Abstract
It is known that the clade A protein phosphatase 2Cs (PP2Cs), including ABI1 and ABI2 and other PP2C members, are key players that function directly downstream of the PYR/PYL/RCAR abscisic acid (ABA) receptors. Here, identification of a crucial site for function of ABI2 protein phosphatase in ABA signalling is reported. It was observed that a calcium-dependent protein kinase (CDPK) phosphorylation site-like motif (CPL) in the ABI2 molecule is required for the interactions of ABI2 with the two members of the ABA receptors PYL5 and PYL9 and with a downstream protein kinase SnRK2.6, and for the catalytic activity of ABI2 in vitro, as well as for the response of ABI2 to the ABA receptors PYL5/PYL9 in relation to the ABA receptor-induced inhibition of the ABI2 phosphatase activity. Further, genetic evidence was provided to demonstrate that this CPL is required for the function of ABI2 to mediate ABA signalling. These data reveal that this CPL is an important site necessary for both the phosphatase activity of ABI2 and the functional interaction between ABI2 and PYL5/9 ABA receptors, providing new information to understand primary events of ABA signal transduction.
Keywords: ABA signalling, ABI2, Arabidopsis thaliana, functional site, protein phosphorylation, type 2C protein phosphatase
Introduction
Abscisic acid (ABA) regulates many aspects of plant developmental processes, such as seed maturation, germination, and post-germination growth, and plays a central role in plant adaptation to environmental challenges (Finkelstein et al., 2002; Adie et al., 2007). Numerous components of ABA signal transduction have been identified during the past decades, which include, for example, phospholipases C/D, G proteins, G-protein-coupled receptors and receptor-like kinases, protein kinases and phosphatases, ubiquitin E3 ligases, and various classes of transcription factors (for reviews, see Finkelstein et al., 2002; Wang, 2002; Himmelbach et al., 2003; Shinozaki et al., 2003; Fan et al., 2004; Hirayama and Shinozaki, 2007; Seki et al., 2007; Cutler et al., 2010). This progress has significantly advanced understanding of ABA signal transduction.
In recent years, two distinct ABA receptor-mediated pathways were identified. A class of START proteins PYR/PYL/RCAR was reported to function as ABA receptors, which mediate a signalling pathway directly upstream of a group of type 2C protein phosphatases (PP2Cs) such as ABI1 and ABI2 (Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009; Santiago et al., 2009a, b; Yin et al., 2009; Cutler et al., 2010). A PYR/PYL/RCAR-mediated ABA signalling pathway from ABA perception to downstream gene expression has been reconstituted in vitro (Fujii et al., 2009; Cutler et al., 2010). The Mg-chelatase H subunit (CHLH/ABAR) was reported to bind ABA and function in ABA signalling as an ABA receptor in Arabidopsis thaliana (Shen et al., 2006; Wu et al., 2009) and also to be a key player to connect the circadian clock with the ABA-mediated plant responses to drought (Legnaioli et al., 2009). A signalling pathway from primary events to downstream gene expression, involving a group of WRKY-domain transcription factors, has been proposed (Shang et al., 2010).
In the PYR/PYL/RCAR-mediated ABA signalling pathway, PP2Cs are key players that relay ABA signal directly from the PYR/PYL/RCAR ABA receptors to their downstream regulators SNF1-related protein kinases (SnRKs), which activate an ABF/AREB/ABI5 clade of bZIP-domain transcription factors via a protein phosphorylation process finally to induce physiological ABA responses (Fujii et al., 2009; Cutler et al., 2010). Thus, reversible protein phosphorylation, mediated by SnRKs and PP2Cs, plays an essential role in the PYR/PYL/RCAR-mediated ABA signalling.
PP2Cs are a class of conserved protein serine/threonine phosphatases, which, in cooperation with other types of protein phosphatases, including protein tyrosine phosphatases and protein serine/threonine phosphatases, PP1s, PP2As, and PP2Bs, regulate many important physiological events including stress responses by catalysing protein dephosphorylation in yeast, plant, and animal cells (Maeda et al., 1994; Sheen, 1996; Gaits et al., 1997; Takekawa et al., 1998; Nguyen and Shiozaki, 1999; Warmka et al., 2001; Meskiene et al., 2003; for a review, see Schweighofer et al., 2004). Plant PP2Cs are encoded by a large multigene family. In Arabidopsis, 76 genes were identified as PP2C candidates, which fall into 10 groups (A–J) (Kerk et al., 2002; Schweighofer et al., 2004). In addition to ABI1 (Leung et al., 1994; Meyer et al., 1994; Gosti et al., 1999) and ABI2 (Leung et al., 1997), HAB1, HAB2 (Leonhardt et al., 2004; Saez et al., 2004, 2006), AHG1 (Nishimura et al., 2007), and PP2CA/AHG3 (Cherel et al., 2002; Kuhn et al., 2006; Yoshida et al., 2006; Lee et al., 2009) were also identified as ABA signalling components, and all six members belong to the clade A PP2Cs (Schweighofer et al., 2004). Recently, two homologous members of clade B PP2Cs, PP2C5 and AP2C1, were reported to be involved in ABA signalling (Brock et al., 2010).
