Abstract
One of the more useful protein tags for a protein in biochemical experiments is biotin, due to its femtomolar dissociation constant with streptavidin or avidin. Robust methodologies have been developed for the in vivo addition of a single biotin to recombinant protein or either in vitro enzymatic or chemical addition of biotin to a protein. Such modified proteins can be used in a variety of experiments, such as affinity selection of phage-displayed peptides or antibodies, pull-down of interacting proteins from cell lysates, or arraying proteins on arrays. We present three complementary approaches for biotinylating proteins in vivo in Escherichia coli, and biotinylating proteins in vitro either chemically or enzymatically that can be scaled up to tag large numbers of proteins in parallel.
Keywords: affinity selection, biotin, biotin ligase, BirA, E. coli, phage-display, protein labeling, protein-protein interactions, streptavidin, streptavidin coated magnetic beads
1. Introduction
Recombinant proteins are typically overexpressed in heterologous hosts (i.e., bacteria, insect cells, mammalian cells, plants) with “fusion tags”, which are short peptides, protein domains, or entire proteins which can be fused to proteins of interest, with the goal of imparting the biochemical properties of the fusion tag to the protein of interest. This is done at the genetic level by fusing the gene of interest to the gene encoding the fusion tag of interest, resulting in the expression of a single protein fused to the tag. In general, the type of fusion tag used is dictated by its application. Short peptide tags (e.g., six-histi-dine, epitopes, StrepTag, calmodulin-binding peptide) regularly serve to permit facile purification of the recombinant protein, permit detection of the fusion protein, or to direct interaction of the recombinant protein with other proteins or inert surfaces. Larger fusion partners, such as protein domains (e.g., chitin-binding domain) or proteins (e.g., cu-tinase, green fluorescent protein (GFP), glutathione-S-transferase (GST), intein, maltose binding protein (MBP), are commonly used to promote folding, solubility, purification, labeling, chemical ligation, or immobilization of the recombinant protein. If desired, the fusion tag can be detached from the protein of interest by cleavage of a linker region with a site-specific protease, which does not cleave the protein of interest.
One popular tag for detecting recombinant and native proteins is the small molecule biotin, which is a component of the vitamin B2 complex. It binds with high affinity to the chicken egg white protein, avidin, and the fungal protein, streptavidin. (Note that a degly-cosylated, recombinant form of avidin, with near-neutral isoelectric point (i.e., pI = 6.3) that minimizes nonspecific interactions, is commercially distributed as "neutravidin".) Avidin and streptavidin are tetrameric proteins that bind four molecules of D-biotin extremely tightly (i.e., dissociation constant of ~10−15 M) (1, 2). Proteins can be modified (i.e., biotinylated) with biotin very easily in vitro with chemical reagents, which are linked to biotin, under relatively mild condition that do not affect protein stability, three-dimensional structure, or function.
However, two drawbacks of in vitro chemically biotinylated target protein is that the number of biotin added is not uniform and the modification of certain lysine residues may lead to inactivation of the binding site(s). In E. coli, the biotin carboxy carrier protein (BCCP) is biotinylated by BirA (3), a biotin ligase which covalently attaches a biotin to the amino group of a lysine residue present within the recognition sequence within BCCP (4). A minimal biotinylation sequence has been found from screens of combinatorial peptide libraries; this 13 amino acid peptide (5), along with a 15 amino acid long variant (6), termed the AviTag™, have been identified as effective in vivo and in vitro substrates for the BirA enzyme. When targets proteins are fused to the AviTag and co-expressed in vivo along with BirA, they can be biotinylated in bacteria (7-9), yeast (10-12), insect (13), or mammalian cells (14, 15). Furthermore, when recombinant proteins are fused to the AviTag and incubated in vitro with purified BirA, they can be bi-otinylated efficiently on the central lysine residue in the AviTag (16, 17).
