Abstract
Bacterial leaf pustule (BLP) disease is caused by Xanthomonas axonopodis pv. glycines (Xag). To investigate the plant basal defence mechanisms induced in response to Xag, differential gene expression in near-isogenic lines (NILs) of BLP-susceptible and BLP-resistant soybean was analysed by RNA-Seq. Of a total of 46 367 genes that were mapped to soybean genome reference sequences, 1978 and 783 genes were found to be up- and down-regulated, respectively, in the BLP-resistant NIL relative to the BLP-susceptible NIL at 0, 6, and 12h after inoculation (hai). Clustering analysis revealed that these genes could be grouped into 10 clusters with different expression patterns. Functional annotation based on gene ontology (GO) categories was carried out. Among the putative soybean defence response genes identified (GO:0006952), 134 exhibited significant differences in expression between the BLP-resistant and -susceptible NILs. In particular, pathogen-associated molecular pattern (PAMP) and damage-associated molecular pattern (DAMP) receptors and the genes induced by these receptors were highly expressed at 0 hai in the BLP-resistant NIL. Additionally, pathogenesis-related (PR)-1 and -14 were highly expressed at 0 hai, and PR-3, -6, and -12 were highly expressed at 12 hai. There were also significant differences in the expression of the core JA-signalling components MYC2 and JASMONATE ZIM-motif. These results indicate that powerful basal defence mechanisms involved in the recognition of PAMPs or DAMPs and a high level of accumulation of defence-related gene products may contribute to BLP resistance in soybean.
Keywords: bacterial leaf pustules, disease resistance, RNA-Seq analysis, soybean
1. Introduction
Bacterial leaf pustule (BLP) disease, caused by Xanthomonas axonopodis pv. glycines (Xag), is one of the most serious diseases in soybean. It reduces grain yield by 15–40% at high temperatures and high humidity, primarily through chlorophyll degradation and premature defoliation.1,2 Early reports3 suggested that BLP resistance in CNS (PI 548445) is controlled by a single recessive gene (rxp) surrounded by two simple sequence repeat (SSR) markers, Satt372 and Satt486, on chromosome 17 (previously linkage group D2).4–7 Based on quantitative trait locus (QTL) mapping, one major and several minor QTLs for resistance to BLP have been identified in six different soybean chromosomes.6 Three loci are closely linked to homeologous rxp regions and their ancestral function was retained in the duplicated rxp loci.8 Despite extensive effort, rxp has yet to be isolated, and the soybean defence mechanisms of response to BLP have not been fully elucidated.
Plants have evolved two defence mechanisms to resist pathogen invasion that involve different strategies of detecting pathogens. On the extracellular face of the host cell, pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs); subsequent stimulation of PRRs leads to PAMP-triggered immunity (PTI).9,10 Although basal immune responses are activated, bacterial effector proteins delivered into host cells via a type III secretion system (TTSS) can suppress PTI.11,12 These pathogen effectors are recognized by specific resistance (R) genes, which encode nucleotide-binding and leucine-rich repeat (LRR) domains. These R gene products activate the second type of immune defence mechanism, effector-triggered immunity (ETI).9,10 Recognition of pathogen attack by PRRs and R genes leads to the activation of defence responses and a type of localized cell death known as the hypersensitive response.13,14
Near-isogenic lines (NILs) are a useful and valuable material for mapping genes. Pairs of NILs differing by the presence or absence of a target gene have been used to isolate markers associated with the target gene and identify markers linked with pathogen resistance.15,16 NILs for BLP resistance have been reported using multiple backcrosses17 and shown to be more suitable for the identification of specific target genes rather than other populations such as recombinant inbred lines.18
Comprehensive transcriptome analysis has provided new insight into developmentally and environmentally induced changes in gene expression. This information can be used to help predict the roles and interactions of individual genes, as well as to elucidate more complex signalling pathways activated in response to external stimuli and uncover potential cross-talk between these pathways. Over the past decade, microarray technology has enabled a more comprehensive understanding of the dynamic composition and regulation of transcripts in many plants, including soybean.19–21 Nonetheless, microarray-based transcript profiling has several limitations, including high background, low sensitivity, and non-specific or cross-hybridization signals, all of which hamper the accurate detection of low abundance transcripts and the discrimination of similar sequences.22,23 To overcome these sensitivity issues with microarrays, hundreds of specific primers for quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis have been designed for the purpose of characterizing the expression of regulatory genes.24 However, both of these methods rely primarily on existing expressed gene sequences for the synthesis of oligonucleotides and for generating qRT–PCR primers.25,26
Next-generation sequencing technologies, such as the widely used Illumina Genome Analyzer,27,28 provide powerful alternative strategies for transcriptome analysis using direct mRNA-sequencing (RNA-Seq). RNA-Seq technology involves generating whole cDNA short reads that are then mapped to genome sequences to obtain the number of ‘mapped reads’ of each gene. Using this approach, one can achieve unprecedented levels of accuracy and specificity in quantifying differentially expressed genes, identifying novel transcribed regions, and identifying alternative splice events.29,30
Using a high-throughput gene expression profiling technique and available complete soybean genome sequences, genes involved in the soybean response to Xag infection were identified by differential expression profiling. The objectives of this study were to identify genes that are differentially expressed in BLP-resistant and BLP-susceptible NILs in response to Xag infection and to gain a better understanding of plant–pathogen interactions using soybean NILs for BLP resistance.
2. Materials and methods
2.1. Plant material and Xag inoculation
The BLP-resistant NILs used in the current study were previously described.17 Briefly, NILs were generated by three cycles of repeated backcrossing of the BLP-resistant line, SS2–2, as the donor parent and the BLP-susceptible line, Taekwangkong, as the recurrent parent. SSR genotyping of the progeny of the backcrosses revealed that more than 93% of the recurrent parent genome was recovered.
For inoculation with Xag, soybean plants were cultivated in growth chambers at 28°C under 12h illumination. Xag strain 8ra was cultured on peptone sucrose agar medium at 28°C for 48h31 prior to inoculation. Bacterial cultures were diluted to a concentration of 1 × 108 colony forming units/ml in 10 mM MgCl2 containing 0.1 ppm rifampicin antibiotic. This suspension was sprayed on the first fully expanded trifoliate leaves of 1-month-old soybean plants using an atomizer. As a control [0h after inoculation (hai)], plants were sprayed with 10 mM MgCl2 buffer. Inoculated plants were grown at 100% relative humidity.
2.2. Sample preparation, read alignment, and sequence analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA purity was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA), 1% formaldehyde gel electrophoresis, and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA from three biological replicates at each time point (0, 6, and 12 hai) was pooled in preparation for next-generation Illumina Genome Analyzer II (GA II) sequencing. All procedures, including mRNA purification, cDNA preparation, end repair of cDNA, adaptor ligation, and cDNA amplification, were carried out according to the manufacturer protocols accompanying the mRNA-Seq Sample Preparation Kit (Cat. RS-930-1001, Illumina Inc., San Diego, CA, USA).
Purified cDNA libraries were dispensed onto an Illumina single-end flow cell composed of eight lanes using the Illumina Cluster Station (Illumina, Inc.). One lane was used per time point for the BLP-susceptible and BLP-resistant NILs; the 6 hai of BLP-resistant NIL was applied to two lanes to test for mechanical reproducibility. The remaining lane was used for an internal control. The 76 bp reads were collected using the Illumina GA II and sequencing-by-synthesis technology.