Eukaryotic PP2Cs carry their conserved catalytic domain at either the N- or C-terminus. The catalytic domain of most (44 out of 76) Arabidopsis PP2Cs, including all the nine members of clade A, is located at the C-terminus (for reviews, see Rodriguez, 1998; Schweighofer et al., 2004). ABI1 and ABI2, like the other PP2Cs, are Mg2+-dependent protein phosphatases, and thus the metal coordinating centre in the catalytic domain should be critical for both their protein phosphatase activity and physiological functions in ABA signalling (for reviews, see Rodriguez, 1998; Schweighofer et al., 2004). The point mutations of both abi1-1 and abi2-1 in ABI1 and ABI2 PP2Cs, respectively, involve substitution of the same amino acid residue (substitution of an aspartic acid residue for a glycine residue) at an equivalent position close to the Mg2+-coordinating centre (G180D for abi1-1 mutation and G168D for abi2-1 mutation) in the catalytic domain, which disrupt the interactions between the PP2Cs and the ABA receptors PYR/PYL/RCAR (Ma et al., 2009; Park et al., 2009), but do not affect the interactions between the PP2Cs and their downstream regulatory components SnRK2s (Umezawa et al., 2009). These mutations reduce PP2C activity of both ABI1 and ABI2 in vitro (Leung et al., 1997; Leube et al., 1998; Gosti et al., 1999), but appear to enhance constitutively the dephosphorylation activity of the PP2Cs for their natural SnRKs in vivo (Umezawa et al., 2009), which explains the dominant mutation of ABA-insensitive phenotypes of the abi1-1 and abi2-1 mutants (Leung et al., 1994, 1997; Meyer et al., 1994; Gosti et al., 1999). Transgenic analysis showed that a mutated form of HAB1 (G260D mutation), which is an equivalent mutation to abi1-1 and abi2-1, resulting in ABA insensitivity in transgenic plants, behaved as a dominant positive or hypermorphic mutation in planta (Robert et al., 2006). In addition to these in planta genetic approaches, biochemical analysis and protoplast transgenic assays also showed that the metal-coordinating motif was important and the N-terminal domain was likely to play a regulatory role in PP2C activity for ABI1 in ABA signalling (Leube et al., 1998; Sheen, 1998). However, PP2Cs have many motifs in their structure (Schweighofer et al., 2004); additional functionally active domains remain to be identified to deepen our understanding of PP2C-mediated ABA signalling.
Here, identification of an important site for function of ABI2 protein phosphatase in ABA signalling is reported. It was observed that a CDPK phosphorylation site-like motif (CPL) in the ABI2 molecule is required for the interactions of ABI2 with two members of the ABA receptors, PYL5 and PYL9, and with a downstream protein kinase, SnRK2.6, and for the catalytic activity of ABI2 in vitro, as well as for the response of ABI2 to the ABA receptors PYL5/PYL9 in relation to the ABA receptor-induced inhibition of the ABI2 phosphatase activity. It was demonstrated that this CPL is required for the function of ABI2 to mediate ABA signalling. These data reveal that this CPL site is necessary for both the phosphatase activity of ABI2 and the functional interaction between ABI2 and PYL5/9 ABA receptors, providing new information to understand primary events of ABA signal transduction.
Materials and methods
Plant materials, generation of transgenic plants, and growth conditions
A T-DNA insertion mutant (SALK_015166, with ecotype Col-0 as background) of the ABI2 gene from the Salk collection of the Arabidopsis Biological Resource Center (ABRC) was isolated. This abi2 mutant allele was named abi2-t1. This mutant allele was also used in previous studies of ABA signalling (Yoshida et al., 2006; Rubio et al., 2009). The molecular characterization of this mutant allele was confirmed. To identify the individual homozygous T-DNA insertion, genomic DNA was obtained and submitted to PCR genotyping using the ABI2-specific primers (forward 5′-AAACTGTTGGGTCTACCTCGG-3′ and reverse 5′-ACCATCCCATATTCTGGTTGG-3′). Sequencing of the T-DNA flanking region in abi2-t1 showed that the insertion was localized 20 nucleotides upstream of the TGA stop codon, and the T-DNA insertion generates a 37 bp deletion from –57 to –21 bp 5′-upstream of the ABI2 translation stop codon. Semi-quantitative RT-PCR amplification was performed for characterization of ABI2 full-length transcript in the abi2-t1 mutant plants. The 4-week-old plants were collected and frozen in liquid nitrogen for RNA extraction. The primers used for the PCR amplifications were: forward primer 5′-ATGGACGAAGTTTCTCCTGCAGTCGCTG-3′ and reverse primer 5′-ATTCAAGGATTTGCTCTTGAATTTCC-3′ for ABI2 (At5g57050), and forward primer 5′-AGGCACCTCTTAACCCTAAAGC-3′ and reverse primer 5′-GGACAACGGAATCTCTCAGC-3′ for β-ACTIN-8. The results indicated that the abi2-t1 is a knock-down allele (see Supplementary Fig. S2A available at JXB online).
For generating transgenic lines, different PCR products were prepared by using the primers listed in Supplementary Table S1 at JXB online. Wild-type ABI2 (ABI2-WT) was amplified by using primers I2-F1and I2-R1 with ABI2 cDNA as template, then fused to the pMD-19-T vector (Takara, Dalian Division, China) and confirmed by sequencing. The point mutations and deletion mutations of the ABI2 gene were generated by overlap PCR using KOD-PLUS- (Toyobo) with ABI2-WT plastid as template. ABI2-3A point mutations were generated as follows: for the point mutation of the first putative CDPK phosphorylation site R-X-X-S/T (RPFT, mutation T16→A) of ABI2 (Furihata et al., 2006), PCR was done with primers I2A1F and I2R1 with ABI2-WT plastid as template, then the product was used as template for PCR with primers I2F1 and I2R1, which resulted in a product A1P. For the mutations of both the first and second putative CDPK phosphorylation sites (RPFT, mutation T16→A; and RTES, mutation T101→A), PCR was performed with primer pairs I2F1 plus I2A2R and I2A2F plus I2R1 by using A1P as template and the products, A2P1 and A2P2, were generated, respectively. Another PCR was performed with I2F1 and I2R1 with A2P1 and A2P2 as templates, which allowed a product A2P to be obtained. For the mutations of all the three putative CDPK phosphorylation sites (RPFT, mutation T16→A; RTES, mutation S101→A; and RGKT, mutation T261→A), primer pairs I2F1 plus I2A3R, I2A3F plus I2R1, and I2F1 plus I2R1 were used for PCR amplification with A2P as template. For ABI2-3D point mutations (RPFT, mutation T16→D; RTES, mutation S101→D; and RGKT, mutation T261→D), similar procedures to those described above for the ABI2-3A mutation were used with primer pairs I2DF plus I2R1, I2F1 plus I2DR, and I2F1 plus I2R1.