To generate biotinylated proteins in the laboratory, one has several options. If the protein is not available but is found to express well in E. coli, then it may be expedient to construct recombinant DNA in which its coding region is fused with the AviTag at its N- or C-terminus. (While there is a single biotin attached per protein molecule, the efficiency of biotinylation typically ranges between 50 and 80%.) Alternatively, if the protein is already available in sufficient amounts, then one can chemically biotinylate the protein prior to affinity selection experiments. (Typically, 100% of the molecules will be labeled, with one or more biotins one of the lysine residues.) Finally, one can construct recombinant DNA with the AviTag fused at the protein's N- or C-terminus, express it and purify it from E. coli, and then biotinylate the protein in vitro with purified BirA. (Typically, 80–100% of the target protein is biotinylated in vitro.) All three approaches are described herein.
2. Materials
2.1. Reagents
Ampicillin (Sigma-Aldrich Chemical Company, St. Louis, MO)
Anti-Fab-HRP antibody (Jackson ImmunoResearch Laboratories, West Grove, PA)
Autoinducing medium (described in (18); can be purchased from Novagen, Madison, WI)
D-biotin (Sigma-Aldrich)
BirA enzyme (Avidity, Boulder, CO)
BugBuster detergent (Novagen)
E. coli strain BL21 (DE3) (Novagen)
EZ-Link® Sulfo-NHS-LC-biotin (Pierce Chemical Company, Rockford, IL; MW = 557n daltons)
Immobilized metal affinity chromatography (IMAC) resin (Qiagen, Valencia, CA)
LB+ampicillin: Luria Broth (10 gm yeast extract, 10 gm peptone, and 5 gm NaCl in one liter of water; autoclaved) plus 100 μg/ml ampicillin
LB+ampicillin+chloramphenicol: LB+ampicillin plus 12.5 μg/mL chloramphenicol
MagnaBind™ Streptavidin Beads (Pierce Chemical Company)
pBirA Cmr biotinylation plasmid (Avidity)
pMCSG16 and pMCSG17 vectors (described in (9); available upon request)
Phosphate Buffered Saline (PBS: 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 1.5 mM KH2PO4)
p-nitrophenyl phosphate (Sigma-Aldrich)
Qiagen Gel Extraction Kit (Qiagen, Valencia, CA)
Slide-A-Lyzer™ dialysis cassettes (Pierce)
Sma I (New England Biolabs, Waverly, MA)
Streptavidin (Sigma-Aldrich Chemical Company, St. Louis, MO)
Streptavidin-alkaline phosphatase (Sigma-Aldrich)
T4 DNA polymerase (Promega Corporation, Madison, WI)
Zebra spin-columns (Pierce)
2.2. Construction of recombinant plasmids encoded protein fusions to the AviTag
To generate biotinylated proteins for affinity selection experiments, one can transfer the open reading frame (ORF) of a protein of interest into plasmids that contain both the AviTag biotinylation sequence and a six-histidine tag, at either the N- or C- terminus of the ORF. The AviTag encodes the peptide sequence, GLNDIFEAQKIEWHE, where the underlined lysine residue is biotinylated by BirA. The two bacterial expression vectors, pMCSG16 and pMCSG17, also contain a ligation independent cloning (LIC) site for efficient cloning of the ORFs, which allows high-throughput cloning, expression, in vivo bi-otinylation, purification, and streptavidin/avidin immobilization of target proteins for affinity selection of phage-displayed libraries (9). Expression of the target-AviTag fusion protein is under the control of the T7 RNA polymerase promoter, which is under the transcriptional control of the LacZ promoter in E. coli strain BL21 (DE3). To produce enough BirA in the bacterial cells, they also contain the pBirA Cmr plasmid (16), which carries resistance to chloramphenicol and a compatible origin of replication. A protocol for generating the recombinants in pMCSG16 and pMCSG17 is briefly described below, with more extensive protocols found elsewhere (9), Dr. Frank Collart's publication in this book).
2.2.1. Preparation of the vector DNA
Digest 5 μg of pMCSG16 and pMCSG17 DNA with the restriction enzyme, Ssp I, which linearizes the plasmid DNA in the center of the LIC site. Check an aliquot for complete digestion by agarose gel electrophoresis.
Purify the linearized DNA by passing it through a YM-100 column (Millipore) for enzyme removal and buffer exchange.
