological replicates. (C–E) Catabolism of 14C-labelled uridine, uracil, and dihydrouracil in PYD1 mutants. Degradation was measured as released [14C]CO2 based on import of the corresponding substrate. Values represent means ±SE of at least five biological replicates. The asterisks indicate significant differences between the wild type and mutants, based on a one-way ANOVA test.
These results show that PYD1 is crucial for pyrimidine degradation. A localization of PYD1 in plastids was demonstrated by use of a GFP fusion protein [Zrenner et al., 2009; and supported by our own analysis using a similar approach (not shown)]. Evidence for a plastid localization of the main uracil salvage activity has also been presented (Mainguet et al., 2009). Therefore, it appears that the plastid stroma is the site where both uracil degradation and salvage are located. It was supposed that PYD1 from plants lacks an N-terminal cofactor-binding site (Zrenner et al., 2009). Therfore, interaction with a component complementing this lack which must be present in the plastid is required. To test this hypothesis, a 21 amino acid N-terminal truncated PYD1 protein was expressed in PYD1-3 and found to reside in the cytosol because of lack of a plastid target sequence. The corresponding plants exhibited no PYD1 activity (not shown), supporting the idea that a plastidic cofactor is required for activity. Although experimental evidence for N-terminal processing of PYD1 after import into the plastid which then leads to a truncation of the protein covering ≥21 amino acids is lacking, it appears less probable that important structural information for the mature protein is present in this region. Furthermore, based on sequence analysis programs for conserved domains (CCDs; Marchler-Bauer et al., 2007), no amino acids involved in cofactor binding in mammalian homologues are present at the far N-terminal part of the proteins.
PYD1 function in seeds and seed germination
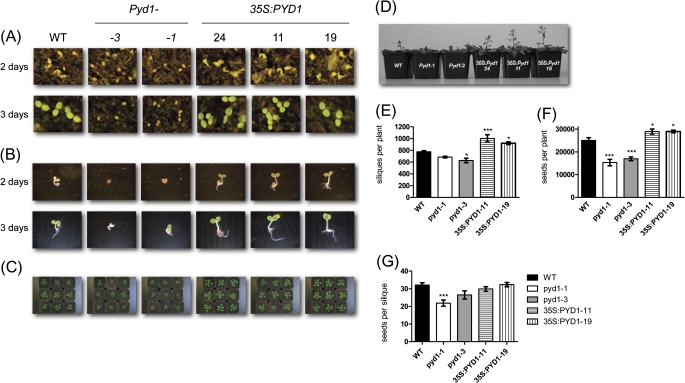
Because of the high PYD1 transcript level in dry seeds, the first 3 d of seed germination after imbibition were monitored. For this, seeds were sown on soil or on wet paper. Under both conditions, a delay in seedling development of ∼2 d became obvious for both PYD1:T-DNA insertion lines (Fig. 3A, B). In contrast, 35S:PYD1 lines grew faster compared with the wild-type controls (Fig. 3A, B). These differences in growth continued subsequently, and were still visible after 6 weeks (Fig. 3C). Flowering also occurred first in the overexpressor lines, followed by the wild type and knockout mutants (Fig. 3D). Thus, a delay throughout the whole plant development of PYD1 knockout lines has to be attested, whereas overexpressor lines showed a faster development. After 10 weeks of growth, wild-type plants developed 780 siliques with 32 seeds each (mean of at least 10 plants, Fig. 3E–G). Under the same growth conditions PYD1:T-DNA mutants developed only 26 and 22 seeds per silique and 629 and 686 siliques per plant, respectively. Overexpressor mutants developed significantly more siliques than wild-type plants. However, the number of seeds per silique was not different from the wild-type situation (Fig. 3E–G).
Fig. 3.