For the first putative CDPK phosphorylation site R-X-X-S/T (RPFT) deletion mutation (amino acid residues 13–16) of ABI2 (dF), PCR was performed with primers I2F2 and I2R1 with ABI2-WT plastid as template, and the PCR product (P1) was gel purified, and then another PCR was perfromed with primers I2F1 and I2R1 using P1 as template. For the second putative CDPK phosphorylation site R-X-X-S/T (RTES) deletion (amino acid residues 98–101) of ABI2 (dS), PCR was performed with primer pairs I2F1 plus I2R3 for isolating the N-terminal product (P2-1) and I2F3 plus I2R1 for isolating the C-terminal product (P2-2) with ABI2-WT as template. The purified PCR products P2-1 and P2-2 were used as templates for the third PCR with the I2F1 plus I2R1 primer pair. For the third putative CDPK phosphorylation site R-X-X-S/T (RGKT) deletion (amino acid residues 258–261) of ABI2 (dT), primer pairs I2F1 plus I2R4, I2F4 plus I2R1, and I2F1 plus I2R1 were used for PCR amplification using similar procedures to those described above for dS mutation. The PCR products harbouring different mutations or deletions, which include ABI2-WT, ABI2-3A, ABI2-3D, dF, dS, dT, and dFS (deletion of both the first and second putative CDPK phosphorylation sites), dFT (deletion of both the first and third putative CDPK phosphorylation sites), dST (deletion of both the second and third putative CDPK phosphorylation sites), and dFST (deletion of all the three putative CDPK phosphorylation sites), were fused to the pMD-19-T vector (Takara, Dalian Division, China) and the nucleotide sequences were confirmed by sequencing the entire insert fragment.
All the above constructs were subcloned into the binary vector pCAMBIA1300 that contains the Cauliflower mosaic virus (CaMV) 35S promoter and a C-terminal green fluorescent protein (GFP) flag. Each vector was confirmed by sequencing. Different constructs were introduced into the GV3101 strain Agrobacterium tumefaciens and transformed into Arabidopsis Col wild type or abi2-t1 mutant plants by the floral dip method as previously described (Clough and Bent, 1998). The homologous T3 generation seeds or plants were used for analysis. At least 10 homozygous overexpression or complementary transgenic lines were obtained with similar ABA-related phenotypes, and, after preliminary analysis, two representative lines were analysed in detail.
Plants were grown in a growth chamber at 19–20 °C on Murashige and Skoog (MS) medium (Sigma-Aldrich) at ∼80 μmol photons m−2 s−1 or in compost soil at ∼120 μmol photons m−2 s−1 over a 16 h photoperiod.
Analysis of protein interaction by the yeast two-hybrid system
Interaction between proteins was carried out using a yeast Gal4-based two-hybrid system 3 (Clontech) according to the manufacturer's instructions. The primers used for cloning the related cDNAs are listed in Supplementary Table S2 at JXB online. The PYL5 or PYL9 cDNA was inserted by the EcoRI (5′ end) and SalI (3′ end) sites into the pGBKT7 plasmid to generate bait plasmids, and the SnRK2.6 cDNA was inserted by the EcoRI (5′ end) and BamHI (3′ end) sites into the pGBKT7 plasmid to generate bait plasmid. The ABI2 variants were cloned by the EcoRI (5′ end) and XhoI (3′ end) sites into pGADT7 plasmid to generate prey plasmids. The transformants were tested on SD screening medium as indicated.
To test the interaction intensity of PYL9 and ABI2 variants in the yeast two-hybrid system, a drop test was used to assay yeast growth. Yeast cells expressing various ABI2 constructs were grown on SD-medium lacking Leu, Trp, His, and Ade overnight, and then transferred to fresh, liquid, Leu-Trp-His-Ade-deficient medium to OD600=0.2. After a further incubation of 4 h, the OD values were measured. The cells were diluted in sterile water, and then a 8 μl aliquot of different concentrations of cells (OD600=0.1, 0.05, 0.01, or 0.001, as indicated) was spotted on the Leu-Trp-His-Ade-deficient medium. The yeast cells were further grown at 30 °C for 2 d or 3 d for observations.
Production of PYL5 and PYL9, ABI2, and mutated ABI2 proteins
The primers used for the protein production are listed in Supplementary Table S3 at JXB online. The full-length open reading frame (ORF) of wild-type ABI2 and the mutated forms of ABI2 were subcloned into pET28a-c(+) with EcoRI and XhoI, and the full-length ORF of PYL5 and PYL9 was subcloned into pET28a-c(+) with EcoRI and SalI. The constructs were expressed in Escherichia coli strain BL21(DE3). The individual proteins were purified according to the manufacturer's instructions (Novagen). It is noteworthy that, for the expression of ABI2s to assay PP2C activity, 5 mM MgCl2·6H2O was added to the E. coli culture medium, and the cells were collected and lysed in a buffer containing 500 mM NaCl, 5 mM MgCl2·6H2O, and 25 mM TRIS, pH 8.0.
Assay of phosphatase activity
The phosphatase activity was measured by the serine-threonine phosphatase assay system (Promega) essentially as previously described (Yin et al., 2009). Each reaction was performed in a 50 μl reaction volume containing 9 μg of ABI2 (equivalent to 3.8 μM), 20 mM HEPES, pH 7.5, 150 mM NaCl, and PYL5 or PYL9 (20 μg, equivalent to 16.9 μM) protein and (+)-ABA (10 μM, Sigma-Aldrich) were added if required. The concentration of PYL5 protein in the reaction medium is as indicated when assaying the PYL5 dose dependence of ABI2 activity inhibition (see Fig. 4B). After incubation with peptide substrate (supplied with the Promega kit) at 30 °C for 30 min, the reaction was stopped by addition of 50 μl of molybdate dye. Absorbance at 630 nm was measured 30 min after the addition of molybdate dye.