Treat the linearized DNA with T4 DNA polymerase (1 unit/μg of DNA) for 2 hours (hr) in the appropriate buffer with 2.5 mM dGTP. Based on the nucleotide sequences adjacent to the Ssp I site in either vector, the proof-reading exonuclease activity of the enzyme will trim back 15 nucleotides from the 3' termini of the linearized DNA.
Purify the treated vector DNA in a 0.5% agarose gel, and recover DNA using the Qi-agen Gel Extraction Kit. Quantify recovery of the DNA spectroscopically and store at -20°C.
2.2.2. Preparation of the insert
Amplify the coding region of the target protein by polymerase chain reaction (PCR). Design the oligonucleotide primers with the sequence 5'-TACTTCCAATCCAATG-GC-3' followed by the nucleotides encoding the target protein. The anti-sense primers should begin with the sequence 5'-TCCACTTCCAATGGA-3' followed by the reverse complement of the 3' end without a stop codon (TAA). The same PCR products, without the TAA codon in the LIC overhang, can be cloned in either pMCSG16 or pMC-SG17 vector, where the AviTag is N- or C-terminal to the cloning site, respectively. (Generally, both vectors are often used to construct recombinant DNA, just in case the N-terminal or C-terminal fusion interferes with protein folding or access to a binding site in the target protein.)
Resolve the PCR product on an agarose gel and purify the desired fragment with the Qiagen Gel Extraction Kit. Quantify recovery of the DNA fragment spectroscopically and store at −20°C.
Incubate the fragment with T4 DNA polymerase in the appropriate buffer with 2.5 mM dCTP for 2 hr, at 37°C.
The T4 polymerase-treated PCR product is purified and stored in 10 mM Tris-HCl (pH 8.0).
2.2.3. Construction of recombinant plasmids
Mix the T4 polymerase-treated PCR products with treated vector, heat to 70°C for 5 min, and allow to cool to room temperature for 30 min. Annealed vector plus insert is then transformed into the E. coli strain BL21 (DE3), which contains the biotin ligase expression plasmid, pBirA Cmr.
Select transformants on Petri plates containing LB+ampicillin+chloramphenicol. Grow overnight at 37°C.
Confirm recombinants by PCR and agarose gel electrophoresis. Under ideal conditions, the rate of recombinants is >90%.
Verify the construction of the recombinant plasmids by DNA sequencing. Store bacterial clones in LB+ampicillin+chloramphenicol with 20% glycerol, at −80°C.
3. Methods
3.1. In vivo enzymatic biotinylation of proteins
To biotinylate of target proteins in E. coli, it is important to grow the expression plasmids in bacteria that contain the pBirA Cmr biotinylation plasmid (16). Without this plasmid, the levels of endogenous BirA enzyme are inadequate to get more than 5% of the overex-pressed protein biotinylated. It is also important to add biotin to the culture medium at the time of overexpression of the recombinant protein, to ensure that sufficient amounts of this molecule are available for post-translational modification of the target protein. A typical protocol for in vivo labeling of AviTagged proteins in bacteria (Fig. 1) consists of the following.
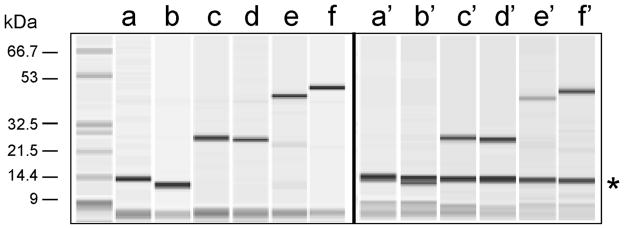
Figure 1. Fractionation of three different AviTagged and His6-tagged proteins.
Three different recombinant proteins in pMCSG16 and pMCSG17 vectors, which contain a His6-tag at the N-terminus, and AviTag at either the N-terminus or C-terminus, respectively, were expressed in E. coli with the pBirA Cmr plasmid present. Total bacterial cell lysates were mixed with streptavidin-coated magnetic beads, tumbled, washed, and bound material resolved by capillary electrophoresis with an Agilent Bioanalyzer 2100. Lanes a-b, c-d, and e-f correspond to human SrcSH3 domain, human superoxide dis-mutase (SOD), and the Bacillus subtilis APC1259 protein with AviTags at their N- and C-termini, respectively, which were purified by IMAC. Lanes a'-f' contain the same protein samples, excepted purified on streptavidin-coated magnetic beads. The asterisk refers to streptavidin monomer released from the streptavidin coated magnetic beads when boiled. This figure is modified from another publication (9).