Sequence reads were aligned using Bowtie (http://bowtie-bio.sourceforge.net), an ultrafast short-read mapping program,32 using the 8× sequence assembly of the soybean genome as a reference (Glyma1.01, http://www.phytozome.net/soybean).33 TopHat (http://tophat.cbcb.umd.edu) was used to identify splice junctions.34 Genes that were differentially expressed in BLP-susceptible and BLP-resistant NILs were identified with the statistical R package DEGseq (http://bioinfo.autsinghua.edu.cn/software/degseq) using an MA-plot-based method and a random sampling model.35 Raw digital gene expression data were normalized as reads per kilobase pair of transcript per million mapped reads (RPKM);27 genes for which the P-value was <0.001 were selected for further analysis. Expression data (reads) were log2-transformed and filtered at a level of 2-fold or greater difference in expression at each time point (0, 6, and 12 hai). Differential patterns of gene expression at the various time points are represented by Venn diagrams.
2.3. Clustering and gene ontology analyses
K-means clustering was performed using TM4: MeV 4.7 software (http://www.tm4.org/mev.html)36 and Pearson's correlation coefficient. Differentially expressed genes were grouped into 10 distinct clusters based on expression patterns. Annotation and assignment of functional categories for the genes in each cluster were assigned based on conserved PFAM domain predictions (http://pfam.sanger.ac.uk/)37 and gene ontology (GO) analysis (http://www.geneontology.org/). Soybean genes identified by GO as containing PFAM domains were classified into three categories:38 biological process, cellular component, and molecular function. Differentially regulated genes were further classified into secondary categories within biological process, cellular component, and molecular function after comparison of BLP-susceptible and BLP-resistant NILs.
2.4. Validations of RNA-Seq data by qRT–PCR
qRT–PCR was performed to validate the RNA-Seq results for nine gene transcripts whose expression differed by more than 2.0-fold between the BLP-susceptible and BLP-resistant NILs after Xag inoculation. Primers for qRT–PCR were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/).39 A Bio-Rad iScript™ cDNA Synthesis Kit (Cat. 170-8891, Hercules, CA, USA) was used to synthesize the cDNAs and real-time quantification was performed using a LightCycler 480 system (Roche Diagnostics, Laval, QC, Canada) and the Bio-Rad iQ™ SYBR Green Supermix Kit (Cat. 170-8882). Expression levels of the selected genes were normalized to tubulin expression. PCR mixtures (final volume, 50 µl) contained 200 ng of cDNA, 500 nM each primer, 18 µl of sterile water, and 25 µl of iQ™ SYBR Green Supermix (Bio-Rad). The conditions for amplification were as follows: 5 min denaturation at 95°C followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 10 s. Following amplification, melting curves were determined in a three-segment cycle of 95°C for 5 s, 65°C for 1 min, and 97°C for 0 s on the continuous acquisition mode. Samples were analysed in triplicate to ensure statistical significance. Data were analysed based on the stable expression level of the reference gene according to method of Livak and Schmittgen.40
3. Results
3.1. BLP symptoms
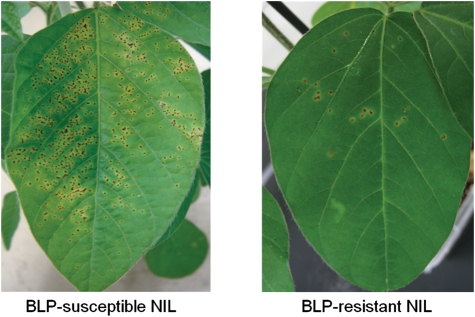
Disease severity in two NILs, one carrying the rxp-susceptible allele and the other carrying the rxp-resistant allele, was assessed 14 days after Xag inoculation (Fig. 1). Pustules surrounded by small yellow haloes were evident in the early stages after inoculation. There were more lesions in the BLP-susceptible NIL, and these lesions subsequently merged to form larger necrotic areas in the late stage of the disease. In the BLP-susceptible NIL, severe haloes and pustules were spread throughout the leaves. In contrast, in the BLP-resistant NIL, BLP disease symptoms were restricted to portions of the leaves (Fig. 1).
Figure 1.
Disease symptoms in BLP-susceptible and BLP-resistant NILs after Xag inoculation. Small yellow to brown lesions with a raised pustule typically formed in the early disease stage, with large necrotic lesions developing later.
3.2. RNA-Seq analysis
Changes in transcript levels between the BLP-susceptible and BLP-resistant NILs at 0, 6, and 12 hai with Xag were analysed by RNA-Seq. A total of 125.7 million reads were generated by 76 bp single-end sequencing from the six cDNA libraries (BLP-susceptible NIL at 0, 6, and 12 hai and BLP-resistant NIL at 0, 6, and 12 hai), constituting 6.3 Gb of cDNA sequence (Table 1). Only the first 50 bp of the reads was used for mapping. Approximately 86% of the sequenced reads (108 million mapped reads) were successfully aligned to the soybean genome reference sequence (Glyma1.01, http://www.phytozome.net/soybean) using Bowtie and TopHat software.32–34 Of 65 781 predicted genes in the soybean genome, the expression levels of 46 367 mapped genes were quantified based on sequence reads. Using the random sampling model in the DEGseq program,35 mapped read counts of each gene with a P-value of < 0.001 were obtained. MA-plots revealed little variation in gene expression patterns for the different time points (Supplementary Fig. S1). A total of 15 678 genes were selected with high confidence based on RPKM values converted from mapped read counts by DEGseq (Supplementary Dataset S1).
Table 1.
Statistics of the Illumina-GA II 76 bp reads and comparison to the G. max reference genome (Glyma 1.01)
| Samples | Time (hours after inoculation) | Number of reads | Number of mapped reads | Mapped reads (%) | Size of nucleotides (bp) |
|---|---|---|---|---|---|
| Susceptible | 0 | 17 834 047 | 15 645 574 | 88 | 891 702 350 |
| NIL | 6 | 17 931 452 | 15 433 901 | 86 | 896 572 600 |
| 12 | 16 408 387 | 13 138 100 | 80 | 820 419 350 | |
| Resistant NIL | 0 | 18 813 182 | 16 446 553 | 87 | 940 659 100 |
| 6a | 18 192 507 | 15 956 847 | 88 | 909 625 350 | |
| 6a | 18 387 123 | 16 173 009 | 88 | 919 356 150 | |
| 12 | 18 048 903 | 15 192 733 | 84 | 902 445 150 | |
| Total | 125 615 601 | 107 986 717 | 6 280 780 050 |
aThese samples were used as technical replicates.
As described by Libault et al.,41 four reference genes, actin (Glyma08g19420), cons4 (Glyma12g02310), cons6 (Glyma12g05510), and tubulin (Glyma08 g01740), were used for the evaluation of gene expression. Eight additional genes containing a tubulin motif were also used as reference genes (Supplementary Table S1). The absolute value of the fold change for the reference genes ranged from 0.0040 (tubulin motif-containing Glyma20g27280 at 0 hai) to 0.7489 (tubulin motif-containing Glyma15g13970 at 0 hai), which indicated that the expression levels of the reference genes were not significantly different across treatment periods in the BLP-susceptible and BLP-resistant NILs. In addition, there was little technical variation, as shown by the similar expression profiles of two replicates of the BLP-resistant NIL at 6 hai (Table 1). These results indicated that the gene transcript data were reliable, and suitable for detection and further transcriptome analysis.