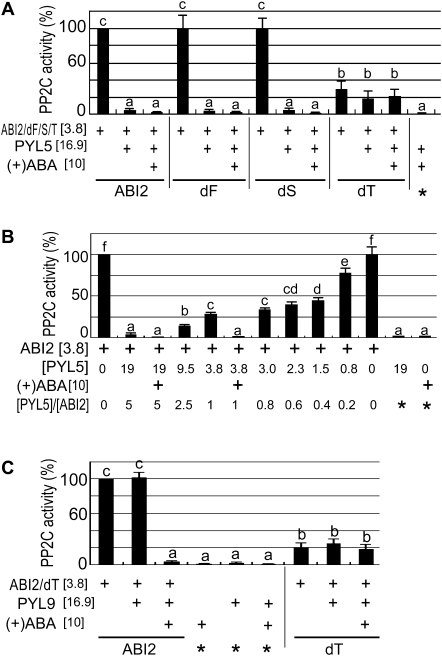
Fig. 4.
Deletion mutation of the third CDPK phosphorylation site-like motif of ABI2 reduces the catalytic activity and abolishes the response of ABI2 to the ABA receptors PYL5 and PYL9 in vitro. (A) Deletion mutation of the third CDPK phosphorylation site-like motif (CPL) of ABI2 (dT), but not the deletion mutations of the first (dF) and second (dS) CPL, reduces the catalytic activity and abolishes the response of ABI2 to the ABA receptor PYL5 in vitro. The concentrations of ABI2/dF/dS/dT protein, PYL5 protein, and (+)ABA in the reaction medium are indicated in square brackets (unit: μM). (B) PYL5 inhibited the ABI2 activity in a dose-dependent manner in the absence of ABA, but ABA can improve the inhibitory effect of PYL5 on ABI2 activity. The PYL5 protein at different concentrations (indicated by [PYL5], unit: μM) was added to the PP2C-assaying medium to investigate the PYL5 dose dependence. The concentration of ABI2 or (+)ABA in the reaction medium is indicated in square brackets (unit: μM). The ratios of the concentrations of PYL5 to ABI2 are shown (indicated by [PYL5]/[ABI2]). (C) ABI2 deletion mutation form dT reduces the catalytic activity and abolishes the response of ABI2 to the ABA receptor PYL9 in vitro. The concentrations of ABI2/dT protein, PYL9 protein, and (+)ABA in the reaction medium are indicated in square brackets (unit: μM). In A–C, wild-type ABI2 protein is indicated by ABI2. Addition of the proteins (or 10 μM ABA) listed on the left to the PP2C-assaying system is indicated by a plus sign (+). Several controls were used and are marked by asterisks (*). Relative PP2C activities (%) were used, which were normalized relative to that of the wild-type ABI2 in the absence of PYL5/9 and ABA, which was taken as 100% with a value of ∼3.3 nmol min−1 mg−1 PP2C. Each value is the mean ±SE of five independent biological determinations, and different letters indicates significant differences at P <0.05 (Duncan's multiple range test).
Phenotypic analysis:
Phenotypic analysis was done essentially as previously described (Shang et al. 2010). Approximately 100 seeds were sterilized and planted in triplicate on MS medium (Sigma, St. Louis, MO, USA; full-strength MS) to assay germination. The medium contained 3% sucrose and 0.7% agar (pH 5.8) and was supplemented or not with different concentrations of (±)-ABA. The seeds were incubated at 4 °C for 3 d before being placed at 20 °C under light conditions, and germination (emergence of radicals) was scored at the indicated times. Seedling growth was assessed by directly planting the seeds in ABA-containing MS medium to investigate the response of seedling growth to ABA after germination.
Accession numbers:
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g57050 (ABI2), At5g05440 (PYL5), and At1g01360 (PYL9).
Results
ABI2 is unlikely to function as a phosphorylated form catalysed by CDPK-mediated phosphorylation in ABA signalling
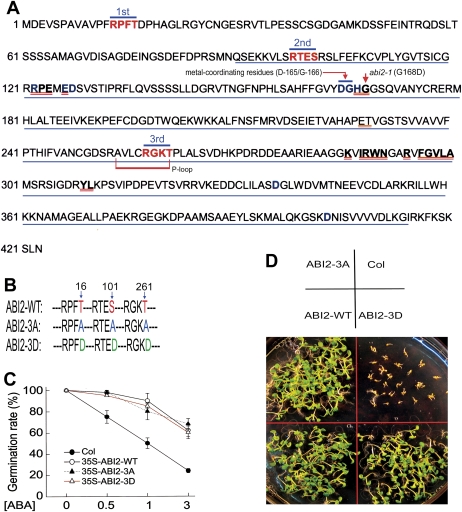
It is known that ABI2, like its homologue ABI1, is a key PP2C involved in ABA signalling (Leung et al., 1994, 1997; Meyer et al., 1994; Gosti et al., 1999). Sequence analysis showed that there are three putative CDPK phosphorylation sites (CPSs) in the ABI2 molecule. The first and second CPSs are conserved in ABI1 and ABI2, but not in other clade 2 PP2Cs such as HAB1 and HAB2, while the third CPS displays sequence similarity among the PP2Cs (Fig. 1A; Supplementary Fig. S1 at JXB online). The first CPS of ABI2 is located in the N-terminus (amino acid residues 13–16, RPFT), and the other two CPSs are located at the PP2C catalytic domain (amino acid residues 98–101, RTES; and amino acid residues 258–261, RGKT) (Fig. 1A). The third CPS falls into a putative ATP/GTP-binding site motif (Fig. 1A; Supplementary Fig. S1).
Fig. 1.
ABI2 is unlikely to function as a phosphorylated form catalysed by CDPK-mediated phosphorylation in ABA signalling. (A) Diagram of some potentially functional domains in the ABI2 molecule. Red letters indicate the three putative CDPK phosphorylation sites, and 1st, 2nd, and 3rd indicate, respectively, the first, second, and third site. Blue letters in bold indicate amino acid residues important for PP2C phosphatase activity. The sequence underlined in blue indicates the PP2C catalytic domain. Letters double-underlined in red denote the amino acid residues essential for interaction between PYL1 and ABI2 as described previously (Miyazono et al., 2009). The ABI2 metal-coordinating residues (D-165/G-166), a potential ATP/GTP-binding site motif or phosphate-binding loop (P-loop: AVLCRGKT), and the point mutation of abi2-1 (G168D) are also indicated. (B) A diagram showing the ABI2-3A (substitution mutation T16A plus S101A plus T261A) and ABI2-3D (substitution mutation T16D plus S101D plus T261D) mutations compared with wild-type ABI2 (ABI2-WT). The numbers of the serine (S)/threonine (T) residues, T16, S101, and T261, which may putatively be phosphorylated by CDPK, are indicated (blue arrows). (C) ABI2-3A and ABI2-3D transgenic lines (with wild-type Col as background) show a similar ABA insensitivity to ABI2-WT transgenic lines in ABA-induced inhibition of seed germination. Each value is the mean ±SE of five independent biological determinations. (D) ABI2-3A and ABI2-3D transgenic lines show a similar ABA insensitivity to ABI2-WT transgenic lines in early seedling growth. The seeds were directly planted in 3 μM ABA-containing medium and the growth status was recorded 12 d after stratification. The experiments were repeated five times with similar results.