Inoculate a 2 mL culture of LB+ampicillin+chloramphenicol with bacteria from an individual colony. Shake overnight at 37°C.
Transfer 1 mL of the stationary phase culture to a flask containing 100 mL of autoin-ducing medium, 100 μg/mL ampicillin, 12.5 μg/mL chloramphenicol, and 20 μM D-biotin. Shake overnight at 37°C.
Pellet the cells, and discard the culture medium.
Lyse the cell pellet with BugBuster detergent, according to the manufacturer's instructions.
Recover the recombinant protein by immobilized metal affinity chromatography (IMAC).
If desired, dialyze away the free imidazole and store the protein in PBS.
Determine the percent of the protein sample that is biotinylated (see Section 3.5).
3.2. In vitro chemical biotinylation of proteins
Dissolve the protein sample in PBS or some amine-free buffer. If the sample is in a Tris or amine-containing buffer such as imidazole, exchange it for an amine-free buffer by dialysis or with the use of a spin column.
-
Calculate the amount of biotin solution to add to the protein sample. The extent of bi-otinylation can be controlled by the molar ratio of biotin to protein. As a general rule, for protein samples at 2-10 mg/mL, a ≥ 12-fold molar excess of biotin should be added. For samples at ≤ 2 mg/mL, a ≥ 20-fold molar excess of biotin should be added. It is important to know the volume (mL), concentration (mg/mL), and molecular weight (Daltons) of the protein sample.
First, determine the millimoles of biotin reagent to add to the protein.
mmol Biotin = mL of protein × mg protein× mmol protein × 12-20 mmol Biotin
Then, calculate the volume of 10 mM biotin solution to add to the reaction.
μL Biotin = mmol Biotin × 1,000,000 μL × L
Before use, remove biotin reagent from freezer and bring to room temperature before opening to prevent excess moisture from entering the vial. Immediately before use, prepare a 10 mM solution of the biotin reagent in water. EZ-Link Sulfo-NHS-LC-Biotin (Sulfosuccinimidyl-6-(biotinamido) hexanoate) is an excellent amine reactive biotinylation reagent with a 22.4 Å long linker.
According to the above calculations, add the appropriate volume of biotin solution to the protein sample.
Incubate on ice for two hr, or at room temperature for 30 minutes.
Stop the biotinylation reaction with the addition of 10 mM glycine (pH 7.2).
Eliminate free biotin (see Protocol 3.4).
Determine the protein concentration by optical absorbance or through SDS-PAGE (see Protocol 3.5).
3.3. In vitro enzymatic biotinylation of proteins
For subsequent BirA biotinylation exchange purified protein to 10 mM Tris, pH 8.0. Glycerol, NaCl. As ammonium sulfate inhibits BirA activity, this chemical should be eliminated or minimized through dialysis.
Incubate 100 μg of protein in 50 mM bicine, pH 8.3, 10 mM ATP, 10 mM Mg(OAc)2, 50 mM biotin with 15 units of BirA for 1 h at 30°C. At protein concentration higher than 40 mM, additional biotin should be supplemented to keep the molar ratio of protein to biotin at ~1:1.
Eliminate free biotin (see Protocol 3.4).
Determine the protein concentration by optical absorbance or through SDS-PAGE.
Determine the percent biotinylation of the protein sample (see Section 3.5; Fig. 2).
