Abstract
Somatic cells are equipped with different silencing mechanisms that protect the genome against retrotransposons. In Drosophila melanogaster, a silencing pathway implicating the argonaute protein PIWI represses retrotransposons in cells surrounding the oocyte, whereas a PIWI-independent pathway is involved in other somatic tissues. Here, we show that these two silencing mechanisms result in distinct chromatin structures. Using sensor transgenes, we found that, in somatic tissues outside of the ovaries, these transgenes adopt a heterochromatic configuration implicating hypermethylation of H3K9 and K27. We identified the Polycomb repressive complexes (PRC1 and 2), but not heterochromatin protein 1 to be necessary factors for silencing. Once established, the compact structure is stably maintained through cell divisions. By contrast, in cells where the silencing is PIWI-dependent, the transgenes display an open and labile chromatin structure. Our data suggest that a post-transcriptional gene silencing (PTGS) mechanism is responsible for the repression in the ovarian somatic cells, whereas a mechanism that couples PTGS to transcriptional gene silencing operates to silence retrotransposons in the other somatic tissues.
Keywords: retrotransposons, Drosophila, heterochromatin, Polycomb
1. Introduction
Due to their ability to transpose to virtually any genomic site, transposable elements (TEs) have the ability to generate deleterious mutations in the host genome. In response, host cells have devised strategies to control TE activity. RNA interference (RNAi), which has a prominent role in this control, is triggered by small RNAs. Several families of small RNAs have now been reported, but it appears that D. melanogaster mainly has two RNA silencing mechanisms that repress transposons expression: endo-siRNAs (endogenous small interfering RNAs) and piRNAs. piRNAs (for PIWI-interacting RNAs) are mainly derived from TE antisense strands and are produced from discrete genomic loci.1 piRNAs are from 26 to 30 nucleotides (nts) in length and have been reported in the reproductive apparatus of Drosophila, mice, rats, and humans.2,3 Endo-siRNAs are ∼21 nts long and associate with the argonaute protein Ago2. Like piRNAs, endo-siRNAs are mainly derived from retrotransposons and other genomic repetitive elements. However, contrasting with piRNAs, endo-siRNAs are likely to be expressed ubiquitously.4–7 It is currently proposed that piRNAs and endo-siRNAs have a germline-specific function and a soma-specific function, respectively, in the establishment of TE silencing.4
Besides these post-transcriptional regulations implicating RNAi silencing pathways, TEs can also be transcriptionally silenced. A range of chromatin modifications suppress their transcription, including histone modifications and altered chromatin packing.8 Furthermore, in both plants and mammals, DNA methylation on cytosine residues is another important signal that represses TE transcription and provides a mechanism for inheritance of TE silencing.
We recently reported that PIWI is necessary for silencing of ZAM and Idefix, two long terminal repeat retrotransposons from Drosophila melanogaster, in the ovarian follicle cells, but is dispensable in other somatic lineages throughout fly development.9 However, it remains unclear whether a specific chromatin structure is deposited on these elements and in which tissues. Furthermore, since DNA methylation is absent from D. melanogaster, nothing is currently known about the mechanisms involved to maintain their silencing through cell divisions. Here, we investigated further the chromatin structures necessary to silence Idefix.
Like most transposable families, many copies of Idefix are detected within the Drosophila genomes. Active copies are generally dispersed on chromosomal arms but many copies corresponding to Idefix vestiges also exist and accumulate in the heterochromatic regions. It was then of prime importance to follow the silencing exerted on a copy known to a target of the RNAi pathways. To this aim, we made use of sensor transgenes carrying a GFP (green fluorescent protein) reporter gene followed by an Idefix fragment that recapitulates the control exerted on active copies of Idefix.9 Analysis of the chromatin structure deposited on the transgene could then be easily explored over the GFP sequence that is unique in the genome.
We show that in somatic cells in contact with the germline, the silencing needs to constantly target the Idefix sequence to be maintained. By contrast, in the other somatic tissues referred to as the soma, the silencing is stably established even when the TE fragment is excised. Our data further indicate that a compact chromatin structure likely associated with Polycomb (PC)-dependent chromatin structures is deposited.
2. Materials and methods
2.1. Drosophila strains and transgenic lines
The following mutant fly stocks were used: ago2414,10 ago251B,11 dcr2[L811fsX],12 PIWI2 and PIWI3,13 r2d21,14 and loqs[f00791].15 Bl279 (+; +/+; MKRS hs-Flip/Tm6) comes from Bloomington stock. pc1 comes from Cavalli's lab.
About 419 bp homologous to the Idefix gag coding region (1003–1422) were inserted either in a sense (tGgIds) or anti-sense orientation (tGgIdas) according to gfp transcription within the pUASt-gfp vector. Three and six independent transgenic lines were analyzed for tGgIds and tGgIdas, respectively (Table 1).
Table 1.