To address the question of whether ABI2 may function in ABA signalling through these CPSs as a phosphorylated form catalysed by CDPK-mediated phosphorylation, transgenic lines expressing two mutated ABI2s were generated, ABI2-3A (substitution mutation T16A plus S101A plus T261A) and ABI2-3D (substitution mutation T16D plus S101D plus T261D) (Fig. 1B). ABI2-3A harbours three loss-of-function mutated CDPK phosphorylation sites, and ABI2-3D has three constitutive active mutated CDPK phosphorylation sites. The results showed that the transgenic lines expressing ABI2-3A or ABI2-3D all displayed ABA-insensitive phenotypes in ABA-induced seed germination inhibition and post-germination growth arrest, and these phenotypes were essentially the same as those of the transgenic lines expressing wild-type ABI2 (Fig. 1C, D). These data suggested that ABI2 may not function as the phosphorylated form catalysed by a CDPK-mediated phosphorylation process in ABA signalling.
The third CDPK phosphorylation site-like motif of ABI2 is required for the interactions of ABI2 both with PYL5/PYL9 and with SnRK2.6
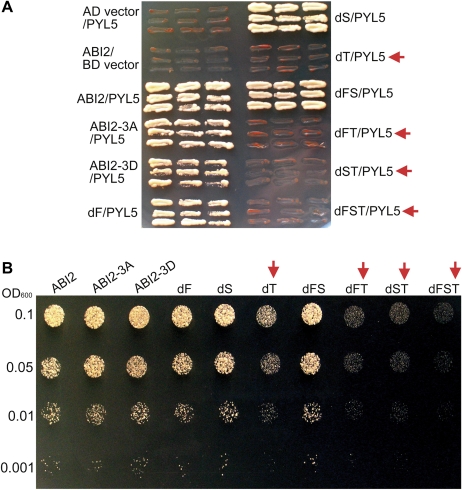
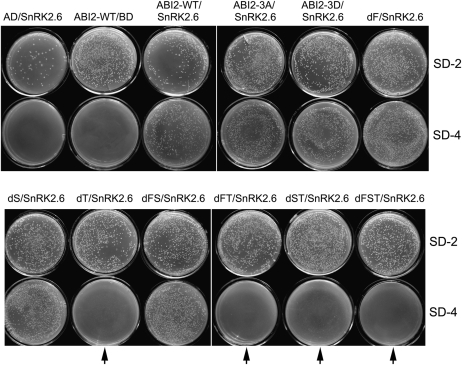
Given that these CPSs in the ABI2 molecule may not function in ABA signalling as true CDPK phosphorylation sites, the putative CPS was renamed the CPS-like motif (abbreviated to CPL) in the following text to describe these motifs only according to their sequence similarity. To assess possible functions of these CPLs, three mutated ABI2s with deletion of the first CPL (four amino acid residues from 13 to 16: RPFT, named dF), the second CPL (four amino acid residues from 98 to 101: RTES, named dS), or the third CPL (four amino acid residues from 258 to 261: RGKT, named dT), respectively, were generated (Figs 1–7). Also, other different combined deletion mutations of ABI2 were created, which include dFS (deletion of both the first and second CPLs), dFT (deletion of both the first and third CPLs), dST (deletion of both the second and third CPLs), and dFST (deletion of all the three CPLs) (Figs 1–7). Using these ABI2 mutations, it was first tested whether any of the CPLs of ABI2 plays a role in the interaction between ABI2 and two members of the START-domain ABA receptors PYL5/RCAR8 and PYL9/RCAR1, which had been genetically better characterized in previous studies (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009b). Additionally, possible roles of these CPLs in the interaction between ABI2 and a downstream protein kinase SnRK2 (the interactions between the mutated ABI2 and the other two ABA signalling components SnRK2.2/2.3 were not tested because of the auto-activation phenomenon in the yeast two-hybrid assay with the protein pairs ABI2–SnRK2.2/2.3), which was identified as a direct downstream component of the clade A PP2Cs like ABI2 (Fujii et al., 2009), were tested. The results showed that deletions of either the first or the second CPL or both CPLs did not affect the interactions between ABI2 and PYL5/PYL9 or between ABI2 and SnRK2.6, but that the deletion of the third CPL disrupted the interactions between ABI2 and PYL5 and between ABI2 and SnRK2.6, and significantly reduced the interaction between ABI2 and PYL9 (Figs 2A, B, 3). Deletions of the third CPL combined with that of the first and second CPLs appeared to have an increasing effect to reduce the interaction between ABI2 and PYL9 (Fig. 2B). These results were not changed by ABA treatments (data not shown), which is consistent with previous observations that the interactions between the HAB1 PP2C and seven PYL members (PYL5–PYL7 and PYL9–PYL12) are independent of ABA treatment (Park et al., 2009). These findings suggested that the third CPL may play an important role in ABA signalling.
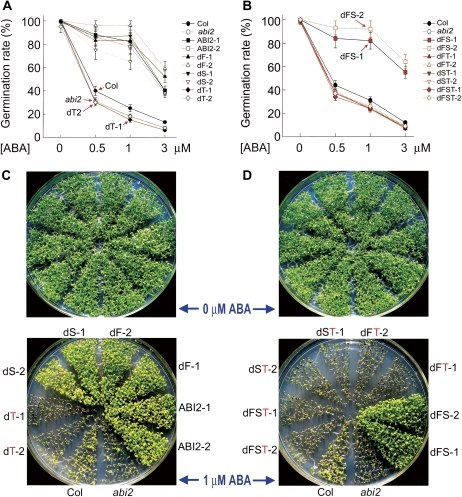
Fig. 7.