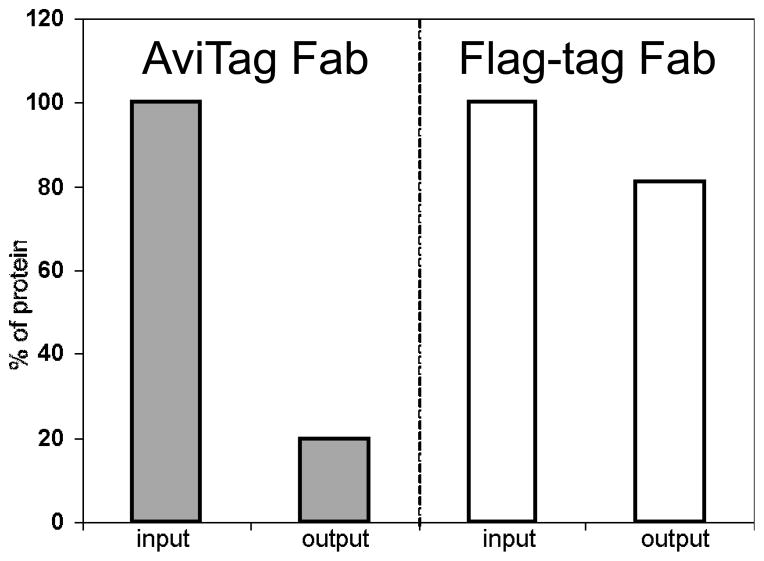
Figure 2. BirA can efficiently attach biotin in vitro to a protein carrying an AviTag.
In one form of the Herceptin Fab, which binds to the ectodomain of the ErbB2 membrane protein (26, 27), the AviTag is at the C-terminus of the light chain, and in another form, an epitope-tag, in which the Flag Tag (28) replaces the AviTag. Both forms of Fab were overexpressed in E. coli, purified by Protein G chromatography, incubated with BirA in vitro, mixed with magnetic streptavidin-coated beads (input), tumbled for one h at 4°C, centrifuged and unbound fractions (output) were collected. (Under these conditions, the AviTagged, but not the Flag-tagged, form of Fab should become biotinylated.) The protein input and output samples were then incubated in microtiter plate wells coated with ErbB2 (gracious gift of Dr. Daniel Leahy, Johns Hopkins University, Baltimore, MD) for 2 hr, and immune complexes in the wells were detected with anti-Fab-HRP antibody. The Y-axis refers to the percent protein bound to antigen in the output relative to the input (100%) fractions, as tested by ELISA. As seen in the histograms, ~80% of the Fab-AviTag fusion could be biotinylated in vitro with little or no negative effect on binding (29, 30), and we interpret the small amount of binding of the non-biotinylated form of the protein (i.e., Fab-Flag tag) to the streptavidin coated magnetic beads to be due to non-specific binding.
3.4. Elimination of free biotin
3.4.1. By dialysis
Pierce has developed a Slide-A-Lyzer™ dialysis cassette that can conveniently remove low molecular weight contaminants and salts. Determine the molecular weight and available volume of your protein to choose an appropriate dialysis cassette size and membrane capacity for the molecular weight cut-off (MWCO). Avoid choosing a membrane with a MWCO that is too close to the size of your protein; this can cause some loss of sample. To protect the physical integrity of the dialysis membrane, use precaution when handling the dialysis cassette and touch only the plastic frame.
As a quality assurance that there is no leakage in the cassette, inject sterile distilled water into the cassette to check for leaks and remove after visual inspection prior to the addition of protein sample.
If the dialysis cassette size requires hydration or a low volume of sample is used, remove the cassette from its pouch, slip it into the accompanying buoy, and immerse in dialysis buffer for 30 sec. Remove the cassette from the buffer and gently tap the edge on a paper towel to remove excess liquid.
Attach a 18-gauge, 1 inch beveled needle (or 21-gauge, 1-inch beveled needle) to the Luer-Lock of the syringe by screwing it into place.
Remove the protective sheath from the needle and draw a small volume of air into the syringe to void the syringe’s dead volume. Immerse the needle into your sample and then slowly draw back on the syringe piston.
Remove the dialysis cassette from the buoy and penetrate the gasket with the needle through one of the syringe ports located at the top corners of the cassette. The needle should penetrate the gasket to a minimal extent to avoid puncturing the membrane. Inject the sample slowly to avoid foaming.
