Abstract
RING finger proteins comprise a large family and play important roles in regulation of growth and development, hormone signalling, and responses to biotic and abiotic stresses in plants. In this study, the identification and functional characterization of a C4C4-type RING finger protein gene from the Chinese wild grapevine Vitis pseudoreticulata (designated VpRFP1) are reported. VpRFP1 was initially identified as an expressed sequence tag (EST) from a cDNA library constructed from leaves of V. pseudoreticulata inoculated with the grapevine powdery mildew Uncinula necator. Sequence analysis of the deduced VpRFP1 protein based on the full-length cDNA revealed an N-terminal nuclear localization signal (NLS) and a C-terminal C4C4-type RING finger motif with the consensus sequence Cys-X2-Cys-X13-Cys-X1-Cys-X4-Cys-X2-Cys-X10-Cys-X2-Cys. Upon inoculation with U. necator, expression of VpRFP1 was rapidly induced to higher levels in mildew-resistant V. pseudoreticulata plants. In contrast, expression of VpRFP1 was down-regulated in mildew-susceptible V. vinifera plants. Western blotting using an antibody raised against VpRFP1 showed that VpRFP1 was also induced to higher levels in V. pseudoreticulata plants at 12–48 hours post-inoculation (hpi). However, there was only slight increase in VpRFP in V. vinifera plants in the same time frame, even though a more significant increase was observed at 96–144 hpi in these plants. Results from transactivation assays in yeast showed that the RING finger motif of VpRFP1 exhibited some activity of transcriptional activation; however, no activity was seen with the full-length VpRFP1. Overexpression of VpRFP1 in Arabidopsis plants was found to enhance resistance to Arabidopsis powdery mildew Golovinomyces cichoracearum, which seemed to be correlated with increased transcript levels of AtPR1 and AtPR2 in the pathogen-infected tissues. In addition, the Arabidopsis transgenic lines showed enhanced resistance to a virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Taken together, the results suggested that VpRFP1 may be a transcriptional activator of defence-related genes in grapevines.
Keywords: C4C4-type RING finger, Chinese wild Vitis pseudoreticulata, disease resistance, powdery mildew, VpRFP1
Introduction
The capacity of plants to protect themselves against pathogens depends on the detection mechanisms that recognize pathogen-derived molecules to activate host defence responses. Sophisticated plant defence responses, such as hypersensitive cell death, oxidative burst, and synthesis of pathogen-related (PR) proteins, are activated upon invasion by pathogens or changes in environmental conditions (Wang et al., 2005; Koga et al., 2006; Shabab et al., 2008). Subsequent to these events, plants may develop a long-lasting and broad-spectrum resistance known as systemic acquired resistance (SAR) (Wang et al., 2005; Park et al., 2007). Plant transcription factors (TFs) play crucial roles in the plant defence response to pathogen infection (Jones et al., 2006; Mozoruk et al., 2006). TFs contribute to the regulation of the plant defence response, including the up-regulation of the PR genes via recognition of specific DNA sequences in the promoter region (Rushton and Somssich, 1998). So far, at least six major families of plant TFs, including R2R3 MYB (Stracke et al., 2001), ERF (Andriankaja et al., 2007), TGA bZIP (Jakoby et al., 2002), WRKY (Zhang and Wang, 2005), NPR1 (Zhang et al., 1999), and Whirly (Desveaux et al., 2004, 2005), have been reported to participate in the regulation of plant defence responses.
Zinc finger proteins are the most abundant proteins in plants, which may be essential for fundamental plant growth and development (Takatsuji, 1998). As a member of TFs, zinc finger proteins can be grouped into the subfamilies of TFIIIA, WRKY, Dof, GATA, RING finger, and PHD (Takatsuji, 1998). RING finger proteins are defined by the consensus sequence containing cysteine (Cys) and histidine (His) residues Cys-X2-Cys-X9–39-Cys-X1–3-His-X2–3-Cys/His-X2-Cys-X4–48-Cys-X2-Cys, where X is any amino acid and the number of X residues varies in different fingers (Borden and Freemont, 1996). Based on the variation in composition of the eight zinc-coordinating Cys (C) and His (H) residues and the distance between the metal ligands, eight types of RING domain have been identified in Arabidopsis thaliana (Stone et al., 2005). The eight types include two canonical RING types: RING-HC (C3HC4) and RING-H2 (C3H2C3), and six modified RING domain types, namely RING-v (C4HC3), RING-C2 (C4C4), RING-D (C3HDC3), RING-S/T (C3H2SC2 or CSCHCTC2), RING-G (C3HGC3) (Albert et al., 2002; Hewitt et al., 2002; Dasgupta et al., 2004; Stone et al., 2005), and C5HC2 (Song et al., 2007). The RING finger domain has also been documented with E3 ligase function, which plays a crucial role in ubiquitin-dependent protein degradation (Smalle and Vierstra, 2004).
Genetic and biochemical studies have identified a number of RING finger proteins that play key roles in various biological processes. Initial functional evaluations have indicated that RING finger proteins regulate photomorphogenesis. For example, the regulatory protein COP1 was found to represses photomorphogenesis in darkness (Deng et al., 1991, 1992; Ang and Deng, 1994). Previous studies also showed that RING finger proteins are associated with plant growth and development (Xu and Li, 2003; Disch et al., 2006; Zhang et al., 2008). Moreover, other studies also demonstrated that some RING finger proteins are directly involved in regulation of hormone signalling pathways by targeting of specific proteins for degradation (Zhang et al., 2005, 2007; Bu et al., 2009). In particular, some reports indicated that an abundance of RING finger proteins play key roles in regulating defence responses against abiotic and biotic stresses. HOS1 is a functional RING finger protein that has ubiquitin E3 ligase activity, is a negative regulator of cold signal transduction, physically interacts with ICE1, and is a TF activating the expression of CBF genes (Lee et al., 2001; Dong et al., 2006). RING finger proteins also seem to be involved in pathogen response or plant defence. Arabidopsis ATL2 is induced rapidly and momentarily by chitin and cellulose treatments (Salinas-Mondragon et al., 1999), and the eca mutants with constitutive expression of the ATL2 gene exhibited up-regulated expression of defence-related genes and salicylic acid (SA)- and jasmonic acid (JA)-responsive genes (Serrano and Guzman, 2004). A T-DNA insertion mutant of Arabidopsis ATL9 results in increased susceptibility to powdery mildew (Ramonell et al., 2005). Arabidopsis RIN2 and RIN3 and tobacco ACRE132 regulate expression of disease resistance genes specifically involved in the hypersensitive response (Kawasaki et al., 2005).
