Abstract
Faithful replication and DNA repair are vital for maintenance of genome integrity. RAD51 is a central protein in homologous recombination repair and during replication, when it protects and restarts stalled replication forks. Aberrant RAD51 expression occurs in glioma, and high expression has been shown to correlate with prolonged survival. Furthermore, genes involved in DNA damage response (DDR) are mutated or deleted in human glioblastomas, corroborating the importance of proper DNA repair to suppress gliomagenesis. We have analyzed DDR and genomic instability in PDGF-B–induced gliomas and investigated the role of RAD51 in glioma development. We show that PDGF-B–induced gliomas display genomic instability and that co-expression of RAD51 can suppress PDGF-B–induced tumorigenesis and prolong survival. Expression of RAD51 inhibited proliferation and genomic instability of tumor cells independent of Arf status. Our results demonstrate that the RAD51 pathway can prevent glioma initiation and maintain genome integrity of induced tumors, suggesting reactivation of the RAD51 pathway as a potential therapeutic avenue.
Keywords: genomic instability, glioma, PDGF-B, RAD51, replicative stress
Cancer is caused by accumulated genetic changes. Failure to protect cells from these may lead to genomic instability, a common feature of nearly all solid tumors.1 Faithful replication is essential to maintain genome integrity. Replication forks frequently encounter obstacles, and a stalled replication fork, if left unresolved, can collapse and may lead to a double-strand break (DSB).2 The major error-free pathway of DSB repair is homologous recombination (HR), in which an identical sister chromatid is used as template during repair. The HR pathway is also involved in replication, repairing errors at replication forks.2,3 Proteins involved in HR are essential for cell survival, because most HR genes cause embryonic lethality when deleted in mice.4–6 Moreover, hereditary mutations in HR genes (eg, BRCA1 and BRCA2) strongly predispose to cancer.7–9 Therefore, functional HR pathways are important for maintenance of genome integrity and prevention of tumor development.
RAD51 is central for HR and necessary for homology search and strand exchange at sites of DSBs. RAD51 nucleoprotein filaments form around single-stranded DNA (ssDNA) at sites of DSBs and will catalyze a DNA strand exchange reaction between the ssDNA and the homologous double-stranded DNA of the sister chromatid.10,11 In addition, it was recently shown that RAD51 and γ-H2AX could participate in restarting stalled replication forks.12,13 Thus, RAD51 is important for both preventing and repairing DSBs.
Elevated levels of RAD51 are common in a wide variety of tumors.14 Large-scale genomic analyses of glioma, breast, and colorectal cancers did not identify any mutations in RAD51, implying it to be a rare event in human cancer.15–17 In glioma most reports support a suppressive role of RAD51. In an array-CGH screen of 42 human gliomas, a minimal deletion comprising RAD51, TP53BP, and FANCG was detected in a subset of the tumors.18 Expression of RAD51 mRNA was determined in 40 astrocytomas of grade II–IV and was found not to differ significantly from normal brain samples.19 However, a study including 68 patients with glioblastoma showed that elevated RAD51 protein expression at initial diagnosis, as well as at recurrence, correlated with significantly increased survival duration.20 Several in vitro studies have shown that overexpression of RAD51 can decrease proliferation and delay cell-cycle progression of human tumor cells,21,22 further corroborating RAD51 as a tumor suppressor. Collectively, these findings argue for a suppressive role of RAD51 in glioma development.
Gliomas are primary central nervous system (CNS) tumors with poor prognosis. They frequently exhibit alterations of the PDGF signaling pathway, with PDGFRA amplifications being the most common.17,23 With use of mouse glioma models, genes involved in HR (Rad51b and Fancc) were tagged in PDGF-B/MMLV–induced gliomas,24 PDGF-B–infected glial progenitor cells from p27Kip1-deficient mice showed genomic instability and impaired RAD51 focus formation,25 and targeted deletions of ATM or Chk2 accelerated PDGF-induced glioma development in Ntv-a mice,26 suggesting involvement of HR and RAD51 in PDGF-driven gliomagenesis.
To investigate the effect of RAD51 on genomic stability and gliomagenesis in vivo, we used the PDGF-B–induced RCAS/TV-A mouse model of glioma development.27,28 We demonstrate that overexpression of PDGF-B in glial stem/progenitor cells caused genomic instability in vitro and in vivo. To modulate DNA repair, RAD51 was transduced together with PDGF-B in neonatal Ntv-a wild-type (wt) and Arf−/− mice.29 Arf encodes p19Arf (p14ARF in human), a positive regulator of p53. Silencing of p14ARF by hypermethylation is common in grade II oligodendrogliomas,30 and because these tumors progress, the INK4a/ARF locus is frequently homozygously deleted.31 We show that high expression of RAD51 could significantly decrease the incidence of PDGF-B–induced glioma, prolong survival, and inhibit aneuploidy of PDGF-driven glioma cells.