3.3. Transcriptome analysis in response to Xag inoculation
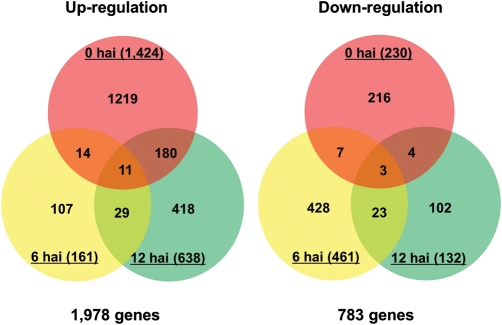
The number of differentially expressed genes in the BLP-resistant NIL compared with the BLP-susceptible NIL at each sampling point was estimated [P < 0.001 and log2 (fold change) >2.0 or <−2.0]. A total of 2415 unique genes were up- or down-regulated in the resistant NIL at various times after inoculation. Of these, 1978 were up-regulated and 783 were down-regulated 0, 6, and/or 12 hai in the BLP-resistant NIL (Fig. 2). Of these, 346 genes were differentially regulated at each time point. For example, Glyma20 g15480 (a homologue of cytochrome P450) was up-regulated at 0 hai but down-regulated at 6 hai in the BLP-resistant NIL. Compared with the BLP-susceptible NIL, 1424 genes were up-regulated and 230 genes were down-regulated at 0 hai. Following Xag inoculation, 161 genes were up-regulated and 461 genes were down-regulated at 6 hai; 638 genes were up-regulated and 132 genes were down-regulated at 12 hai. A subset of genes was either up- or down-regulated at both the 6 and 12h time points (234 and 37, respectively) in the BLP-resistant NIL compared with the BLP-susceptible NIL. Of the 2415 genes that were differentially expressed, only 11 were up-regulated and 3 were down-regulated at all time points (Supplementary Datasets S2 and S3). Most of the genes that were up-regulated in the BLP-resistant NIL were expressed at 0 hai (Fig. 2, left); most of the genes that were down-regulated were expressed at 6 hai (Fig. 2, right).
Figure 2.
Number of gene transcripts in the BLP-resistant NIL that were up- and down-regulated [P < 0.001 and log2 (fold change) >2.0 or<−2.0] compared with the BLP-susceptible NIL. The number inside the parentheses indicates the number of genes expressed at the hours after Xag inoculation (hai). The total number of gene transcripts is at the bottom of each Venn diagram.
3.4. Clustering and GO analyses
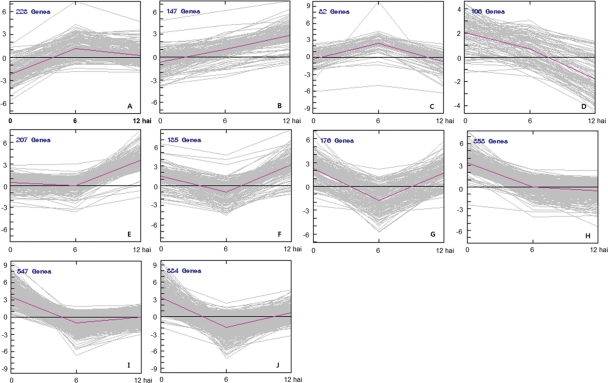
Clustering analysis was used to group the 2415 differentially expressed genes (P< 0.001 and fold change >2 or <−2) into clusters based on common expression patterns. Ten distinct clusters emerged reflecting the general trends and key transitional states in the BLP-resistant NIL following Xag inoculation (Fig. 3). The genes in each cluster are listed in Supplementary Dataset S4. Clusters A and C contained genes that were up-regulated from 0 to 6 hai, and then down-regulated from 6 to 12 hai (Fig. 3). The 147 genes in cluster B were consistently up-regulated from 0 to 12 hai, while the 106 genes in cluster D and 353 genes in cluster H were constitutively down-regulated. The genes in cluster D exhibited a more dramatic decrease after 6 hai compared with those in cluster H, which showed only a slight decrease after 6 hai. Genes grouped into the remaining five clusters E, F, G, I, and J were down-regulated from 0 to 6 hai, but up-regulated from 6 to 12 hai (Fig. 3).
Figure 3.
Cluster analysis of 2415 genes differentially expressed following Xag inoculation. The genes were classified based on similarity of expression pattern over the time course of infection. Ten clusters were identified by K-means clustering. The pink lines indicate representative transcriptional regulators; x- and y-axes represent hours after Xag inoculation (hai) and log2 fold change [log2 (BLP-resistant NIL/BLP-susceptible NIL)], respectively.
Annotation of the 1978 up-regulated and 783 down-regulated genes was carried out based on the identification of conserved PFAM domains (Supplementary Dataset S4). Within the group of 1978 up-regulated genes, there were 2268 PFAMs assigned; within the 783 down-regulated genes, 824 PFAMs were identified. PFAM domains were converted into GO-identities (IDs) using mapping to GO (http://www.geneontolgy.org). Since some of the genes were assigned to multiple PFAMs, the total number of GO IDs was greater than that of PFAM assignments, and these GO IDs could be assigned to multiple GO terms (Table 2). Based on GO terms,38 there were 3494 up-regulated and annotated genes in the BLP-resistant NIL following Xag inoculation. The genes were classified as follows: 1152 genes mapped to biological process terms; 311 genes mapped to cellular component terms; and 2031 genes mapped to molecular function terms (Table 2). Among the 1047 down-regulated and annotated genes, 326 mapped to biological process terms; 105 mapped to cellular component terms; and 616 mapped to molecular function terms. In addition to metabolic processes, genes related to biological regulation, establishment of localization, response to stimulus, and signalling process were highly expressed within the biological process category. Most of the genes categorized under molecular function were involved in binding and catalytic activity (Table 2). Since many of the genes that mapped to biological process terms, particularly ‘response to stimulus’, were differentially expressed, plant resistance genes involved in responses to stimuli were closely evaluated using the database.9,10 The ‘response to stimulus’ group (GO:0050896) comprised 7.90 and 3.07% of all of the up-regulated and down-regulated genes, respectively, within the biological process category (Table 2).
Table 2.