Deletion mutation of the third CDPK phosphorylation site-like motif of ABI2 disrupts the ABI2 function. The genetic background of all the transgenic lines was abi2-t1 (indicated by abi2) mutant plants. (A and B) Test of ABA-induced inhibition of seed germination in different transgenic lines. The germination rates were recorded 36 h after stratification. Each value is the mean ±SE of three biological determinations. (C and D) Test of ABA-induced post-germination growth arrest in different transgenic lines. The seeds were planted in ABA-free (top panels) or 1 μM ABA-containing medium for 3 d stratification, and growth status was recorded 15 d after stratification. The experiments were repeated three times with similar results. All the abbreviations are the same as used in Figs 2 and 6, and the suffix numbers (1, 2) indicate the numbers of the transgenic lines.
Fig. 2.
Deletion mutation of the third CDPK phosphorylation site-like motif of ABI2 disrupts ABI2–PYL5 interaction or reduces ABI2–PYL9 interaction. The test was done in a yeast two-hybrid system. (A) Yeast cells were transformed with different construct pairs harbouring ABI2/mutant ABI2 and PYL5 and investigated for cell growth. Red arrows indicate the yeast cells transformed with a construct harbouring a deletion mutation of the third putative CDPK phosphorylation site (T mutation). AD and BD vectors were used as negative controls. The experiments were repeated three times with the same results. (B) Semi-quantitative drop test: yeast cells were transformed with different construct pairs harbouring ABI2/mutant ABI2 and PYL9 and investigated for cell growth. Red arrows indicate the yeast cells transformed with a construct harbouring a deletion mutation of the third putative CDPK phosphorylation site (T mutation). The OD600 values (0.1, 0.05, 0.01, and 0.001) were used to indicate the yeast concentrations as described in the Materials and methods. The experiments were repeated three times with similar results. For the abbreviations ABI2-WT, ABI2-3A, and ABI2-3D, see Fig. 1B–D. dF, mutant ABI2 with deletion of the first CDPK phosphorylation site-like motif (CPL); dS, mutant ABI2 with deletion of the second CPL; dT, mutant ABI2 with deletion of the third CPL; dFS, mutant ABI2 with deletion of both the first and second CPLs; dFT, mutant ABI2 with deletion of both the first and third CPLs; dST, mutant ABI2 with deletion of both the second and third CPLs; dFST, mutant ABI2 with deletion of all the three CPLs.
Fig. 3.
Deletion mutation of the third CDPK phosphorylation site-like motif of ABI2 substantially disrupts ABI2–SnRK2.6 interaction. The test was done in a yeast two-hybrid system. Yeast cells were transformed with different construct pairs harbouring ABI2/mutant ABI2 and SnRK2.6, and investigated for cell growth. Arrows indicate the yeast cells transformed with a construct harbouring a deletion mutation of the third putative CDPK phosphorylation site. AD and BD vectors were used as negative controls. The experiments were repeated three times with the same results. SD-2 indicates SD medium lacking Leu and Trp, and SD-4 indicates Leu-Trp-His-Ade-deficient SD medium. All the other abbreviations are the same as used in Fig. 2.
The third CDPK phosphorylation site-like motif of ABI2 is required for the catalytic activity of ABI2 in vitro
It was further tested whether the CPL deletion mutations of ABI2 affect the PP2C phosphatase activity in vitro by using a phosphor-peptide as a substrate. It was confirmed that PYL9-induced inhibition of the ABI2-PP2C activity depends on ABA (Fig. 4C; Ma et al., 2009; Szostkiewicz et al., 2010). It was observed, however, that PYL5 could inhibit the PP2C activity of ABI2 in the absence of ABA, and that this inhibition of ABI2 activity was PYL5 dose dependent (Fig. 4A, B). ABA could improve the inhibiting effect of PYL5 on ABI2 activity when the added amounts of the PYL5 protein were low in the assaying system (Fig. 4B). Previous reports showed ABA dependence of the PYL5-induced inhibition of ABI2-PP2C activity (Ma et al., 2009; Melcher et al., 2010), which may be due to a low level of the PYL5 protein present in their PP2C assay system (Melcher et al., 2010: 0.6–2 μM PYL5 protein in 0.2 μM ABI2-containing medium; Ma et al. 2009: 0.6 μM PYL5 protein in 0.04 –M ABI2-containing medium).
It was shown that neither the first nor the second CPL deletion mutations affected the PP2C phosphatase activity of ABI2, and that these two mutated forms of ABI2 showed the same PYL5-induced inhibition of their PP2C activity and the same ABA independence of this PYL5-induced PP2C inhibition as the wild-type ABI2 (Fig. 4A). However, the third CPL deletion significantly reduced the phosphatase activity of ABI2, and abolished the response of ABI2 to the ABA receptors PYL5 and PYL9 in vitro: this reduced phosphatase activity could not be further inhibited by either PYL5 or PYL9 in vitro (Fig. 4A, C).