Material and Methods
DNA Constructs
The RCAS-RAD51 construct was created by inserting the human RAD51 coding sequences (1062 bp) into pBS (Bluescript) to add correct restriction sites for subsequent ligation into the retroviral RCAS-Y vector. The pBS-RAD51 constructs were digested with NotI/ClaI, creating a fragment of ∼1.1 kb, which was cloned into the RCAS-Y vector. RCAS-RAD51 carries the wt coding sequence of human RAD51. The RCAS-PDGF-B-eGFP (called RCAS-PDGF-B in the article) construct has been described elsewhere.27
RNA Extraction and Detection of RCAS-mediated Gene Expression
Total RNA was extracted using TRIzol reagent (Invitrogen). Complementary DNA (cDNA) was made through reverse transcription of 0.5–1 µg of total RNA using random primers (S12545; New England Biolabs) and M-MuLV reverse transcriptase (New England Biolabs). One hundredng of cDNA was used for polymerase chain reaction (PCR) detection of PDGF-B of human origin from the RCAS virus using the PuReTaq Ready-To-Go PCR Beads (GE Healthcare). RNA from the U-343MGa glioma cell line was used as positive control, and RNA from primary uninfected Ntv-a wt cells was used as negative control. Primers used specific for human PDGF-B were forward primer (5′-TGCTGCTACCTGCGTCTGGTC) and reverse primer (5′-TTCTTCCACGACCCAAGCTCT) yielding a 208-bp fragment. PCR setup was 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, ending at 72°C for 10 min. PCR products were visualized on an agarose gel, and pictures were taken using the Gel Foto System 1000 (Techtum Lab).
Cell Cultures, Transfection of DF-1 Cells, and Isolation and Infection of Primary Brain Cells
To produce RCAS retroviruses, DF-1 chicken fibroblasts were transfected with the various RCAS constructs using FuGene6 (Roche Diagnostics). The transient transfection caused production of RCAS retroviruses and a subsequent viral infection of the cells. The transfected cells were cultured for at least 2 weeks before they were used in in vitro or in vivo experiments to allow for efficient viral spread throughout the culture.
Primary brain cell cultures were prepared from neonatal mouse brains of Ntv-a wt and Ntv-a Arf−/− genotype. The whole brain was aseptically dissected out, and the cerebellum was removed. The cerebrum was mechanically dissociated, by using 18- and 22-gauge needles, in DMEM supplemented with 10% fetal bovine serum, 4 mM of L-glutamine, and 1% penicillin/streptomycin. The cells were collected by centrifugation and re-suspended in N2 media (DMEM/F12 supplemented with 2.5 mM of L-glutamine, 1% penicillin/streptomycin, 8.6 mM of glucose, 1.5 mg/mL NaHCO3, 3.7 mg/mL HEPES, 0.1 mg/mL transferrin, 25 µg/mL insulin, 100 µg/mL putrescine, 30 nM of Na-selenite, and 20 nM progesterone; pH was adjusted to 7.2). The cells were plated on tissue culture dishes coated with 10 µg/mL polyornithine hydrobromide and 1 µg/mL fibronectin. The media were changed every second day, and 10 ng/mL FGF2 was added to the cells every day to select for neural progenitor cells.
Primary cells were infected with supernatants from DF-1 cells producing RCAS-PDGF-B, RCAS-RAD51, or empty RCAS (RCAS-X). Conditioned media from the respective retrovirus-producing cells were collected after 24 h, sterile filtered through 0.45-mM filters, and added to the primary Ntv-a cell cultures together with 10 ng/mL FGF2. This was repeated every day for 7 days. After RCAS-X + RCAS-PDGF-B infection, immortalized Ntv-a wt cells were cultured in DMEM with addition of 12% fetal bovine serum, 4 mM of L-glutamine, and 1% penicillin/streptomycin for >20 passages before a second round of infection was done.
Tumor cells isolated from Ntv-a wt and Arf−/− mice infected with RCAS-PDGF-B were cultured in DMEM with addition of 10% fetal bovine serum, 4 mM of L-glutamine, and 1% penicillin/streptomycin. BrdU incorporation was done by adding BrdU at a final concentration of 10 µg/mL and incubating the product for 16 h.
Proliferation Assay
Primary Ntv-a Arf−/− cells (passage 3) were infected for 14 days with different viral supernatants before seeding 2.5 × 104 cells/35-mm dish (duplicates) for proliferation assay. The number of cells per dish was determined at day 5 after plating using a Coulter counter (Coulter Electronics).
Immunocytochemical Analyses
Cells were plated on coverslips and fixed in 4% paraformaldehyde for 15 min or in ice-cold methanol-acetone (ratio, 1:1) for PCNA stainings. Cells subjected to BrdU staining were treated with 2M of HCl for 30 min before staining to allow for denaturation of DNA. After washing in PBS, cells were treated with 0.5% TritonX-100 in PBS and blocked for 1 h in PBS, 2% BSA. Incubation was done at room temperature with primary antibody, polyclonal anti-RAD51 (H-92; Santa Cruz Biotechnology), monoclonal anti-PCNA (Cell Signaling Technology), monoclonal anti-Ki67 (DAKO), or monoclonal anti-BrdU (Abcam) for 1 h. Coverslips were washed in PBS and incubated with the secondary antibody, goat polyclonal anti-rabbit fluorescein isothiocyanate (FITC; Abcam), anti-rat Alexa 555, anti-rabbit Alexa 555, anti-rabbit Alexa 488, and anti-mouse Alexa 488 (Invitrogen) in the dark for 1 h at room temperature; washed in PBS; and mounted in Vectashield with DAPI (Vector Labs) or ImmuMount (Shandon) mounting media with DAPI.
Coronary sections of formalin-fixed, paraffin-embedded mouse brains were deparaffinized by standard protocol using xylene. Heat-induced epitope retrieval in citrate buffer (pH 6.0) was used for all antibodies. Sections were blocked for 1 h in PBS 0.05% Tween (PBS-T) and 5% nonfat dry milk and incubated with primary antibody, polyclonal rabbit anti-RAD51 (H-92, Santa Cruz Biotechnology), monoclonal mouse anti-PCNA (Cell Signaling Technology), rabbit anti-phospho-histone H2A.X (Ser139; Cell Signaling Technology), and monoclonal rabbit anti-Ki67 (DAKO) at 4°C overnight. Sections were washed in PBS-T; incubated with secondary antibody, goat polyclonal anti-rabbit FITC (Abcam), and anti-mouse Rhodamine (Abcam) in the dark for 1 h at room temperature; washed in PBS-T; and mounted in Vectashield with DAPI (Vector Labs).