GO functional categorization of PFAM domain-containing soybean genes differentially regulated in the BLP-resistant NIL
| Category | Up-regulation |
Down-regulation |
||
|---|---|---|---|---|
| Number of genes | Proportion (%) | Number of genes | Proportion (%) | |
| Biological process | ||||
| Biological regulation | 197 | 17.10 | 44 | 13.50 |
| Carbon utilization | 3 | 0.26 | 0 | 0.00 |
| Cell proliferation | 1 | 0.09 | 0 | 0.00 |
| Cell wall organization or biogenesis | 10 | 0.87 | 6 | 1.84 |
| Cellular component organization | 10 | 0.87 | 1 | 0.31 |
| Cellular process | 69 | 5.99 | 22 | 6.75 |
| Developmental process | 2 | 0.17 | 0 | 0.00 |
| Establishment of localization | 110 | 9.55 | 26 | 7.98 |
| Metabolic process | 633 | 54.95 | 210 | 64.42 |
| Multicellular organismal process | 1 | 0.09 | 0 | 0.00 |
| Response to stimulus | 91 | 7.90 | 10 | 3.07 |
| Signalling process | 25 | 2.17 | 6 | 1.84 |
| Viral reproduction | 0 | 0.00 | 1 | 0.31 |
| Subtotal | 1152 | 100.00 | 326 | 100.00 |
| Cellular component | ||||
| Cell part | 212 | 68.17 | 62 | 59.05 |
| Extracellular region | 32 | 10.29 | 15 | 14.29 |
| Macromolecular complex | 32 | 10.29 | 5 | 4.76 |
| Organelle | 35 | 11.25 | 23 | 21.90 |
| Subtotal | 311 | 100.00 | 105 | 100.00 |
| Molecular function | ||||
| Antioxidant activity | 26 | 1.28 | 2 | 0.32 |
| Binding | 965 | 47.51 | 261 | 42.37 |
| Catalytic activity | 811 | 39.93 | 282 | 45.78 |
| Electron carrier activity | 70 | 3.45 | 27 | 4.38 |
| Enzyme regulator activity | 24 | 1.18 | 11 | 1.79 |
| Molecular transducer activity | 35 | 1.72 | 7 | 1.14 |
| Nutrient reservoir activity | 0 | 0.00 | 1 | 0.16 |
| Structural molecule activity | 5 | 0.25 | 2 | 0.32 |
| Transcription regulator activity | 34 | 1.67 | 6 | 0.97 |
| Transporter activity | 61 | 3.00 | 17 | 2.76 |
| Subtotal | 2031 | 100.00 | 616 | 100.00 |
| Total | 3494 | 1047 | ||
3.5. Expression of plant resistance genes
Because of the limited information available for mapping Glyma PFAMs to GO terms, defence response genes (GO:0006952) were further analysed by sequence comparisons with the corresponding Arabidopsis protein sequences using a threshold of <e−100. Based on BLASTP analysis of the defence response genes, 134 putative defence response genes in Glycine max exhibited significant differences in expression between the BLP-susceptible and BLP-resistant NILs following Xag inoculation (Supplementary Dataset S5). Interactions between plant and pathogen were probed by analysing the transcript levels of PRRs, ETI receptors, and jasmonic acid (JA)- and salicylic acid (SA)-related proteins that were differentially expressed in the BLP-susceptible and BLP-resistant NILs.
3.5.1. Expression of PTI-related genes
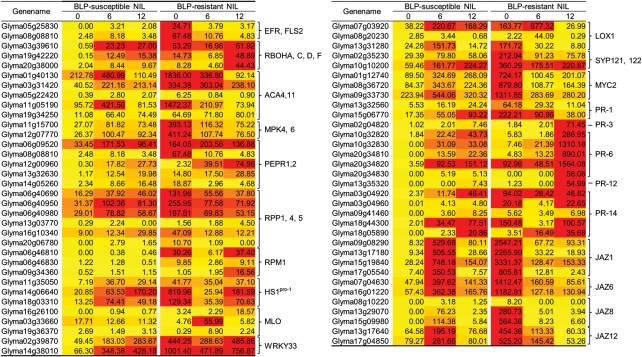
In plants, there are two types of immunity to bacterial pathogens, PTI and ETI (Fig. 4). Stimulation of PRRs is a key step in the early stages of PTI. Based on BLASTP searches, Glyma05g25830 and Glyma08g08810, close homologues of flagellin sensing 2 (FLS2) and EF-Tu receptor (EFR) in G. max, were identified as highly expressed at 0 hai in the BLP-resistant NIL compared with the BLP-susceptible NIL (Fig. 5 and Supplementary Fig. S2A).
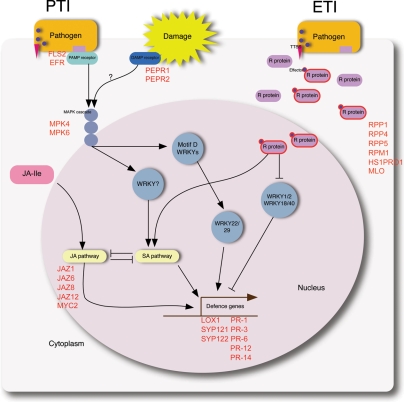
Figure 4.
Schematic diagram of plant immunity to bacterial pathogens, adapted from Dodds and Rathjen.10 Plants use two strategies to respond to pathogen attacks: PTI and ETI. Ultimately, through several branched and multi-component pathways, defence-related genes are transcribed. Genes that were up-regulated in the BLP-resistant NIL are represented as reddish bold characters.
Figure 5.
Heat maps of gene transcripts in BLP-susceptible and BLP-resistant NILs after Xag infection [P < 0.001 and log2 (fold change) >2.0 or<−2.0]. PTI-, ETI-, JA-, and defence-related soybean genes whose expression differed significantly are represented. Darker colours indicate higher transcript levels.
Respiratory burst oxidase homologue (RBOH) is an important factor in the production of reactive oxygen species during the plant response to abiotic and biotic stresses.42,43 Changes in the expression levels of G. max RBOHA-, C-, D-, and F-like genes in response to Xag inoculation were assessed (Fig. 5 and Supplementary Fig. S2B). Three soybean genes (Glyma03g39610, Glyma19g42220, and Glyma20g38000) with homology to RBOH exhibited low level expression in the BLP-susceptible NIL at 0 hai. In the BLP-resistant NIL, transcript levels were relatively higher, with a slight reduction at 6 hai; the level of these three transcripts was markedly increased at 12 hai.
Calcium is an essential second messenger in the signal transduction pathways that regulate plant responses.42,43 The expression levels of G. max Ca2+-ATPase 4 (ACA4)- and ACA11-like genes were much higher than any other defence-related genes (Fig. 5 and Supplementary Fig. S2C). In particular, the expression levels of Glyma01g40130 and Glyma11g05190 in the BLP-resistant NIL were extremely high at 0 hai relative to the BLP-susceptible NIL.
Genes involved in the mitogen-activated protein kinase (MAPK) signalling cascade are downstream components in PTI42,43 (Fig. 4). Interestingly, the expression of G. max MAP kinase (MPK) 4- and MPK6-like genes was higher in the BLP-resistant NIL at 0 hai (Fig. 5 and Supplementary Fig. S2D). High expression of damage-associated molecular pattern (DAMP)-related plasma membrane LRR receptor kinase 1 (PEPR1) and PEPR2 homologues in G. max was observed in the BLP-resistant NIL at 0 hai (Fig. 5 and Supplementary Fig. S2E), suggesting that DAMPs for eliciting PAMP downstream may also be involved in BLP resistance.
3.5.2. ETI-related genes
The second type of stress perception in plants involves recognition of pathogen virulence molecules called effectors by specific plant intracellular receptors, or ETI. To investigate the role of ETI in BLP resistance, R gene sequences were collected from the Plant Resistance Gene database (PRGdb, http://prgdb.cbm.fvg.it).44 Expression of several corresponding G. max genes, including RPP1-, 4- and 5-, RPM1-, beet cyst nematode Heterodera schachtii Schmidt resistance gene (HS1PRO-1)- and mildew-resistance locus O (MLO)-like genes was induced following Xag inoculation (Fig. 5 and Supplementary Fig. S3). Of the six RPP-like genes surveyed, Glyma06g40690, Glyma06g40950, and Glyma06g40980 exhibited significantly higher expression levels than three other genes regardless of time points or NILs. Furthermore, these three genes were the most highly expressed in the resistant NIL at 0 hai (Fig. 5 and Supplementary Fig. S3A). Expression of Glyma16g10340 in the susceptible NIL increased steadily after Xag inoculation, whereas in the resistant NIL, expression was reduced, with the highest level of expression observed at 0 hai.