Characteristics of tGgIds and tGgIdas lines in somatic tissues and their homologous piRNA and endo-siRNAs (esiRNAs) reported in the databases4,31
| Number of lines | Silencing in follicle cells | Silencing in larvae | Number of sense piRNAs | Number of antisense piRNAs | Number of sense esiRNAs | Number of antisense esiRNAs | |
|---|---|---|---|---|---|---|---|
| tGgId sense | 3 | On | Off | 0 | 194 | 0 | 0 |
| tGgId antisense | 6 | On | On | 34 | 0 | 3 | 0 |
2.2. Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) was performed using the standard procedures, which was modified as follows: for each immunoprecipitation, three OD260 of chromatin in ChIP buffer was pre-incubated in the presence of 80 μl of Protein A-Agarose (PAA) beads (Millipore) for 30 min at 4°C. PAA were removed, antibodies were added (a control in the absence of antibody named Input was included), and samples were incubated overnight at 4°C in a rotating wheel. Then, 60 μl of PAA were added and incubation was continued for 1h at 4°C. Samples were washed in saline concentration buffer. Chromatin was eluted from PAA in 500μl of elution buffer (sodium dodecyl sulfate and NaHCO3) at room temperature for 15 min. The eluate was incubated overnight at 65°C to reverse cross-links and treated by proteinase K for 1h at 50°C. Samples were phenol–chloroform extracted and ethanol precipitated. DNA was resuspended in 40 μl of 10 mM Tris (pH 7.5).
qPCR (quantitative polymerase chain reaction) (ABI 7300) from at least two biological-independent extractions and three technical replicate was performed using SYBR Green (Sigma) and Platinum Taq DNA Polymerase (Invitrogen). Enrichment was calculated relative to RpL32 and values were normalized to input measurements; the error is indicated by the standard error of mean (SEM). Antibodies used include: H3K9-di-methylation #07-441, H3K9-tri-methylation #07-523, H3K9/K14-acetylation #06-599, H3K27-di-methylation #06-421, H3K27-tri-methylation #06-449, H4K20-mono-methylation #07-440, H2A-ubi #05-678 from Millipore, heterochromatin protein 1a (HP1a) (C1A9) from DSHB, and PC and PH antibodies were provided by G. Cavalli Lab. The following primers were used uptssF: CCAAGCTTTGCGTACTCG and uptssR: CCGTGGGGTTTGAATTAAC for the upstream gfp TSS; gfpF: ACCATTACCTGTCCACACAA and gfpR: CCAGCAGCTGTTACAAACTC for the gfp gene; and RpL32F: CCGCTTCAAGGGACAGTATC and RpL32R: GACAATCTCCTTGCGCTTCT for the control gene RpL32.
2.3. Fluorescent staining and microscopy
Ovaries were dissected and fixed in 5% formaldehyde in phosphate buffer solution for 15 min each hour after the heat shock. GFP was viewed in whole-mount ovaries using the 488 nm filter set of the ZEISS LSM 510 confocal microscope.
2.4. In situ hybridization
FISH (fluorescence in situ hybridization) was performed on larval imaginal discs, as described previously.16 Detailed coordinates of the PCR fragments used to produce the probes can be provided upon request. Three-dimensional images were acquired on a Leica SP5 confocal microscope using a 63X objective. The colocalization was searched for automatically by using a script written on Imaris XT software (Supplementary Fig. S2). Three hundred and five and 179 nuclei were examined from seven tGgIdas and seven tGgDId larval discs, respectively.
3. Results
3.1. Idefix sequence is continuously required for its silencing in the follicle cells but not in other somatic tissues
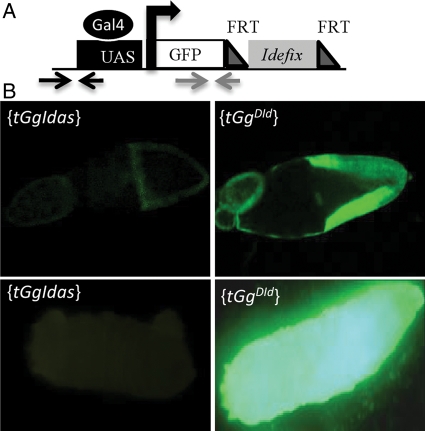
To characterize further the silencing targeting Idefix in the somatic tissues, we used a sensor transgene called tGgIdas. In this transgene, the gfp reporter gene driven by the UASt promoter was linked to a fragment of the gag gene of Idefix (Fig. 1A).9 When tGgIdas is driven by a ubiquitous driver, Actine-Gal4, a GFP signal is detected neither in the follicle cells nor in other somatic tissues throughout development: larvae, pupae, and adults (Fig. 1B). When Idefix is excised and a new line established with the sensor transgene lacking Idefix sequence, then GFP is fully recovered (see tGgDId line, Fig. 1B). Since tGgIdas is repressed in all the somatic tissues within and outside of the ovaries because of the Idefix sequence, we used it to test whether its silencing might switch to an active state as soon as the targeted fragment of Idefix has been excised.
Figure 1.