Infection of Ntv-a Transgenic Mice and Tumor Surveillance
DF-1 cells producing RCAS-PDGF-B, RCAS-RAD51, or RCAS-X were injected alone or in combination into the right cerebral hemisphere of neonatal Ntv-a wt and Arf−/− mice.32 Infected mice were monitored every second day and euthanized when they showed signs of illness or at 12 weeks of age. Experiments were performed in accordance with the rules and regulations of the local animal ethics committee (C18/6 and C158/8 to LU).
Histopathology and Immunohistochemical Analyses
Mouse brains were fixed in formalin and cut into 5 coronary pieces, dehydrated according to standard protocol, embedded in paraffin, coronary sectioned, and analyzed for the presence of tumors by hematoxylin and eosin staining. Immunostainings were performed using the Ventana Discovery Automated Stainer (Ventana Medical Systems). Deparaffinization was done in the machine. Heat-induced epitope retrieval in Tris Borate EDTA buffer (pH 8.0) was used for all antibodies. A streptavidin-horseradish peroxidase–based DAB kit provided by Ventana was used for detection, and slides were counterstained with hematoxylin. The antibodies used were rabbit anti-human PDGF-B (Lab Vision), rabbit polyclonal anti-Rad51 (H-92, Santa Cruz Biotechnology), rabbit polyclonal anti-PDGFRα (Cell Signaling Technology), rabbit monoclonal anti-PDGFRβ (Cell Signaling Technology), mouse monoclonal anti-PCNA (Cell Signaling Technology), and rabbit monoclonal anti-Ki67 (DAKO).
Flow Cytometry
The cells were prepared as described by Castro et al.33 and analyzed using a LSR II flow cytometer (Becton Dickinson). The data were analyzed using ModFit LT 3.2 (Verity Software House).
Feulgen Staining
Measurement of DNA content in the tumors was done using Feulgen photocytometry.34 Optical density of at least 100 nuclei per tumor was measured, and a DNA index was calculated and displayed as a histogram.11 Normal tissue and diploid tumors display a major peak at 2.0 c, and near-diploid tumors have a major peak ≤2.5c.35 Aneuploid tumors display a broader distribution of peaks spanning 2.5–6 c. The modal value represents the highest peak in each tumor and is presented as the DNA content for that tumor. A tumor-free area in the corresponding tissue section was for each tumor used as a reference for normal DNA content and set to 2.0 c.
Image Acquisition
Pictures of immunohistochemical stains were acquired using a Leica DFC320 with FireCam 1.3 at 2088 × 2558 pixels and 16 bpp. Immunofluorescence pictures for Figs 1 and 3 were taken using an AxioCamHR at 1300 × 1030 pixels, pixel dimensions of 100 × 100 × 1000 nm, and 8 bpp. Pictures for Fig. 4 were taken using a Zeiss 510 Meta Confocal microscope at 2048 × 2048 pixels, pixel dimension of 150 × 150 × 1200 nm (C, E) or 160 × 160 × 1200 nm (B, D), and 8 bpp. All images were processed using Adobe Photoshop.
Fig. 1.
Autocrine PDGF stimulation and expression of DNA damage response proteins in PDGF-B–induced mouse gliomas. (A) Glioma incidence and malignancy grade in PDGF-B–induced gliomas from Ntv-a wt and Arf−/− mice. Fischer's exact test was used to analyze the difference in incidence between wild-type and Arf−/− mice. (B–F) Representative pictures of PDGF-B–induced mouse gliomas stained for (B) viral transduced human PDGF-B, (C) PDGFRα, (D) PCNA and Ki67 (arrows indicate tumor cells positive for PCNA and negative for Ki67), (E) γH2AX, and (F) RAD51. Scale bars = 50 μm.
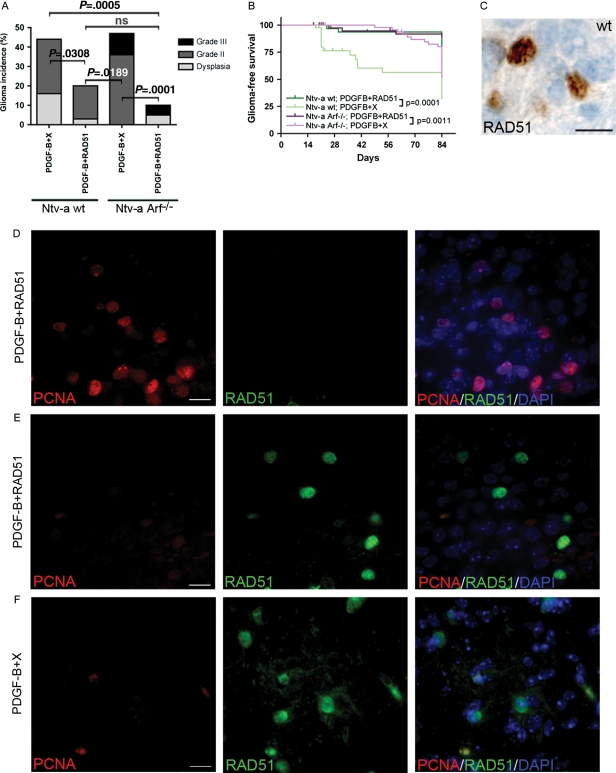
Fig. 3.