Before removing the needle from the cassette, draw up on the syringe piston to remove air from the cassette cavity. This ensures the sample solution contacts the greatest amount of surface area. At the same time, avoid contacting the needle to the membrane. Mark the corner of the cassette with a permanent marker to identify the port injected.
Reattach the buoy and float in a beaker with 200-500 times the volume of your sample of dialysis buffer. Dialyze at room temperature for 2 h, change the dialysis buffer and dialyze for another two hr. Finally, change the dialysis buffer and dialyze overnight at 4°C. Keeping the solution in constant stirring can speed up the dialysis process.
To remove the sample after dialysis, use a new syringe and needle and fill the syringe with air at least equal to the volume of your sample size. Penetrate the gasket at an unused syringe port. Discharge the air into the cavity to separate the membrane to prevent from piercing the membrane. Insert only the tip of the needle through the syringe port.
Rotate the cassette so that the syringe and the sample are on the bottom and slowly draw back on the syringe piston to capture the dialyzed sample. Remove the needle and discard the dialysis cassette. Expel the sample into a new tube.
3.4.2. With a microcon device
Determine the molecular weight of your protein to choose an appropriate filter membrane capacity for the nominal molecular weight limit (NMWL). Avoid choosing a membrane with a NMWL that is too close to the size of your protein; this can cause some loss of sample. It should be noted, that generally some of the protein sample is lost due to sticking to the membrane.
Assemble the microcon centrifugal filter device by inserting the sample reservoir into the microfuge tube.
Pipette sample solution into sample reservoir. Do not add more than 500 μL into the sample reservoir. Avoid touching the membrane at the bottom of the sample reservoir with pipette tip. Close the tube with attached cap.
Place assembled filter device with sample into a compatible centrifuge aligning the cap strap toward the center of the rotor.
Centrifuge using appropriate spin times according to the NMWL at 4°C. Repeat two times adding more solvent to 500 μL and discarding the flow through before centrifugation.
Remove assembled filter device from centrifuge and separate the sample reservoir from the tube. Carefully invert the sample reservoir into a new tube.
Centrifuge for 3 min at 1000 × g to collect concentrated sample. Notice that the attached cap will not close when the sample reservoir is inverted. To prevent the cap from breaking off, align the cap to the center of the rotor so that it is flush against the rotor.
3.5. Verification of protein biotinylation
Biotinylation of proteins can be monitored by three different means:
An enzyme linked immunosorbent assay (ELISA) format is a quick method for demonstrating that the target protein is biotinylated. First, biotinylated proteins are immobilized onto polystyrene microtiter plates, and after washing away the free protein, non-specific binding of proteins is blocked with excess BSA, and the wells incubated with streptavidin alkaline-phosphatase (1 μg/mL in PBST, one h incubation). The amount of streptavidin-enzyme conjugate retained in the wells is detected using p-nitrophenyl phosphate, with the yellow color (measured at 405 nm wavelength) proportional to the amount of biotinylated protein adsorbed to the microtiter plate well surface.
The number of biotins per protein molecule can be determined using a kit (Pierce Chemical Co., # 28005). In the kit, the 4′-hydroxyazobenzene-2-carboxylic acid (HABA) dye binds to avidin to produce a yellow-orange color, which absorbs at 500 nm wavelength. Biotin conjugated to a protein will displace the dye and cause the absorbance to decrease; a standard curve can be established using the free biotin, permitting the number of moles of biotin incorporated (after biotinylating a protein) to be estimated. A similar kit, but more sensitive (based on the displacement of a ligand tagged with a quencher dye from the biotin-binding sites of Biotective™ Green reagent), is available from Invitrogen (# F30751).
The presence of non-biotinylated proteins in a sample can be estimated by following the binding of the sample to streptavidin coated magnetic beads by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A sample can be incubated with streptavidin coated magnetic beads for 30 minutes and the amount of non-bound protein can be compared to input (Fig. 3). In a successful biotinylation experiment, 100% of the biotinylated protein should be removed from sample, assuming that the streptavidin coated magnetic beads are in excess to the amount of biotin attached to the protein, compared to a much lower value for non-biotinylated protein, which is non-specifically stuck to the beads.
