Abstract
Normalization of quantitative gene expression data with a suitable reference gene is essential for accurate and reliable results. However, the availability and choice of most suitable reference gene(s) showing uniform expression across all the experimental conditions remain a drawback. We have developed a web server, PlantRGS (http://www.nipgr.res.in/PlantRGS), for the identification of most suitable candidate reference gene(s) at the whole-genome level using microarray data for quantitative gene expression studies in plants. Microarray data from more than 11 000 tissue samples for nine plant species have been included in the PlantRGS for meta-analysis. The web server provides a user-friendly graphical user interface-based analysis tool for the identification of most suitable reference genes in the selected plant species under user-defined experimental conditions. Various parameter options and output formats will help users to investigate desired number of most suitable reference genes with wide range of expression levels. Validation of results revealed that novel reference genes identified by the PlantRGS outperforms the traditionally used reference genes in terms of expression stability. We anticipate that the PlantRGS will provide a platform for the identification of most suitable reference gene(s) under given experimental conditions and facilitate quantitative gene expression studies in plants.
Keywords: gene expression, reference gene, microarray, normalization, web server
1. Introduction
Gene expression studies are very important to gain insights into the function of gene(s). Two commonly used techniques for measurement of gene expression levels in complementation to each other are DNA microarray and reverse-transcription–quantitative PCR (RT–qPCR) analysis.1 The RT–qPCR has very high sensitivity and is often used to validate the results of DNA microarray studies.2–4 Normalization is considered as the key factor, which affects the reliability of RT–qPCR results.5 The normalization is done with a suitable reference gene, which is ubiquitously expressed at stable level in different biological contexts. Until recently, a variety of housekeeping genes, such as actin, tubulin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), elongation factor 1-α (EF1-α), polyubiquitin and ribosomal RNA, were being used as reference gene(s), which were supposed to express stably in different biological contexts because of their known role in basic cellular processes. However, it has been reported in many studies that the expression of these genes are also regulated and therefore, their transcript level varies in different cell types and/or experimental conditions, making them unsuitable for use as reference genes in normalization of gene expression results.6–11 The expression stability of reference genes showing highly stable expression under one set of experimental conditions may vary under other set of experimental conditions, and thus, their choice remains a drawback. Furthermore, the set of most suitable reference genes may differ in various species even under the identical experimental conditions. Therefore, for a particular study, a species-specific set of most suitable reference genes needs to be identified for accurate normalization of gene expression data.
Several studies have been performed to identify the most suitable housekeeping gene(s) under specific environmental conditions in various organisms, including plants, to use them as reference genes for normalization.12–15 However, only a few studies have been performed to identify the novel reference genes.9,16–18 Using microarray data, expression of all the genes can be examined simultaneously to identify the novel genes showing uniform expression under selected experimental conditions or tissue types. A plethora of microarray data have been generated for various plant species in different biological contexts and are available in the public databases, which can be utilized for this purpose. Only a few studies have been carried out using microarray data of Arabidopsis, Glycine max (soybean) and rice to predict the most suitable reference genes under specific experimental conditions followed by their validation using RT–qPCR and/or statistical algorithms.9,16,17,19
In this study, we have developed a web server, PlantRGS, which identifies the most suitable candidate reference genes for quantitative gene expression studies in plants based on the microarray data. PlantRGS provides a user-friendly graphical user interface (GUI) that allows automatic analysis of thousands of arrays simultaneously and generates a list of most suitable candidate reference genes with their gene description, expression statistics [mean signal value, standard deviation and coefficient of variance (CV)] and graph showing relative expression as output. The server will be very valuable for the scientific community to identify the most suitable reference gene(s) to be used for normalization in gene expression studies.
2. Materials and methods
2.1. Microarray data download and pre-processing
Microarray data based on Affymetrix platform for the nine different plant species, including Arabidopsis thaliana (Arabidopsis), G. max (soybean), Hordeum vulgare (barley), Oryza sativa (rice), Populus (poplar), Solanum lycopersicum (tomato), Triticum aestivum (wheat), Vitis vinifera (grape vine) and Zea mays (maize), available at the Gene Expression Omnibus (GEO) database of NCBI comprised of 11 187 arrays, were downloaded. All the downloaded CEL files were subjected to normalization using MAS5 algorithm using Affymetrix Expression Console software (http://www.affymetrix.com) at default parameters. Signal intensities and absolute calls (present, marginal or absent) measured for each probeset in all the arrays using Expression Console were recorded. The probesets having detection P-value < 0.05 were assigned as present call, those with detection P-value ≥0.065 were assigned as absent call and the remaining as marginal call.