Grapevine (Vitis vinifera L.) is economically the most important fruit species worldwide. Vitis vinifera is currently the major species cultivated for its high quality in producing wine, juice, table grapes, and dried fruit. However, it is susceptible to many fungal diseases including powdery mildew [Uncinula necator (Schw.) Burr. or Erysiphe cichoracearum], anthracnose [Elsinoe ampelina (de Bary) Shear], and downy mildew [Plasmopara viticola (Berk. & Curt.) Berl. & De Toni ] (Pavlousek, 2007). Fungal pathogens are a major problem in grapevine globally; they cause huge losses in yield and significant reduction in berry and wine quality. China is one of the origins of Vitis species, and some Chinese wild Vitis species possess desirable disease resistance to various pathogens, such as extremely high resistance to anthracnose and ripe rot [Glomerella cingulata (Ston.) Spauld et Schrenk], high resistance to powdery mildew, and resistance to crown gall (Agrobacterium vitis) (Chai et al., 1997; He, 1999). Baihe-35-1 is a unique accession of Chinese wild Vitis pseudoreticulata W. T. Wang, which possesses a high resistance to multiple fungi, particularly to U. necator (Wang et al., 1995).
To reveal the molecular mechanisms involved in the defence response to pathogen infection in Chinese wild V. pseudoreticulata, pathogen-induced genes have previously been isolated from a cDNA library of V. pseudoreticulata leaves inoculated with U. necator (Xu et al., 2009). Among them, one was predicted to encode a putative RING finger protein, and its full-length cDNA (GenBank accession no. FJ356672) was obtained based on one expressed sequence tag (EST) sequence (GenBank accession no. DQ354158). This gene was designated as VpRFP1 (V. pseudoreticulata RING-finger protein 1). In this study, the VpRFP1 gene from Chinese wild V. pseudoreticulata accession Baihe-35-1 was cloned and its expression patterns were investigated. It was shown that ectopic overexpression of VpRFP1 in transgenic Arabidopsis resulted in enhanced resistance against both the powdery mildew pathogen Golovinomyces cichoracearum and the bacterial pathogen Pseudomonas syringae pv. tomato DC3000.
Materials and methods
Plant materials
Grapevine (Chinese wild V. pseudoreticulata accession Baihe-35-1, V. vinifera cv. Carignane) leaves were obtained from the Grape Repository of Northwest A&F University, Yangling, Shaanxi, PR China. Arabidopsis thaliana plants (ecotype Columbia, Col-0) were grown in a soil mix of peat moss, perlite and vermiculite (3:1:1, v/v/v) under a 12/12 h day/night cycle at 24 °C with 60% humidity.
Pathogen inoculations
The pathogen U. necator, collected from leaves of field-grown V. vinifera cv. Cabernet Sauvignon, was maintained in greenhouse-grown V. vinifera cv. Carignane plantlets. The pathogens were collected and suspended in sterile water with a concentration of 5×105 sporangia ml−1. The spore suspension were sprayed onto the abaxial leaf surface of attached grapevine leaves. The inoculated leaves were enclosed in plastic bags to maintain high humidity. After inoculation for 0, 12, 24, 48, 72, 96, 120, and 144 h, leaves were sampled, immediately frozen in liquid nitrogen, and stored at –80 °C for use.
The disease assay with G. cichoracearum isolate UCSC1 was obtained from Dingzhong Tang and conducted as previously described (Wilson et al., 2001). VpRFP1-overexpressing Arabidopsis were inoculated by spraying leaves with the pathogen conidial suspension (5×105 conidia ml−1). The inoculated plants were placed in a plant-growth box (25 °C with 16 h of illumination per day and 100% relative humidity). After inoculation for 0, 24, 48, 72, and 96 h, the leaves were sampled.
Pseudomonas syringae pv. tomato DC3000 was provided by Dingzhong Tang, and grown at 28 °C in King's B medium (supplemented with 100 mg l−1 rifampicin) overnight, then diluted to ∼107 cfu ml−1 with 10 mM MgCl2 solution. Approximately 10 μl of bacterial suspension was infiltrated into the abaxial side of 4–5 leaves per plant using a 1 ml needleless syringe. Quantification of bacterial growth was performed as described (Wang et al., 2007). Trypan blue staining was used to detect cell death as described (Yang et al., 2009).
DNA sequencing and sequence analysis
The open reading frame (ORF) of VpRFP1 was cloned into the pMD-19 vector (TaKaRa, Dalian, China) and sequenced (SunBiotech Company, Beijing, China). DNA sequences were analysed using BLASTN and BLASTX in the National Center for Biotechnology Information (NCBI) databases (http://www.ncbi.nlm.nih.gov). The protein conserved domain was analysed using Smart (http://smart.emblheidelberg.de/smart/change_mode.pl) and ExPASy (http://au.expasy org /tools/). The deduced amino acid sequence was aligned and the phylogenetic tree was generated by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Preparation of fusion protein and polyclonal antibodies
The VpRFP1 gene from Chinese wild V. pseudoreticulata was PCR cloned in-frame into the pGEX-4T-1 vector. The construct was verified by DNA sequencing and transformed into Escherichia coli BL21. Expression of the fusion protein glutathione S-transferase (GST)–VpRFP1 was induced by isopropyl-β-D-thiogalactopyranoside (IPTG). The purification of the fusion protein was performed by electrodialysis assay, and New Zealand rabbits were immunized with the purificied product to obtain polyclonal antibodies. The polyclonal antisera were used for western blot and subcellular immunogold labelling.