Structure and silencing of tGgIdas. (A) Structure of tGgIdas: the gfp reporter gene is depicted by a white rectangle and its TSS by an arrow. The Idefix portion (grey) are inserted between two FRT sites (dark grey triangles). Position of the qPCR primer sets taken in the gfp sequence and upstream of the TSS are indicated by arrows. (B) An example of GFP silencing targeting tGgIdas in the follicle cells (above) and in larvae (below) is presented on the left panels; expression of the reference line carrying a sensor transgene, tGgDId, in which the Idefix fragment has been excised, is presented on the right.
We made use of the Flp/FRT (Flip/FLP recognition target) system and its selective induction by the heat shock Flp (HS-Flp) driver. Heat shocks were performed for 1 h on third instar larvae. If tGgIdas silencing is stably established in somatic tissues, GFP repression should persist through cell divisions even after the Idefix fragment has been flipped out. Alternatively, if the silencing is labile and depends on a regulatory pathway that needs to constantly target Idefix sequence to establish the repression, then the silencing should be lost and the GFP expression recovered during the life of the fly as soon as the Idefix sequence is flipped out (Fig. 2A).
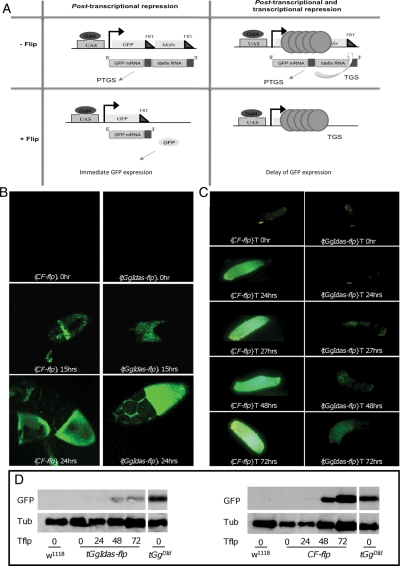
Figure 2.
The silencing exerted on tGgIdas is lost in the follicle cells as soon as the targeted retrotransposon sequences are excised, but remains in the soma. (A) Tested hypothesis: (left) if tGgIdas silencing is labile and depends on a regulatory pathway that needs to constantly target Idefix sequence to establish the repression, then the silencing should be lost and the GFP expression recovered even during the life of the fly as soon as the Idefix sequences is flipped out; (right) if the silencing is stably established in tissues due to a compact heterochromatic structure able to maintain a cellular memory, GFP repression should persist through cell divisions even after the Idefix fragment has been flipped out. (B) (left column) When the CF line [HS-Flp, Actin>FRT>yellow>FRT>Gal4, UASt-gfp] is submitted to a heat shock treatment, GFP expression is detected in the follicle cells as soon as 15 h after treatment (second panel) and a full expression at 24h (third panel). (right column) When the tGgIdas line is submitted to the heat shock, GFP is detected as soon as 15h after treatment and a full expression at 24h compared with tGgDid (first panel). (C) (left column) When CF larvae are submitted to a heat shock treatment, GFP expression is detected 24h after treatment; GFP is fully recovered after 48h (fifth panel). (right column) After heat shock, GFP expression remains faint in tGgIdas larval tissues even 72h after heat shock. (D) Western blot analysis for GFP expression in larvae submitted to heat shock kinetics. Western blotting was performed with an antibody that specifically detects GFP. The w1118 line with no gfp transgene is used as a negative control and tGgDId is used as a positive control for GFP expression. The transgenic lines tGgIdas (left panel) and the test line CF (right panel) are analyzed at time intervals presented below the figure: 0, 24, 48 and 72h. Tub is a loading control.
In a first set of experiments, we determined the time necessary for the FLP recombinase to excise a DNA fragment flanked by FRT sequences after induction by heat shock. We used a triple transgenic line designated as CF (for control Flp). A first transgene is the Flp recombinase under the control of the heat shock promoter (HS-Flp). A second transgene (Actin>FRT-yellow-FRT>Gal4) carries the ubiquitous actin promoter upstream of a yellow gene flanked by FRT sequences and a Gal4 reporter gene placed downstream. When yellow is present, Gal4 is not expressed. The third transgene of the CF line carries the UASt promoter placed upstream of the gfp reporter gene. If a heat shock is performed in this triple transgenic line, the yellow gene flanked by FRT sites is excised. The Gal4 protein is then expressed, and in turn, activates the UAS-gfp transgene giving rise to fluorescence. A kinetic of GFP appearance in both the follicle and larval cells at time intervals of 1h after heat shock was performed, and the time necessary to recover GFP expression was determined in both tissues. We estimated that 15 and 24h was the beginning of expression of the UAS-gfp reporter gene in the follicle cells and the larval tissues, respectively, and a complete expression was recovered after 24h in the follicle cells and 48h in larvae (Fig. 2B and C, left columns).
Then, we determined how long the silencing of the tGgIdas transgene persists after Idefix has been flipped out. A heat shock was performed on flies or larvae with the genetic backgrounds [Actin-Gal4/+; tGgIdas/HS-Flp] or [Actin-Gal4/+; tGgIdas/TM6].