RAD51 could inhibit PDGF-B–induced glioma development in Ntv-a wt and Arf−/− mice. (A) Incidence and distribution of grade from PDGF-B + X and PDGF-B + RAD51 tumors in wt and Arf−/− mice. Fischer's exact test was used to compare incidence rates. ns indicates nonsignificant. (B) Kaplan-Meier graph showing glioma-free survival. Censored objects are depicted with a vertical bar. Survival curves were compared using the log-rank test. (C) A representative immunohistochemical staining for RAD51 in a PDGF-B + RAD51 tumor. Scale bar = 25 μm. (D and E) Representative pictures of double immunofluorescence stainings for PCNA and RAD51 in tumor tissue. Different areas of the same PDGF-B + RAD51 tumor showing one area with high PCNA expression and no RAD51 expression (D) and another area with intense nuclear RAD51 staining and no PCNA-positive cells (E). (F) A representative PDGF-B + X tumor showing high endogenous RAD51 expression and no overlap with PCNA. Scale bar = 50 μm.
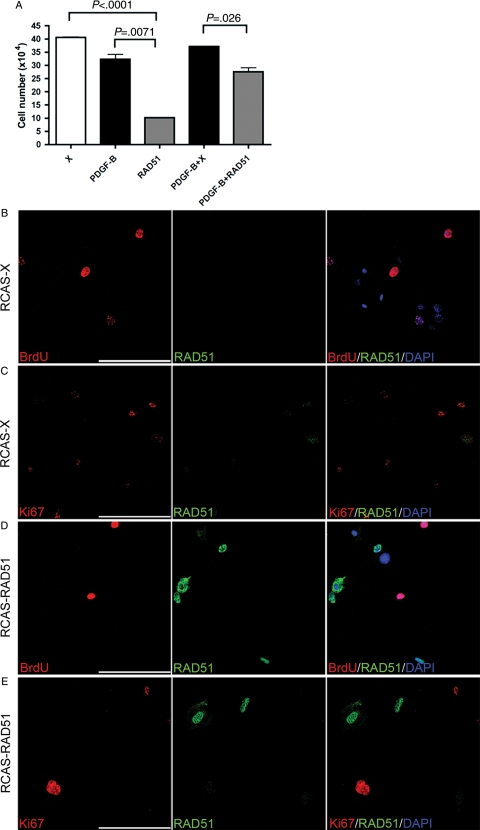
Fig. 4.
Inhibitory effect by RAD51 on in vitro proliferation of PDGF-B–infected Ntv-a wt and Arf−/− cells. (A) Total cell numbers at day 5 after infection of primary Ntv-a Arf−/− cells with the depicted viruses. Cell numbers were compared using the unpaired t test. (B–E) Double immunofluorescence stainings for RAD51 and BrdU or Ki67 on IC wt cells infected with RCAS-X (B and C) or RCAS-RAD51 (D and E). Scale bar = 100 μm.
Statistical Analyses of Tumor Numbers
Statistical analyses were performed with the GraphPad Software Prism, version 4.0a, using Fischer's exact test for the comparison of incidence rates and DNA content, χ2 test for comparisons of malignancy, log-rank test applied to Kaplan-Meier graphs, and unpaired t test for cell proliferation.
Results
PDGF-B–induced Hyperplasias and Tumors Displayed Autocrine PDGF Stimulation, Activation of DDR, and Genomic Instability
In a hyperplasia xenograft model, a combination of FGF2 (fibroblast growth factor-2), SCF (stem cell factor), and endothelin-3 have been shown to induce genomic instability in the hyperplastic lesions.36 To analyze whether genomic instability also could be induced by PDGF-B, we analyzed brain tumors induced in neonatal Ntv-a wt and Arf−/− mice with RCAS-PDGF-B+ RCAS-X (PDGF-B + X), where RCAS-X is a control virus with no exogenous gene inserted. Tumor lesions were divided into 3 groups on the basis of histopathological findings: hyperplasias, low-grade gliomas (grade II), and anaplastic gliomas (grade III). Tumor incidence and malignancy were comparable to findings from previous studies27,29 (Fig. 1A, Table 1). Hyperplasias were typically found in mice that had hydrocephalus within 5 weeks after injection and consisted of small, atypical, hyper-proliferative, Olig2-positive (data not shown) intra/peri-ventricular lesions not yet infiltrating the brain parenchyma that were considered to be early neoplastic lesions. Hyperplasias and tumors expressed PDGF-B (Fig. 1B) and PDGFRA (Fig. 1C), supporting the presence of autocrine PDGF stimulation, and co-expression of Ki67 and PCNA (Fig. 1D) suggested that PDGF-driven proliferation occurred. However, there were also cells positive for PCNA only (Fig. 1D, indicated by arrows) proposing induction of DDR in these cells.
Table 1.