Three gene transcripts encoding RPM1-like genes, Glyma06g46810 (3e−136), Glyma06g46830 (5e−138), and Glyma09g34360 (9e−118), were identified by BLASTP searches using Arabidopsis RPM1. These genes had the lowest e-values, and the differences in expression of these three genes between the NILs was considerable (Fig. 5 and Supplementary Fig. S3B). In the susceptible NIL, there were very low levels of Glyma06g46810, Glyma06g46830, and Glyma09g34360, whereas transcript levels were quite high in the resistant NIL, particularly for Glyma06g46810 at 0 and 12 hai. The transcript levels of Glyma06g46830 and Glyma09g34360 were lower than that of Glyma06g46810, but the pattern of expression of Glyma06 g46830 was similar to Glyma06g46810. The expression of Glyma09g34360 increased hours after Xag infection in the resistant NIL (Supplementary Fig. S3B).
Three G. max genes (Glyma11g35050, Glyma14g06640, and Glyma18g03310) with homology to HS1PRO-1 were identified after a BLASTP search against the Arabidopsis database (Fig. 5 and Supplementary Fig. S3C). The expression of these three genes was lower in the susceptible NIL compared with the resistant NIL. Glyma14g06640 expression was highest in the resistant NIL at 0 hai, then was decreased at 6 hai and increased again at 12 hai. The pattern of expression of Glyma18g03310 was similar to that of Glyma14g06640, but it was expressed to lower levels compared with Glyma14g06640 (Supplementary Fig. S3C).
Of particular interest, Glyma03g33660, Glyma16g26100, and Glyma19g36370 were highly similar to MLO, which indicated that the functions of plant resistance genes may be conserved across species (Fig. 5 and Supplementary Fig. S3D). Glyma03g33660 was dramatically increased in the resistant NIL at 6 hai compared with 0 hai, but was subsequently decreased at 12 hai. Although the transcript levels of Glyma19g36370 were much lower than Glyma03g33660, the pattern of expression of the two genes was similar. Glyma16g26100 remained elevated by at least 3-fold from 6 to 12 hai in the resistant NIL. The levels of expression of Glyma16g26100 and Glyma19g36370 were very low in the susceptible NIL (Supplementary Fig. S3D).
3.5.3. Defence-related genes
WRKY33, LOX1, SYP121, SYP122, MYC2, and pathogenesis-related (PR) genes have been implicated in plant immune responses to pathogen attacks. WRKY transcription factors (TFs) play important roles in plant immune systems in response to abiotic and biotic stress,45,46 and are involved at various points in the signalling pathways that regulate these responses (Fig. 4). Glycine max genes containing WRKY domains were selected in the search for conserved PFAM domain (PF03106). The expression of two WRKY33 TFs, Glyma02g39870 and Glyma14g38010, increased steadily following Xag inoculation in the BLP-susceptible NIL, and both genes were expressed at higher levels in the resistant NIL compared with the susceptible NIL at all time points. There was a large difference in expression between the NILs at 0 hai, with the highest level of Glyma14g38010 observed at 0 hai in the BLP-resistant NIL (Fig. 5 and Supplementary Fig. S4A).
Three G. max homologues of lipoxygenase 1 (LOX1) were identified (Fig. 5 and Supplementary Fig. S4B). In the BLP-susceptible NIL, transcript levels of Glyma07g03920 and Glyma13g31280 were low at 0 hai, increased at 6 hai and then subsequently decreased at 12 hai. The expression of Glyma07g03920 in the BLP-resistant NIL was shown a 4-fold increase in transcript levels at 6 hai relative to 0 hai and then a subsequent decrease at 12 hai. Although the expression level of Glyma08g20230 was much lower than the other two genes (Glyma07g03920 and Glyma13g31280), transcript levels were increased 6 hai in the BLP-resistant NIL (Supplementary Fig. S4B).
Two soybean genes (Glyma02g35230 and Glyma10g10200) were identified by BLASTP searches for SYP121 and SYP122 homology. Expression of Glyma02g35230 and Glyma10g10200 was elevated in the BLP-resistant NIL only at 0 hai relative to the BLP-susceptible NIL (Fig. 5 and Supplementary Fig. S4C).
MYC2, a bHLH TF, plays a crucial role in the JA-signalling pathway.47 Four G. max genes retrieved using Phytozome were identified: Glyma01g12740, Glyma07g05740, Glyma08g36720, and Glyma09g33730. Three of these (Glyma01g12740, Glyma08g36720, and Glyma09g33730) were highly expressed in the BLP-resistant NIL at 0 hai compared with any other time point in either NIL (Fig. 5 and Supplementary Fig. S4D).
Several soybean genes related to Arabidopsis PR genes48 were retrieved using Phytozome,33 including soybean homologues to PR-1, -3, -4, -6, -12, and -14. Glyma13g32560 and Glyma15g06770 (PR-1-like G. max genes) were highly expressed in the BLP-resistant NIL at 0 hai (Fig. 5 and Supplementary Fig. S5A). Of the basic chitinase-like genes identified in G. max (PR-3 and -4 homologues), the PR-3 homologue Glyma02g04820 was highly expressed in the BLP-resistant NIL at 12 hai (Fig. 5 and Supplementary Fig. S5B and C). Of seven PR-6 G. max genes, five homologues of the proteinase inhibitor PR-6 (Glyma10g32820, Glyma10g32830, Glyma20g34810, Glyma20g34820, and Glyma20g34830) were significantly induced in the BLP-resistant NIL at 12 hai (Fig. 5 and Supplementary Fig. S5D). Expression of Glyma13g35320, a homologue of the plant defensing PR-12, was elevated 2-fold in the BLP-resistant NIL compared with the BLP-susceptible NIL at 12 hai (Fig. 5 and Supplementary Fig. S5E). Of the 11 homologues of PR-14, a lipid transfer protein, only Glyma03g04920 and Glyma18g44300 at 0 hai, Glyma18g05890 at 6 hai, and Glyma03g04960 at 0 and 12 hai were highly expressed relative to the BLP-susceptible NIL (Fig. 5 and Supplementary Fig. S5F).
3.5.4. Plant hormones-related genes
The SA and JA-ethylene (ET) phytohormone pathways are important regulators of defence gene expression.49,50 These pathways function downstream in PTI and ETI, and are well characterized relative to other steps in the process51–53 (Fig. 4). We examined the expression of two non-expressor of PR gene 1 (NPR1)-like genes in G. max, Glyma09g02430 and Glyma15g13320, since NPR1 is an important regulatory component in SA signalling (Fig. 5 and Supplementary Fig. S6A). In the BLP-susceptible NIL, expression of Glyma09g02430 and Glyma15g13320 started out high at 0 hai, and then increased at 6 hai before returning to pre-inoculation levels 12 hai. There were no significant differences in the expression of Glyma09g02430 and Glyma15g13320 between the NILs.
Isochorismate synthase 1 (ICS1) is a key enzyme responsible for increasing SA accumulation and pathogen resistance.53,54 Overall, the expression levels of the ICS1-like G. max genes Glyma01g25690 and Glyma03g17420 were not significantly different between the two NILs (Supplementary Fig. S6B). The expression of Glyma01g25690 was increased following inoculation in the BLP-susceptible NIL, but was decreased at 6 hai in the BLP-resistant NIL relative to 0 and 12 hai. In both NILs, the expression of Glyma03g17420 was similar at 0 and 6 hai, and then increased at 12 hai.
Enhanced disease susceptibility 1 (EDS1) and EDS5 were also evaluated. The expression of EDS1-like genes (Glyma04g34800, Glyma06g19920, Glyma06g19890, and Glyma06g19900) and EDS5-like genes (Glyma11g11970 and Glyma11g11990) was similar in both NILs (Supplementary Fig. S6C and D). Thus, there were no major differences in the expression of SA signalling genes between the BLP-susceptible and BLP-resistant NILs.