Gene expression analysis by real-time quantitative PCR and semi-quantitative PCR
Total RNA was extracted from grapevine leaves as previously described (Asif et al., 2000). First-strand cDNA was synthesized from 2 μg of DNase-treated total RNA using PrimeScript™ RTase (TaKaRa). Semi-quantitative RT-PCR was performed by using a PrimeScript™ RT-PCR Kit (TaKaRa). Control reactions to normalize RT-PCR were done with primers derived from grapevine GAPDH (glyceraldehyde phosphate dehydrogenase) sequences. PCRs on serial dilutions of cDNA were performed at 57 °C for 29 cycles to define semi-quantitative conditions that resulted in amplification linear to RNA amounts. RT-PCR products were separated on 1.2% agarose gels. The experiments were performed three times with similar results. The special primers used for PCR are listed in Supplementary Table S2 available at JXB online.
For real-time PCR, reactions were carried out on the Bio-Rad IQ5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). PCRs were carried out in triplicate in a reaction buffer containing 1× SYBR® Premix Ex Taq™ (TaKaRa), 0.2 μM of forward and reverse primers, and 10 ng of reverse-transcribed RNA in a final volume of 25 μl. Thermal cycling conditions were: 30 s at 95 °C followed by 40 cycles of 15 s at 94 °C, 30 s at 58 °C, and 30 s at 72 °C. The specificity of the individual PCR amplifications was checked using a heat dissociation curve from 55 °C to 95 °C following the final cycle of the PCR. The results obtained for each gene of interest at each time point were normalized against the grapevine GAPDH gene, which was used as a reference gene. Mean values and SDs were obtained from three duplicate experiments and are representative of three independent experiments. Primers used for real-time quantitative PCR are listed in Supplementary Table S2 at JXB online.
Plant protein extraction and protein gel blot analysis
Total protein was extracted from infected leaves as described (Wang et al., 2003, 2006). The concentration of the protein extract was determined with the Bradford protein assay (Bradford, 1976). SDS–PAGE was carried out according to standard procedures with 25 μg of total proteins. Proteins were electrotransferred onto PVDF membranes (Bio-Rad). The membrane was blocked for 3 h in blocking buffer [phosphate-buffered saline (PBS), 0.1% (w/v) Tween-20, and 5% (w/v) skim milk] before incubating overnight in a 1:1000 dilution of polyclonal antiserum prepared in blocking buffer. Detection of VpRFP1 was achieved with goat anti-rabbit IgG secondary antibody and the BCIP/NBT chemiluminescent system according to the manufacturer's instructions (Sigma-Aldrich, Beijing, China). The experiment was performed twice with similar results.
Analysis of VpRFP1 transcriptional activity
The deletion constructs of VpRFP1 for the transactivation assay were generated by fusing the VpRFP1 gene downstream of the GAL4 DNA-binding domain (BD). Fifteen VpRFP1 gene regions were PCR amplified to introduce a BglII site at the 5′ end and a PstI site at the 3′ end. The fragments were digested with BglII and PstI, and cloned into the same sites of pGBKT7. The gene-specific primers are listed in Supplementary Table S1 at JXB online. The pGBKT7 vector which expressed GAL4 BD alone served as the negative control, and the GAL4-VpERF1 fusion construct (designated as pGBKT7-VpERF1) was used as a positive control.
The 14 deletion constructs of VpRFP1 gene fused to the vector pGBKT7 for the yeast GAL4 BD were generated. The fusion constructs were transformed into the Saccharomyces cerevisiae strain AH109 and cultured on YPDA plates at 28 °C for 3 d. The well-isolated colonies were transferred from YPDA plates to SD/-Ade /-His/-Trp/X-α-Gal plates to test the transcriptional activation activity according to their growth and the activity of β-galactosidase by observing the blue colour produced.
Subcellular immunogold labelling
Ultrathin sections were prepared from Lowicryl K4M-embedded specimens according to the method previously described (Peng et al., 2003). The ultrathin sections were first blocked for 30 min at room temperature by floating the grids on droplets of PBS containing 8 mM Na2HPO4, 1.5 mM KH2PO4, 3 mM KCl, and 500 mM NaCl (pH 7.4) supplemented with 50 mM glycine and continuously blocked with PBS supplemented with 0.1% (w/v) gelatin, 0.5% (w/v) bovine serum albumin (BSA), and 0.1% (v/v) Tween-20 (pH 7.4, PBGT). Without rinsing, the sections were incubated with New Zealand rabbit antiserum directed against VpRFP1 (all diluted 1:100 in PBGT buffer) overnight at 4 °C. Following extensive washes with PBGT buffer, the sections were incubated with secondary antibody (goat anti-rabbit IgG antibody conjugated with 10 nm gold) at a 1:100 dilution in PBGT buffer for 2 h at room temperature. The sections were rinsed consecutively with PBGT and double-distilled water, followed by staining with 2% uranyl acetate in 50% ethanol for 25 min at 25 °C and with alkaline lead citrate for 15 min. After washing extensively with double-distilled water, the ultrathin sections were examined with an electron microscope.
The specificity and reliability of the immunogold labelling were tested by two negative controls. In the first one, the antiserum was omitted to test possible unspecific labelling of the goat anti-rabbit IgG antibody–gold conjugate. In the second, rabbit pre-immune serum was used instead of the rabbit antiserum before immunogold labelling to test the specificity of the antiserum. At least three repetitions for each control were conducted for each sample.
Plant expression vector construction and Arabidopsis transformation
To generate the VpRFP1-overexpressing gene construct, the VpRFP1 coding region including the termination codon was amplified using the following primers: VpRFP1 forward primer: 5′-GGGGTCGACATGATTACCGATTCGATCAC-3′ (SalI site underlined), and VpRFP1 reverse primer: 5′-GGGGGTACCCTAAGACCTTGCAATCATGC-3′ (KpnI site underlined). The PCR products were inserted in the pMD-19-T vector (TaKaRa) and identified by DNA sequencing. The digested VpRFP1 fragments were subcloned in the same sites of the plant expression vector pWRII (Lei et al., 2009) in which the VpRFP1 was driven by the ehanced Cauliflower mosaic virus (CaMV) 35S promoter and possesses hygromycin resistance in plants. The recombinant construct harbouring VpRFP1 was introduced into Agrobacterium tumefaciens strain EHA105 via electroporation. Arabidopsis transformation of the VpRFP1 gene was carried out according to the floral dipping method (Clough and Bent, 1998). For selection of Arabidopsis transgenic lines, seeds were surface-sterilized with 70% ethanol for 2 min, then 2% sodium hypochlorite (NaOCl) solution for 6 min and rinsed five times in sterile water. Seeds were plated on basal MS salt medium containing 1% sucrose and 20 mg l−1 hygromycin. Plates were kept at 4 °C for 2–4 d in darkness to synchronize germination, then transferred to growth chambers, and maintained under the environmental conditions described above for plant growth in pots. Fourteen-day-old seedlings were transferred to the soil mix. Wild-type plants and transgenic Arabidopsis T3 lines were used for further analysis.