Tumor incidence in Ntv-a wt and Arf−/− mice injected with combinations of RCAS-RAD51 (RAD51), RCAS-PDGF-B (PDGF-B), and RCAS-X (X)
| Arf status | RCAS | No. of mice | No. of tumors | Incidence, % |
|---|---|---|---|---|
| wt | RAD51 | 39 | 0 | 0 |
| wt | PDGF-B + X | 43 | 19 | 44 |
| wt | PDGF-B + RAD51 | 35 | 7 | 20 |
| Arf−/− | RAD51 | 41 | 0 | 0 |
| Arf−/− | PDGF-B + X | 47 | 22 | 47 |
| Arf−/− | PDGF-B + RAD51 | 42 | 4 | 9 |
To further analyze whether DDR activation occurred in PDGF-B–induced tumors, we performed immunostainings for DDR markers γ-H2AX and RAD51. The majority of hyperplasias and tumors had areas positive for γ-H2AX (Fig. 1E) and RAD51 (Fig. 1F), whereas adjacent normal brain tissue samples tested negative for these proteins (data not shown). This result suggested that overexpression of PDGF-B activated DDR early in tumor development. RAD51 expression was present in tumors from both Ntv-a wt and Arf−/− mice, but it occurred more frequently and intensely in Ntv-a Arf−/− tumors. A putative explanation could be an inability of p53 to repress RAD51 expression in Arf−/− mice.37
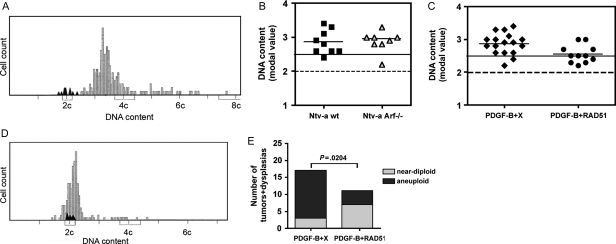
Genomic stability of PDGF-B–induced tumors was analyzed by Feulgen staining.34 This is a recognized technique used in the clinic to assess ploidy in formalin-fixed, paraffin-embedded human tumor material. Feulgen staining enabled us to estimate the DNA content on a cell-to-cell basis in tumor tissue, and the result for each tumor was presented as a histogram (Fig. 2A). Analysis of modal values of PDGF-B + X induced hyperplasias (n = 2) and tumors (n = 15) in Ntv-a wt and Arf−/− mice showed that all samples but 2 had an aneuploid DNA content (Fig. 2B). Two tumors had a near-diploid DNA content (1 each from wt and Arf−/−). Interestingly, both hyperplasias were aneuploid. This indicated that PDGF-B could induce aneuploidy in experimental gliomas and implied that it is an early event in tumorigenesis.
Fig. 2.
Assessment of genomic instability by Feulgen analysis on hyperplasias and tumors. (A) A typical histogram for a PDGF-B + X tumor. Black peaks depict the reference DNA content of normal brain cells in the same tissue section. (B) Modal values for PDGF-B + X hyperplasias and tumors (depicted by squares and triangles) and mean for each group (depicted by short black lines) are indicated. The dotted line indicates the DNA content of normal diploid cells, and the solid line indicates the maximum value for near-diploid cells. (C) Modal values for PDGF-B + X (diamonds) and PDGF-B + RAD51 (circles) hyperplasias and tumors where the results from wild-type and Arf−/− mice have been combined. The dotted line indicates the DNA content of normal diploid cells, and the solid line indicates the maximum value for near-diploid cells. (D) A histogram of a near-diploid PDGF-B + RAD51 tumor with a modal value of 2.2c. Black peaks represent normal diploid cells analyzed in a tumor-free area of the same tissue section. (E) Fischer's exact test shows a significant difference in distribution of near-diploid and aneuploid dysplasias + tumors between the PDGF-B + X and PDGF-B + RAD51 groups.
PDGF-B Caused Genomic Instability in Cultured Tumor Cells and Primary Glial Cells
Because, to our knowledge, PDGF-B has not previously been shown to induce genomic instability, we wished to further substantiate our findings by analyzing the DNA content of cultured tumor cells derived from PDGF-B–induced mouse gliomas. We used tumor cells from both Ntv-a wt (cultures # 2164 and 2185) and Ntv-a Arf−/− (cultures # 1769 and 2167) mice. To verify that cultured cells were tumor-derived, expression of human PDGF-B mRNA in the tumor cell cultures was confirmed (Supplementary Fig. 1A). DNA FACS analysis was performed on uninfected primary Ntv-a wt cells and on early passage tumor cell cultures. Cultures 1769 and 2167 were found to be diploid (data not shown) when compared to primary Ntv-a wt cells (diploid control, Supplementary Fig. 1C). However, culture 2164 showed clear aberrant DNA content (Supplementary Fig. 1C), and culture 2185 showed an increased peak at 4 c and a small extra population of cells at 8 c, indicating presence of tetraploid cells in this cell population (Supplementary Fig. 1D). Tetraploidy has been suggested to trigger chromosomal instability and eventually aneuploidy.38 Taken together, our data from cultured tumor cells support the hypothesis that oncogenic PDGF-B can induce genomic instability.
The result from PDGF-B–induced hyperplasias and tumors suggested that the effect by PDGF-B on genomic instability would be an early event in tumorigenesis. We therefore investigated the effect of PDGF-B on genomic instability in primary glial cells from newborn Ntv-a wt and Arf−/− mice. Early passage cells from Ntv-a wt and Arf−/− mice were infected with RCAS-PDGF-B, referred to as IC (infected cells) wt or IC Arf−/− cells, and expression of the virally transduced human PDGF-B mRNA was confirmed (Supplementary Fig. 1A). DNA FACS analysis showed that IC wt cells displayed genomic instability, compared with uninfected wt cells (Supplementary Fig. 1E), with a DNA content that changed over time. At passage 18, a fraction of cells retained a diploid DNA content, whereas the majority of the cells were tetraploid (Supplementary Fig. 1F). At later passages, genomic instability had progressed and several aneuploid peaks had appeared (Supplementary Fig. 1G), suggesting tetraploidization to be a first step toward aneuploidy in these cells.