Ovaries were dissected every hour from the heat-shocked flies and GFP expression examined in the follicle cells. We found that fluorescence was detected in some patches 15h after the heat shock (Fig. 2B, right column, middle panel) and a strong green signal in the entire follicular epithelium was recovered after 24h (Fig. 2B, right column, lower panel). These data indicate that the repression exerted in the follicle cells is reversible as soon as the targeted retrotransposon sequence is excised.
When examined in larvae, GFP expression remained undetectable 24 s after heat shock induction in flies [Actin-Gal4/+; tGgIdas/HS-Flp]. Fluorescence was hardly detected 48h after treatment and remained very faint even after 72h, indicating that a full expression was not recovered (Fig. 2C, right column).
As controls, we verified first that the Idefix fragment has been indeed excised after the heat shock treatment (PCR amplifications not shown), and second, that GFP expression is not due to the heat shock treatment, but indeed to Idefix excision since no GFP was detected in cells with the [Actin-Gal4/+; tGgIdas/TM6] genetic background after heat shock.
Then, proteins were extracted from the treated larvae at different time intervals from 0 to 72h and western blot experiments were performed with an antibody raised against GFP. The reference line established with the tGgDId transgene with no Idefix sequence and the w1118 line with no gfp transgene were used as positive and negative controls of GFP expression, respectively. In accordance with results described above, western blots revealed that in [Actin/+; tGgIdas/hs-flp] larval tissues (denoted tGgIdas-flp on Fig. 2D), the level of GFP was hardly detected by 48 or 72 as opposed to the level of GFP expressed from the CF line 48 or 72h after heat shock (Fig. 2D, left and right panels, respectively).
Overall, experiments performed on larval tissues indicate that in these somatic tissues, the silencing persists even after the Idefix sequence has been excised what contrasts with the silencing exerted in follicle cells.
3.2. Histone methylation marks cover silenced transgenes in somatic tissues outside ovaries
We asked whether specific chromatin structures could mark tGgIdas within and/or outside of the ovaries. In a first set of experiments, post-translational histone modifications including lysine acetylation and methylation were analyzed. Whereas acetylation is generally linked with gene activity, methylation of histone H3 on Lys9 (H3K9) and Lys27 (H3K27) correlates with gene silencing. We performed ChIP assays using antibodies against H3K9/K14 acetylation, H3K9me3, H3K27me2, H3K27me3, and H4K20me1, followed by a real-time PCR performed with a primer set targeting gfp from both tGgIdas and tGgDId transgenes (Fig. 1A).
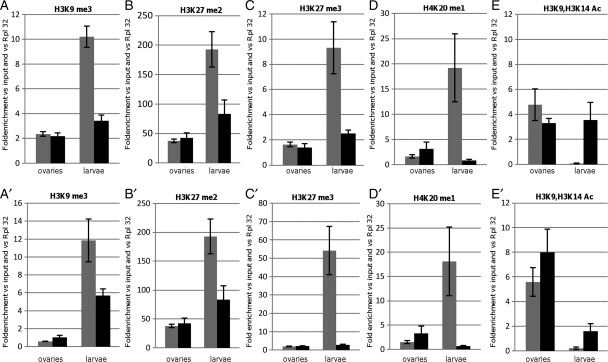
In the ovaries, the H3K9me3, H3K27me2, or H3K27me3 marks were found absent from tGgIdas and tGgDId transgenes (Fig. 3A–C). As expected, when the repressive marks are absent, both of these transgenes displayed an enrichment of H3K9 acetylation (Fig. 3E).
Figure 3.
Histone post-translational modifications associated with the silenced sensor transgene in ovaries and larvae: the histograms show relative amounts of H3K9me3 (A and A′), H3K27me2 (B and B′), H3K27me3 (C and C′), H4K20me1 (D and D′), and H3K9/H3K14Ac (E and E′). Each panel displays the results obtained from ovaries (left) and larvae (right) of transgenic lines tGgIdas (gray bar) and tGgDId (black bar). ChIP was performed on the gfp reporter gene (A–D) and upstream of its TTS (A′–D′) (see primer sets in Materials and methods). Measurements were normalized to the RpL32 gene. Error bars, SEM.
By contrast, a very strong enrichment of the methylated marks was detected on tGgIdas in larval tissues (Fig. 3A–C). These repressive chromatin hallmarks depend on the presence of Idefix because they are enriched >3–10-fold when the Idefix sequence is present compared with tGgDId. They also depend on tGgIdas transcription since they are found absent when it is not activated by the Gal4 driver (data not shown). In contrast, histone marks such as H3K9 acetylation were found completely absent from tGgIdas compared with tGgDId (Fig. 3E).
We further verified that the H4K20me1 mark, generally associated with transcriptional repression and chromosomal condensation, is also enriched on tGgIdas when compared with tGgDId in larval tissues, whereas it is absent in ovaries (Fig. 3D). Finally, we also analyzed the H3K9me2 modification and found no enrichment on tGgIdas compared with tGgDId (Supplementary Fig. S1).