2.2. Microarray data processing and output
The microarray data processing method implemented in the PlantRGS for the identification of most suitable reference genes is based on the previous studies.9,17 User-selected microarray experiments/arrays and parameters from the server application are taken as an input in the Perl processing module. The absolute call (present/marginal/absent) information about all probesets for the selected arrays and signal intensity (linear or log transformed) values only for the probesets having present call in at least given percentage (user-defined) of the selected arrays are obtained from the MySQL database. These probesets are further filtered on the basis of signal intensity range defined by the user for selected arrays. Mean signal intensity, standard deviation and CV (mean/standard deviation) for the filtered probesets are calculated and the given number of probesets showing least CV with their statistics is displayed along with the corresponding gene locus id and description (if available). In addition, user-specified probesets (if provided) are also searched in the list of filtered probesets and their statistics is also displayed. The gene description for Arabidopsis, rice and soybean probesets was used from TAIR (http://www.arabidopsis.org/), Rice Oligonucleotide Array Database (http://www.ricearray.org/) and SoyBase (http://soybase.org/), respectively. However, for other plants, gene descriptions were derived from the significant (E-value ≤ 1e-5) best hit of the target sequences of the probesets with Arabidopsis or Uniprot proteins in BLASTX search.
2.3. Statistical assessment of gene expression stability
To assess the gene expression stability of the reference genes predicted by the server and traditional reference genes, we used geNorm v3.5 software.20 The signal values were converted into relative quantities for analysis using geNorm. geNorm calculates the average expression stability (M) value for each gene being analysed based on the mean pairwise variation in the expression of a gene from all other genes under investigation.20
2.4. Web server architecture and implementation
PlantRGS adopts a four-tier architecture (Java/Java/Perl/MySQL) as depicted in Supplementary Fig. S1A. The server has been presently hosted on a Linux workstation with two quad-core 3.33 GHz Intel Xeon processors and 12 GB of RAM. The server application has been implemented in Java and the client GUI was developed using Java 2 Swing technology. The pre-processed microarray data have been stored into MySQL database on the server. Processing of the data has been employed in Perl programming language. The client-side GUI can be launched either by using web browser-dependent applet or web browser-independent Java Web Start technology (using .jnlp file available for download). The server can accept requests from multiple clients simultaneously using a specified port.
3. Results
3.1. Data acquisition and web server
Microarray data from Affymetrix platform comprised of a total of 593 experiments including 11 187 arrays from nine different plant species were used in the current version of PlantRGS. Microarray data generated using Affymetrix platform show high degree of reproducibility and homogeneity across different laboratories when compared with other platforms and thus provide better opportunity to identify the genes with uniform expression across various experimental conditions. Largest data set was available for Arabidopsis (432 experiments and 5689 arrays) followed by soybean (23 experiments and 3109 arrays) and least for tomato (7 experiments and 173 arrays). The microarray data represented a wide variety of experimental conditions, including developmental stages, abiotic and biotic stress conditions, light/hormone/chemical treatments and genotypes/mutants/transgenics. The number of experiments and arrays for all the nine plant species included in the PlantRGS is given in Supplementary Table S1. A complete list of all the experiments along with their accession number and description available at GEO has been provided in the download option of the web server. Other plant species, for which very less microarray data are available at GEO, has not been included in the current version. We intend to include the microarray data for more plants in the subsequent releases of PlantRGS, when sufficient data for them become available in the public databases.
All the microarray experiments were curated either manually or using curated data source PLEXdb21 to assign them to broad experimental factor ontology term(s). The microarray experiments were classified broadly into development/tissue-type, abiotic stress, biotic stress, hormone, other stimuli and genetic line/cultivar, ontology term(s). The experiments which could not be classified into these ontology terms were kept in miscellaneous category. This classification will help the users to select all the microarray experiments for a specific environmental condition under study, without putting extra effort in going through their description.