Growth analysis of wild type (WT), PYD1 knockout, and PYD1 overexpressor plants. (A) Germination was monitored on soil after 2 d and 3 d (B) Germination was monitored for 2 d and 3 d on wet paper. (C) Six-week-old plants grown on soil. (D) Typical appearance of PYD1 mutants at the time of bolting. (E–G) Seeds and siliques were harvested from plants grown for 4 weeks under short day conditions and subsequently for 6 weeks under long day conditions. Values represent means ±SE of at least 10 biological replicates. The asterisks indicate significant differences between the WT and mutants, based on a one-way ANOVA test.
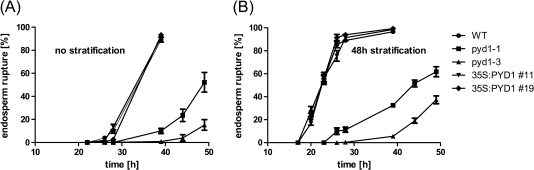
As the next step, germination itself, which begins with water uptake of the seed and ends when the radicle has protruded through all covering layers (Müller et al., 2006), was looked at more closely. Successful germination can be monitored as endosperm rupture. Endosperm rupture was followed over time for the wild type and PYD1 mutants with and without prior stratification, as described in Müller et al. (2006).
Differences in the germination behaviour between the various PYD1 mutants became obvious. Whereas wild-type and 35S:PYD1 overexpressors showed 50% germinated seeds after ∼33 h without stratification and after ∼22 h with stratification, both knockout mutants germinated markedly more slowly (Fig. 4A, B). PYD1-1 mutants showed 50% germinated seeds after 48 h without stratification and after 43 h with stratification (Fig. 4A, B). Even after 49 h the PYD1-3 mutants germinated to 15% without stratification and 37% with stratification (Fig. 4A B). The seeds used were 9 months after harvest, and yet there was still a small effect of stratification in all populations. However, this is typical of other observations on fully after-ripened seeds (Müller et al., 2006). The wild type and PYD1 overexpressor mutants showed no difference in endosperm rupture, thus it can be concluded that in this case post-germination effects account for the faster development of the corresponding plants. In contrast, PYD1-1 and PYD1-3 mutants were markedly delayed in germination. ABA is known to act in an inhibitory manner on dormancy and thus seed germination, while GA (gibberellic acid) is an antagonist of ABA. A current model of how endosperm rupture (seed germination) is regulated is presented in Linkies et al. (2009). While water uptake and testa rupture are not affected by ABA, the procedure of endosperm cap weakening, a prerequisite of endosperm rupture, is inhibited by ABA. GA, as an embryo signal, can induce the cap weakening process. In the late phase of germination, ethylene biosynthesis comes into play and interferes with ABA signalling (Linkies et al., 2009).
Fig. 4.
Germination of seeds was monitored and shown as endosperm rupture. (A) Seeds were incubated for the indicated times and monitored for endosperm rupture without stratification. (B) Seeds were stratified for 48 h in the dark at 4 °C. Five plates with 50 seeds each were analysed and the mean values ±SE were calculated.
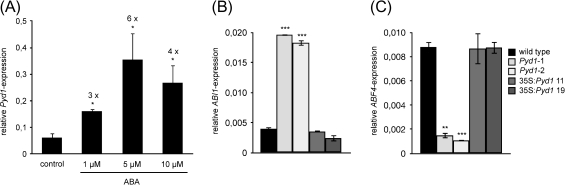
Therefore, a possible role for ABA in the delay of germination of PYD1 knockout mutants was analysed. When 9-day-old seedlings grown in liquid culture were subjected to ABA treatment, a 3- to 6-fold increase in PYD1 transcript was observed, dependent on the ABA concentration (Fig. 5A). These results are in accordance with data available through the eFP browser database (Winter et al., 2007). This pointed to an interaction of PYD1 function and ABA metabolism. To proceed further in this analysis, ABA-responsive genes were analysed with respect to their expression in the PYD1:T-DNA mutants. ABI1 is known to be a regulator of ABA signalling during germination and shows increased transcript levels as a response to ABA treatment (Hoth et al., 2002). PYD1:T-DNA seeds, 2 d after imbibition, showed a 5-fold increased ABI1 expression compared with wild-type seeds of the same age (Fig. 5B). As a further marker for the ABA status, ABF4 was monitored. ABF4 is an ABA-responsive element-binding factor and member of the bZIP transcription factors, up-regulated by ABA (Choi et al., 2000). ABF4 showed the opposite response compared with ABI1 in the present experiments; the relative expression in PYD1:T-DNA mutants accounted for 15% of the wild-type level (Fig. 5C). Although the differential expression pattern of ABI1 and ABF4 in the PYD1 knockout mutants cannot be explained, the fact that both are deregulated and the observed delayed germination point to an interaction of PYD1 with ABA signalling during germination.