Results
Isolation and sequence analysis of a novel RING finger protein (VpRFP1) gene from Chinese wild V. pseudoreticulata
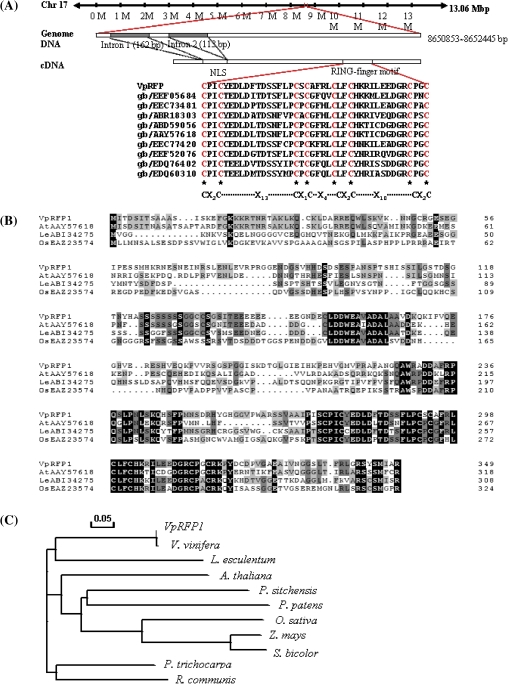
Based on the EST sequence (DQ354158) obtained previously (Xu et al., 2009), the full-length cDNA sequence (FJ356672) has now been obtained. The VpRFP1 cDNA of 1415 nt contains a 170 nt untranslated region (UTR) at the 5' end and a 192 nt 3'-UTR. The genomic sequence of the VpRFP1 gene was also cloned directly from the V. pseudoreticulata genomic DNA (GeneBank accession no. GU446678), VpRFP1 is located on chromosome 17 of the published Pinot Noir whole genome (Jaillon et al., 2007). Alignment of the genomic DNA sequence with the VpRFP1 cDNA indicated that the VpRFP1 gene contained two introns and three exons. The predicted ORF encodes a RING finger protein of 350 amino acids, which has a theoretical pI value of 5.83 and a deduced molecular mass of 38 005 Da. The deduced amino acid sequence of VpRFP1 contained a nuclear localization signal (NLS) at its N-terminus and the RING finger motif at its C-terminus. Compared with other species, the RING finger motifs belong to a novel RING finger variant of the C4C4 type, with the consensus sequence Cys-X2-Cys-X13-Cys-X1-Cys-X4-Cys-X2-Cys-X10-Cys-X2-Cys (X indicates any amino acid, and subscript number indicate the number of amino acids) (Fig. 1A). AtPEX10 is a typical C3HC4-type RING finger (Schumann et al., 2007) which possesses the conserved sequence C-X2-C-X9–39-C-X1–3-H-X2–3-C-X2-C-X4–48-C-X2-C (Stone et al., 2005) (Fig. 1A). Comparative analysis of these two motifs revealed that both are remarkably similar, but with the histidine of the C3HC4 type replaced by a cysteine residue.
Fig. 1.
Sequence analysis of the VpRFP1. (A) Schematic representation of the primary structure of the putative VpRFP1 gene and major motif. (B) Amino acid sequence alignment of VpRFP1 and closely related proteins from Arabidopsis thaliana (GenBank accession no. AAY57618), Solanum lycopersicum (formerly Lycopersion esculentum; GenBank accession no. ABI34275), and Oryza sativa (GenBank accession no. EAZ23574). Identical amino acids are shaded in black, and conserved amino acid changes are shaded in grey. (C) Phylogenetic analysis of VpRFP1 and closely related proteins. The tree was generated based on the deduced amino acids from 10 plant species using the ClustalW method of the MegAlign program: Arabidopsis thaliana (GenBank accession no. AAY57618), Lycopersion esculentum (GenBank accession no. ABI34275), Picea sitchensis (GenBank accession no. ABR18303), Oryza sativa (GenBank accession no. EAZ23574), Zea mays (GenBank accession no. NP_001143878), Populus trichocarpa (GenBank accession no. XP_002321557), Sorghum bicolor (GenBank accession no. XP_002447976), Ricinus communis (GenBank accession no. XP_002511474), Physcomitrella patens (GenBank accession no. XP_001758896), and Vitis vinifera (GenBank accession no. XP_002281744). (This figure is available in colour at JXB online.)
Comparisons of the amino acid sequences among VpRFP1 and other putative proteins are presented in Fig. 1B. The VpRFP1 protein shared 50% identical amino acids with a Solanum lycopersicum (formerly Lycopersicon esculentum) hypothetical protein (accession no. ABI34275), 46% homology with an A. thaliana RING finger family protein (accession no. AAY57618), and 38% homology with an Oryza sativa hypothetical protein (accession no. EAZ23574). Phylogenetic tree analysis showed that they were classified into three subfamilies. Vitis pseudoreticulata (accession no. FJ356672), V. vinifera (accession no. XP_002281744), and S. lycopersium (accession no. ABI34275) were part of the same subfamily (Fig. 1C).