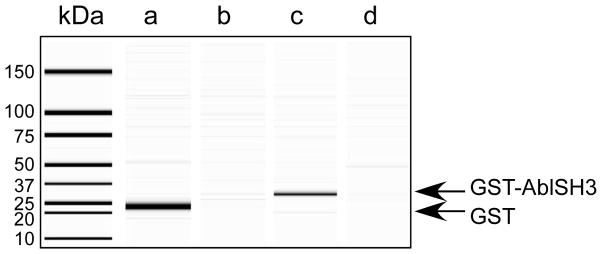
Figure 3. Chemical biotinylation of two proteins in vitro and confirmation that they bind to streptavidin coated magnetic beads.
Purified Glutathione-S-transferase (GST) and GST-Abl SH3 domain fusion proteins were chemically biotinylated with EZ-Link Sul-fo-NHS-LC-Biotin (Sulfosuccinimidyl-6-(biotinamido) hexanoate). After quenching the reagent with excess glycine and spinning the sample in a microconcentrator to remove free biotin, the proteins were separately mixed with streptavidin coated magnetic beads. Input and bound material was then resolved by capillary electrophoresis using a BioRad Experion. The left hand lane contains molecular weight standards listed in kilodaltons (kDa). Lanes a-d resolves (a) biotinylated GST, (b) biotinylated GST that did not bind to the streptavidin coated magnetic beads (i.e., after it was tumbled with streptavidin-coated beads), (c) GST-AblSH3 domain fusion protein, and (d) GST-AblSH3 domain fusion protein that did not bind to streptavidin-coated magnetic beads.
4. Notes
From our experience, we find we can achieve 50-80% biotinylation of our in vivo expressed AviTagged proteins. Generally, this level of biotinylation is sufficient for our needs, as one can conveniently capture the biotinylated target protein from lysed E. coli directly on streptavidin- or avidin-coated microtiter plate wells or streptavidin coated magnetic beads, without prior purification (9). If 100% biotinylation of a particular protein is required, we recommend either enzymatic biotinylation of an AviTagged protein or chemical biotinylation, although the number of biotins attached to the protein will differ.
For labeling purposes, it is possible to incorporate different forms of biotin or biotin containing labeling agents. For example, the biotin analog, desthiobiotin, which retains the uredio ring of biotin, but lacks the 5-member sulfur-containing thiophene ring, can be a substrate for efficient in vitro desthiobiotinylation by BirA (17).The use of desthio-biotin offers a gentle method for affinity purifying desthiobiotinylated proteins, as they can be eluted off avidin or streptavidin matrices by competition with biotin (19). There are also several analogs of biotinylation reagents available from Pierce Chemical Company available for use in chemical biotinylation, such as EZ-Link NHS-Chro-mogenic Biotin, which contains a chromophore that absorbs strongly at 354 nanome-ters and allows accurate measurement of the number of biotins attached to a protein sample, and EZ-Link NHS-SS-Biotin, which allows release of the biotin tag with exposure to reducing agents. Finally, it should be noted that there are a variety of other strategies for adding biotin to a protein (reviewed in (20)), including translation with bi-otinylated puromycin (21, 22), intein-mediated site-specific biotinylation (23), and use of novel peptide tags (24) or protein domains (25), which are substrates for post-translational modification with biotin-derivatized compounds.
Acknowledgments
The authors acknowledge intellectual contributions by Mr. Michael Scholle and Dr. Frank Collart, and financial support from the National Institutes of Health (1R01 GM079096, P01 GM075913, 1U54 CA119343).