JA functions as an antagonist of SA in plant hormone pathways, and several Arabidopsis genes involved in JA signalling pathways have been identified.55 Soybean homologues of jasmonate resistant 1 (JAR1), an enzyme that catalyses the conjugation of JA to isoleucine,56 and coronate insensitive 1 (COI1), a jasmonate receptor,57 were not significantly expressed between the NILs (Supplementary Fig. S7A and B). Jasmonate ZIM-motif (JAZ) proteins bind to TFs such as MYC2 and control a variety of jasmonate-mediated responses, including defence gene expression and growth responses to wounding.58,59 The expression of JAZ3- and JAZ4-like genes was unchanged at both time points after inoculation (Fig. 5 and Supplementary Fig. S7D and E). Expression of four other JAZ-like proteins, JAZ1, JAZ6, JAZ8, and JAZ12, was significantly elevated at 0 hai in the BLP-resistant NIL; of these genes, JAZ1 was the most highly expressed (Fig. 5 and Supplementary Fig. S7C, F–H).
3.6. Validation of RNA-Seq results by qRT–PCR
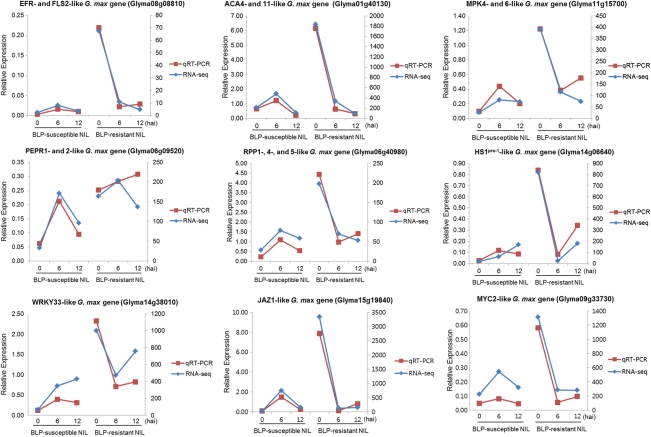
qRT–PCR was used to validate several of the differentially expressed genes identified by RNA-Seq in the BLP-susceptible and BLP-resistant NILs. Nine genes involved in defence mechanisms, including PTI and ETI (Fig. 6), were evaluated: Glyma08g08810 (EFR and FLS2), Glyma01g40130 (ACA4 and 11), Glyma11g15700 (MPK4 and 6), Glyma06g09520 (PEPR1 and 2), Glyma06g40980 (RPP1, 4 and 5), Glyma14g06640 (HS1PRO-1), Glyma14g38010 (WRKY33), Glyma15g19840 (JAZ1), and Glyma09g33730 (MYC2), all of which were shown by RNA-Seq analysis to be up-regulated at 0 hai in the BLP-resistant NIL compared with the BLP-susceptible NIL. The primer sets used for qRT–PCR are listed in Supplementary Table S2. The validation results were consistent with the gene expression patterns identified by RNA-Seq. The expression levels of Glyma11g15700 and Glyma06g09520 at 12 hai in the resistant NIL were slightly higher based on qRT–PCR than RNA-Seq. These results highlighted the fidelity and reproducibility of the RNA-Seq analysis used in the present study.
Figure 6.
Validation of RNA-Seq data by qRT–PCR. Nine PTI-, ETI-, and defence-related genes were selected for validation.
4. Discussion
Plants have two main defence mechanisms that can be activated in response to microbial plant pathogens, PTI and ETI9,10 (Fig. 4). PAMPs trigger an influx of calcium ions and an oxidative burst in the early stage of infection. This is followed by activation of MAPK and calcium-dependent protein kinase, and stomatal closure and transcriptional reprogramming in the intermediate stage. Finally, SA accumulation and callose deposition occur in the late stage of infection.42,43 Plant pathogenic bacteria, including Xanthomonas, deliver effector proteins into host cells via a TTSS to promote parasitism.11,12 These bacterial effectors are recognized directly or indirectly by specific R genes, leading to the gene-for-gene theory of ETI.60 Specific recognition of pathogen effectors leads to the induction of disease resistance and HR.13,14
In the current study, plant immune responses triggered by the interaction of soybean with the plant pathogen Xag were investigated. The expression patterns of PTI- and ETI-related genes in response to Xag infection in BLP-susceptible and BLP-resistant NILs were compared using RNA-Seq. Most of the genes related to PTI were up-regulated at 0 hai in the BLP-resistant NIL relative to the BLP-susceptible NIL (Figs 2 and 5). EFR- and FLS2-like genes, as putative PAMP receptors, were highly expressed at 0 hai (Fig. 5 and Supplementary Fig. S2A). Of note, key PTI genes triggered by PAMPs, such as ACA4, ACA11, MPK4, MPK6, and RBOH genes, were highly expressed in the BLP-resistant NIL at 0 hai (Fig. 5 and Supplementary Fig. S2B–D). Recently, the first DAMP receptors (PEPR1 and PEPR2) were identified by virtue of their ability to recognize AtPep1,61 DAMPs are involved in the activation of plant immune responses, and include polysaccharides released from the cell wall and cuticular fragments, as well as some endogenous peptides.62,63 The expression of PEPR1 and 2 in soybean differed significantly in the BLP-resistant and BLP-susceptible NILs at 0 hai (Fig. 5 and Supplementary Fig. S2E). Based on expression patterns, the PTI-related genes (EFR-, FLS2-, RBOH-, ACA4-, ACA11-, MPK4-, and MPK6-like G. max genes) and DAMP receptors (PEPR1- and 2-like G. max genes) were grouped into clusters H and I (Fig. 3 and Supplementary Dataset 4). The up-regulation of PTI-related genes at 0 hai could potentiate the immune reaction to pathogen attack. Genes expressed in early stage responses, such as 0 hai, might be more important for BLP resistance than those expressed at later stages (i.e. 6 hai). This is supported by the observation that sufficient accumulation of R proteins in the initial stage of response influences effective resistance against biotrophic pathogens, demonstrated by the mildew-resistance locus A R gene in barley.64 Thus, the gene response at 0 hai may be involved in BLP resistance to Xag in the BLP-resistant NIL.
Among the R genes obtained from the Plant Resistance Genes database (PRGdb),44 RPP1-, 4-, and 5-like; RPM1-like; HS1PRO-1-like; and MLO-like genes were significantly different in expression between the BLP-susceptible and BLP-resistant NILs (Fig. 5 and Supplementary Fig. S3). RPP genes trigger localized cell death in Arabidopsis upon recognition of downy mildew avirulence genes.65 The transcript levels of the three RPP genes (Glyma06g40690, Glyma06g40950, and Glyma40980) were much higher at 0 hai in the BLP-resistant NIL relative to the BLP-susceptible NIL. In Arabidopsis, RPM1 encodes an intracellular immune sensor that is induced in response to Pseudomonas syringae, and attack by this pathogen leads to the expression of RPM1 disease resistance proteins.66,67 In the BLP-resistant NIL, the expression of soybean RPM1 homologues was elevated. In sugar beet, the HS1PRO-1 gene reportedly confers resistance to the beet cyst nematode.68 Soybean homologues of HS1PRO-1 were also expressed at high levels at 0 hai in the BLP-resistant NIL. These patterns suggest that a strong surveillance system exists in the BLP-resistant NIL, and that these genes play important roles in the modulation of defence responses to biotic and abiotic stress stimuli. In contrast, MLO, which plays an important role in defence responses to biotic and abiotic stresses,69 exhibited a different pattern. The expression of G. max MLO increased hours after Xag infection. Thus, MLO might be involved downstream of the other R genes in defence response signalling pathways.