PlantRGS GUI provides various tools and options to select the plant species and experiments/arrays, process the data and analyse, manage, export and load the results. The signal intensity values of each probeset for all arrays after normalization with MAS5 algorithm have been stored in MySQL database of the web server. The user-friendly GUI provides different tools and parameter options to investigate most suitable reference genes without the requirement of high computational resources and technical expertise. The user interface outputs the most suitable reference genes listed in the ascending order of CV. A lower value of CV indicates higher expression stability of that gene across the selected set of arrays or experimental conditions. The server may be accessed by multiple users simultaneously without compromising the speed and accuracy of prediction. The results were generated in couple of minutes on the server, when tested for the data set of soybean, with default parameters for simultaneous requests from three different clients.
3.2. Identification of most suitable reference genes using PlantRGS
3.2.1. PlantRGS workflow
The workflow for using PlantRGS has been illustrated in Supplementary Fig. S1B. After launching the application, selection of a plant species to be studied loads all the microarray data available for it on the GUI. Microarray data representing different experimental conditions can be selected from the hierarchical tree of experiments/arrays or ontology terms. A set of parameters, including log transformation, per cent present call, signal intensity range and number of reference genes to be identified may be specified. In addition, probeset ids may be provided for comparison of their expression stability with those predicted by the server. After selecting experiments/arrays and suitable parameters, run the analysis and results will be displayed in result area of GUI. The results can be exported in tab-delimited text file (tables) and images (graph).
3.2.2. Key parameters of PlantRGS
The PlantRGS web server highlights several key parameters for identifying most suitable candidate reference genes as described below.
Selection of microarray experiments. The microarray experiments for a plant species can be selected depending on the experimental conditions to be analysed. The broad ontology terms may be used to select the microarray data for desired experimental conditions. More is the number of microarray experiments selected, better will be the reliability of the reference genes identified by the server. In case of non-availability of the microarray data for the identical experimental conditions being analysed, microarray experiments representing broadly similar conditions may be used for the analysis. Although it is not recommended to use the microarray data of a closely related species to identify the reference genes, candidate reference genes may be shortlisted. It is strongly recommended that expression stability of orthologues of such candidate reference genes should be validated experimentally before their use in the gene expression studies.
Log transformation and present call. Log-transformed or linear signal intensity values of the probesets can be used for the analysis. Log transformation of the microarray data has been recommended to make the distribution more symmetric and Gaussian-like. We also suggest the analysis of microarray data in the log-transformed state. In addition, the option of per cent present calls filters the probesets showing present call (detection P-value <0.05) in at least given number (%) of arrays from the total number of selected arrays. For example, the default option of 90% ensures that the expression of each probeset is detected with a present call in at least 90% of the arrays selected for the analysis.
Signal intensity range. A good reference gene should have comparable expression level to that of the gene(s) of interest to be analysed under given experimental conditions. There might be cases where the expression level of gene(s) to be analysed is very low, for example, transcription factor-encoding genes.2 For such studies, the highly stable but lowly expressed reference gene(s) might be the best choice for normalization of quantitative gene expression data.9 By defining the signal intensity range in the parameters, reference genes with desired expression level suitable for the given study can be identified.
User-defined probeset ids. This option helps users to compare the expression stability of the traditional reference genes or genes of their choice with that of predicted by the PlantRGS, in the selected experiments/arrays. Defining user-defined probesets lists their expression statistics along with those predicted by the server for a comparative analysis of their expression stabilities to identify the most suitable one. This option also helps to identify the most suitable reference gene showing the expression level comparable to the gene(s) of interest.
3.2.3. PlantRGS output
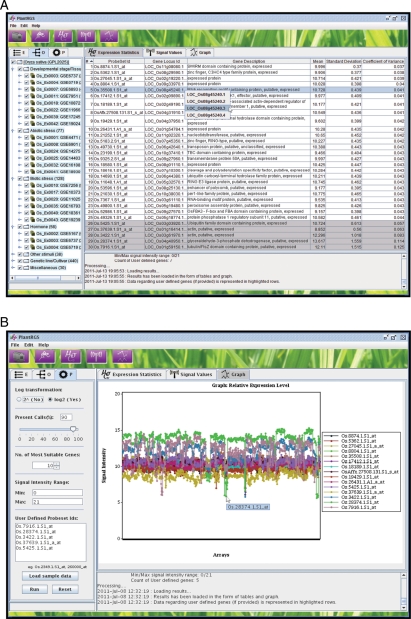
After the identification of most suitable reference genes by the server, GUI displays results in the form of tables (Fig. 1A) and graph (Fig. 1B). Two tables are generated in the result area, one showing the description (probeset id, gene locus id and gene description) and statistics (mean signal intensity, standard deviation and CV) for the given number of most suitable reference gene predicted by server and user-defined probeset ids (if provided). The detailed information about these genes can be investigated on their respective genome project web sites in web browser through the links provided on the corresponding gene IDs. In the second table, the signal values of the predicted reference genes and user-defined probeset ids (if provided) in all the arrays used for the analysis are displayed, which are used to generate graph for the relative expression levels of the selected probesets. Figure 1A and B shows the screenshots of the 25 most suitable reference genes identified by the PlantRGS from the whole data set of rice along with five user-defined probesets (highlighted in grey) and graph showing their relative expression levels in all the arrays, respectively.