Because primary cells from Ntv-a Arf−/− mice are immortal,39 we first investigated whether mere loss of p19Arf would cause genomic instability. DNA FACS analysis showed that uninfected Arf−/− cells were diploid (Supplementary Fig. 1H). Upon PDGF-B infection, a subset of IC Arf−/− cells was tetraploid, as shown by an enlarged peak at 4c and an extra G2 peak at 8c (Supplementary Fig. 1I). Together, our data from PDGF-B–induced tumors and tumor cells and PDGF-B–infected primary glial cells support the hypothesis that aberrant PDGF signaling can cause genomic instability.
RAD51 Inhibited PDGF-B–induced Glioma Formation and Genomic Instability
The fact that RAD51 was endogenously expressed in the PDGF-B–induced gliomas indicated that DDR was induced. This induction, however, was not sufficient to suppress the genomic instability seen in almost all PDGF-B–induced tumors. To analyze whether forced expression of RAD51 could affect PDGF-induced genomic instability and tumorigenesis in our model, we co-injected RCAS-RAD51 + RCAS-PDGF-B or RCAS-RAD51 + RCAS-X (as a control) intracerebrally in neonatal Ntv-a wt and Arf−/− mice. In the RAD51 + X group, some mice developed hydrocephalus and RAD51 could be detected in the peri-ventricular areas of the brain (data not shown), but no tumors or dysplastic lesions were found in any of the mice by 12 weeks of age (Table 1). This showed that RAD51 could be virally transduced and highly expressed by glial cells in vivo but was not oncogenic by itself.
In the RAD51 + PDGF-B–injected group, there was a significant decrease in tumor incidence versus the PDGF-B + X–injected mice (Table 1, Fig. 3A). In addition, cross-wise comparisons (wt; PDGF-B + X vs. Arf−/−; PDGF-B + RAD51 and Arf−/−; PDGF-B + X vs. wt; PDGF-B + RAD51) generated significant decreases in incidence (Fig. 3A), supporting the hypothesis that the effect was caused by RAD51 and independent of Arf loss. Accordingly, co-expression of RAD51 significantly prolonged survival in both Ntv-a wt and Arf−/− mice (Fig. 3B).
To investigate whether RAD51 expression could hamper PDGF-induced genomic instability, we performed Feulgen stainings of PDGF-B + RAD51 induced dysplasias (n = 3) and tumors (n = 8) from wt and Arf−/− mice. No tumors or dysplasias induced by PDGF-B + RAD51 had a truly diploid DNA content, but the majority of lesions contained a near-diploid DNA content (Fig. 2C). An example of a near-diploid PDGF-B + RAD51 tumor is shown in Fig. 2D. Analysis of the combined numbers in wt and Arf−/− mice of near-diploid and aneuploid tumors and dysplasias showed a significant increase in near-diploid lesions in the PDGF-B + RAD51 group, compared with the PDGF-B + X group (Fig. 2E). These data strongly support the hypothesis that RAD51 overexpression could protect tumor cells from genomic instability.
RAD51 Inhibited Proliferation of Tumor Cells in vivo and Primary Glial Cells in vitro
To further investigate the mechanism by which RAD51 could inhibit glioma development and genomic instability, we analyzed the expression of RAD51 in relation to PCNA in the tumor tissue, because previous studies showed that overexpression of RAD51 in vitro could lead to decreased proliferation and delay in cell cycle progression.21,22 RAD51 was nuclearly expressed in distinct areas of most PDGF-B + RAD51–induced hyperplasias and tumors (Fig. 3C), and in some tumors, it also displayed cytoplasmic staining proximal to the nucleus (data not shown), which has been described for cells overexpressing RAD51 for an extended period of time.40 There was no overlap with PCNA in either hyperplasias or tumors. In fact, most tumors displayed high expression of PCNA and low expression of RAD51 (Fig. 3D), indicating increased proliferation and/or DDR. However, in areas with high RAD51 expression, PCNA-positive cells were notably less common (Fig. 3E), and we could never find cells that co-expressed nuclear RAD51 and PCNA (Fig. 3D and E), suggesting that RAD51 overexpression results in lower proliferation and/or DDR. Interestingly, also in PDGF-B + X tumors that had distinct expression of endogenous nuclear RAD51, the vast majority of cells did not co-express PCNA (Fig. 3F).
To analyze the effect of RAD51 on proliferation, we used primary glial cells from Ntv-a Arf−/− mice infected with RCAS-PDGF-B and/or RCAS-RAD51. Expression of RAD51 resulted in significantly lower cell numbers than in corresponding controls (Fig. 4A) and there was no increase in apoptosis (data not shown). The effect by RAD51 on IC wt cells was also studied. RAD51 and control infected cells were analyzed for RAD51 expression and proliferation as measured by BrdU incorporation and Ki67 staining. Control cells expressed low levels of endogenous RAD51 (Fig. 4B and C) while most RAD51 infected cells showed high expression of RAD51 (Fig. 4D and E). In line with the data from tumor tissue, cells with high nuclear RAD51 expression were never BrdU (Fig. 4D) or Ki67 (Fig. 4E) positive. Together, our data from tumor tissue and primary cells strongly indicated that nuclear expression of RAD51 could inhibit PDGF-B–stimulated proliferation.
Discussion
Genomic instability is a hallmark of human cancer and is acquired by most solid tumors. It is suggested to be an early event in the tumorigenic process, facilitating additional genetic aberrations and tumor progression.1 Activation of oncogenes and growth-promoting pathways has been shown to induce genomic instability both in vitro and in vivo.36,41 Genomic instability often precedes mutations of so-called caretaker genes, such as TP53,36,42 supported by data showing that deletion of TP53 in vitro and in vivo did not lead to aneuploidy.43 In our study, we used a PDGF-B–induced mouse model of gliomagenesis that mimics the proneural subtype of human glioma.44 We found that the experimental gliomas displayed expression of DDR proteins and genomic instability and that these features likely were due to the aberrant PDGF signaling and occurred early in the tumorigenic process. As for TP53, loss of p19Arf did not predispose for genomic instability.