Fig. 5.
Relative transcript level of PYD1 in response to ABA treatment and relative expression of ABI and ABF4 in 2-day-old PYD1 mutants. Expression was monitored by quantitative RT-PCR using EF1α as the reference gene. Values represent means ±SE of at least three biological replicates. The asterisks indicate significant differences between the wild type and mutants, based on a one-way ANOVA test.
To exclude the possibility that an altered composition of seed storage products might be responsible for the observed differences in germination and development of the mutants, total carbon and nitrogen in dry seeds was determined by elemental analysis and, in addition, total protein and lipid were extracted and quantified from seeds. No significant differences were detectable between all mutants and wild-type plants with respect to all storage compounds (Supplementary Table S2 at JXB online). Obviously, PYD1 activity does not influence seed development with respect to the main storage compounds but acts from the time of germination onwards until new seed is produced.
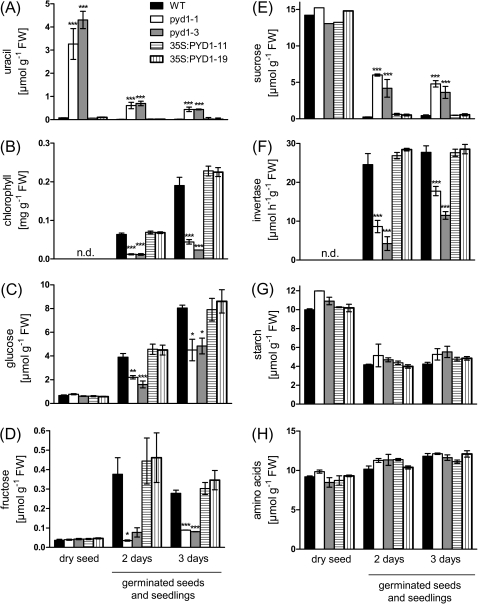
Due to the limited ability of PYD1 knockout lines to degrade uracil, markedly increased uracil levels up to 50-fold compared with the wild type were found in dry seeds of PYD1 knockouts (Fig. 5A). The uracil level remains high during the first days of germination, although the amount on a fresh weight basis declines as a consequence of hydration and cell expansion of the seeds (Fig. 6A). Also other plant tissues such as senescent leaves, stems, and siliques are characterized by markedly increased uracil levels (Supplementary Fig. S4A–C). As already indicated (see Fig. 3), development of PYD1:T-DNA lines is delayed, which is illustrated by the markedly lowered chlorophyll contents during the first 3 d after imbibition (Fig. 6B). A deeper analysis of the metabolic changes during early germination reveals lower monosaccharide contents in T-DNA mutants, accompanied by higher sucrose contents, compared with the wild type, whereas overexpressors show no significant differences (Fig. 6C–E). Sucrose and monosaccharides are connected by the activity of neutral invertase and it was shown that this enzyme activity is indispensable for normal growth of Arabidopsis (Barratt et al., 2009). Neutral invertase activity accounted for 24 μmol g FW−1 h−1 in 2-day-old seedlings of the wild type and had similar activity in 3-day-old seedlings (Fig. 6F). In contrast, the invertase activity in PYD1-1 and PYD1-2 was reduced to ∼25% in 2-day-old and ∼50% in 3-day-old seedlings compared with the wild type (the activity in overexpressor mutants was similar to that in the wild type; Fig. 6F). The amounts ofig. 6G, H). Therefore, it appears that invertase activity is a key regulator of growth affected by PYD1 during the early phase of Arabidopsis development.