Defence-related genes were analysed using the GO term ‘defence response’ (GO:0006952) (Fig. 5 and Supplementary Fig. S4). The expression of LOX1-, SYP121-, SYP122-, WRKY33-, and MYC2-like genes differed significantly between NILs. With the exception of LOX1, these genes were all grouped into cluster H or I, which consisted of genes that were up-regulated at 0 hai, down-regulated at 6 hai, and then up- or down-regulated slightly at 12 hai in the BLP-resistant NIL relative to the BLP-susceptible NIL (Fig. 3 and Supplementary Dataset 4). LOX1-mediated pathways are crucial for lipid peroxidation during plant defence responses to pathogen infection.70 An important gene for defence, SYP121 (closest homologue, PEN1), is required for the timely formation of cell wall appositions.71 These defence-related genes were highly expressed in the BLP-resistant NIL. WRKY33 expression levels were also much higher in the resistant NIL (Supplementary Fig. S4A). WRKY TFs have also been shown to be involved in plant immune responses to bacterial pathogens.45,46
Plant resistance to biotrophic pathogens is thought to be controlled largely by SA-mediated signalling pathways. In contrast, resistance to necrotrophic pathogens is mediated by the JA and ET signalling pathways.54 Even though Xanthomonas is a known biotrophic pathogen, we did not observe any significant difference in the expression of SA-related genes in the NILs (Supplementary Fig. S6). However, the core JA-signalling components JAZ (JAZ1, JAZ6, and JAZ8) and MYC2 were highly expressed in the BLP-resistant NIL at 0 hai, and the expression patterns of all of these genes were very similar (Fig. 5 and Supplementary Figs S4D and S7). Up-regulation of JAZs and MYC2 related to JA-signalling was shown in the BLP-resistant NIL at 0 hai. One possibility for the elevated expression of JA signalling components may be that spraying by the atomizer was perceived as wounding. Alternatively, endogenous levels of JA may be elevated in the BLP-resistant NIL compared with the BLP-susceptible NIL. According to Chini et al.,58 JAZ genes are constitutively overexpressed in MYC2 transgenic plants in the absence of jasmonate treatment. Furthermore, JAZ genes were found to be transcriptional targets of MYC2 via negative regulation, leading to the self-repression of MYC2 in response to increased MYC2 expression levels.58 Binding of the active form of JA, JA-isoleucine, to COI1 and subsequent interaction of the complex with JAZ proteins is followed by ubiquitination by SCFCOI1 and proteasomal degradation, allowing the release of MYC2 and activation of JA responses.72 Our results are consistent with these previous studies of JA signalling pathways.
Previously, QTL mapping was used to place rxp along with eight SSRs and two SNPs in the centre of chromosome 17.7 Three candidate genes for rxp, Glyma17g09770, Glyma17g09780, and Glyma17g09790, were also proposed as putative BLP resistance genes. The chromosomal position of rxp was confirmed using the same BLP-susceptible and BLP-resistant NILs used in the current study. Of note, no significant differences were observed in the expression levels of the three rxp candidate genes (Supplementary Table S3). These results could be explained by either a dilution effect or to differences in sampling time between the two studies. A strong dilution effect of mRNA from un-inoculated regions of the leaves could be avoided by laser capture microdissection (LCM), which allows the selective isolation of targeted cells from a tissue section, thereby avoiding contamination from adjacent non-infected tissue or cells.73,74 In both studies, the leaves were cut manually, not by LCM, introducing potential inconsistencies in the area of the excised leaf tissue, which may in turn result in small differences in expression levels of the candidate resistance genes. Compared with the previous QTL mapping study,7 the current study employed a different sampling schedule after Xag inoculation. There are a variety of reasons why there might be differences between the results of transcriptome analysis just hours after Xag inoculation and QTL mapping based on a BLP resistance phenotype a week after Xag inoculation. Since a change or alteration of causal genes for resistance does not necessarily mean changes at the transcript level, the instability of the translation product might be a factor. Thus, translational changes may be responsible for BLP resistance, although there were no significant differences in transcript accumulation of the candidate genes between the NILs.
In conclusion, 2415 differentially expressed genes were identified in BLP-resistant and BLP-susceptible NILs following Xag inoculation. PTI-, ETI-, defence-, and hormone-related genes were identified and analysed for their potential role in plant immune reactions using the methods of clustering and GO analysis. The PAMP receptors EFR and FLS2 and PAMP-induced genes, including RBOH, ACA4, ACA11, MPK4, and MPK6, were all highly expressed in the BLP-resistant NIL at 0 hai. Furthermore, the DAMP receptors PEPR1 and PEPR2 were also highly expressed at 0 hai, supporting the idea that DAMP-induced genes are involved in BLP-resistance. These results indicate that the plant surveillance system for detecting pathogen invasion is well-established in the BLP-resistant NIL. PAMP and DAMP receptors share common pathways from the initial steps of the defence mechanism, and these two factors may be crucial in mediating BLP resistance. No significant differences were observed for several components of SA-signalling pathways, whereas core JA-signalling components such as JAZ proteins and MYC2 TFs exhibited significant differences in expression between the NILs. Although JA participates to a greater extent than SA in the BLP defence mechanism, both the JA and SA pathways are associated with PR genes and PR-mediated signalling mechanisms. Thus, the defence mechanisms involved in BLP resistance are likely to be quite complex. The current findings contribute a better understanding of plant pathogen defence responses, plant–pathogen interactions, and signal transduction pathways. TFs such as WRKY33 and MYC2 were highly expressed in the BLP-resistant NIL at 0 hai. These differentially expressed TF genes open up an interesting avenue of study into the cis- or trans-elements involved in BLP resistance using other advanced technologies such as Chip-Seq.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This research was supported by a grant from the Next-Generation BioGreen 21 Program (no. PJ0081172011), Rural Development Administration, Republic of Korea.