Deletion of the third CDPK phosphorylation site-like motif of ABI2 disrupts the function of ABI2 in ABA signalling
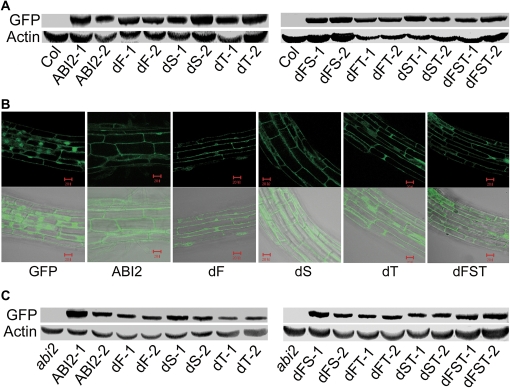
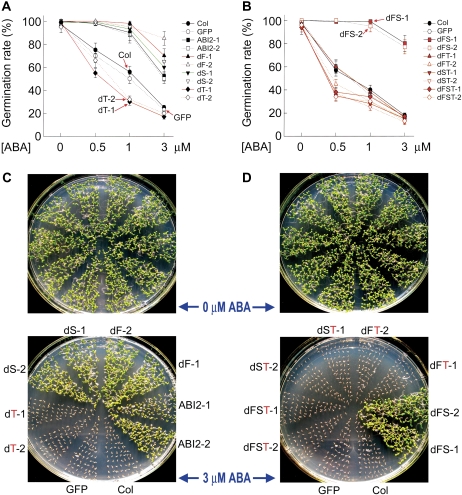
Genetic approaches were used to investigate whether the various deletion mutations of ABI2 affect ABA signalling. All the mutated ABI2s were shown to be expressed correctly in the transgenic lines, and the mutations did not affect the cytosolic and nuclear subcellular localization (Fig. 5A–C). It was observed that the wild-type ABI2 transgenic lines in wild-type Col plants as the transgenic background displayed ABA-insensitive phenotypes in ABA-induced inhibition of seed germination and post-germination growth arrest, and the transgenic plants expressing the mutated ABI2s harbouring the first or second single CPL deletion or first and second double CPL deletion showed substantially the same ABA-related phenotypes as the wild-type ABI2 transgenic lines (Fig. 6A–D). However, the transgenic lines expressing various mutated forms of ABI2 harbouring the third CPL deletion mutation showed wild-type ABA-related phenotypes in ABA-induced inhibition of seed germination and post-germination growth arrest (Fig. 6A–D), and even weak ABA hypersensitive phenotypes were observed in ABA-induced inhibition of seed germination (Fig. 6A, B).
Fig. 5.
Characterization of the various transgenic lines. (A) Immunoblotting analysis of the GFP-tagged ABI2 and mutant ABI2 in the transgenic lines with Col wild-type plants as the background: GFP was immunodetected to assess the expression levels of the ABI2 and mutant ABI2 proteins. Actin was used as a loading control. The experiments were repeated three times with similar results. Col, wild-type (a control); ABI2, wild-type ABI2. The other abbreviations for construct pairs are the same as described in Figs 1 and 2. The suffix numbers (1, 2) indicate the numbers of the transgenic lines. (B) The subcellular localization of the transgenic ABI2 and mutant ABI2. The roots of the transgenic lines were investigated with a confocal microscope. Top panel, GFP fluorescence; and bottom, merged images of the GFP fluorescence and bright field. The abbreviations are the same as in A. The experiments were repeated three times with similar results. (C) Immunoblotting analysis of the GFP-tagged ABI2 and mutant ABI2 in the transgenic/complementation lines with the abi2-t1 mutant (indicated by abi2) as the background. The same experiments were done and the same abbreviations are used as described in A. The experiments were repeated three times with similar results.
Fig. 6.
Deletion mutation of the third CDPK phosphorylation site-like motif of ABI2 disrupts the ABI2 function. The genetic background of all the transgenic lines was wild-type Col plants. (A and B) Test of ABA-induced inhibition of seed germination in different transgenic lines. The germination rates were recorded 36 h after stratification. Each value is the mean ±SE of three biological determinations. (C and D) Test of ABA-induced post-germination growth arrest in different transgenic lines. The seeds were planted in ABA-free (top panels) or 3 μM ABA-containing medium for 3 d stratification, and growth status was recorded 9 d after stratification. The experiments were repeated three times with similar results. All the abbreviations are the same as used in Fig. 2, and the suffix numbers (1, 2) indicate the numbers of the transgenic lines.
To assess further the function of these CPLs, a T-DNA insertion mutant (SALK_015166) was isolated, which is a knock-down allele and named abi2-t1 (Supplementary Fig. S2A at JXB online). This abi2 mutant allele displayed an ABA-hypersensitive phenotypes in ABA-induced seed germination inhibition and post-germination growth arrest, and enhanced drought tolerance (Supplementary Fig. S2A–C). It is noteworthy that these observations are consistent with those of a previous report (Yoshida et al., 2006), but another group did not observe ABA-hypersensitive phenotypes of this abi2 mutant allele (Rubio et al., 2009). This differerence may be caused by differences in environmental conditions where the plants grow.
Transgenic lines expressing the same constructs as mentioned above were generated using the abi2 knock-down allele abi2-t1 as background, and it was observed that the transgenic lines expressing either the wild-type ABI2 or mutated ABI2s harbouring the first or second single CPL deletion or first and second double CPL deletion mutation displayed ABA-insensitive phenotypes in ABA-induced inhibition of seed germination and post-germination growth arrest, whereas overexpression of various mutated forms of ABI2 harbouring the third CPL deletion mutation could not rescue ABA-hypersensitive phenotypes in seed germination and post-germination growth (Fig. 7A–D). All these genetic findings showed clearly that the deletion of the third CPL from the ABI2 molecule disrupts its function in ABA signalling.