References
- 1.Wilchek M, Bayer EA, Livnah O. Essentials of biorecognition: the (strept)avidin-biotin system as a model for protein-protein and protein-ligand interaction. Immunol Lett. 2006;103:27–32. doi: 10.1016/j.imlet.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Laitinen OH, Nordlund HR, Hytonen VP, Kulomaa MS. Brave new (strept)avidins in biotechnology. Trends Biotechnol. 2007;25:269–277. doi: 10.1016/j.tibtech.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Chapman-Smith A, Cronan JEJ. Molecular biology of biotin attachment to proteins. J Nutr. 1999;129:477S–484S. doi: 10.1093/jn/129.2.477S. [DOI] [PubMed] [Google Scholar]
- 4.Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26:1414–1420. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatz P. Use of peptide library to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specified biotinylation in Escherichia coli. Bio/Tech. 1993;11:1138–1142. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 6.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashraf SS, Benson RE, Payne ES, Halbleib CM, Gron H. A novel multi-affinity tag system to produce high levels of soluble and biotinylated proteins in Escherichia coli. Protein Expr Purif. 2004;33:238–245. doi: 10.1016/j.pep.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Penalva LO, Keene JD. Biotinylated tags for recovery and characterization of ribonucleoprotein complexes. BioTechniques. 2004;37:604, 606, 608–10. doi: 10.2144/04374ST05. [DOI] [PubMed] [Google Scholar]
- 9.Scholle MD, Collart FR, Kay BK. In vivo biotinylated proteins as targets for phage-display selection experiments. Protein Expr Purif. 2004;37:243–252. doi: 10.1016/j.pep.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Athavankar S, Peterson BR. Control of gene expression with small molecules: biotin-mediated acylation of targeted lysine residues in recombinant yeast. Chem Biol. 2003;10:1245–1253. doi: 10.1016/j.chembiol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Parthasarathy R, Bajaj J, Boder ET. An immobilized biotin ligase: surface display of Escherichia coli BirA on Saccharomyces cerevisiae. Biotechnol Prog. 2005;21:1627–1631. doi: 10.1021/bp050279t. [DOI] [PubMed] [Google Scholar]
- 12.Scholler N, Garvik B, Quarles T, Jiang S, Urban N. Method for generation of in vivo biotinylated recombinant antibodies by yeast mating. J Immunol Methods. 2006 doi: 10.1016/j.jim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 14.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viens A, Mechold U, Lehrmann H, Harel-Bellan A, Ogryzko V. Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal Biochem. 2004;325:68–76. doi: 10.1016/j.ab.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- 17.Wu SC, Wong SL. Development of an enzymatic method for site-specific incorporation of desthiobiotin to recombinant proteins in vitro. Anal Biochem. 2004;331:340–348. doi: 10.1016/j.ab.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 18.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal Biochem. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]
- 20.Chattopadhaya S, Tan LP, Yao SQ. Strategies for site-specific protein biotinylation using in vitro, in vivo and cell-free systems: toward functional protein arrays. Nat Protoc. 2006;1:2386–2398. doi: 10.1038/nprot.2006.338. [DOI] [PubMed] [Google Scholar]
- 21.Starck SR, Green HM, Alberola-Ila J, Roberts RW. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem Biol. 2004;11:999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Agafonov DE, Rabe KS, Grote M, Voertler CS, Sprinzl M. C-terminal modifications of a protein by UAG-encoded incorporation of puromycin during in vitro protein synthesis in the absence of release factor 1. Chembiochem. 2006;7:330–336. doi: 10.1002/cbic.200500358. [DOI] [PubMed] [Google Scholar]
- 23.Tan LP, Lue RY, Chen GY, Yao SQ. Improving the intein-mediated, site-specific protein biotinylation strategies both in vitro and in vivo. Bioorg Med Chem Lett. 2004;14:6067–6070. doi: 10.1016/j.bmcl.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 24.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 25.Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 26.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 27.Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J Mol Biol. 2002;320:415–428. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 28.Einhauer A, Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods. 2001;49:455–465. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 29.Saviranta P, Haavisto T, Rappu P, Karp M, Lovgren T. In vitro enzymatic biotinylation of recombinant fab fragments through a peptide acceptor tail. Bioconjug Chem. 1998;9:725–735. doi: 10.1021/bc9800217. [DOI] [PubMed] [Google Scholar]
- 30.Sibler AP, Kempf E, Glacet A, Orfanoudakis G, Bourel D, Weiss E. In vivo biotinylated recombinant antibodies: high efficiency of labelling and application to the cloning of active anti-human IgG1 Fab fragments. J Immunol Methods. 1999;224:129–140. doi: 10.1016/s0022-1759(99)00016-2. [DOI] [PubMed] [Google Scholar]