Expression patterns of VpRFP1 in grapevine resistance responses
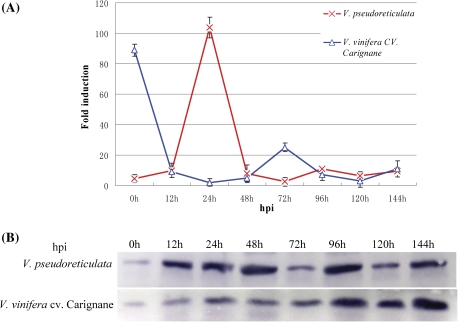
To investigate whether the expression pattern of RFP1 was similar among different Vitis species, the expression levels of RFP1 in a resistant grapevine (accession Baihe-35-1 of V. pseudoreticulata) and a susceptible variety (V. vinifera cv. Carignane) inoculated with U. necator were determined using real-time PCR and western blot. The results showed that the expression of RFP1 was induced by U. necator at the transcriptional level. For resistant grapevine accession Baihe-35-1 of V. pseudoreticulata, the up-regulated expression of VpRFP1 was detected at 12 hours post-inoculation (hpi) (Fig. 2A), peaked at 24 hpi, and returned to low baseline levels at 48 hpi. In contrast, the susceptible V. vinifera cv. Carignane displayed a high transcription level at 0 h, but this decreased immediately after inoculation, and reached the lowest levels at 24 hpi and 48 hpi, respectively.
Fig. 2.
Expression of RFP1 in grapevine leaves that were inoculated with the pathogen U. necator. Leaves of plantlets of V. pseudoreticulata accession Baihe-35-1 and V. vinifera cv. Carignane were inoculated with U. necator (5×105 spores ml−1). Control leaves were sprayed with sterile water. Leaves were collected at different time points as indicated. Hpi, hours post-inoculation with fungal spores. (A) Analysis of VpRFP1 expression at the transcript level by real-time PCR. VpGAPDH was used as the internal control. Data are the mean ±SE (n=3) of a representative experiment. (B) Analysis of VpRFP1 expression at the translational level by western blot. A 28 μg aliquot of total protein was used for each line, separated by SDS–PAGE, transferred onto a PVDF membrane, and immunodetected with a VpRFP1-specific antiserum. (This figure is available in colour at JXB online.)
To further determine whether RFP1 was induced by the pathogen at the translational level, western blot assay was carried out using a specific antiserum. The result showed that the expression of RFP1 at the translational level was induced by U. necator both in the resistant accession of V. pseudoreticulata and in the susceptible V. vinifera cv. Carignane (Fig. 2B). The expression of RFP1 in V. vinifera cv. Carignane presented a gradually increasing trend after inoculation, and reached maximum expression at 144 hpi. In contrast, the expression of VpRFP1 in the resistant accession Baihe-35-1 of V. pseudoreticulata was transiently changeable, and high peaks occurred at 48 hpi and 96 hpi, and low peaks t 72 hpi and 120 hpi, respectively.
RFP1 is induced by U. necator in both V. pseudoreticulata and V. vinifera cv. Carignane; the expression pattern of RFP1, however, was different between the two grapevine genotypes. This prompted the further elucidation of the function of VpRFP1 in response to pathogen attack.
Immunogold labelling localization of VpRFP1
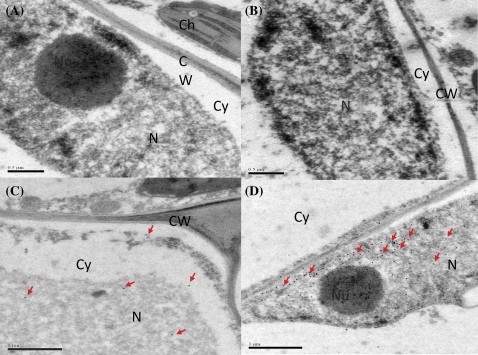
VpRFP1 fused with a GST tag was induced in E. coli BL21 (Supplementary Fig. S1A at JXB online), and then purified by an electrodialysis procedure (Supplementary Fig. S1B). The purified protein was used to immunize New Zealand rabbits to obtain polyclonal antiserum. Immunogold labelling with VpRFP1 antiserum showed that gold particles were mainly localized in the nucleus (Fig. 3). No substantial signal was detected in controls (Fig 3A, B), confirming the specificity of the immunolocalization. Non-infected leaves treated with the specific antiserum against VpRFP1 showed few gold particles in the nucleus and fewer in the cytoplasm (Fig. 3C). At 24 hpi, pathogen-infected samples showed a high density of gold particles in the nucleus (Fig. 3D), while similar signals were found in the cytoplasm of non-infected samples.
Fig. 3.
Immunogold localization of VpRFP1 in Vitis pseudoreticulata leaves. (A) With use of the rabbit pre-immune serum instead of the specific rabbit antiserum as control, no substantial signal was detected. (B) The absence of the antiserum to test the non-specific labelling of goat anti-rabbit IgG antibody–gold conjugate; no substantial signal was detected. (C) Before inoculation with pathogen, a few gold particles were detected in the nucleus and cytoplasm. (D) At 24 h after inoculation with pathogen, more gold particles were detected in the nucleus. N, nucleus; Nu, nucleolus; Cy, cytoplasm; CW, cell wall. (This figure is available in colour at JXB online.)
Transcriptional activation activity of VpRFP1 in yeast
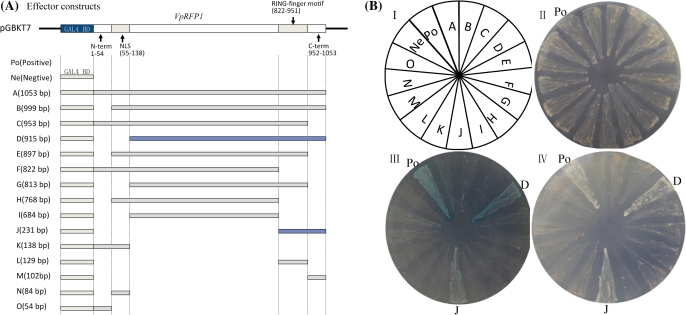
To examine whether VpRFP1 protein could function as a transcriptional activator in yeast, a series of deletion constructs of the VpRFP1 gene were fused to the GAL4 BD domain. The constructs were then transformed into yeast (strain AH109), and screened on SD medium. The VpERF1 cloned from Chinese wild grapevine has been previously confirmed to be active in yeast (unpublished data). Therefore, VpERF1::GAL4 was used as the positive control, and the empty vector pGBKT7 as the negative control.