Supplementary Material
References
- 1.Kennedy B.W., Tachibana H. Bacterial diseases. In: Caldwell B.E., editor. Soybeans: Improvement, Production, and Uses. Madison, WI: American Society of Agronomy; 1973. pp. 491–504. [Google Scholar]
- 2.Takamiya K., Tsuchiya T., Ohta H. Degradation pathway(s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci. 2000;5:426–31. doi: 10.1016/s1360-1385(00)01735-0. [DOI] [PubMed] [Google Scholar]
- 3.Hartwig E., Lehman S. Inheritance of resistance to the bacterial pustule disease in soybeans. Agron. J. 1951;43:226–9. [Google Scholar]
- 4.Palmer R.G., Lim S.M., Hedges B.R. Testing for linkage between the Rxp locus and 9 isozyme loci in soybean. Crop Sci. 1992;32:681–3. [Google Scholar]
- 5.Narvel J.M., Jakkula L.R., Phillips D.V., et al. Molecular mapping of Rxp conditioning reaction to bacterial pustule in soybean. J. Hered. 2001;92:267–70. doi: 10.1093/jhered/92.3.267. [DOI] [PubMed] [Google Scholar]
- 6.Van K., Ha B.-K., Kim M.Y., et al. SSR mapping of genes conditioning soybean resistance to six isolates of Xanthomonas axonopodis pv. glycines. Kor. J. Genet. 2004;26:47–54. [Google Scholar]
- 7.Kim D.H., Kim K.H., Van K., et al. Fine mapping of a resistance gene to bacterial leaf pustule in soybean. Theor. Appl. Genet. 2010;120:1443–50. doi: 10.1007/s00122-010-1266-0. [DOI] [PubMed] [Google Scholar]
- 8.Kim K.D., Shin J.H., Van K., et al. Dynamic rearrangements determine genome organization and useful traits in soybean. Plant Physiol. 2009;151:1066–76. doi: 10.1104/pp.109.141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones J., Dangl J. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11:539–48. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 11.Lahaye T., Bonas U. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 2001;6:479–85. doi: 10.1016/s1360-1385(01)02083-0. [DOI] [PubMed] [Google Scholar]
- 12.Grant S., Fisher E., Chang J., et al. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Microbiology. 2006;60:425–49. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 13.Lam E., Kato N., Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–53. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- 14.Shirasu K., Schulze-Lefert P. Regulators of cell death in disease resistance. Plant Mol. Biol. 2000;44:371–85. doi: 10.1023/a:1026552827716. [DOI] [PubMed] [Google Scholar]
- 15.Young N., Zamir D., Ganal M., et al. Use of isogenic lines and simultaneous probing to identify DNA markers tightly linked to the Tm-2a gene in tomato. Genetics. 1988;120:579–85. doi: 10.1093/genetics/120.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wight C., O'Donoughue L., Chong J., et al. Discovery, localization, and sequence characterization of molecular markers for the crown rust resistance genes Pc38, Pc39, and Pc48 in cultivated oat (Avena sativa L.) Mol. Breed. 2005;14:349–61. [Google Scholar]
- 17.Kim K.H., Kim M.Y., Van K., et al. Marker-assisted foreground and background selection of near isogenic lines for bacterial leaf pustule resistant gene in soybean. J. Crop Sci. Biotechnol. 2008;11:263–8. [Google Scholar]
- 18.Keurentjes J.J.B., Bentsink L., Alonso-Blanco C., et al. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J.J., Rodriguez-Zas S., Aldea M., et al. Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol. Plant Microbe Interact. 2005;18:1161–74. doi: 10.1094/MPMI-18-1161. [DOI] [PubMed] [Google Scholar]
- 20.Libault M., Wan J.R., Czechowski T., et al. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 2007;20:900–11. doi: 10.1094/MPMI-20-8-0900. [DOI] [PubMed] [Google Scholar]
- 21.Brechenimacher L., Kim M.Y., Benitez M., et al. Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2008;21:631–45. doi: 10.1094/MPMI-21-5-0631. [DOI] [PubMed] [Google Scholar]
- 22.Sultan M., Schulz M.H., Richard H., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Bloom J.S., Khan Z., Kruglyak L., et al. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libault M., Joshi T., Takahashi K., et al. Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol. 2009;151:1207–20. doi: 10.1104/pp.109.144030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marioni J.C., Mason C.E., Mane S.M., et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Vliet A.H.M. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 2010;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 27.Mortazavi A., Williams B.A., Mccue K., et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Wilhelm B., Landry J. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–57. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Blencowe B.J., Ahmad S., Lee L.J. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Gene Dev. 2009;23:1379–86. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh C., Heu S., Choi Y. Sensitive and pathovar-specific detection of Xanthomonas campestris pv. glycines by DNA hybridization and polymerase chain reaction analysis. Plant Pathol. J. 1999;15:57–61. [Google Scholar]
- 32.Langmead B., Trapnell C., Pop M., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 34.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L.K., Feng Z.X., Wang X., et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 36.Saeed A.L., Sharov V., White J., Liang W., Bhagabati N., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 37.Sonnhammer E.L.L., Eddy S.R., Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins: Struct. Funct. Genet. 1997;28:405–20. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 38.Berardini T., Mundodi S., Reiser L., et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135:745–55. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 40.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Libault M., Bilgin S., Radwan D., et al. Identification of four soybean reference genes for gene expression normalization. Plant Genome. 2008;1:44–54. [Google Scholar]
- 42.Zipfel C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009;12:414–20. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Clay N., Adio A., Denoux C., et al. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanseverino W., Roma G., De Simone M., et al. PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2010;38:D814–21. doi: 10.1093/nar/gkp978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey S., Somssich I. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–55. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eulgem T., Somssich I. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–71. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Chini A., Boter M., Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid signalling module. FEBS J. 2009;276:4682–92. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- 48.Sels J., Mathys J., De Coninck B., Cammue B., De Bolle M.F.C. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 2008;46:941–50. doi: 10.1016/j.plaphy.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 49.Dong X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998;1:316–23. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 50.Vlot A., Dempsey D.A., Klessig D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 51.Aarts N., Metz M., Holub E., et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Plant Biol. 1998;95:10306–11. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkel B.N., Brooks D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–31. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 53.Loake G., Grant M. Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 2007;10:466–72. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Phytopathology. 2005;43:205–27. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 55.Gfeller A., Liechti R., Farmer E. Arabidopsis jasmonate signaling pathway. Sci. Signal. 2010;3:cm4. doi: 10.1126/scisignal.3109cm4. [DOI] [PubMed] [Google Scholar]
- 56.Suza W.P., Staswick P.E. The role of JAR1 in jasmonoyl-l-isoleucine production during Arabidopsis wound response. Planta. 2008;227:1221–32. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 57.Yan J., Zhang C., Gu M., et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–36. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chini A., Fonseca S., Fernandez G., et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 59.Thines B., Katsir L., Melotto M., et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 60.Flor H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971;9:275–96. [Google Scholar]
- 61.Krol E., Mentzel T., Chinchilla D., et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 2010;285:13471–9. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huckelhoven R. Cell wall-associated mechanisms of disease resistance and susceptibility. Phytopathology. 2007;45:101–27. doi: 10.1146/annurev.phyto.45.062806.094325. [DOI] [PubMed] [Google Scholar]
- 63.Lotze M., Zeh H., Rubartelli A., et al. The grateful dead: damage associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 64.Bieri S., Mauch S., Shen Q.-H., et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–95. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rehmany A., Gordon A., Rose L., et al. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–50. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mackey D., Holt B.F., Wiig A., et al. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–54. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 67.Serrano M., Hubert D., Dangl J., et al. A chemical screen for suppressors of the avrRpm1-RPM1-dependent hypersensitive cell death response in Arabidopsis thaliana. Planta. 2010;231:1013–23. doi: 10.1007/s00425-010-1105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thurau T., Kifle S., Jung C., et al. The promoter of the nematode resistance gene Hs1 pro-1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol. Biol. 2003;52:643–60. doi: 10.1023/a:1024887516581. [DOI] [PubMed] [Google Scholar]
- 69.Piffanelli P., Zhou F., Casais C., et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–85. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casey R., Hughes R. Recombinant lipoxygenases and oxylipin metabolism in relation to food quality. Food Biotechnol. 2005;18:135–70. [Google Scholar]
- 71.Assaad F., Qiu J., Youngs H., et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol. Cell. 2004;15:5118–29. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trujillo M., Shirasu K. Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 2010;13:402–8. doi: 10.1016/j.pbi.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Santi S., Schmidt W. Laser microdissection-assisted analysis of the functional fate of iron deficiency-induced root hairs in cucumber. J. Exp. Bot. 2008;59:697–704. doi: 10.1093/jxb/erm351. [DOI] [PubMed] [Google Scholar]
- 74.Barcala M., Garcia A., Cabrera J., et al. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010;61:698–712. doi: 10.1111/j.1365-313X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.