Fig. 6.
Metabolite levels and invertase activity (as indicated) determined in dry seeds, and 2- and 3-day-old seedlings. Seeds were sown on wet paper in trays and incubated for 48 h in the dark at 4 °C for imbibition. The trays were then transferred to a growth chamber (see Materials and methods) and plant material was harvested after 48 h or 72 h (4 h after lights on). Values represent means ±SE of at least five biological replicates. The asterisks indicate significant differences between the wild type and mutants, based on a one-way ANOVA test.
Is the uracil level the critical factor for the observed delay in development? To test this hypothesis, Arabidopsis seedlings were grown on agar plates in the presence of up to 10 mM uracil without observing any growth phenotype. However, it is possible that this experiment does not resemble the situation in PYD1:T-DNA mutants as nothing is known about the cell type-specific or subcellular accumulation of uracil in these plants.
The complete degradation of uracil leads to the formation of ammonia and β-alanine. Whereas release of ammonia due to pyrimidine degradation increases under conditions of nitrogen limitation and thus attenuates the effects of nitrogen starvation (Zrenner et al., 2009; own unpublished observations), much less is known about the role of β-alanine in plant metabolism. However, β-alanine represents a precursor of CoA biosynthesis and one prominent biosynthetic reaction requiring CoA is fatty acid synthesis. However, a major contribution of uracil catabolism to pantothenate synthesis is unlikely based on incorporation studies with radiolabelled uracil (for discussion, see Zrenner et al., 2009).
PYD1 function in senescence
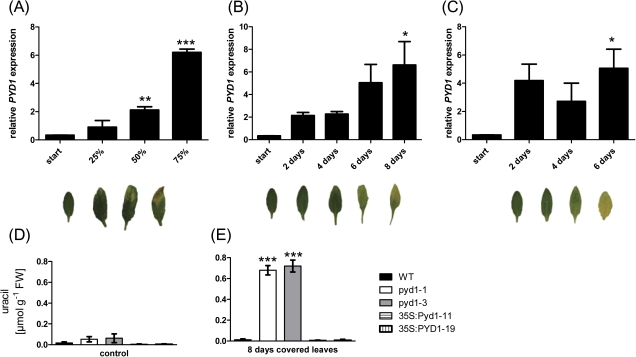
At the end of their development, plants enter a programmed ageing process known as senescence. This has been well studied especially in leaves. For this kind of analysis, different experimental approaches are used. Leaves of different ages can be harvested from ‘old’ plants (natural senescence), leaves can be covered to induce senescence (dark-induced senescence), or leaves can be detached and incubated in the dark (van der Graaff et al., 2006). Under all conditions described, a marked increase of PYD1 transcript up to 20-fold above controls was observed (Fig. 7A–C). In line with these observations was an increase in uracil accumulation in leaves from PYD1-1 and PYD1-2 plants after covering the leaves for 8 d (Fig. 7D). The already elevated uracil content of untreated PYD1-1 and PYD1-2 mutant leaves compared with those of the wild type was further increased after covering leaves up to 0.77 and 0.67 μmol g FW−1, respectively. Darkened leaves of wild-type plants still only accumulated 0.02 μmol uracil (Fig. 7D). This is indicative of an up-regulation of pyrimidine degradation in senescent tissues to liberate nitrogen bound in nucleic acids and nucleotides and make these available for other tissues, such as developing seeds. It has been shown that accelerated nitrogen remobilization from senescing leaves, achieved by overexpression of pyruvate phosphate dikinase, positively affected seed weight (Taylor et al., 2010). Furthermore, nitrogen limitation is a condition in which uracil degradation is markedly elevated (Zrenner et al., 2009). Therefore, it is likely that altering the efficiency of uracil catabolism affects seed yield (Fig. 3F) through altered nitrogen supply.