AH109 competent cells harbouring the recombinant plasmid or the empty vector pGBKT7 were able to grow on SD/-Trp medium, indicating that all recombinant plasmids or the empty vector pGBKT7 were transferred into AH109 (Fig. 4BII). All yeast cells carrying constructs pGBKT7-A, pGB KT7-B, pGBKT7-C, pGBKT7-E, pGBKT7-F, pGBKT7-G, pGBKT7-H, pGBKT7-I, pGBKT7-K, pGBKT7-L, pGB KT7-M, pGBKT7-N, and pGBKT7-O could not grow on the SD/-Trp/-Ade/-His medium, or yield the colour reaction on the SD/-Trp/-Ade/-His/+X-α-gal medium. Cells expressing the constructs pGBKT7-D and pGBKT7-J, and the positive control caused yeast growth on the SD/-Trp/-Ade/-His medium, and induced the blue colour on the SD/-Trp/-Ade/-His/+X-α-gal medium (Fig. 4BIII, IV), indicating that portions D and J of VpRFP1 have potential transcriptional activity in yeast. The effector constructs pGBKT7-D and pGBKT7-J (Fig. 4A) shared the C-terminal region with the RING finger motif, and displayed obvious transcriptional activities. This result suggested that the C-terminal region with the RING finger motif plays a significant role in the transcriptional activity. Interestingly, the full-length VpRFP1 has no transcriptional activity in yeast.
Fig. 4.
Functional analysis of the potential transcriptional activation of the VpRFP1 gene in yeast. (A) Diagram of the effector constructs. (B) Transcriptional activation assay of the different fragments of the VpRFP1 gene in yeast. The yeast strain AH109 harbouring plasmids that encoded GAL4-VpRFP1 and different fragments were grown on: II, SD/-Trp medium; III, SD/-Trp/-Ade/-His medium; IV, SD/-Trp/-Ade/-His/+X-α-gal medium. Plates were incubated at 28 °C for 3 d. For the positive control, yeast cells harbouring pGBK-T7-VpERF1 was used as a positive control. The empty vector pGBKT7 expressing GAL4 BD alone served as a negative control. (This figure is available in colour at JXB online.)
Overexpression of VpRFP1 in Arabidopsis enhanced resistance to pathogens
To further study the biological role of VpRFP1 in defence responses, functional analyses of VpRFP1 were performed in transgenic Arabidopsis plants transformed with the overexpression construct pWRII/VpRFP1 (Fig. 5A). Upon inoculation, VpRFP1 transgenic plants were more resistant against the pathogen than the wild type (Fig. 5B, C). At 8 days post-inoculation (dpi) with G. cichoracearum, wild-type leaves showed some disease symptoms, while transgenic plants remained disease free with no visible fungal colonies. The microscopic images showed that the transgenic Arabidopsis plant inhibited the spread of G. cichoracearum (Fig. 5D). At 12 dpi, the disease symptoms in wild-type leaves became more severe than in the transgenic plants, and only a few symptoms occurred in transgenic plant leaves (Fig. 5C). As is shown (Fig. 5E), VpRFP1 was constitutively expressed in the T3 generation, but was also affected by G. cichoracearum inoculation. Interestingly, PR1 and PR2 displayed the same expression pattern as VpRFP1, whose expression increased gradually, and began to decrease after reaching a peak at 72 hpi. In contrast, the transcripts of PR3 and PDF1.2 were suppressed at 24 hpi, and reached maximum expression at 96 hpi.
Fig. 5.
Enhanced disease resistance and elevated expression levels of defence-related genes in transgenic Arabidopsis plants. (A) The schematic representation of the pWRII/VpRFP1 construct used for Arabidopsis transformation. RB, right border; LB, left border; ED 35S, CaMV 35S promoter; omega, the 5′-leader sequence of tobacco mosaic virus (TMV); NOS-T, nopaline synthase terminator; 35S, CaMV 35S promoter; HPTII, hygromycin phosphotransferase; 3'UTR, 3'-untranslated region. (B) Disease symptoms developed on the leaves of wild-type and transgenic lines 8 d after inoculation with fungal spores. (C) Disease symptoms developed on the leaves of wild-type and transgenic lines 12 d after inoculation. (D) Microscopic visualization of fungal growth on leaves of 3-week-old plants at 8 dpi with G. cichoracearum. Leaves were stained with trypan blue. Scale bar=100 μm. Three independent experiments showed similar results. (E) RT-PCR analysis of defence-related genes in transgenic T3 lines, and transgenic plant #1 is indicated. Amplification of actin is shown for the equal loading of RNA. Hpi, hours post-inoculation. (This figure is available in colour at JXB online.)
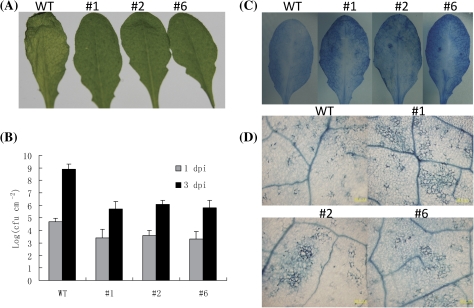
Experiments were also carried out to examine whether or not the transgenic plants were resistant to P. syringae pv. tomato DC3000 infection. The result indicated that none of the VpRFP1 transgenic plants exhibited spreading maceration in the infected leaves, and displayed clearly enhanced resistance (Fig. 6A). The growth of P. syringae pv. tomato DC3000 in inoculated plants was measured and it was found that the bacterial titres in transgenic lines were significantly lower than in the wild-type plants at 3 dpi (Fig. 6B). Macroscopic and microscopic analyses were also performed to detect the cell death. The results indicated that at the macroscopic level, cell death in infected mature leaves significantly increased in the transgenic lines compared with the wild-type plants (Fig. 6C), and at the microsopic level, the extent of visible lesions also increased significantly in the transgenic lines (Fig. 6D).
Fig. 6.