Figure 1.
Screenshots of PlantRGS showing results. (A) Screenshot of the results in expression statistics tab showing the list of the most suitable reference genes (probeset ids) predicted by PlantRGS and user-defined probesets (highlighted in grey colour) with their gene locus ID, gene description, mean signal value, standard deviation and CV. (B) Screenshot of the graph tab showing the relative expression levels of all the reference genes predicted by PlantRGS and user-defined probesets in all the arrays (samples/experimental conditions).
3.3. Validation of the PlantRGS results
We identified most suitable reference genes for all the nine plants using whole microarray data sets available for each species. A list of top 100 most suitable candidate reference genes identified by PlantRGS is available as Supplementary Table S2. Their gene description shows that most of these represent novel reference genes involved in various cellular processes. A few case studies were performed to validate the results obtained from PlantRGS. The results were assessed using a widely used statistical algorithm, geNorm,20 which calculates the M value for each gene being analysed. The gene with lowest M value is considered to have most stable expression and rank higher. We identified most suitable reference genes for the whole data sets of rice and Arabidopsis using PlantRGS and compared their expression statistics with that of traditional reference genes followed by validation of their expression stability using geNorm. In addition, we also compared the results obtained from PlantRGS and those reported in the previous studies using smaller data sets.
3.3.1. Validation of most suitable reference genes from whole data set of rice
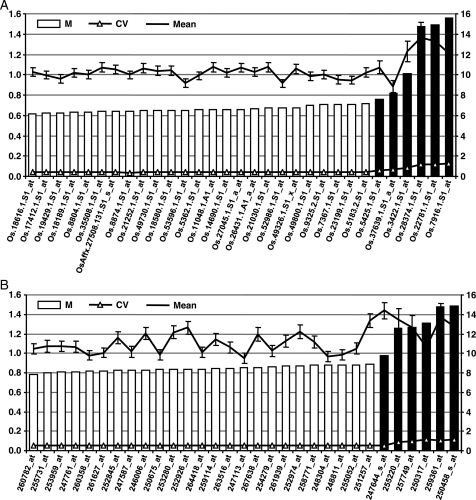
Using the whole microarray data set of rice comprised of 688 arrays, we identified top 25 most suitable candidate reference genes using default parameters and specifying six user-defined probesets for traditionally used reference genes (representing ubiquitin, actin, GAPDH and tubulin) using PlantRGS. All the reference genes predicted by PlantRGS ranked higher in terms of their expression stability (lower CV) than the traditional reference genes (Fig. 2A). Further, the assessment by geNorm also showed lower M value and hence more expression stability of the reference genes identified by the PlantRGS in comparison to the traditional reference genes (Fig. 2A). Although one traditional reference gene encoding for ubiquitin (Os.5425.1.S1_at) was comparable with novel reference genes in terms of M value, it ranked lower due to marginally higher M value and CV. Mean signal intensity was higher for most of the traditional reference genes with higher standard deviation, indicating their higher and variable expression across the experimental conditions. The most suitable reference genes identified from the whole data set of rice may be used for normalization of quantitative gene expression data from a wide variety of experimental conditions.
Figure 2.
Validation of the results obtained by PlantRGS for whole data set of rice (A) and Arabidopsis (B). The expression stability (M, represented by bars) of top 25 novel reference genes predicted by PlantRGS (open bars) and five traditionally used reference genes (black bars) were calculated by geNorm in all the arrays/experiments available for rice and Arabidopsis. A lower value of M (scale on left axis) indicates higher expression stability. The line with triangles indicates CV for each gene (scale on left axis), and the line with points indicates mean signal intensity (scale on right axis) with standard deviation (error bars).