Fig. 7.
Relative PYD1 transcript accumulation monitored at different stages of leaf sensescence and uracil levels in senescent leaves from PYD1 mutants. (A) Samples were taken from plants undergoing natural senescence, (B) plants with covered leaves, and (C) detached leaves. Expression was monitored by quantitative RT-PCR using EF1α as the reference gene. (D) Uracil was determined in leaves covered for 8 d. Values represent means ±SE of at least three biological replicates. The asterisks indicate significant differences between the wild type and mutants, based on a one-way ANOVA test
In summary, a delayed germination, seedling development, and altered seed production were observed when PYD1 is missing from Arabidopsis plants. One of the earliest metabolic alterations in these mutants is an accumulation of sucrose due to decreased cytosolic invertase activity, in addition to constitutively high uracil levels. The delayed endosperm rupture and altered expression of ABA-dependent genes point to an interaction of PYD1 with ABA signalling which might also influence invertase activity directly. In the later phase of development, PYD1 is involved in nucleic acid breakdown and possibly in remobilization of nitrogen supply to developing seeds.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Genetic characterization of PYD1 T-DNA insertion mutants.
Figure S2. Growth of the wild type and PYD1 mutants in the presence and absence of toxic 5-fluorouracil.
Figure S3. Catabolism of 14C-labelled thymine in PYD1 mutants.
Figure S4. Uracil contents of the wild-type and PYD1 mutants.
Table S1. PCR primers used in this study.
Table S2. Contents of storage compounds in wild-type and PYD1 mutant seeds.
Acknowledgments
This work was supported by DFG-grant MO 1032/3-1. We gratefully acknowledge support of this work by Professor H. E. Neuhaus.
References
- Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proceedings of the National Academy of Sciences, USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt R, Zrenner R. Purine and pyrimidine biosynthesis in higher plants. Physiologia Plantarum. 2003;117:297–304. doi: 10.1034/j.1399-3054.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. Journal of Biological Chemistry. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coxon KM, Chakauya E, Ottenhof HH, Whitney HM, Blundell TL, Abell C, Smith AG. Pantothenate biosynthesis in higher plants. Biochemical Society Transactions. 2005;33:743–746. doi: 10.1042/BST0330743. [DOI] [PubMed] [Google Scholar]
- Curie C, Liboz T, Bardet C, Gander E, Medale C, Axelos M, Lescure B. Cis and trans-acting elements involved in the activation of Arabidopsis thaliana A1 gene encoding the translation elongation factor EF-1 alpha. Nucleic Acids Research. 1991;19:1305–1310. doi: 10.1093/nar/19.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt MM, Carr JM, Williamson CL, Slocum RD. Effects of phosphate limitation on expression of genes involved in pyrimidine synthesis and salvaging in Arabidopsis. Plant Physiology and Biochemistry. 2005;43:91–99. doi: 10.1016/j.plaphy.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. Journal of Cell Science. 2002;115:4891–4900. doi: 10.1242/jcs.00175. [DOI] [PubMed] [Google Scholar]
- Islam MR, Kim H, Kang SW, Kim JS, Jeong YM, Hwang HJ, Lee SY, Woo JC, Kim SG. Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Molecular Biology. 2007;63:465–477. doi: 10.1007/s11103-006-9101-3. [DOI] [PubMed] [Google Scholar]
- Jung B, Flörchinger M, Kunz HH, Traub M, Wartenberg R, Jeblick W, Neuhaus HE, Möhlmann T. Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. The Plant Cell. 2009;21:876–891. doi: 10.1105/tpc.108.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Hoffmann C, Möhlmann T. Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. Plant Journal. 2011;65:703–711. doi: 10.1111/j.1365-313X.2010.04455.x. [DOI] [PubMed] [Google Scholar]
- Kafer C, Zhou L, Santoso D, Guirgis A, Weers B, Park S, Thornburg R. Regulation of pyrimidine metabolism in plants. Frontiers in Bioscience. 2004;9:1611–1625. doi: 10.2741/1349. [DOI] [PubMed] [Google Scholar]