Reactions of the VpRFP1 transgenic Arabidopsis plants to P. syringae pv. tomato DC3000 infection. (A) Disease symptoms developed on the leaves of wild-type and transgenic lines 2 days post-inoculation with P. syringae pv. tomato DC3000. (B) The numbers of bacterial cells in the leaf tissues were measured at different time points after infiltration with bacterial suspension. Data are the means ±SD from three independent experiments. (C) The transgenic Arabidopsis and the wild-type leaves stained with trypan blue 2 d post-inoculation with P. syringae pv. tomato DC3000; (D) Trypan blue-stained leaves were viewed under a light microscope for plant cell death 2 d after inoculation with P. syringae pv. tomato DC3000. (This figure is available in colour at JXB online.)
Discussion
Based on previous findings from studies of other plant C4C4-type RING finger (RING finger-C2) genes (Albert et al., 2002; Stone et al., 2005), it was hypothesized that VpRFP1 isolated from the resistant accession Baihe-35-1 of Chinese wild V. pseudoreticulata may be a novel C4C4-type RING finger protein involed in grapevine–powdery mildew interaction. The gene was found to show strong evidence for transcriptional change in the susceptible V. vinifera and a significantly different response in the disease-resistant V. pseudoreticulata (Fig. 2). This rapid and strong transcriptome response to U. necator infection is consistent with recent findings of the C3HC4-type RING finger protein in pepper–pathogen interaction (Hong et al., 2007). It is possible that this induced expression pattern of defence-related transcripts plays a critical role in the regulation of early defence singal pathways in the leaves of disease-resistant V. pseudoreticulata. Further investigations will be required to provide evidence for the relationship between the expression pattern of this gene and U. necator resistance.
The translocation of TFs from the cytoplasm into the nucleus is considered to be an important step in post-translational control (Laskey and Dingwall, 1993), and requires an NLS for selective transportation into the nucleus (Raikhel, 1992). The present data suggest that the VpRFP1 amino acid sequence from residues 18 to 46 contained a putative NLS, thus suggesting that VpRFP1 can function as a TF and may be targeted to the nucleus. Evidence from immunolocalization analyses indicated that VpRFP1 was not only located in the nucleus (Fig. 3), but was also induced by the pathogen. It is interesting to note that the full-length VpRFP1 and most of the deletion mutants showed no transcriptional activation activity, whereas the RING finger motif connected to the C-terminal region of VpRFP1 (pGBKT7-D and pGBKT7-J) presented transactivation activity in yeast (Fig. 4). A possible explanation is that the transcriptional activation activity of VpRFP1 may require an environmental stimulus to become activated, such as pathogen induction. This induced activation seems to be coordinated with the phosphorylation state of the TF gene involved in response to extracellular signals via the modulation of the activities of protein kinases and protein phosphatases (Whitmarsh and Davis, 2000). It has also been documented that the activation function of TFs in planta was considerably different from that in yeast systems (Sprenger-Haussels and Weisshaar, 2000; Heinekamp et al., 2002). Therefore, another possible explanation is that the novel VpRFP1 could function as a transcriptional repressor in yeast, but as a transcriptional activator in grapevine.
Functional analyses revealed that the expression of the novel VpRFP1 was regulated by U. necator at both the transcriptional and translational levels, and also enhanced the resistance to G. cichoracearum and P. syringae pv. tomato DC3000 in transgenic Arabidopsis plants. Further functional characterization of VpRFP1 in interacting with other proteins, such as ubiquitin ligases activity in vitro and the degradation substrate, is required to provide more comprehensive information about the disease-resistant specifities of the novel C4C4-type RING finger protein VpRFP1, and will also be helpful in understanding the molecular mechanism of disease resistance in Chinese wild V. pseudoreticulata species.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. VpRFP1 expression in E. coli and purification of GST–VpRFP1 fusion protein.
Table S1. Primers used for amplifying the target gene.
Table S2. Primers used for RT-PCR and real-time PCR analysis of target gene expression.
Acknowledgments
We would like to thank Dr Wenping Qiu (Division of Plant Sciences, Missouri State University), Dr Hily Jean-michel (Cornell University College of Agriculture and Life Sciences), and Dr. Shunyuan Xiao for critical review and comments on the manuscript. This study was supported by National Natural Science Foundation of China (grant nos 30771493 and 30971972).
Glossary
Abbreviations
- EST
expressed sequence tag
- GST
glutathione-S-transferase
- IPTG
isopropyl-β-D-thiogalactopyranoside
- NLS
nuclear localization signal
- PR
pathogenesis-related
- RING
really interesting new gene
- RT-PCR
reverse transcription-PCR
- SAR
systemic acquired resistance
- SD
synthetic dextrose
- YPAD
yeast extracts peptone adenine dextrose
References
- Albert T, Hanzawa H, Legtenberg Y, De Ruwe M, van den Heuvel F, Collart M, Boelens R, Timmers H. Identification of a ubiquitin C protein ligase subunit within the CCR4 CNOT transcription repressor complex. EMBO Journal. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker D, de Carvalho-Niebel F. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. The Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L, Deng X. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. The Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M, Dhawan P, Nath P. A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Molecular Biology Reporter. 2000;18:109–115. [Google Scholar]
- Borden K, Freemont P. The RING finger domain: a recent example of a sequence–structure family. Current Opinion in Structural Biology. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, et al. The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiology. 2009;150:463–481. doi: 10.1104/pp.109.135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, He P, Cheng L, Cui Y. The resistance of wild Vitis species in China to. Agrobacterium tumefaciens. Acta Horticulturae Sinica. 1997;24:129–132. [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of. Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Ramsey K, Smith J, Auble D. Sir Antagonist 1 (San1) is a ubiquitin ligase. Journal of Biological Chemistry. 2004;279:26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- Deng X, Caspar T, Quail P. copl: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes and Development. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Deng X, Matsui M, Wei N, Wagner D, Chu A, Feldmann K, Quail P. COP1, an arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G [beta] homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Desveaux D, Marechal A, Brisson N. Whirly transcription factors: defense gene regulation and beyond. Trends in Plant Science. 2005;10:95–102. doi: 10.1016/j.tplants.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Despres C, Mess J, Levesque C, Fobert P, Dangl J, Brisson N. A ‘Whirly’ transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Developmental Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma V, Laux T, Fletcher J, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Current Biology. 2006;16:272–279. doi: 10.1016/j.cub.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Dong C, Agarwal M, Zhang Y, Xie Q, Zhu J. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences, USA. 2006;21:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P. Viticulture. Beijing: China Agriculture Press; 1999. [Google Scholar]