3.3.2. Validation of most suitable reference genes from whole data set of Arabidopsis
A total of 5689 arrays of Arabidopsis representing wide variety of experimental conditions and tissue types were analysed for the identification of most suitable reference genes using default parameters. All the top 25 reference genes predicted by the server were superior in terms of expression stability exhibiting lower CV and M value when compared with the six traditionally used reference genes, including polyubiquitin, actin, tubulin, EF1-α and GAPDH (Fig. 2B). The M value was significantly higher for most of the traditional reference genes indicating their lower expression stability.
3.3.3. Comparison with previously identified reference genes
We have also compared the results of PlantRGS with those reported in previous studies using a subset of microarray data for rice and Arabidopsis. We used the same microarray data set(s) used in previous studies and identified most suitable reference genes using our server and compared the results. The analysis revealed that among the top five reference genes identified by Narsai et al.19 using microarray data for different stress conditions in rice; four were included in the top 25 genes identified by the PlantRGS with a comparable M value and CV (Supplementary Fig. S2A). A comparison with the reference genes identified for different developmental stages in rice17 showed that the top 25 genes predicted by the server outperformed the previously identified genes (Supplementary Fig. S2B). Likewise, we also compared the results for various developmental stages and abiotic stress conditions in Arabidopsis reported by Czechowski et al.9 using the same microarray data sets. In both data sets, we found the reference genes predicted by the PlantRGS performed better or comparable to the previously identified reference genes (Supplementary Fig. S2C and D). It is noteworthy, that in all the above case studies, the reference genes predicted by the server outperformed the traditionally used reference genes in terms of expression stability.
4. Discussion
RT–qPCR is a very powerful tool for the accurate and reliable quantification of gene expression, if implicated without any systematic errors. The most common source of error in the RT–qPCR results is the inappropriate choice of reference gene(s) for normalization, which may lead to erroneous results. Therefore, the identification of most suitable reference genes for a particular set of experimental conditions and their systematic validation is crucial for quantitative gene expression studies. Although efforts to identify the most suitable reference gene(s) for a particular study followed by validation before their use have been increasing, unfortunately, however, its importance is still being disregarded. This problem is mainly due to the poor availability of user-friendly tools and resources, and requirement of substantial time and efforts. The web server, PlantRGS, developed in this study, provides user-friendly, interactive and platform-independent GUI tool for the identification of most suitable candidate reference gene(s), which adopts Java 2 Swing technology for automatic and very fast processing of a large amount of microarray data with minimal technical skills.
A few previous studies have suggested the use of microarray data for the identification of better candidate reference gene(s) for RT–qPCR data normalization.9,17,19 Czechowski et al.9 reported the first use of microarray data for the identification of superior reference genes. Recently, using microarray data sets encompassing organ, development, biotic and abiotic conditions, 151 genes with relatively stable expression under all conditions were identified in rice.19 The microarray data analysis-based strategy has been implemented in PlantRGS also, for the identification of most suitable candidate reference gene(s) under given experimental conditions. PlantRGS utilizes the whole microarray data set available for a plant species or for a specific biological context (e.g. developmental stage/tissue-type, abiotic stress, biotic stress and hormone treatment etc.), for the identification of best candidate reference gene(s) with minimum expression variance. The selection of signal intensity range is a very important feature of the PlantRGS, which facilitates the identification of a reference gene(s) having expression levels similar to the gene(s) of interest. The geNorm-based statistical analysis validated the better performance of the novel candidate reference genes identified by PlantRGS when compared with traditionally used housekeeping genes. However, it is recommended that the stable expression of candidate reference genes identified by PlantRGS should further be validated experimentally before using for normalization. Further, in case the orthologues of reference genes identified for one plant species are being used for normalization in other species, the validation of expression stability is must. Recently, Narsai et al.19 showed that among the 15 orthologues of novel Arabidopsis reference genes,9 only one exhibited stable expression in rice, suggesting that caution must be taken while selecting reference genes from other plant species.
Recently, next-generation sequencing (NGS) technologies are being increasingly used for the quantification of gene expression. Although these technologies provide high throughput and absolute abundance of the transcripts, their high cost and less data availability remain a drawback when compared with the microarray studies as of now for global gene expression profiling. In addition, the choice of statistical methods for normalization and expression analysis using NGS data is a challenge.22 A few studies have been performed using sequence-based approach for the identification of suitable reference genes.23,24 We propose that the concept/method used in PlantRGS may be used for the identification of candidate reference genes from NGS data in future with the availability of more data and better statistical methods for the analysis.