Given the evidence that heterochromatin structures can spread along the chromatin fiber, we wondered whether histone marks associated with the sensor transgenes in the larval tissues could be detected outside of the transcription start site (TSS). We performed ChIP experiments using primers spanning a region from −440 to −330 bp upstream of the TSS (Fig. 1A). We observed similar results as those obtained with primer targeting gfp. On one hand, there is an enrichment of H3K9me3, H3K27me2, H3K27me3, or H4K20me1 on tGgIdas in the larval tissues. On the other hand, these repressive marks were found absent from the sensor transgene in the ovaries, whereas an H3K9 acetylation similar to tGgDId was observed (Fig. 4A′–E′).
Figure 4.
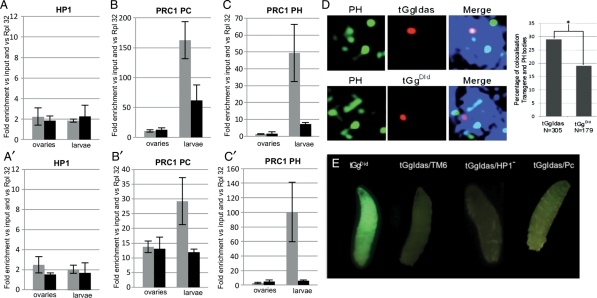
PC and PH, not HP1, are required to silence the sensor transgene in larvae: the histograms show relative amounts of HP1 in A and A′, the PC protein in B and B′, and PH protein in C and C′. Each panel displays the results obtained from ovaries (left) and larvae (right) of transgenic lines tGgIdas (gray bar) and tGgDId (black bar). ChIP was performed on the gfp reporter gene (A–C) and upstream of its TTS (A′–C′). Measurements were normalized to the RpL32 gene. Error bars, SEM. (D) The silenced tGgIdas transgenes colocalize with Pc-G bodies: characteristic individual nuclei analyzed by FISH-I in larval imaginal discs: Hoechst staining, the Pc-G bodies, the transgene insertion site and the merge of the three channels are shown. The scale bar represents 1 µm. On the right, quantification of the percentage of nuclei in which the transgene, either tGgIdas or tGgDId, is present in a Pc-G body. Genotype and the total number of nuclei analyzed are indicated below each bar. Asterisk denotes that the colocalization with PH was statistically different between tGgDId and tGgIdas (P < 0.02). (E) GFP expression from tGgIdas in an heterozygous Su(var)2055 (noted HP1−) and pc1 mutant background (last two larvae on the right) compared with a non-silenced transgene tGgDId when Idefix is excised from tGgIdas (first larvae on the left) and to a strong silenced transgene tGgIdas in a Pc and HP1 wild-type background (second larvae from the left).
We conclude that distinct chromatin structures cover tGgIdas in larval versus ovarian tissues. Post-translational histone modifications generally linked to repression are only observed in non ovarian somatic tissues.
3.3. PC binding, and not HP1, correlates with histone methylation at silenced transgenes
HP1 has been implicated in TE silencing in ovaries.17 Thus, we were interested to test whether HP1 could be detected on the silenced transgene either in ovaries or larvae. ChIP experiments revealed that tGgIdas could not be immunoprecipitated with an anti-HP1 antibody at levels above background (Fig. 4A and A′). ChIP experiments performed on a region targeted by HP1 (the heterochromatic flam/COM locus) was used as a positive control for the ChIP experiment (Supplementary Fig. S1). These results indicate that HP1 does not cover tGgIdas in somatic cells.
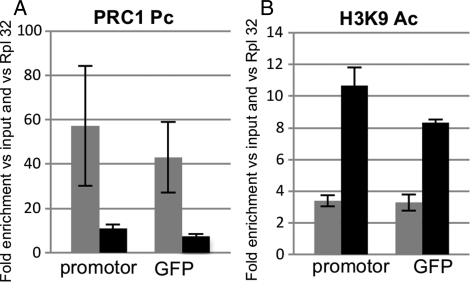
Evidence of H3K27 methylation in larval tissues suggested that Polycomb group (Pc-G) proteins might be implicated in the transgene silencing. Indeed, the methylation of H3K27 is a hallmark of the silencing mediated by PRC2, since it is carried out by the Histone Methyl Transferase Enhancer of Zeste, E(z), the catalytic subunit of PRC2. The chromodomain of the Drosophila PC protein from the PC repressive complex 1 (PRC1) binds to the H3 tail peptide trimethylated at K9 and more strongly at K27me2/3. This prompted us to investigate Pc-G proteins occupancy. ChIP assays were performed with antibodies against PC and Polyhomeotic (PH). Again, two distinct patterns of enrichment were observed. A very strong enrichment of both these proteins was detected on the gfp gene in larval tissues, whereas neither PC nor PH proteins could be detected on the gfp gene in the ovarian follicle cells (Fig. 4B and C).
Given the evidence that heterochromatin structures can spread along the chromatin fiber, ChIP assays were performed using primers taken upstream of the TSS of tGgIdas (Fig. 1). Results indicated that an enrichment of PC and PH was also detected in larval tissues outside of the TSS (Fig. 4B′ and C′). A comparably low amount of PC and PH marks was detected on tGgIdas and tGgDId in the follicle cells (Fig. 4B′ and C′).