The oncogene-induced DNA replication stress model is based on the hypothesis that replicative stress as a result of oncogenic signaling will contribute to genomic instability early in tumor development.45 Activation of DDR in response to stalled replication forks is suggested to work as an anti-cancer barrier in pre-cancerous lesions.36,42 In mammalian cells, RAD51 was recently shown to protect and restart stalled replication forks even before DSBs had formed,12,13 demonstrating the central role of RAD51 to protect against genomic instability. Here, we demonstrate in vivo that forced expression of RAD51 can decrease the incidence of oncogene-induced glioma, prolong survival, and reduce genomic instability in tumors in a p19Arf-independent manner. The fact that endogenous RAD51 expression was up-regulated in PDGF-B + X tumors indicated that oncogene-induced replicative stress and/or DNA damage occurred in response to PDGF but that the response was insufficient to protect cells from genomic instability and malignant transformation. A recent study showed that loss of function of DNA repair pathway proteins in combination with PDGF-B stimulation caused increased glioma incidence and malignancy,26 supporting our findings of a tumor suppressive function by DNA repair proteins.
The fact that RAD51 could reduce genomic instability may be of clinical importance, because it has been shown that patients who have near-diploid tumors have a significantly better prognosis, compared with those who have aneuploid tumors.35,46 Moreover, high RAD51 protein expression in glioblastoma has been shown to correlate with increased survival of patients.20 Our investigation endorses a putative therapeutic role for activation of the HR pathway through increased RAD51 expression. The results from tumor tissue, tumor cells, and PDGF-B–infected normal cells strongly implied that high nuclear expression of RAD51 could inhibit PDGF-driven proliferation, and the absence of BrdU incorporation in these cells indicated that they were halted in the G1 phase of the cell cycle. The finding that the RAD51-mediated tumor inhibitory and genome stabilization effects occurred in both wt and Arf−/− mice strongly implied that the suppressive effects by RAD51 were independent of p19Arf. This is essential, because approximately one-half of human glioblastomas carry mutations or homozygous deletions of CDKN2A that will affect p14ARF, and it suggests that these patients may still benefit from reactivating the RAD51 pathway.
Supplementary Material
Conflict of interest statement. None declared.
Funding
The Swedish Cancer Society, the Swedish Research Council, the Association for International Cancer Research, the Swedish Childhood Cancer Foundation, and the Medical Faculty of Uppsala University to L.U. Robert Lundberg's Foundation to U.K.W. The Swedish Childhood Cancer Foundation, Karolinska Institutet, Karolinska University Hospital, and the Cancer Society in Stockholm to M.N.
Supplementary Material
Acknowledgments
We thank Marianne Kastemar, Kerstin Nystrom-Nord, and Juan Castro for technical support.
References
- 1.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. doi: 10.1038/nrm2858. doi:10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 2.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 3.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11(11):S52–S59. doi: 10.1016/s0962-8924(01)02149-3. doi:10.1016/S0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 4.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16(12):7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12(2):191–194. doi: 10.1038/ng0296-191. doi:10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 6.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19(24):6675–6685. doi: 10.1093/emboj/19.24.6675. doi:10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins N, McManus R, Wooster R, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12–13. Oncogene. 1995;10(8):1673–1675. [PubMed] [Google Scholar]
- 8.Castilla LH, Couch FJ, Erdos MR, et al. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat Genet. 1994;8(4):387–391. doi: 10.1038/ng1294-387. doi:10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 9.Reliene R, Bishop AJ, Schiestl RH. Involvement of homologous recombination in carcinogenesis. Adv Genet. 2007;58:67–87. doi: 10.1016/S0065-2660(06)58003-4. doi:10.1016/S0065-2660(06)58003-4. [DOI] [PubMed] [Google Scholar]
- 10.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87(4):757–766. doi: 10.1016/s0092-8674(00)81394-x. doi:10.1016/S0092-8674(00)81394-X. [DOI] [PubMed] [Google Scholar]
- 11.Kronenwett U, Huwendiek S, Ostring C, et al. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64(3):904–909. doi: 10.1158/0008-5472.can-03-2451. doi:10.1158/0008-5472.CAN-03-2451. [DOI] [PubMed] [Google Scholar]
- 12.Lundin C, Schultz N, Arnaudeau C, Mohindra A, Hansen LT, Helleday T. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J Mol Biol. 2003;328(3):521–535. doi: 10.1016/s0022-2836(03)00313-9. doi:10.1016/S0022-2836(03)00313-9. [DOI] [PubMed] [Google Scholar]