5. Conclusion
In this study, we have developed a user-friendly web server tool, PlantRGS, for the identification of most suitable reference genes under selected experimental conditions using microarray data for various plant species. Various features implemented in PlantRGS allow the identification of best suitable reference gene(s) for a given study. Validation studies showed that the novel reference genes identified by PlantRGS outperform the traditionally used reference genes and are better candidates for gene expression normalization. We anticipate that the availability of this tool will facilitate immensely the reliability and accuracy of the quantitative gene expression studies in plants.
6. Availability
The PlantRGS web server is publicly available at http://www.nipgr.res.in/PlantRGS to all the users without any login/subscription requirement.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported financially by the Department of Biotechnology, Government of India, New Delhi, India, under the Next Generation Challenge Programme on Chickpea Genomics and IYBA scheme, and core grant from the NIPGR.
Supplementary Material
References
- 1.Clarke J.D., Zhu T. Microarray analysis of the transcriptome as a stepping stone towards understanding biological systems: practical considerations and perspectives. Plant J. 2006;45:630–50. doi: 10.1111/j.1365-313X.2006.02668.x. [DOI] [PubMed] [Google Scholar]
- 2.Czechowski T., Bari R.P., Stitt M., Scheible W.R., Udvardi M.K. Real-time RT–PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38:366–79. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- 3.Gachon C., Mingam A., Charrier B. Real-time PCR: what relevance to plant studies? J. Exp. Bot. 2004;55:1445–54. doi: 10.1093/jxb/erh181. [DOI] [PubMed] [Google Scholar]
- 4.Garg R., Jhanwar S., Tyagi A.K., Jain M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010;17:353–67. doi: 10.1093/dnares/dsq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT–PCR. Nat. Protoc. 2006;1:1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 6.Thellin O., Zorzi W., Lakaye B., et al. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 1999;75:291–5. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 7.Schmittgen T.D., Zakrajsek B.A. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT–PCR. J. Biochem. Biophys. Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee P.D., Sladek R., Greenwood C.M., Hudson T.J. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12:292–7. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubie C., Kempf K., Hans J., et al. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol. Cell. Probes. 2005;19:101–9. doi: 10.1016/j.mcp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez L., Mauriat M., Guenin S., et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription–polymerase chain reaction (RT–PCR) analysis in plants. Plant Biotechnol. J. 2008;6:609–18. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 12.Nicot N., Hausman J.F., Hoffmann L., Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005;56:2907–14. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- 13.Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006;345:646–51. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 14.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. Normalisation of real-time RT–PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–9. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 15.Garg R., Sahoo A., Tyagi A.K., Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.) Biochem. Biophys. Res. Commun. 2010;396:283–8. doi: 10.1016/j.bbrc.2010.04.079. [DOI] [PubMed] [Google Scholar]
- 16.Libault M., Thibivilliers S., Bilgin D.D., et al. Identification of four soybean reference genes for gene expression normalization. Plant Genome. 2008;1:44–54. [Google Scholar]
- 17.Jain M. Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Sci. 2009;176:702–6. [Google Scholar]
- 18.Kwon M.J., Oh E., Lee S., et al. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PLoS One. 2009;4:e6162. doi: 10.1371/journal.pone.0006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narsai R., Ivanova A., Ng S., Whelan J. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010;10:56. doi: 10.1186/1471-2229-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandesompele J., De Preter K., Pattyn F., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wise R.P., Caldo R.A., Hong L., Shen L., Cannon E., Dickerson J.A. BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. Methods Mol. Biol. 2007;406:347–63. doi: 10.1007/978-1-59745-535-0_17. [DOI] [PubMed] [Google Scholar]
- 22.Bullard J.H., Purdom E., Hansen K.D., Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coker J.S., Davies E. Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques. 2003;35:740–8. doi: 10.2144/03354st04. [DOI] [PubMed] [Google Scholar]
- 24.Chari R., Lonergan K.M., Pikor L.A., et al. A sequence-based approach to identify reference genes for gene expression analysis. BMC Med. Genomics. 2010;3:32. doi: 10.1186/1755-8794-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.