To further verify that the binding of Pc-G protein is indeed a hallmark of the chromatin structure associated with the repressed transgene in the somatic tissues outside of the ovaries, we performed three additional tests. First, we investigated whether a colocalization of Pc-G bodies and the targeted transgene might be visualized in larval imaginal discs. A combination of 3-D FISH and immunostaining technique (FISH-I) was used, allowing the detection of the relative localization of the transgene with Pc-G bodies visualized using an anti-PH antibody. The tGgIdas transgene used in this study is integrated on chromosome 3R at the cytological position 25591560, 937 bp upstream of the Kay gene. This site is not a Pc-G binding site reported by Negre et al.18 We found that the colocalizations of tGgDId on one side, and tGgIdas on the other, with Pc-G bodies are statistically different, 19 versus 29%, respectively (P < 0.02; Fig. 4D). Thus, insertion of tGgIdas results in a higher incidence of this locus to colocalize with Pc-G bodies than the same transgene without the Idefix sequence. Second, we performed similar ChIP assays on tissues extracted from adult carcasses and verified that indeed the same enrichment in Pc-G proteins is observed in these somatic tissues (Fig. 5A) compared with a low H3K9 acetylation (Fig. 5B). Third, we verified that H2A ubiquitination of lysine 119, which is performed by the PRC1 E3 ubiquitin ligase Sce/RING, is also a histone mark linked to PC silencing of tGgIdas19,20 (Supplementary Fig. S1).
Figure 5.
H3K9 acetylation and PC association with the silenced sensor transgene in adults: the histograms show relative amounts of PC (A) and H3K9ac (B) from carcasses of the transgenic lines tGgIdas (gray bar) and tGgDId (black bar). Each panel displays the results obtained when ChIPs were performed with primer sets taken at the GFP reporter gene (right) and upstream of the TSS (left) (see primer sets in Materials and methods). Measurements were normalized to the RpL32 gene. Error bars, SEM.
Finally, to definitively implicate Pc-G protein in the silencing, we aim at analyzing the effect of PC mutant on the silencing exerted on tGgIdas. However, described mutants of PC are recessive lethals. Moreover, a clonal analysis was not possible due to the FRT sequences present in the transgene. Nevertheless, since Pc alleles are found regularly but with low expressivity as dominant phenotypes, transgenic flies tGgIdas were crossed to pc1/TM6 mutant backgrounds. GFP expression was examined in the larval progeny [tGgIdas/+; pc1/+] and compared with [tGgDId/+; +/+] larvae as a control for GFP expression when the Idefix sequences has been flipped out and to [tGgIdas/+; +/+] as a positive control for GFP silencing. As exemplified Fig. 4E, a faint but significative expression of GFP was recovered in the Pc heterozygous mutant background (Fig. 4E, right). Finally, to verify that HP1 is not implicated in the silencing of tGgIdas, we examined GFP expression in larvae heterozygous for an HP1 mutant allele, [tGgIdas/+; Su(var)2055/+]. No GFP expression was recovered in these larvae (Fig. 4E).
Overall, the data indicate that a repressive structure implicating Pc-G protein is associated with the sensor transgene tGgIdas in somatic tissues outside of the ovaries.
3.4. Which signal is required in the soma to target Pc-G proteins to the sensor transgene?
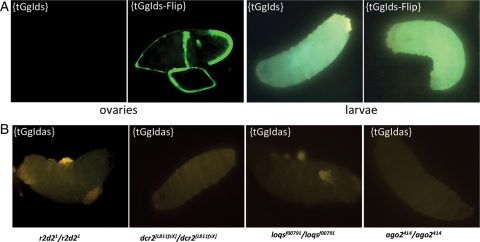
We wondered whether the silencing observed in the somatic cells has the capacity to discriminate one strand of the sensor transgene from the other. To this end, we established new transgenic lines carrying a transgene, tGgIds, similar to tGgIdas except that the same Idefix fragment has been inserted in a sense orientation. Three independent transgenic lines were established. When the expression of tGgIds and tGgIdas was examined in the follicle cells where the Piwi pathway has been clearly implicated, we found that both of them were repressed in this tissue (Fig. 6A). However, we found that, unlike tGgIdas, tGgIds is not targeted to silencing in the soma where a full GFP expression is observed (Fig. 6A) and it is not enriched in repressive chromatin hallmarks such as H3K27me3 as illustrated in Supplementary Fig. S1.
Figure 6.
Characteristics of the silencing targeting tGgIds (A) and tGgIdas (B) transgenes. (A) The silencing targeting the sensor transgene tGgIds is active in the ovaries and inactive in the larvae (first and third panels from the left, respectively). Expression of tGgIds is driven by the ubiquitous actin-Gal4 driver. Expression of the transgene after flp-recombinase action (tGgIds-Flip) is presented as a positive expression of the GFP reporter gene in the second and last panels. (B) Mutations of genes involved in the endo-siRNA pathway do not release the silencing exerted on the tGgIdas sensor transgene. GFP expression is never recovered in larvae from homozygous mutants for r2d2, dcr2, loq, and ago2.