- Kimura M, Nambara E. Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibitions. Plant Molecular Biology. 2010;73:119–129. doi: 10.1007/s11103-010-9603-x. [DOI] [PubMed] [Google Scholar]
- Linkies A, Muller K, Morris K, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. The Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguet SE, Gakiere B, Majira A, Pelletier S, Bringel F, Guerard F, Caboche M, Berthome R, Renou JP. Uracil salvage is necessary for early Arabidopsis development. The Plant Journal. 2009;60:280–291. doi: 10.1111/j.1365-313X.2009.03963.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Research. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat BA, Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Physiologists; 2002. Online publication DOI/111199/tab0018- http://aspborg/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- Piskur J, Schnackerz KD, Andersen G, Bjornberg O. Comparative genomics reveals novel biochemical pathways. Trends in Genetics. 2007;23:369–372. doi: 10.1016/j.tig.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Rawls JM. Analysis of pyrimidine catabolism in Drosophila melanogaster using epistatic interactions with mutations of pyrimidine biosynthesis and beta-alanine metabolism. Genetics. 2006;172:1665–1674. doi: 10.1534/genetics.105.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold T, Alawady A, Grimm B, et al. Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. The Plant Journal. 2007;50:293–304. doi: 10.1111/j.1365-313X.2007.03049.x. [DOI] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE. Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiology. 2004;136:3524–3536. doi: 10.1104/pp.104.049502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler H, Geserick C, Zrenner R. Arabidopsis thaliana nucleosidase mutants provide new insights into nucleoside degradation. New Phytologist. 2011;191:349–359. doi: 10.1111/j.1469-8137.2011.03711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population GABI-Kat. for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Su YH, Kunze R, Warner S, Hewitt M, Slocum RD, Ludewig U, Frommer WB, Desimone M. UPS1 and UPS2 from Arabidopsis mediate high affinity transport of uracil and 5-fluorouracil. Journal of Biological Chemistry. 2004;279:44817–44824. doi: 10.1074/jbc.M405433200. [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiology. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. A high-throughput Arabidopsis reverse genetics system. The Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C, Katahira R, Thorpe TA, Ashihara H. Purine and pyrimidine nucleotide metabolism in higher plants. Journal of Plant Physiology. 2003;160:1271–1295. doi: 10.1078/0176-1617-01169. [DOI] [PubMed] [Google Scholar]
- Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM. Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. The Plant Journal. 2010;62:641–652. doi: 10.1111/j.1365-313X.2010.04179.x. [DOI] [PubMed] [Google Scholar]
- van der Graaf GE, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G. First off the mark: early seed germination. Journal of Experimental Botany. 2011;62:3289–3309. doi: 10.1093/jxb/err030. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoSOne. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Riegler H, Marquard CR, Lange PR, Geserick C, Bartosz CE, Chen CT, Slocum RD. A functional analysis of the pyrimidine catabolic pathway in Arabidopsis. New Phytologist. 2009;183:117–132. doi: 10.1111/j.1469-8137.2009.02843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annual Review of Plant Biology. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
- Zuther E, Kwart M, Willmitzer L, Heyer AG. Expression of a yeast-derived invertase in companion cells results in long-distance transport of a trisaccharide in an apoplastic loader and influences sucrose transport. Planta. 2004;218:759–766. doi: 10.1007/s00425-003-1148-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.