Discussion
Structural analysis has revealed several essential sites for interaction between ABI2 and a member of the START-domain ABA receptors PYL1 (Fig. 1; Miyazono et al., 2009). Consistently, the abi1-1 and abi2-1 mutations correspond to one of the interaction sites close to the Mg-coordination motif (see Fig. 1), both of which disrupt ABI1/2-PYR/PYL/RCAR interactions and reduce the phosphatase activity of the PP2Cs in vitro (Leung et al., 1997; Leube et al., 1998; Gosti et al., 1999; Ma et al., 2009; Park et al., 2009). In the present study, the third CPL in the ABI2 molecule was identified as a novel, important site for interactions between ABI2 and PYL5/PYL9 (Fig. 2). Given the previous structural observation that the third CPL is not the direct interaction site of ABI2 with PYL1, it is suggested that the ABI2 molecule may not interact directly with PYL5/PYL9 through this site. However, this site may be essential for an appropriate conformation for ABI2 to interact with the PYL5/PYL9. It was shown that the third CPL is required for the catalytic activity of ABI2 in vitro and, importantly, this site is also essential for the response of ABI2 to the ABA receptors PYL5/PYL9 in relation to the ABA receptor-induced inhibition of the ABI2 phosphatase activity (Fig. 4), which is consistent with the important role of the third CPL in the ABI2–PYL5/9 interaction (Fig. 2). Further, genetic evidence is provided to demonstrate that the third CPL is required for the function of ABI2 to mediate ABA signalling (Figs 5–7). These data reveal that the third CPL is an important site necessary for both the phosphatase activity of ABI2 and the functional interaction between ABI2 and PYL5/9 ABA receptors. Additionally, disruption of the interaction in vitro between ABI2 and SnRK2.6 by deletion of the third CPL suggests that the third CPL may also be important for relay of ABA signalling from ABI2 to downstream signalling components SnRK2s, which is consistent with the loss-of-function property of ABI2 harbouring this CPL deletion mutation, as evidenced by transgenic genetic approaches (Figs 6, 7). It is particularly noteworthy that this CPL deletion mutation of ABI2 is different from the point mutations of abi1-1 and abi2-1 in ABI1 and ABI2 PP2Cs, which disrupt the interactions between the PP2Cs and the ABA receptors PYR/PYL/RCAR (Ma et al., 2009; Park et al., 2009), but does not affect the interactions between the PP2Cs and SnRK2s, and appear to enhance constitutively the dephosphorylation activity of PP2Cs for SnRKs (Umezawa et al., 2009).
The third CPL is a part of a putative ATP/GTP-binding site motif (Schweighofer et al., 2004). A previous report suggested that the putative ATP/GTP-binding site motif may not be required for ABI1 PP2C activity in vitro (Leube et al., 1998). However, biochemical and genetic evidence is provided that deletion of the third CPL affects both the phosphatase activity and function of ABI2 to regulate ABA signalling (Figs 2–7), suggesting that the putative ATP/GTP-binding site motif may be required for the function of ABI2, though further biochemical and genetic approaches will be necessary to characterize the ATP/GTP binding properties of the putative ATP/GTP-binding site motif and to determine the functional significance of the possible ATP/GTP binding ability. Given that the third CPL is substantially conserved among ABI1, ABI2, HAB1, and HAB2 of clade A PP2Cs (Supplementary Fig. S1 at JXB online), this CPL site may play important role in the clade A PP2C-mediated ABA signalling.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Alignment of four members of the Arabidopsis PP2C family, ABI1, ABI2, HAB1, and HAB2, which are involved in ABA signalling.
Figure S2. Characterization of the abi2-t1 T-DNA insertion mutant.
Table S1. Primers used to create ABI2 mutations/deletions.
Table S2. Primers used for yeast two-hybrid constructs.
Table S3. Primers used for production of PYL5, PYL 9, ABI2, and mutated ABI2 proteins in E. coli.
Acknowledgments
The abi2-t1 mutant seeds were provided by the Arabidopsis Biological Resource Center (ABRC). We thank Dr Peng Li (Tsinghua University, Beijing, China) for advice and help with PP2C measurement. This work was supported by grants from the National Natural Science Foundation of China (Grants 90817104) and the Ministry of Agriculture of China (Grant 2008ZX08009-003).
References
- Adie BAT, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solanoa R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nurnberger T, Gust AA. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiology. 2010;153:1098–1111. doi: 10.1104/pp.110.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB. Physical and functional interaction of the Arabidopsis K channel AKT2 and phosphatase AtPP2CA. The Plant Cell. 2002;14:1133–1146. doi: 10.1105/tpc.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Fan LM, Zhao Z, Assmann SM. Guard cells: a dynamic signaling model. Current Opinion in Plant Biology. 2004;7:537–546. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala S, Rock C. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(suppl.):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signaling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences, USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaits F, Shiozaki K, Russell P. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. Journal of Biological Chemistry. 1997;272:17873–17879. doi: 10.1074/jbc.272.28.17873. [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudata J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell. 1999;11:1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends in Plant Science. 2007;12:343–350. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiology. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis and effects of abh1 on AtPP2CA mRNA. Plant Physiology. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences, USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO Journal. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. The Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube MP, Grill E, Amrhein N. ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Letters. 1998;424:100–104. doi: 10.1016/s0014-5793(98)00149-5. [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACD-INSENSITIVE2 (ABI2) and ABI1 encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. The Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christman A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptor. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Xu Y, Ng LM, et al. Identification and mechanism of ABA receptor antagonism. Nature Structural and Molecular Biology. 2010;17:1102–1109. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. Journal of Biological Chemistry. 2003;278:18945–18952. doi: 10.1074/jbc.M300878200. [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, et al. Structural basis of abscisic acid signaling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Nguyen AN, Shiozaki K. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes and Development. 1999;13:1653–1663. doi: 10.1101/gad.13.13.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signalling in Arabidopsis seed. The Plant Journal. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, Merlot S, N'Guyen V, Boisson-Dernier A, Schroeder JI. A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Letters. 2006;580:4691–4696. doi: 10.1016/j.febslet.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL. Protein phosphatase 2C (PP2C) function in high plants. Plant Molecular Biology. 1998;38:919–927. doi: 10.1023/a:1006054607850. [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiology. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. The Plant Journal. 2004;37:354–369. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiology. 2006;141:1389–1399. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009a;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. The Plant Journal. 2009b;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Current Opinion in Plant Biology. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. The Mg-chelatase H subunit antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell. 2010;22:1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proceedings of the National Academy of Sciences, USA. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad AF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. The Plant Journal. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- Takekawa M, Maeda T, Saito H. Protein phosphatase 2C alpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO Journal. 1998;17:4744–4752. doi: 10.1093/emboj/17.16.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Phospholipase D in hormonal and stress signaling. Current Opinion in Plant Biology. 2002;5:408–414. doi: 10.1016/s1369-5266(02)00283-2. [DOI] [PubMed] [Google Scholar]
- Warmka J, Hanneman J, Lee J, Amin D, Ota I. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Molecular and Cellular Biology. 2001;21:51–60. doi: 10.1128/MCB.21.1.51-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, et al. The Mg-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiology. 2009;150:1940–1954. doi: 10.1104/pp.109.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nature Structural and Molecular Biology. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiology. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.