This result indicated that the silencing mechanism involved in the soma is likely to be an RNA silencing pathway able to discriminate between the two mRNA strands produced by the transgenes. Since the silencing of TEs is presumed to be directed by the endo-siRNA pathway in the soma, we investigated whether proteins involved in the endo-siRNA pathway were necessary actors for the silencing exerted on tGgIdas. Therefore, we compared GFP expression in homozygous mutant and wild-type flies for dcr2, ago2, loq, and R2D2. We found no release of GFP silencing in either of these mutant backgrounds (Fig. 6B).
4. Discussion
In this study, we tested whether the silencing targeting a TE sequence in somatic lineages constantly needs the presence of the target or alternatively, once established, the silencing is maintained even though the targeted sequence is lost. We further correlated differences observed in the ovarian follicle cells versus other somatic tissues with differences in the chromatin structures established on the target.
We found that, in vivo, the Idefix sequence is continuously required for its ongoing repression in the follicle cells. Its repression is not heritably maintained through cell divisions. Accordingly, neither the hallmarks of histone post-translational modifications that generally correlate with compact chromatin structure, nor HP1 or Pc-G proteins are linked to the silenced sensor transgenes. This result argues against the current model in which piRNA–PIWI protein complexes silence transposons in ovarian tissues by directing assembly of heterochromatin-like domains.17 Interestingly, using ovarian somatic stem cells transfected with double-strand RNAs corresponding to Piwi, Haase et al.21 reported recently that the integrity of the piRNA pathway is essential for the ongoing repression of mobile elements. Consistent with our results, they further provided evidence that, once set by the action of Piwi proteins on chromatin, a transposon silencing cannot be autonomously maintained.
Considered together, these studies promote a model in which, in the follicle cells, TEs are submitted to a post-transcriptional gene silencing (PTGS) which is not coupled to an epigenetic silencing state implicating an HP1-dependent heterochromatin structure.
By contrast, the assembly of a compact chromatin structure marked by the methylation of H3K9 and H3K27 targets TEs in the somatic cells outside of the ovaries. Furthermore, our study identifies Pc-G proteins as major actors. The two PRCs, PRC1 and PRC2, are involved in this structure seeing that H3K27me3 mediated by the histone methyl transferase E(z), belonging to PRC2, is detected as well as PC and PH proteins which belong to PRC1. We further demonstrate that this compact chromatin structure has two main consequences on the targeted locus. First, it can spread along the chromatin fiber and invade the genome upstream of the TSS of the sensor transgene. Second, once established, repression of the sensor transgene is then stably maintained throughout cell divisions and does not need the TE sequence to constantly direct the silencing.
Exploiting the mechanism of PC-mediated silencing to repress endogenous retroelements has also been described by several studies performed on mice.22,23 Furthermore, in a recent study, the E(z) homolog, EZH, in the green algae Chlamydomonas reinhardtii, has been implicated in the repression of both (hetero)euchromatic transgenes and dispersed retrotransposons.24 Therefore, consistent with the present study, all these data point PRC1 and PRC2 as evolutionarily conserved epigenetic factors employed to silence TE and convey the silencing from one generation of cells to the next.
Interestingly, our study further points out that HP1 is not a partner of the chromatin structure associated with tGgIdas either in the ovaries or in the soma. Although HP1 has been frequently reported as associated with TE,17,25 our data are in line with recent data published by the Drosophila modENCODE project.26 This project has generated data sets that profile histone modifications, chromosomal proteins, transcription factors, and nucleosome properties across a developmental time course and in multiple cell lines. Their data show that only TE located in heterochromatin are marked by HP1. By contrast, TE inserted in euchromatic regions are not covered by HP1 as found for tGgIdas. Thus, one can assume that HP1 might be a signature of TE vestiges because of their heterochromatic localization, whereas HP1-independent silencing pathways might act within euchromatic regions to specifically silence full-length TE and prevent their mobilization.
Overall, if our data implicate PRC1 and PRC2 as epigenetic factors employed to silence TE, they fail to explain how the TE sequence is recognized. In our effort to identify the effector mechanism directing Pc-G proteins on the targeted sensor transgene, we found that the silencing machinery active in the soma has the capacity to discriminate between both orientations of the Idefix sequence present within the sensor transgene. Indeed, tGgIdas is targeted to silencing, whereas tGgIds is not. This result is consistent with a recognition occurring at the RNA, and not the DNA, level.