- 13.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. doi:10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7(5):686–693. doi: 10.1016/j.dnarep.2007.12.008. doi:10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. doi:10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 16.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bredel M, Bredel C, Juric D, et al. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res. 2005;65(10):4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. doi:10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Z, Hu J, Li X, Jiang Y, Zhou W, Lu D. Expression analyses of 27 DNA repair genes in astrocytoma by TaqMan low-density array. Neurosci Lett. 2006;409(2):112–117. doi: 10.1016/j.neulet.2006.09.038. doi:10.1016/j.neulet.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Welsh JW, Ellsworth RK, Kumar R, et al. Rad51 protein expression and survival in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2009;74(4):1251–1255. doi: 10.1016/j.ijrobp.2009.03.018. doi:10.1016/j.ijrobp.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Raderschall E, Bazarov A, Cao J, et al. Formation of higher-order nuclear Rad51 structures is functionally linked to p21 expression and protection from DNA damage-induced apoptosis. J Cell Sci. 2002;115(Pt 1):153–164. doi: 10.1242/jcs.115.1.153. [DOI] [PubMed] [Google Scholar]
- 22.Flygare J, Falt S, Ottervald J, et al. Effects of HsRad51 overexpression on cell proliferation, cell cycle progression, and apoptosis. Exp Cell Res. 2001;268(1):61–69. doi: 10.1006/excr.2001.5265. doi:10.1006/excr.2001.5265. [DOI] [PubMed] [Google Scholar]
- 23.Ozawa T, Brennan CW, Wang L, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24(19):2205–2218. doi: 10.1101/gad.1972310. doi:10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson FK, Brodd J, Eklof C, et al. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc Natl Acad Sci USA. 2004;101(31):11334–11337. doi: 10.1073/pnas.0402716101. doi:10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.See WL, Miller JP, Squatrito M, Holland E, Resh MD, Koff A. Defective DNA double-strand break repair underlies enhanced tumorigenesis and chromosomal instability in p27-deficient mice with growth factor-induced oligodendrogliomas. Oncogene. 2010;29(12):1720–1731. doi: 10.1038/onc.2009.465. doi:10.1038/onc.2009.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18(6):619–629. doi: 10.1016/j.ccr.2010.10.034. doi:10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. doi:10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland EC, Hively WP, Gallo V, Varmus HE. Modeling mutations in the G1 arrest pathway in human gliomas: overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev. 1998;12(23):3644–3649. doi: 10.1101/gad.12.23.3644. doi:10.1101/gad.12.23.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchougounova E, Kastemar M, Brasater D, Holland EC, Westermark B, Uhrbom L. Loss of Arf causes tumor progression of PDGFB-induced oligodendroglioma. Oncogene. 2007;26(43):6289–6296. doi: 10.1038/sj.onc.1210455. doi:10.1038/sj.onc.1210455. [DOI] [PubMed] [Google Scholar]
- 30.Collins VP. Brain tumours: classification and genes. J Neurol Neurosurg Psychiatry. 2004;75(suppl 2):ii2–ii11. doi: 10.1136/jnnp.2004.040337. doi:10.1136/jnnp.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe T, Katayama Y, Yoshino A, et al. Aberrant hypermethylation of p14ARF and O6-methylguanine-DNA methyltransferase genes in astrocytoma progression. Brain Pathol. 2007;17(1):5–10. doi: 10.1111/j.1750-3639.2006.00030.x. doi:10.1111/j.1750-3639.2006.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhrbom L, Kastemar M, Johansson FK, Westermark B, Holland EC. Cell type-specific tumor suppression by ink4a and arf in kras-induced mouse gliomagenesis. Cancer Res. 2005;65(6):2065–2069. doi: 10.1158/0008-5472.CAN-04-3588. doi:10.1158/0008-5472.CAN-04-3588. [DOI] [PubMed] [Google Scholar]
- 33.Castro J, Heiden T, Wang N, Tribukait B. Preparation of cell nuclei from fresh tissues for high-quality DNA flow cytometry. Cytometry. 1993;14(7):793–804. doi: 10.1002/cyto.990140712. doi:10.1002/cyto.990140712. [DOI] [PubMed] [Google Scholar]
- 34.Forsslund G, Nilsson B, Zetterberg A. Near tetraploid prostate carcinoma. Methodologic and prognostic aspects. Cancer. 1996;78(8):1748–1755. doi:10.1002/(SICI)1097-0142(19961015)78:8<1748::AID-CNCR15>3.0.CO;2-Y. [PubMed] [Google Scholar]
- 35.Forsslund G, Zetterberg A. Ploidy level determinations in high-grade and low-grade malignant variants of prostatic carcinoma. Cancer Res. 1990;50(14):4281–4285. [PubMed] [Google Scholar]
- 36.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. doi:10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 37.Arias-Lopez C, Lazaro-Trueba I, Kerr P, et al. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7(2):219–224. doi: 10.1038/sj.embor.7400587. doi:10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121(Pt 23):3859–3866. doi: 10.1242/jcs.039537. doi:10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 39.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. doi:10.1016/S0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 40.Orre LM, Falt S, Szeles A, Lewensohn R, Wennborg A, Flygare J. Rad51-related changes in global gene expression. Biochem Biophys Res Commun. 2006;341(2):334–342. doi: 10.1016/j.bbrc.2005.12.185. doi:10.1016/j.bbrc.2005.12.185. [DOI] [PubMed] [Google Scholar]
- 41.Woo RA, Poon RY. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 2004;18(11):1317–1330. doi: 10.1101/gad.1165204. doi:10.1101/gad.1165204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. doi:10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 43.Bunz F, Fauth C, Speicher MR, et al. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62(4):1129–1133. [PubMed] [Google Scholar]
- 44.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. doi:10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 46.Pollack A, Zagars GK, el-Naggar AK, Gauwitz MD, Terry NH. Near-diploidy: a new prognostic factor for clinically localized prostate cancer treated with external beam radiation therapy. Cancer. 1994;73(7):1895–1903. doi: 10.1002/1097-0142(19940401)73:7<1895::aid-cncr2820730721>3.0.co;2-0. doi:10.1002/1097-0142(19940401)73:7<1895::AID-CNCR2820730721>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.