- Heinekamp T, Kuhlmann M, Lenk A, Strathmann A, Dröge-Laser W. The tobacco bZIP transcription factor BZI-1 binds to G-box elements in the promoters of phenylpropanoid pathway genes in vitro, but it is not involved in their regulation in vivo. Molecular Genetics and Genomics. 2002;267:16–26. doi: 10.1007/s00438-001-0636-3. [DOI] [PubMed] [Google Scholar]
- Hewitt E, Duncan L, Mufti D, Baker J, Stevenson P, Lehner P. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO Journal. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Choi H, Hwang I, Hwang B. Role of a novel pathogen-induced pepper C3-H-C4 type RING-finger protein gene, CaRFP1, in disease susceptibility and osmotic stress tolerance. Plant Molecular Biology. 2007;63:571–588. doi: 10.1007/s11103-006-9110-2. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends in Plant Science. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jones A, Thomas V, Bennett M, Mansfield J, Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with. Pseudomonas syringae. Plant Physiology. 2006;142:1603–1620. doi: 10.1104/pp.106.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Nam J, Boyes D, Holt B, III, Hubert D, Wiig A, Dangl J. A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. The Plant Journal. 2005;44:258–270. doi: 10.1111/j.1365-313X.2005.02525.x. [DOI] [PubMed] [Google Scholar]
- Koga J, Kubota H, Gomi S, Umemura K, Ohnishi M, Kono T. Cholic acid, a bile acid elicitor of hypersensitive cell death, pathogenesis-related protein synthesis, and phytoalexin accumulation in rice. Plant Physiology. 2006;140:1475–1483. doi: 10.1104/pp.105.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R, Dingwall C. Nuclear shuttling: the default pathway for nuclear proteins? Cell. 1993;74:585–586. doi: 10.1016/0092-8674(93)90505-k. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu J. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes and Development. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Wang Y, Xu W, Zhang F, Yu H, Wang X. Overexpression in Arabidopsis thaliana with a novel gene involved in ALDH from Chinese wild Vitis pseudoreticulata. Journal of Fruit Science. 2009;26:37–42. [Google Scholar]
- Mozoruk J, Hunnicutt L, Cave R, Hunter W, Bausher M. Profiling transcriptional changes in Citrus sinensis (L.) Osbeck challenged by herbivory from the xylem-feeding leafhopper Homalodisca coagulata (Say) by cDNA macroarray analysis. Plant Science. 2006;170:1068–1080. [Google Scholar]
- Park S, Kaimoyo E, Kumar D, Mosher S, Klessig D. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- Pavlousek P. Evaluation of resistance to powdery mildew in grapevine genetic resources. Journal of Central European Agriculture. 2007;8:C105–C114. [Google Scholar]
- Peng YB, Lu YF, Zhang DP. Abscisic acid activates ATPase in developing apple fruit especially in fruit phloem cells. Plant, Cell and Environment. 2003;26:1329–1342. [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiology. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen. Erysiphe cichoracearum. Plant Physiology. 2005;138:1027–1036. doi: 10.1104/pp.105.060947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P, Somssich I. Transcriptional control of plant genes responsive to pathogens. Current Opinion in Plant Biology. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragon R, Garciduenas-Pina C, Guzman P. Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in. Arabidopsis thaliana. Plant Molecular Biology. 1999;40:579–590. doi: 10.1023/a:1006267201855. [DOI] [PubMed] [Google Scholar]
- Schumann U, Prestele J, O'Geen H, Brueggeman R, Wanner G, Gietl C. Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proceedings of the National Academy of Sciences, USA. 2007;104:1069–1074. doi: 10.1073/pnas.0610402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Guzman P. Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics. 2004;167:919–929. doi: 10.1534/genetics.104.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, Harzen A, Colby T, Kamoun S, van der Hoorn R. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. The Plant Cell. 2008;20:1169–1183. doi: 10.1105/tpc.107.056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra R. The ubiquitin 26S proteasome proteolytic pathway. Plant Biology. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Song X, Huang W, Shi M, Zhu M, Lin H. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Sprenger-Haussels M, Weisshaar B. Transactivation properties of parsley proline-rich bZIP transcription factors. The Plant Journal. 2000;22:1–8. doi: 10.1046/j.1365-313x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- Stone S, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of. Arabidopsis. Plant Physiology. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger transcription factors in plants. Cellular and Molecular Life Sciences. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver N, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Wang W, Devoto A, Turner J, Xiao S. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Molecular Plant-Microbe Interactions. 2007;20:966–976. doi: 10.1094/MPMI-20-8-0966. [DOI] [PubMed] [Google Scholar]
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresti M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003;24:2369–2375. doi: 10.1002/elps.200305500. [DOI] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27:2782–2786. doi: 10.1002/elps.200500722. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, He P, Chen J, Lamikanra O, Lu J. Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis. 1995;34:159–164. [Google Scholar]
- Whitmarsh A, Davis R. Regulation of transcription factor function by phosphorylation. Cellular and Molecular Life Sciences. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I, Schiff C, Hughes D, Somerville S. Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics. 2001;158:1301–1309. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Li Q. A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Molecular Biology. 2003;53:37–50. doi: 10.1023/b:plan.0000009256.01620.a6. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu Z, Xiao Y, Wang Y. Construction of a cDNA library of Vitis pseudoreticulata native to China inoculated with Uncinula necator and the analysis of potential defence-related expressed sequence tags (ESTs) South African Journal for Enology and Viticulture. 2009;30:65–71. [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner J, Xiao S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. The Plant Journal. 2009;60:539–550. doi: 10.1111/j.1365-313X.2009.03978.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua N. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes and Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proceedings of the National Academy of Sciences, USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng X. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. The Plant Cell. 2008;20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. The Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.