However, PRC1 and PRC2 employed in this silencing do not have specificity for only one strand and not the other. Therefore, one has to assume that their binding occurs under the presence of a primary signal which brings the sequence specificity. In our study, we were unable to identify this signal. Kanhere et al.27 reported that short RNA from 50 to 200 nts transcribed from the 5′ ends of PC target genes play a role in the association of PRC2. Such short RNAs complementary to Idefix sequences could potentially explain the targeting. Another likely possibility is that the recognition might occur via homologous siRNAs produced by the cell. Epigenetic complexes made of small RNAs together with PRC1 or PRC2 might potentially explain their recruitment to homologous genomic sequences.28–30
Given the evidence that TEs are mainly silenced in the soma via the endo-siRNA pathway, genetic mutants affecting these pathways should have resulted in a loss of the sensor transgene repression. However, no variation in the GFP expression could be detected in these mutant backgrounds. A plausible explanation is that, once established on the transgene, the silent epigenetic state cannot be released even though the targeted fragment is excised or the silencing pathway necessary for the target recognition mutated. Nevertheless, our study pointed out that, like in the follicle cells where piRNAs are involved (Table 1),9,31 the primary effectors acting in the soma are likely to be single-stranded molecules since only one strand of the mRNA produced by the sensor transgene is targeted. This is comforted by the list of endo-siRNAs reported in the databases (Table 1). Endo-siRNAs able to target and silence tGgIdas are reported in the databases, whereas none is complementary to the Idefix fragment inserted in tGgIds.
Overall, in the soma, we propose that the silencing of TE is initiated by a PTGS mechanism which is successively coupled to transcriptional gene silencing. This latter displays all the molecular imprint underlying cell memory and epigenetic inheritance. This epigenetic state, absent in the ovarian follicle cells, accounts for the molecular mechanism controlling maintenance of TE silencing during development.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC 1139). J.D. received a graduate grant from the Ministère de l'Enseignement Supérieur et de la Recherche (MESR). E.B. and P.P. received a grant from the Région Auvergne.
Supplementary Material
Acknowledgements
We are grateful to F. Bantignies for helpful discussions and PC and PH antibodies. We thank S. Ronsseray for mutant flies. We thank P. George and C. Fritsch for critical review of the manuscript. We thank F. Pellissier for technical assistance and Flyfacility for transgenic lines (www.Fly-facility.com).
References
- 1.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. doi:10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 3.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 4.Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–81. doi: 10.1126/science.1157396. doi:10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung W.J., Okamura K., Martin R., Lai E.C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. doi:10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. doi:10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., et al. Drosophila endogenous small RNAs bind to argonaute 2 in somatic cells. Nature. 2008;453:793–97. doi: 10.1038/nature06938. doi:10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 8.Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–85. doi: 10.1038/nrg2072. doi:10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 9.Desset S., Buchon N., Meignin C., Coiffet M., Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. doi:10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–22. doi: 10.1101/gad.1454806. doi:10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu K., Bogert B.A., Li W., Su K., Lee A., Gao F.B. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr. Biol. 2004;14:1025–34. doi: 10.1016/j.cub.2004.05.055. doi:10.1016/j.cub.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., et al. Distinct roles for Drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. doi:10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 13.Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–76. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway , Science. 2003;301:1921–25. doi: 10.1126/science.1088710. doi:10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 15.Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., et al. Normal microRNA maturation and germ-line stem cell maintenance requires loquacious, a double-stranded RNA-binding domain protein , PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. doi:10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brasset E., Bantignies F., Court F., Cheresiz S., Conte C., Vaury C. Idefix insulator activity can be modulated by nearby regulatory elements. Nucleic Acids Res. 2007;35:2661–70. doi: 10.1093/nar/gkm140. doi:10.1093/nar/gkm140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klenov M.S., Lavrov S.A., Stolyarenko A.D., Ryazansky S.S., Aravin A.A., Tuschl T., et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–38. doi: 10.1093/nar/gkm576. doi:10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Negre N., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. doi:10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritsch C., Beuchle D., Muller J. Molecular and genetic analysis of the Polycomb group gene sex combs extra/ring in Drosophila. Mech. Dev. 2003;120:949–54. doi: 10.1016/s0925-4773(03)00083-2. doi:10.1016/S0925-4773(03)00083-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–78. doi: 10.1038/nature02985. doi:10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 21.Haase A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. doi:10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golding M.C., Zhang L., Mann M.R. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell. 2010;6:457–67. doi: 10.1016/j.stem.2010.03.014. doi:10.1016/j.stem.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K., Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–76. doi: 10.1101/gad.544410. doi:10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaver S., Casas-Mollano J.A., Cerny R.L., Cerutti H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics. 2010;5:301–12. doi: 10.4161/epi.5.4.11608. doi:10.4161/epi.5.4.11608. [DOI] [PubMed] [Google Scholar]
- 25.Haynes K.A., Caudy A.A., Collins L., Elgin S.C. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 2006;16:2222–27. doi: 10.1016/j.cub.2006.09.035. doi:10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riddle N.C., Minoda A., Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Tolstorukov M.Y., et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21:147–63. doi: 10.1101/gr.110098.110. doi:10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanhere A., Viiri K., Araujo C.C., Rasaiyaah J., Bouwman R.D., Whyte W.A., et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. doi:10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimaud C., Negre N., Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–75. doi: 10.1007/s10577-006-1069-y. doi:10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Christoforou A., Aravind L., Emmons S.W., van den Heuvel S., Haber D.A. The C. elegans Polycomb gene SOP-2 encodes an RNA binding protein. Mol. Cell. 2004;14:841–47. doi: 10.1016/j.molcel.2004.06.001. doi:10.1016/j.molcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–56. doi: 10.1126/science.1163045. doi:10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. doi:10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.